Abstract
Copy number variations (CNVs) have attracted increasing evidences to represent their roles as cancer susceptibility regulators. However, little is known about the role of CNV in epithelia ovarian cancer (EOC). Recently, the CNV-67048 of WW domain-containing oxidoreductase (WWOX) was reported to alter cancer risks. Considering that WWOX also plays a role in EOC, we hypothesized that the CNV-67048 was associated with EOC risk. In a case-control study of 549 EOC patients and 571 age (±5 years) matched cancer-free controls, we found that the low copy number of CNV-67048 (1-copy and 0-copy) conferred a significantly increased risk of EOC (OR = 1.346, 95% CI = 1.037–1.747) and it determined the risk by means of copy number-dependent dosage effect (P = 0.009). Data from TCGA also confirmed the abovementioned association as the frequency of low copies in EOC group was 3.68 times more than that in healthy group (P = 0.023). The CNV also negatively interacted with oral contraceptive use on EOC risk (P = 0.042). Functional analyses further showed a lower mRNA level of WWOX in tissues with the 0-copy or 1-copy than that in those with the 2-copy (P = 0.045). Our data suggested the CNV-67048 to be a risk factor of EOC in Chinese women.
1. Introduction
Ovarian cancer (OC) is one of the most common tumors of genital system and causes of cancer-related death among females. In America, it was estimated that there were 21,290 new OC patients and 14,180 deaths caused by it in 2015 [1]. In China, OC ranked the tenth cancer incidence with a rate of 6.89/105 in 2011 [2]. Despite the fact that treatments for OC have greatly improved these years, high mortality caused by it was still observed as above data presented. The poor prognosis depends on a late diagnosis as more than 50% of patients were diagnosed with advanced OC. Prevention is the first-rank policy to decrease OC detriment such as etiological intervention, early detection, and early diagnosis, of which the foremost step is to discriminate high risk group of OC with respect to these risk factors of OC that included both environmental and genetic factors.
To date, multiple association studies have established several genetic factors, most of which are single nucleotide polymorphisms (SNPs), to be susceptible loci for OC [3–7]. For example, three genome-wide association studies (GWASs) had revealed seven SNPs at loci 9p22, 8q24, 2q31, 19p13, 3q25, and 17q21 to be risk alleles of OC. However, as the relatively small increments in risk are exerted, these SNPs can only represent a small proportion of OC heritability, which would cause low accuracy on predicting individuals' onset risk of OC in a population [8]. Thus, to reveal missing heritability of OC is impending. Missing heritability can be explained by other variants in genomics of human, such as copy number variants (CNVs), del/ins, and DNA transversion. Among them, CNV is the second major category of genetic variants after SNP, which makes up 10%~13% of human variants [9]. More and more evidences had indicated that CNVs are associated with various cancer risks [10–14]. However, little is known about CNV on OC risk.
Recently, two studies had reported that one germline CNV-67048 that is located in a tumor suppressor gene WW domain-containing oxidoreductase (WWOX) contributed increased risks of lung cancer and gliomas in the Chinese [15, 16]. Also, CNVs across WWOX were reported to have potential contributions to breast cancer initiation and progression [17]. Functioning as a proapoptotic molecular and RNA-binding protein, WWOX protein plays a suppressor role in OC development [18–20]. As reported, WWOX can inhibit OC stem cells proliferation by downregulating expression of cell cycle proteins cyclin E-CDK2 and cyclin D1-CDK4 and promote cell apoptosis by upregulating expression of Wnt-5α, JNK, and caspase-3 [20]. WWOX also involves epithelial-mesenchymal transition of human OC stem cells [18]. Moreover, loss of WWOX expression was observed in OC, especially epithelial ovarian cancer (EOC) [21, 22]. Because previous studies showed that CNV-67048 influences WWOX expression in tumor tissues, considering the function and abnormal expression of WWOX in EOC, we hypothesized that the CNV-67048 contributes to EOC development. To test this hypothesis, we performed a case-control study including 549 EOC patients and 571 age (±5 years) matched cancer-free controls among Chinese women. We also performed functional assays to reveal the function of the CNV in EOC.
2. Materials and Methods
2.1. Study Subject
We conducted a case-control study in Guangzhou city in China. During September 2011 and July 2015, 549 EOC patients and 571 age (±5 years) matched cancer-free controls were recruited from the Guangdong Provincial Maternity and Child Care Center. EOC diagnosis is performed according to the International Federation of Gynecology and Obstetrics (FIGO). Individuals with tumor history were excluded. After a written informed consent was obtained, each participant was asked to denote 3 mL peripheral blood sample and complete a questionnaire to collect their data on sociodemographic, smoking status, alcohol consumption, menstrual and reproductive histories, and contraceptive use. This study was approved by the institutional review boards of Guangdong Provincial Maternity and Child Care Center.
2.2. Genotyping
Genomic DNA was extracted from 3 mL peripheral blood sample. Each DNA was then diluted into a concentration of 50 ng/μL. The copy number of CNV-67048 was tested with the TaqMan® Copy Number Assay according to the protocol of Applied Biosystems by Life Technologies. Briefly, a 10 μL reaction system including 1 μL DNA, 5 μL TaqMan Master Mix, 1 μL special probes and primers for CNV-67048 (cat# Hs03922779, Applied Biosystems), 0.5 μL control RNase P probe (Applied Biosystems), and 2.5 μL deionized water for each subject was prepared and run on the ABI 7900 system. Then the copy number was directly calculated by the CopyCaller® Software 2.1. 5% of the samples were randomly selected to repeat genotyping and the results were 98% in agreement.
2.3. Bioinformatics Analysis
To validate the association between the CNV and EOC risk as well as the possible effect of the CNV on WWOX expression, we downloaded available EOC germline CNVs data and WWOX expression value of 20 Asian EOC individuals from TCGA (https://tcga-data.nci.nih.gov/tcga/). The data of Asian common CNVs in healthy controls were also downloaded from one previously published study [23].
2.4. WWOX mRNA Level Estimation
A total of 31 EOC and 22 normal ovarian tissues were collected from the Guangdong Provincial Maternity and Child Care Center. Total RNA was extracted using the RNAiso reagent (Takara, Japan) and then reverse-transcribed into cDNA with the PrimeScript RT Master Mix (Takara, Japan). The SYBR-Green real-time PCR was used to assess the mRNA level of WWOX in the abovementioned tissues with the primers as suggested by previously published study, 5′-TGG GTT TAC TAC GCC AAT C-3′ (forward) and 5′-GTC CGT TCT CAT CAG TTT CT-3′ (reverse) [16]. The β-actin was used as an internal reference. Method of 2ΔCt was used to demonstrate the mRNA level of WWOX. All analyses were performed in a blinded fashion with the laboratory persons unaware of genotyping data and each assay was done in triplicate.
2.5. Statistical Analysis
The chi-square test was used to assess differences in the distributions of CNV-67048 copy number between EOC cases and controls. The unconditional logistic regression model with or without adjustment for surrounding factors including age, age at menarche, number of births, menstrual history, oral contraceptive use, family history of cancer, smoking status, and alcohol intake was used to infer odds ratio (OR) and 95% confidence interval (95% CI) for each association between the CNV-67048 and EOC risk. The multiplicative interaction model was used to assess the possible interaction between the CNV-67048 and selected variables on cancer risk. The Kruskal-Wallis test was used to evaluate the effect of CNV-67048 on WWOX expression in tissues. All tests were two-sided by using the IBM SPSS software (version 22.0). P < 0.05 was considered to be statistically significant.
3. Results
3.1. Characteristics of the Study Subjects
As shown in Table 1, age matched well between EOC cases and controls with no significant difference (P = 0.382). Also, there was no significant difference in frequency distribution of menstrual history, family history of cancer, smoking status, and alcohol intake between the two groups (P > 0.05 for all). However, significantly higher frequency of menarche age less than 15 years, births number no less than 4, and reported null oral contraceptive use were observed in EOC cases than in controls (P values are 0.039, <0.001, and 0.039 in turn). Moreover, EOC cases were more likely to be heavy smokers with no less than 20 pack-years smoked than controls (P = 0.029).
Table 1.
Frequency distributions of selected variables among EOC cases and controls.
| Variables | Case (n = 549) n (%) |
Control (n = 571) n (%) |
P a |
|---|---|---|---|
| Age (years) | |||
| ≤55 | 277 (50.46) | 303 (53.06) | 0.382 |
| >55 | 272 (49.54) | 268 (46.94) | |
| Age at menarche (years) | |||
| <15 | 211 (53.55) | 257 (46.73) | 0.039∗ |
| ≥15 | 183 (46.45) | 293 (53.27) | |
| Unclear | 155 (—) | 21 (—) | |
| Number of births | |||
| 0 | 16 (2.91) | 11 (1.93) | <0.001 |
| 1–3 | 409 (74.50) | 490 (85.81) | |
| ≥4 | 124 (22.59) | 70 (12.26) | |
| Menstrual history | |||
| Premenopause | 152 (27.69) | 131 (22.94) | 0.068 |
| Menopause | 397 (72.31) | 440 (77.06) | |
| Oral contraceptive use | |||
| Never | 414 (82.31) | 399 (75.86) | 0.039∗ |
| Seldom | 56 (11.13) | 78 (14.83) | |
| Often | 33 (6.56) | 49 (9.32) | |
| Unclear | 46 (—) | 45 (—) | |
| Family history of cancer | |||
| Yes | 40 (7.29) | 38 (6.65) | 0.678 |
| No | 509 (92.71) | 533 (93.35) | |
| Smoking status | |||
| Ever | 68 (12.39) | 54 (9.46) | 0.116 |
| Never | 481 (87.61) | 517 (90.54) | |
| Pack-years smoked | |||
| ≥20 | 36 (6.56) | 18 (3.15) | 0.029 |
| <20 | 32 (5.83) | 36 (6.30) | |
| 0 | 481 (87.61) | 517 (90.54) | |
| Alcohol intake | |||
| Ever | 34 (6.19) | 49 (8.58) | 0.127 |
| Never | 515 (93.81) | 522 (91.42) | |
| Staging | |||
| I + II | 43 (7.84) | ||
| III | 420 (76.50) | ||
| IV | 86 (15.66) |
a P values for a χ2 test.
∗Statistical analysis excluded subjects with unclear or unknown data.
3.2. Contribution of WWOX CNV-67048 to EOC Development
As shown in Table 2, three types of copy number of CNV-67048, which are 2-copy, 1-copy, and 0-copy, were detected. The alter frequency of loss allele in the current study (10.3%) was equivalent to that in Asian individuals as reported (10.0%) [23]. The CNV was related to EOC susceptibility as its frequency distributions of copy number were significantly different between EOC cases and controls (P = 0.005). Results from the unconditional logistic regression model without adjustment for surrounding factors presented significant increases in EOC risk in both 1-copy carriers (OR = 1.325, 95% CI = 1.024–1.714) and 0-copy carriers (OR = 2.425, 95% CI = 1.261–4.665) compared to 2-copy carriers. A tendency for an increased EOC risk was further observed accompanied by decreased copy number (P = 0.002). Moreover, after adjustment for surrounding factors, the 0-copy (OR = 2.198, 95% CI = 1.111–4.348) and a combination of 1-copy and 0-copy (OR = 1.346, 95% CI = 1.037–1.747) still conferred significantly increased risk of EOC. The tendency is also significant (P = 0.009). However, we did not find any statistically significant associations between CNV-67048 and EOC stages in case only study (Table 2). In addition, in order to validate our result, we compared the Asian germline CNVs data from TCGA database of EOC and Asian common CNVs data of health population as published [23]; the frequency of low copy number in EOC group was 3.68 times more than that in healthy group with statistical significance (P = 0.023).
Table 2.
Associations between WWOX CNV-67048 copy numbers and EOC risk and stages.
| CNV-67048 genotypes | n (%) | n (%) | P | Crude OR (95% CI) |
Adjusted OR (95% CI)a |
|---|---|---|---|---|---|
| Case-control study | Patients | Controls | |||
| Total number | 549 | 571 | |||
| 2-copy | 339 (61.75) | 397 (69.53) | 0.005 | 1.000 (ref.) | 1.000 (ref.) |
| 1-copy | 181 (32.97) | 160 (28.02) | 1.325 (1.024–1.714) | 1.268 (0.967–1.663) | |
| 0-copy | 29 (5.28) | 14 (2.45) | 2.425 (1.261–4.665) | 2.198 (1.111–4.348) | |
| 1 + 0-copy | 210 (38.25) | 174 (30.47) | 1.413 (1.103–1.811) | 1.346 (1.037–1.747) | |
| Trend test P value | 0.002 | 0.009 | |||
|
| |||||
| Case only study | Stages III + IV | Stages I + II | |||
| Total number | 506 | 43 | 0.308 | ||
| 2-copy | 309 (61.07) | 30 (69.77) | 1.000 (ref.) | 1.000 (ref.) | |
| 1-copy | 168 (33.20) | 13 (30.23) | 1.255 (0.637–2.470) | 1.330 (0.655–2.701) | |
| 0-copy | 29 (5.73) | 0 (0.00) | — | — | |
| 1 + 0-copy | 197 (38.93) | 13 (30.23) | 1.471 (0.749–2.888) | 1.554 (0.771–3.129) | |
| Trend test P value | 0.129 | 0.112 | |||
aAdjusted in a logistic regression model that included age, age at menarche, number of births, menstrual history, oral contraceptive use, family history of cancer, smoking status, and alcohol intake.
3.3. Associations between CNV-67048 and EOC Risk Stratified by Selected Variables
As shown in Table 3, the contributions of CNV-67048 on EOC risk were only significant in subgroups of 1 to 3 births' number, null oral contraceptive use, no family history of cancer, never smokers, heavy smokers, and never drinkers. However, the nonsignificant effect in the corresponding subgroups may be due to the limited sample size. Interestingly, the interaction analysis showed that the CNV-67048 significantly interacted with oral contraceptive use on EOC risk. As the CNV exerted a risk effect and oral contraceptive use exhibited a protective effect on EOC, their interactions are negative (P = 0.042). We further used the additive interaction model to show detailed interaction effect between them in Figure 1. As shown, compared to subjects who carried 2-copy CNV-67048 and never used oral contraceptive, those who carried 1-copy or 0-copy and never used oral contraceptive harbored the highest EOC risk (OR = 1.786, 95% CI = 1.317–2.423).
Table 3.
Associations between CNV-67048 and EOC risk stratified by selected variables.
| EOC Patients (n = 549) | Controls (n = 571) | Adjusted OR (95% CI)a |
P inter b | |||
|---|---|---|---|---|---|---|
| 2-copy n (%) |
1 + 0-copy n (%) |
2-copy n (%) |
1 + 0-copy n (%) |
1 + 0-copy versus 2-copy | ||
| Age (years) | ||||||
| ≤55 | 177 (63.90) | 100 (36.10) | 206 (67.99) | 97 (32.01) | 1.175 (0.817–1.692) | 0.396 |
| >55 | 162 (59.56) | 110 (40.44) | 191 (71.27) | 77 (28.73) | 1.474 (0.990–2.194) | |
| Age at menarche (years) | ||||||
| <15 | 133 (63.03) | 78 (36.97) | 177 (68.87) | 80 (31.13) | 1.315 (0.878–1.971) | 0.564 |
| ≥15 | 124 (67.76) | 59 (32.24) | 207 (70.65) | 86 (29.35) | 1.193 (0.775–1.839) | |
| Number of births | ||||||
| 0 | 10 (62.50) | 6 (43.75) | 7 (63.64) | 4 (36.36) | 0.414 (0.043–4.002) | 0.075 |
| 1–3 | 238 (58.19) | 171 (41.81) | 342 (69.80) | 148 (30.20) | 1.556 (1.164–2.081) | |
| ≥4 | 91 (73.39) | 33 (26.61) | 48 (68.57) | 22 (31.43) | 0.873 (0.442–1.722) | |
| Menstrual history | ||||||
| Premenopause | 89 (58.55) | 63 (41.45) | 92 (70.23) | 39 (29.77) | 1.451 (0.788–2.671) | 0.563 |
| Menopause | 250 (62.97) | 147 (37.03) | 305 (69.32) | 135 (30.68) | 1.262 (0.931–1.710) | |
| Oral contraceptive use | ||||||
| Never | 239 (57.73) | 175 (42.27) | 283 (70.93) | 116 (29.07) | 1.825 (1.348–2.471) | 0.042 |
| Seldom | 35 (62.50) | 21 (37.50) | 51 (65.38) | 27 (34.62) | 1.019 (0.454–2.291) | |
| Often | 23 (69.70) | 10 (30.30) | 33 (67.35) | 16 (32.65) | 0.464 (0.110–1.953) | |
| Family history of cancer | ||||||
| Yes | 23 (57.50) | 17 (42.50) | 25 (65.79) | 13 (34.21) | 2.440 (0.826–7.208) | 0.654 |
| No | 316 (62.08) | 193 (37.92) | 372 (69.79) | 161 (30.21) | 1.333 (1.016–1.748) | |
| Smoking status | ||||||
| Ever | 43 (63.24) | 25 (36.76) | 37 (68.52) | 17 (31.48) | 1.353 (0.513–3.573) | 0.488 |
| Never | 296 (61.54) | 185 (38.46) | 360 (69.63) | 157 (30.37) | 1.528 (1.165–2.003) | |
| Pack-years smoked | ||||||
| ≥20 | 23 (63.89) | 13 (36.11) | 14 (77.78) | 4 (22.22) | 10.067 (1.110–91.293) | 0.695 |
| <20 | 20 (62.50) | 12 (37.50) | 23 (63.89) | 13 (36.11) | 0.676 (0.175–2.616) | |
| 0 | 296 (61.54) | 185 (38.46) | 360 (69.63) | 157 (30.37) | 1.528 (1.165–2.003) | |
| Alcohol intake | ||||||
| Ever | 24 (70.59) | 10 (29.41) | 36 (73.47) | 13 (26.53) | 1.548 (0.383–6.248) | 0.455 |
| Never | 315 (61.17) | 200 (38.83) | 361 (69.16) | 161 (30.84) | 1.401 (1.070–1.835) | |
aAdjusted in a logistic regression model that included age, age at menarche, number of births, menstrual history, oral contraceptive use, family history of cancer, smoking status, and alcohol intake.
b P value from a multiple interaction analysis.
Figure 1.
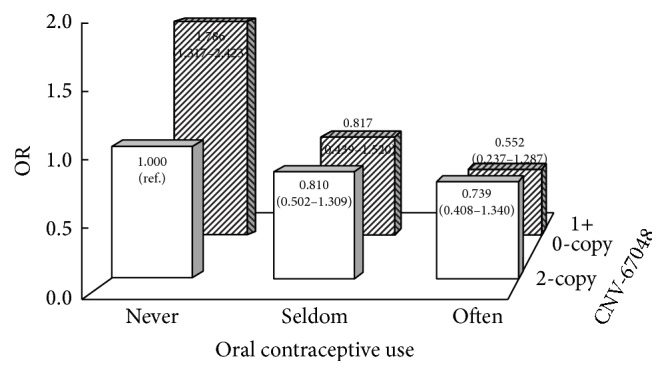
Negative interaction between the CNV-67048 and oral contraceptive use on EOC risk. The subjects who carried 2-copy CNV-67048 and never used oral contraceptive were defined as reference. The loss copy of CNV-67048 significantly interacted with null oral contraceptive use on EOC risk.
3.4. Effect of the CNV-67048 on WWOX Expression
As shown in Figure 2, in the total 53 cases of ovarian tissues, the mRNA levels of WWOX were significantly lower in tissues with the 0-copy (median: 0.0342) or 1-copy (median: 0.0347) of CNV-67048 than in those with the 2-copy (median: 0.0617; P = 0.045). We also queried the TCGA database and downloaded the 20 cases of Asians gene expression data; the gene expression of WWOX was also higher in tissues with 2-copy than in those with 0-copy or 1-copy. The difference between them is 0.082 (2-copy, 0.725 ± 0.100, versus 0-copy and 1-copy, 0.643 ± 0.053).
Figure 2.
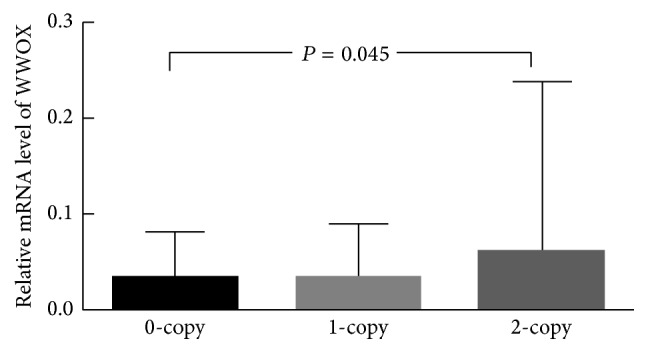
Effect of the CNV-67048 on WWOX mRNA expression in ovary tissues. The Kruskal-Wallis test was used to evaluate the effect of CNV-67048 on WWOX expression in tissues.
4. Discussion
In the current study, we found that the CNV-67048 of WWOX was significantly related to the risk of EOC in Chinese women. The CNV also negatively interacted with oral contraceptive use, because it could significantly tarnish the protective role of oral contraceptive use on EOC development. However, this study did not show any significant association between the CNV-67048 and EOC stages.
The WWOX protein is a kind of broad-spectrum tumor suppressor involving many kinds of human cancers [24]. Functional suppression of WWOX prevents apoptotic cell death induced by a variety of stress stimuli, such as tumor necrosis factor, UV radiation, and chemotherapeutic drug treatment [25]. Through protein-protein interaction, WWOX could directly bond onto a lot of well-known cancer-related molecules such as the p53, p73, Jun, and ErbB4 to enhance apoptosis [26]. WWOX also participates in the cellular metabolism and affects tumor metabolism and thus inhibits tumorigenesis [27]. Alteration of WWOX has been observed in many tumors, including breast [28], ovarian [22], prostate [29], lung [16], hepatocellular [30], gastric [31], and other cancers [32–35], and loss or reduction of its expression is reported to be correlated with worse clinical prognosis such as breast and ovarian cancer [28, 36]. Similarly in EOC, lower WWOX expression was found in tumors compared to normal ovaries [22]. Previous studies had showed that the 0-copy or 1-copy could cause a lower WWOX expression in human tissues than the 2-copy; thus individuals carrying low copy might be more predisposed to develop EOC in response to carcinogenic stimulation than individuals carrying 2-copy because of their innate differences in WWOX expression.
We also found a negative interaction between the CNV-67048 and oral contraceptive use on EOC risk. It is well-established that oral contraceptive plays a protective role in EOC risk [37]. Oral contraceptive can inhibit ovulation frequency and thus decrease the risk of EOC, because chronic stimulus caused by several ovulations can result in abnormal cell proliferation and repair and further tumorigenesis. As mentioned above, WWOX participates in the cell process in response to such stimulus and plays a role in cell repair. Thus, it is possible that the low expression of WWOX driven by low copy number of CNV-67048 enhances the cellular malignant change and suppresses the protective effect of oral contraceptive use.
WWOX CNV-67048 has been reported to be associated with lung cancer and gliomas risk in previous studies [15, 16]. Here, consistently it was found to be related to EOC risk. However, although loss of WWOX expression correlates with advanced EOC stages as reported, we did not find that the CNV was correlated with the stages. By now, only four studies have tested the effect of CNV on EOC development in large populations. Gonzalez Bosquet et al. used the integrating high-throughput data from TCGA including CNVs to construct a molecular signature to predict chemoresponse in EOC [38]. Kamieniak et al. presented a CNV hallmark of BRCA1 and BRCA2 EOC [39]. Huang et al. identified special copy number landscapes for EOC histotypes [40]. Fridley et al. tested genome-wide CNVs but found no association between inherited CNVs and ovarian cancer survival [41]. It is to be observed that these above studies did not study the effect of CNV on EOC risk. To the best of our knowledge, our study is the first study to show that a CNV is related to the risk of EOC.
Although this case-control study presented a significant association between the WWOX CNV-67048 and EOC risk and got TCGA support to be reliable and functional, there were some unavoidable limitations in the current study. As a hospital-based case-control study, there must be a selection bias such as Berkson bias and an information bias including recall bias. Meanwhile, our study sample size is relatively small. Additionally, for those interviews completed, data on some variables were not obtained or were unclear. This may cause bias on the estimation of the abovementioned association. Thus, further study with large sample size in other ethnics is warranted.
In conclusion, our data revealed the CNV-67048 and its interaction with oral contraceptive use to be in association with EOC risk in Chinese women. This CNV might be a genetic risk factor of EOC in Chinese women.
Acknowledgments
This study was supported by the Guangdong Medical Scientific Research Grants no. A2010068 (Xiping Luo) and Scientific Research Grants of Guangdong Provincial Population Family Planning Commission no. 2010241 (Xiping Luo).
Competing Interests
The authors declare that they have no competing interests.
Authors' Contributions
Yongxiu Chen and Xiaochang Tan performed the experiments, collected the data, and wrote the paper; Bi Mai, Xiaowen Huang, and Guiying Hu performed the investigation and collected the samples; Xiping Luo conceived the study and supervised all aspects of the study. Yongxiu Chen and Xiaochang Tan contributed equally to this work.
References
- 1.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2015. CA: A Cancer Journal for Clinicians. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Zeng H., Zhang S. The updated incidences and mortalities of major cancers in China, 2011. Chinese Journal of Cancer. 2015;34:p. 53. doi: 10.1186/s40880-015-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelemen L. E., Lawrenson K., Tyrer J., et al. Genome-wide significant risk associations for mucinous ovarian carcinoma. Nature Genetics. 2015;47(8):888–897. doi: 10.1038/ng.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuchenbaecker K. B., Ramus S. J., Tyrer J., et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nature Genetics. 2015;47(2):164–171. doi: 10.1038/ng.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen K., Ma H., Li L., et al. Genome-wide association study identifies new susceptibility loci for epithelial ovarian cancer in Han Chinese women. Nature Communications. 2014;5, article 4682 doi: 10.1038/ncomms5682. [DOI] [PubMed] [Google Scholar]
- 6.Couch F. J., Wang X., McGuffog L., et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genetics. 2013;9(3) doi: 10.1371/journal.pgen.1003212.e1003212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pharoah P. D., Tsai Y.-Y., Ramus S. J., et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nature Genetics. 2013;45(4):362–370. doi: 10.1038/ng.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gusev A., Bhatia G., Zaitlen N., et al. Quantifying missing heritability at known GWAS loci. PLoS Genetics. 2013;9(12) doi: 10.1371/journal.pgen.1003993.e1003993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stankiewicz P., Lupski J. R. Structural variation in the human genome and its role in disease. Annual Review of Medicine. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 10.Song Y., Li L., Ou Y., et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;508(7498):91–95. doi: 10.1038/nature13176. [DOI] [PubMed] [Google Scholar]
- 11.Yang L., Lu X., Qiu F., et al. Duplicated copy of CHRNA7 increases risk and worsens prognosis of COPD and lung cancer. European Journal of Human Genetics. 2015;23(8):1019–1024. doi: 10.1038/ejhg.2014.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L., Liu B., Qiu F., et al. The effect of functional MAPKAPK2 copy number variation CNV-30450 on elevating nasopharyngeal carcinoma risk is modulated by EBV infection. Carcinogenesis. 2014;35(1):46–52. doi: 10.1093/carcin/bgt314. [DOI] [PubMed] [Google Scholar]
- 13.Liu B., Yang L., Huang B., et al. A functional copy-number variation in MAPKAPK2 predicts risk and prognosis of lung cancer. American Journal of Human Genetics. 2012;91(2):384–390. doi: 10.1016/j.ajhg.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X., Chen X., Hu G., et al. Combined analysis with copy number variation identifies risk loci in lung cancer. BioMed Research International. 2014;2014:9. doi: 10.1155/2014/469103.469103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu K., Fan J., Ding X., et al. Association study of a functional copy number variation in the WWOX gene with risk of gliomas among Chinese people. International Journal of Cancer. 2014;135(7):1687–1691. doi: 10.1002/ijc.28815. [DOI] [PubMed] [Google Scholar]
- 16.Yang L., Liu B., Huang B., et al. A functional copy number variation in the WWOX gene is associated with lung cancer risk in Chinese. Human Molecular Genetics. 2013;22(9):1886–1894. doi: 10.1093/hmg/ddt019. [DOI] [PubMed] [Google Scholar]
- 17.Masson A. L., Talseth-Palmer B. A., Evans T.-J., Grice D. M., Hannan G. N., Scott R. J. Expanding the genetic basis of copy number variation in familial breast cancer. Hereditary Cancer in Clinical Practice. 2014;12(1, article 15) doi: 10.1186/1897-4287-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan H., Sun Y. Evaluation of the mechanism of epithelial-mesenchymal transition in human ovarian cancer stem cells transfected with a WW domain-containing oxidoreductase gene. Oncology Letters. 2014;8(1):426–430. doi: 10.3892/ol.2014.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gourley C., Paige A. J. W., Taylor K. J., et al. WWOX gene expression abolishes ovarian cancer tumorigenicity in vivo and decreases attachment to fibronectin via integrin α3. Cancer Research. 2009;69(11):4835–4842. doi: 10.1158/0008-5472.CAN-08-2974. [DOI] [PubMed] [Google Scholar]
- 20.Yan H., Tong J., Lin X., Han Q., Huang H. Effect of the WWOX gene on the regulation of the cell cycle and apoptosis in human ovarian cancer stem cells. Molecular Medicine Reports. 2015;12(2):1783–1788. doi: 10.3892/mmr.2015.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan H., Sun J. Methylation status of WWOX gene promoter CpG islands in epithelial ovarian cancer and its clinical significance. Biomedical Reports. 2013;1(3):375–378. doi: 10.3892/br.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lan C., Chenggang W., Yulan B., Xiaohui D., Junhui Z., Xiao W. Aberrant expression of WWOX protein in epithelial ovarian cancer: a clinicopathologic and immunohistochemical study. International Journal of Gynecological Pathology. 2012;31(2):125–132. doi: 10.1097/pgp.0b013e3182297fd2. [DOI] [PubMed] [Google Scholar]
- 23.Park H., Kim J.-I., Ju Y. S., et al. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nature Genetics. 2010;42(5):400–405. doi: 10.1038/ng.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewandowska U., Zelazowski M., Seta K., Byczewska M., Pluciennik E., Bednarek A. K. WWOX, the tumour suppressor gene affected in multiple cancers. Journal of Physiology and Pharmacology. 2009;60(supplement 1):47–56. [PubMed] [Google Scholar]
- 25.Lo J.-Y., Chou Y.-T., Lai F.-J., Hsu L.-J. Regulation of cell signaling and apoptosis by tumor suppressor WWOX. Experimental Biology and Medicine. 2015;240(3):383–391. doi: 10.1177/1535370214566747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salah Z., Aqeilan R., Huebner K. WWOX gene and gene product: tumor suppression through specific protein interactions. Future Oncology. 2010;6(2):249–259. doi: 10.2217/fon.09.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abu-Remaileh M., Aqeilan R. I. The tumor suppressor WW domain-containing oxidoreductase modulates cell metabolism. Experimental Biology and Medicine. 2015;240(3):345–350. doi: 10.1177/1535370214561956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nunez M. I., Ludes-Meyers J., Abba M. C., et al. Frequent loss of WWOX expression in breast cancer: correlation with estrogen receptor status. Breast Cancer Research and Treatment. 2005;89(2):99–105. doi: 10.1007/s10549-004-1474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin H. R., Iliopoulos D., Semba S., et al. A role for the WWOX gene in prostate cancer. Cancer Research. 2006;66(13):6477–6481. doi: 10.1158/0008-5472.CAN-06-0956. [DOI] [PubMed] [Google Scholar]
- 30.Huang C., Tian Y., Peng R., et al. Association of downregulation of WWOX with poor prognosis in patients with intrahepatic cholangiocarcinoma after curative resection. Journal of Gastroenterology and Hepatology (Australia) 2015;30(2):421–433. doi: 10.1111/jgh.12722. [DOI] [PubMed] [Google Scholar]
- 31.Maeda N., Semba S., Nakayama S., Yanagihara K., Yokozaki H. Loss of WW domain-containing oxidoreductase expression in the progression and development of gastric carcinoma: clinical and histopathologic correlations. Virchows Archiv. 2010;457(4):423–432. doi: 10.1007/s00428-010-0956-y. [DOI] [PubMed] [Google Scholar]
- 32.Yang J., Cogdell D., Yang D., et al. Deletion of the WWOX gene and frequent loss of its protein expression in human osteosarcoma. Cancer Letters. 2010;291(1):31–38. doi: 10.1016/j.canlet.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 33.Diniz M. G., Borges E. R., Pimenta F. J., et al. Evidence of molecular alterations in the tumour suppressor gene WWOX in benign and malignant bone related lesions of the jaws. Oncology Reports. 2011;25(2):499–502. doi: 10.3892/or.2010.1094. [DOI] [PubMed] [Google Scholar]
- 34.Chiang M.-F., Chen S.-T., Lo C.-P., Sze C.-I., Chang N.-S., Chen Y.-J. Expression of WW domain-containing oxidoreductase WOX1 in human nervous system tumors. Analytical Cellular Pathology. 2013;36(5-6):133–147. doi: 10.3233/acp-140087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowakowska M., Płuciennik E., Wujcicka W. I., et al. The correlation analysis of WWOX expression and cancer related genes in neuroblastoma- a real time RT-PCR study. Acta Biochimica Polonica. 2014;61(1):91–97. [PubMed] [Google Scholar]
- 36.Nunez M. I., Rosen D. G., Ludes-Meyers J. H., et al. WWOX protein expression varies among ovarian carcinoma histotypes and correlates with less favorable outcome. BMC Cancer. 2005;5, article 64 doi: 10.1186/1471-2407-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greer J. B., Modugno F., Allen G. O., Ness R. B. Short-term oral contraceptive use and the risk of epithelial ovarian cancer. American Journal of Epidemiology. 2005;162(1):66–72. doi: 10.1093/aje/kwi162. [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez Bosquet J., Marchion D. C., Chon H., Lancaster J. M., Chanock S. Analysis of chemotherapeutic response in ovarian cancers using publicly available high-throughput data. Cancer Research. 2014;74(14):3902–3912. doi: 10.1158/0008-5472.can-14-0186. [DOI] [PubMed] [Google Scholar]
- 39.Kamieniak M. M., Muñoz-Repeto I., Rico D., et al. DNA copy number profiling reveals extensive genomic loss in hereditary BRCA1 and BRCA2 ovarian carcinomas. British Journal of Cancer. 2013;108(8):1732–1742. doi: 10.1038/bjc.2013.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang R. Y., Chen G. B., Matsumura N., et al. Histotype-specific copy-number alterations in ovarian cancer. BMC Medical Genomics. 2012;5, article 47 doi: 10.1186/1755-8794-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fridley B. L., Chalise P., Tsai Y.-Y., et al. Germline copy number variation and ovarian cancer survival. Frontiers in Genetics. 2012;3, article 142 doi: 10.3389/fgene.2012.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]


