Abstract
Scalable production of recombinant adeno-associated virus vectors (rAAV) in baculovirus-infected Sf9 cells yields high burst sizes but variable infectivity rates per packaged AAV vector genome depending on the chosen serotype. Infectivity rates are particularly low for rAAV5 vectors, based on the genetically most divergent AAV serotype. In this study we describe key improvements of the OneBac system for the generation of rAAV5 vectors, whose manufacturing has been unsatisfactory in all current insect cell-based production systems. The Sf9 cell-based expression strategy for AAV5 capsid proteins was modified to enhance relative AAV5 VP1 levels. This resulted in a 100-fold boost of infectivity per genomic AAV5 particle with undiminished burst sizes per producer cell. Furthermore, the issue of collateral packaging of helper DNA into AAV capsids was approached. By modifications of the AAV rep and cap expression constructs used for the generation of stable Sf9 cell lines, collateral packaging of helper DNA sequences during rAAV vector production was dramatically reduced down to 0.001% of packaged rAAV genomes, while AAV5 burst sizes and infectivity rates were maintained. OneBac 2.0 represents the first insect cell-based scalable production system for high per-particle AAV5 infectivity rates combined with minimal collateral packaging of helper DNA, allowing the manufacturing of safe AAV5-based gene therapies for clinical application.
Introduction
Adeno-associated virus vectors (rAAV) for gene therapy are becoming increasingly successful for a wide spectrum of genetic and acquired diseases. The availability of currently 12 naturally occurring AAV serotypes and numerous natural and engineered variants thereof allow in vivo transduction of almost any cell type or tissue. Successful clinical trials have so far relied on the natural serotypes AAV1, AAV2, or AAV8.1–3 The prevalence of preexisting neutralizing antibodies for serotypes AAV1 and AAV2 is high (up to 80%) in the human population, largely precluding repeated AAV vector applications. Neutralizing antibodies are less frequent for AAV5 (around 40%), the evolutionary most divergent member of the AAV family.4 AAV5 capsids show less than 60% amino acid homology to any other AAV serotype.5 The targeting profile of AAV5 in liver,6 lung,7 and retinal pigment epithelium8 makes rAAV5 vectors attractive candidates for gene therapy.
rAAV5 vectors with high transduction efficiencies can be produced by plasmid cotransfection in adherent HEK 293 cells.9 In this system, the AAV5 capsid gene (cap) is transcribed from the authentic AAV2 p40 promoter. The combination of alternative splicing and the usage of noncanonical start codons result in a VP1:VP2:VP3 stoichiometry of approximately 1:1:10. For scale-up production in insect Sf9 cells, AAV rep and cap, as well as the vector backbone, were initially introduced through infection with three or two recombinant baculoviruses.10,11 Whereas VP expression was adequate for AAV2 and several additional serotypes, rAAV5 production suffered from low infectivity-per-particle ratios that correlated with suboptimal VP1 levels.12,13 The N-terminus unique for VP1 harbors the phospholipase domain that is required for AAV infectivity.14
We have recently developed the OneBac system to allow scalable, high-titer production of the full range of rAAV serotypes by infection with a single baculovirus. OneBac uses stable insect Sf9 cell lines harboring silent copies of AAV rep and cap genes for production of the different serotypes (rAAV1–12).15 Rep and cap expression is induced upon infection with a baculovirus that carries the rAAV genome for packaging. Upon infection the integrated rep and cap genes are highly amplified for efficient gene expression.16 For AAV cap expression in insect cells, the individual VP proteins are translated from a single transcript. VP1 is initiated at a mutated start codon (ACG), while VP2 and VP3 are translated from their authentic start codons to yield the prototypic 1:1:10 ratio. Most rAAV serotype vectors generated with the OneBac system displayed transduction efficiencies comparable or higher than 293 cell-derived AAVs. The AAV5 producer Sf9 cell line also yielded high AAV5 titers but with reduced infectivity rates compared with other serotypes. The reduced infectivity of rAAV5 vector particles correlated with low VP1 expression levels in the stable Sf9 cell lines.
In this report we advanced the OneBac system by introduction of an artificial intron in AAV5 cap that enhances VP1 expression and boosts rAAV5 per-particle infectivity rates. Furthermore, while maintaining the achieved high AAV5 yields, collateral packaging of foreign DNA could be drastically reduced by deletion of the Rep binding element (RBE) previously postulated to be required for template amplification.
Materials and Methods
Plasmids
Plasmid pIR-rep78-hr2-RBE was described previously.16 The AAV5 cap coding sequence was amplified by PCR. An artificial intron of 230 bp containing the baculovirus polyhedrin (polh) promoter17 was inserted into the cap gene between nucleotides 2231 and 2232 of the AAV5 genome (accession number AF085716.1). AAV2 cap of pIR-VP-hr2-RBE was replaced by the intron-containing AAV5 cap gene. For the generation of the ΔRBE rep- and cap-encoding plasmids, the two BglII sites directly flanking the RBE were used for its excision.
Cell culture and construction of stable Sf9 cell lines
HEK 293- or HeLa-derived C12 cells were cultivated as described.15 Sf9 cells and cell lines derived thereof were maintained either as adherent monolayers or in suspension culture at 27°C under constant agitation in serum-free medium. Sf9-derived cell lines for rAAV5 production were generated and screened by infection with Bac-rAAV-GFP as described.15
Recombinant baculovirus
Recombinant baculoviruses carrying the rAAV cassette for GFP expression under the control of the chicken β-actin-CMV hybrid (CBA) promoter (Bac-rAAV-GFP), or the empty transfer plasmid pFBDM (Bac-control) were generated by the MultiBac system.18 Baculovirus stocks were prepared and titers were quantified by plaque assays on monolayer Sf9 cells. For short-term storage, stocks were kept at 4°C in the dark. For long-term storage, baculovirus-infected Sf9 cells were stored in liquid nitrogen.
rAAV5 production, purification, and quantification
rAAV vectors generated by two plasmid transfection in HEK 293 cells were produced as described.9 For vector production in insect cells, Sf9-derived AAV rep/cap-expressing cell lines were continuously held in suspension culture within the logarithmic growth phase before infection with a recombinant baculovirus Bac-rAAV-GFP, or Bac-control (MOI=5). Infected cells were incubated for 72 hr at 27°C under constant agitation. Vectors were purified by one-step AVB sepharose affinity chromatography. Genomic DNA titers were analyzed by quantitative Light Cycler PCR as described.15
Protein analysis and evaluation of transduction efficiency
Western blot analysis, silver-staining gels, and rAAV5 infectivity tests on HeLa C12 cells followed by FACS analysis for quantification of transduced cells were described before.15
Total DNA isolation and quantification of gene copy number
Total DNA from approximately 1×107 Sf9 cells was isolated by the QIAamp DNA Blood Mini Kit (Qiagen). For copy number determination of rep, cap, and blasticidin resistance gene (bsd) or hr2-0.9 in Sf9 cells, 100 ng of total genomic DNA (equals 1.1×105 cells) was analyzed by quantitative Light Cycler PCR with gene-specific primers (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/hum). The same gene-specific primers were also used for determination of AAV-packaged foreign genes. Aliquots of purified AAV vector preparation were digested with Proteinase K (Roth) to release packaged DNA.
Southern blot analysis
Samples of 10 μg of total genomic DNA, isolated from Sf9 cells, were digested with SwaI, with a combination of AvrII, Bsu36I, and MfeI, or with AseI and XbaI for 12 hr and separated on a 0.7% agarose gel. Separated DNA was visualized by Midori Green staining (Nippon Genetics). The separated DNA was depurinated with 0.25 M HCl, denatured with 0.5 M NaOH and 1.5 M NaCl, neutralized with 0.5 M Tris-HCl pH 7.0 and 1.5 M NaCl, and transferred overnight to a nylon membrane (Hybond-N; GE Healthcare) by capillary blotting in 25 mM phosphate buffer pH 6.8. The transferred DNA was fixated by UV cross-linking (Strata-Linker). The membrane was prehybridized for 1 hr at 42°C in 5% SDS, 125 mM phosphate buffer pH 7.2, 0.25 M NaCl, 1 mM EDTA, and 45% formamide. For the detection of AAV5 cap sequences, a 1.4 kb fragment (SmaI-SmaI) from the pIR-VP1-intron-AAV5-cap plasmid was used, which was labeled with biotin-11-dUTP, according to the manual of the DecaLabel DNA Labeling Kit (Fermentas). The probe was added to hybridization solution and incubated at 42°C on the membrane for 24 hr. After repeated washing with 2× SSC and 0.1% SDS at 42°C and with 0.1× SSC and 0.1% SDS at 55°C, the membrane was incubated twice with 5% SDS and 5% milk in PBS for 10 min at room temperature before the addition of horseradish peroxidase-conjugated streptavidin (1:10,000), which was incubated for 20 min at room temperature. After repeated washing of the membrane with 1% SDS in PBS and 0.1% Tween-20 in PBS, signal detection was performed with ECL Western Blotting Substrate (Pierce).
Results
Generation of optimized rAAV5 insect producer cell lines with increased VP1 expression
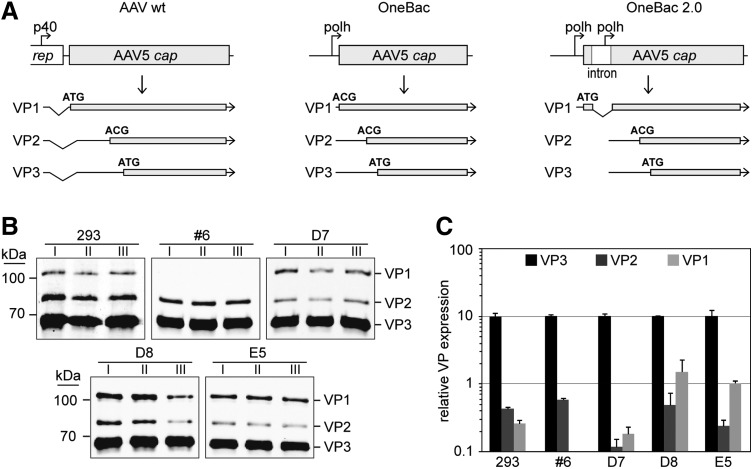
Under the assumption that insufficient VP1 expression was the reason for the reduced infectivity-to-particle ratio of AAV5 vectors produced with the OneBac system,15 the VP1 expression strategy was modified as follows: The original ATG start codon for VP1 was reintroduced into the constructs for stable expression in Sf9 cells (Fig. 1A). In order to prevent VP1 overexpression at the cost of VP2 and VP3 expression levels, an artificial intron was inserted at the AAV major splice acceptor site between the VP1 and VP2 translation initiation codons.17 The inserted intron of 230 bp comprises the consensus sequences for splicing and the baculovirus polyhedrin promoter for expression of a second unspliced transcript to express VP2 and VP3 (Fig. 1A, right panel). As described for the original OneBac system,15 this construct for AAV5 cap was co-transfected in Sf9 cells with the described AAV2 rep construct. Sf9 cell clones were selected with blasticidin and analyzed for cap expression induced by baculovirus infection (Bac-rAAV-GFP). Of over 70 cell lines tested, all VP-positive cell lines showed strong VP1 bands in Western blots (data not shown). Cell lines were screened by small-scale rAAV5 vector productions (30 ml) for high infectivity-to-genomic particle ratios. Cell clones denoted as D7, D8, and E5 were chosen for further analysis.
Figure 1.
AAV5 capsid expression pattern in stable Sf9 cell lines in comparison to transiently transfected 293 cells. (A) Depiction of different cap expression systems for production of rAAV5. (Left panel) Regulation of capsid protein expression in AAV wt. (Center) Capsid protein expression strategy in the OneBac system with an ACG start codon for VP1. (Right) Capsid protein expression in OneBac 2.0 with an artificial intron containing a second polh promoter inserted into the cap gene. The promoters and their transcripts are displayed. The start codons for the translation of the individual VP proteins are indicated. (B) AAV5 Cap expression was analyzed by Western blot analysis of cell extracts from three independent rAAV5 packaging experiments (I–III). Cell extracts from 293 cells were analyzed 72 hr posttransfection. Cell extracts from rep/cap-expressing Sf9 cells were prepared 72 hr postinfection with recombinant Bac-rAAV at MOI=5. VP proteins expressed were detected with mAb B1. (C) Quantitative analysis of the Western blots shown in (B) to compare the relative ratios of VP1, VP2, and VP3 expression in 293 or Sf9 cells, respectively. A secondary antibody emitting near-infrared fluorescence was used and its fluorescence was quantified by an Odyssey imager. Expression levels of VP3 were arbitrarily set to 10 and expression levels of VP2 and VP1 were calculated as relative values thereof. The results of three experiments are given as mean±standard deviation. Please note the logarithmic scale of the y axis. AAV, adeno-associated virus vectors; rAAV, recombinant AAV.
Comparative analysis of the capsid expression profile
The AAV5 cell lines D7, D8, and E5 were compared side by side to the previously described rAAV5 producer cell line (clone #6)15 and to rAAV5 production in HEK 293 cells. While baculovirus-induced expression of Rep was indistinguishable (data not shown), the expression profile of the capsid proteins was clearly different. In contrast to cell line #6, the three new cell lines showed a strongly enhanced proportion of VP1 (Fig. 1B), comparable to the HEK 293-based production system.
A quantitative analysis of the relative amounts of the different capsid proteins (Fig. 1C) showed VP1 expression levels in the range of 10% of VP3 expression in the cell lines D8 and E5. For the previously described Sf9 cell line for rAAV5 production, clone #6, VP1 expression levels were below 1% of VP3.
Analysis of the AAV5 vector yields per cell
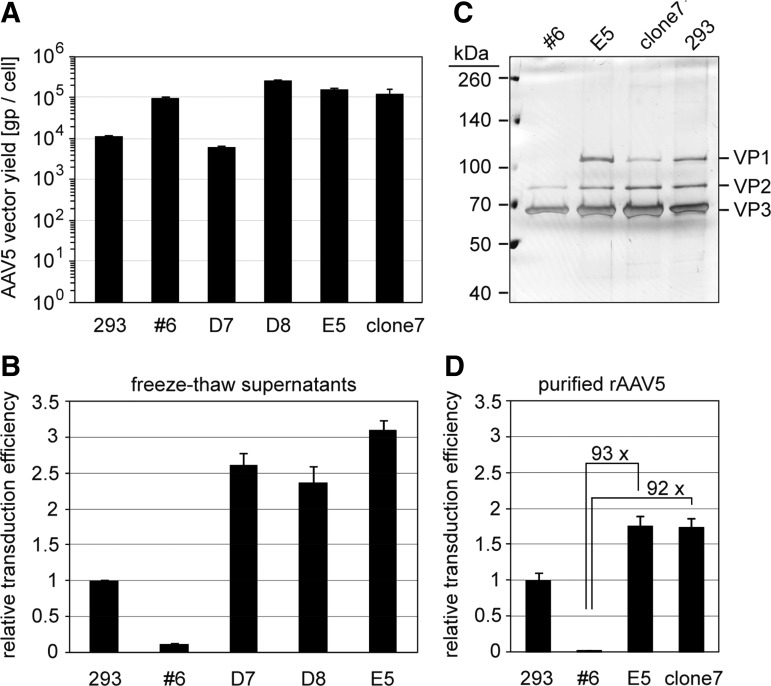
High rAAV yields per cell constitute the prime requirement for the production of high-titer AAV vector preparations. In a side-by-side analysis, the burst sizes of the rAAV5 producer cell lines were determined in triplicates of 30 ml cultures infected with Bac-rAAV-GFP. Yields of approximately 1×105 rAAV5-GFP genomic particles (gp) per cell were attained for cell lines #6, D8, and E5 (Fig. 2A). The burst size of cell line D7 was roughly 10-fold lower comparable to the HEK 293 two-plasmid transfection system.9
Figure 2.
Comparative analysis of the burst size and cell transduction efficiency of the rAAV5 vectors produced in 293 or Sf9 cells. (A) The yields of rAAV5 preparations were quantified as benzonase-resistant genomic particles (gp) per cell by qPCR using rAAV genome-specific primers. rAAV burst sizes of three experiments are displayed as mean±standard deviation. Please note the logarithmic scale of the y axis. (B) Analysis of the transduction efficiencies of rAAV5 vectors derived from freeze–thaw supernatants as determined in HeLa C12 cells (MOI: 10,000). Transduction efficiencies of rAAV5s prepared in 293 were arbitrarily set to 1.0, while those of rAAV5 vectors derived from Sf9 cells are displayed as percentage thereof. The experiments were performed in triplicates and are displayed as mean±standard deviation. (C) Silver staining gel analysis of AVB-chromatography-purified rAAV5 preparations separated on 8% SDS-PAGE with 5×109 gp loaded per lane. (D) Analysis of rAAV5 transduction as described in (B), except that highly purified rAAV5 vector preparations were used (MOI: 1000). Data on clone 7 (ΔRBE) are added for comparison, and details are discussed in the context of Fig. 3. RBE, Rep binding element.
Comparative analysis of the transduction efficiency
Transduction efficiencies were compared among rAAV5 vectors produced in HEK 293 cells, the published Sf9 cell line #6 (low VP1), and the new rAAV5 producer cell lines expressing higher ratios of VP1. Identical amounts of genomic AAV particles from cleared freeze–thaw supernatants were used to infect HeLa C12 cells followed by quantification of GFP expression through FACS analysis. The transduction efficiency of 293-derived AAV5 vectors was used as a reference and was arbitrarily set to 1.0. As observed previously,15 rAAV5 vectors from the Sf9 cell line #6 showed a roughly 10-fold reduced transduction efficiency compared with 293 cell-derived rAAV5 vectors (Fig. 2B). In contrast, vectors produced in the new Sf9 cell lines D7, D8, and E5 showed even higher relative transduction efficiencies than those derived from HEK 293 cells. The enhancement of transduction rates of rAAV5 produced in the new Sf9 cell lines ranged between 20- and 30-fold in comparison to the initially published rAAV5 producer cell line. To confirm these results with highly purified vector preparations, 400 ml suspension cultures of Sf9 cell lines #6 (low VP1) and E5 (high VP1) were prepared in parallel and infected with Bac-rAAV-GFP. In parallel, rAAV5-GFP was produced from plasmid-transfected HEK 293 cells. All rAAV5 preparations were purified by HPLC-AVB column chromatography. AAV purity was evaluated in silver-stained SDS gels. Distinct and pure VP bands of the expected sizes were detected for all HPLC-purified vector preparations (Fig. 2C). Relative transduction efficiencies of the purified AAV5 preparations were determined that largely reproduced the results obtained with the freeze–thaw supernatants (Fig. 2D, compare with Fig. 2B). In comparison to the original AAV5 producer cell line, the transduction efficiency of AAV5 vectors produced by the new Sf9 cell line optimized for VP1 expression was increased almost 100-fold (p<0.001).
Analysis of helper gene-related DNA impurities in purified AAV vector preparations
For the previously described OneBac system,15 purified rAAV vector preparation had initially been analyzed for copackaged AAV rep-cap DNA sequences viewed as a potential source of replication-competent AAVs. Only low rates of combined rep/cap sequences were found in this analysis. Fusion of rep and cap gene sequences from two separate plasmids is a rare event, but a prerequisite for the generation of wtAAV. To further analyze this issue, quantitative PCRs for collateral packaging of subfragments of candidate helper sequences were performed. In AVB sepharose-purified rAAV vector preparations, significantly higher copy numbers of isolated rep or cap gene sequences per vector genome were detected for most AAV serotypes (data not shown).
Generation of rAAV5 Sf9 producer cell lines without Rep binding elements
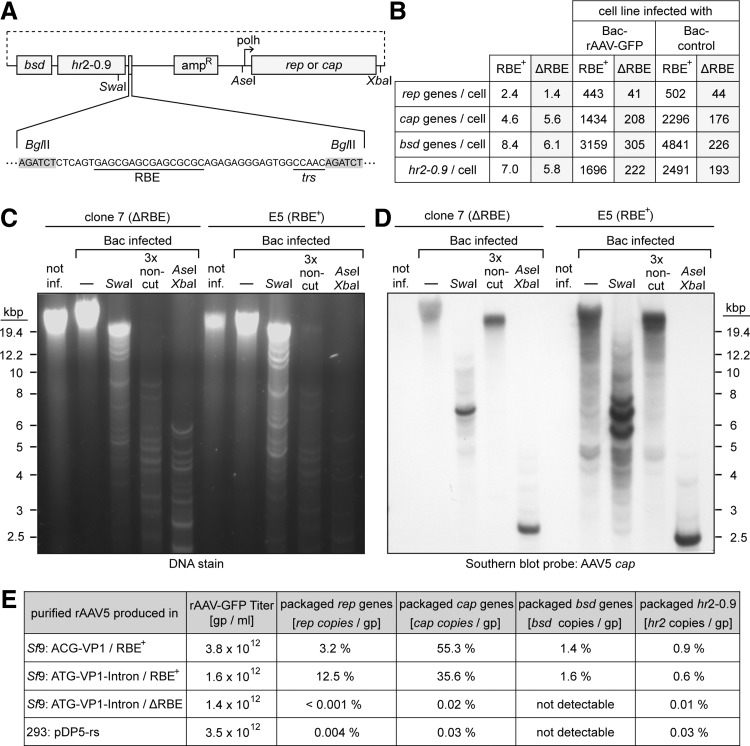
Previously, the AAV2 RBE derived from the inverted terminal repeat (ITR) was inserted into the rep and cap expression constructs for the generation of rAAV producer cell lines.16 It was postulated that the RBE would mediate Rep-dependent rescue and template amplification required for sufficient Rep and VP expression as a prerequisite for high-titer AAV production.
The RBEs are parts of the AAV ITRs and also serve as packaging signals. This led to the assumption that the RBE embedded in the constructs might be also responsible for collateral rep and cap gene packaging as observed in the OneBac system. To explore this issue, the RBE was excised from the AAV2 rep expression plasmid and from the here-described AAV5 cap expression plasmid with the artificial intron (Fig. 3A). The resulting plasmids were cotransfected in Sf9 cells to generate producer cell lines for the production of highly infectious rAAV5 vectors with reduced AAV rep and cap gene packaging. Of the derived rAAV5 producer cell lines, clone 7 was further analyzed. Upon infection with Bac-rAAV-GFP, a vector yield of 1.3×105 gp/cell was achieved, comparable to the other RBE-containing Sf9 AAV5 producer cell lines (Fig. 2A). The transduction efficiency of AVB sepharose-purified AAV5 vectors produced in clone 7 (Fig. 2C) was determined on HeLa C12 cells. The ratio of infectious to genomic rAAV5 was identical to rAAV5 vectors produced in cell clone E5 (Fig. 2D). These results clearly show that the presence or absence of RBE has little effect on rAAV vector yields or transduction efficiency.
Figure 3.
Comparative analysis of the RBE+ and ΔRBE rAAV5 producer cell lines. (A) Depiction of the constructs for the expression of rep and cap. Rep and cap expression is regulated by a polyhedrin promoter (polh) and the enhancer element hr2-0.9. Further elements on these construct include an ampicillin resistance gene (ampR), the blasticidin S deaminase gene (bsd), and the RBE with the terminal resolution site (trs). (B) Number of rep, cap, and bsd gene or hr2-0.9 copies in uninfected or baculovirus-infected AAV5 cell lines E5 (RBE+) or clone 7 (ΔRBE). In case of infected cells, total DNA was isolated 72 hr after baculovirus infection. Gene copy numbers were determined by quantitative Light Cycler PCR with gene-specific primers. (C) Analysis of total genomic DNA by agarose gel (0.7%) electrophoresis. An amount of 10 μg of total DNA from two different rAAV5 producer cell lines (clone 7 and E5) was loaded per lane. For baculovirus-infected cell lines, DNA was additionally digested with different sets of restriction enzymes as indicated. SwaI represents a single cutter for the cap expression constructs that were transfected in Sf9 cells for the generation of the cell lines. AvrII, Bsu36I, and MfeI were chosen as noncutting enzymes (3× noncut) for the cap expression constructs. The restriction enzymes AseI and XbaI flank the AAV5 cap gene resulting in a 2650 bp fragment. (D) For the Southern blot a hybridization probe was used that detects the AAV5 cap gene (AAV5 genome: nt 2232–3603). (E) Analysis of copackaging of rep, cap, bsd, and hr2-0.9 sequences into AAV5 capsids during vector production. Aliquots from AVB sepharose-purified AAV vector preparation were digested with proteinase K to release the packaged DNA from the capsids. The samples were analyzed by quantitative Light Cycler PCR with primers specific for the vector genome or the indicated genes, respectively. The titers of rAAV5 particles containing the vector genome are shown as absolute values, whereas the portions of rep-, cap-, or bsd-gene and hr2-0.9 element-containing particles are displayed as percentages thereof.
Comparative analysis of RBE+ and ΔRBE rAAV5 producer cell lines
For the generation of stable insect cell lines, the rep and cap constructs were transfected in molecular ratios of 1:2.5. In the resulting rAAV5 producer cell lines, approximately two gene copies of rep and five gene copies of cap per cell were detected, irrespective of the presence of the RBE (Fig. 3B). Upon baculovirus infection, the numbers of rep and cap genes were highly amplified, even in the absence of the RBE. However, the degree of rep and cap gene amplification was about 10-fold higher in RBE+ compared with ΔRBE cell lines (Fig. 3B). The result shows that the lower level of gene amplification is obviously sufficient for high-titer rAAV5 production as shown in Fig. 2A.
Furthermore, our data show that the amplification mechanism in the cell lines is not dependent on baculovirus-transduced rAAV genomes. AAV rep and cap are similarly amplified in presence of a “control” baculovirus with an empty transfer plasmid. The state of the amplified cap genes was further analyzed by restriction digests of total genomic DNAs separated on agarose gels. The high-molecular-weight bands of undigested, genomic DNA only marginally changed upon digestion with SwaI (Fig. 3C). SwaI cuts statistically every 65 kb and represents a single-cut enzyme for the transfected expression cassettes. Digests with a combination of three noncut enzymes for the transfected expression cassettes, or two restriction enzymes that cut at flanking sites of the rep or cap genes almost entirely digested high-molecular-weight genomic DNA. On the corresponding Southern blot probed with AAV5 cap sequences, high-molecular-weight bands were predominant in the case of undigested, baculovirus-infected cell DNA (Fig. 3D). SwaI-digested DNA of the ΔRBE cell lines produced a band of approximately 6.8 kb that corresponded to the linearized transfected cap expression construct (6,788 bp). The RBE+ cell line showed stronger signals and additional off-size bands. Digests with a combination of three noncut enzymes left the high-molecular-weight bands intact, although the genomic DNA was entirely digested. Genomic DNA cut with AseI and XbaI produced a band of approximately 2.6 kb that corresponds to the size of the AseI-XbaI fragment of the transfected cap expression construct (2650 bp).
The combination of an agarose gel and corresponding Southern blot probed with AAV5 cap documented that the rep and cap expression cassettes were amplified to high-molecular-weight, predominantly head-to-tail concatemers, irrespective of the presence of an RBE. The additional off-size bands observed in the presence of the RBE (Fig. 3D) appear to reflect the overall higher level of amplification (Fig. 3B). In addition, the blasticidin resistance gene (bsd) and the hr2-0.9 element, which are a parts of either expression construct, were similarly amplified (Fig. 3B). The data document that baculovirus-induced AAV rep/cap template amplification is partially independent of the Rep-RBE interaction.
Comparative analysis of collateral packaging of helper gene in rAAV5 producer cell lines
AVB sepharose-purified AAV5 vector preparations were produced for the three generations of Sf9 rAAV5 producer cell lines and for HEK 293 cell-produced rAAV5 (Fig. 2C). Packaged genomes were released and analyzed by quantitative Light Cycler PCR. While the titers of vector genome-containing particles were comparable, the percentage of capsids containing rep, cap, or bsd genes varied dramatically (Fig. 3E). In the two Sf9 AAV5 RBE+ producer cell lines, considerable amounts of packaged rep, cap, and bsd genes or hr2-0.9 sequences were detectable in highly purified AAV preparations. In contrast, in rAAV5 vector preparations derived from the ΔRBE cell line, the copy numbers of copackaged sequences were reduced to less than 0.001%. The proportion was even lower than that of AAV5 vectors generated in HEK 293 cells. These results clearly show that the presence of the RBE in the constructs for Sf9 cell generation is responsible for increased collateral packaging of rep and cap in rAAV vector preparations.
Discussion
In this study we improved the OneBac system by solving two inherent problems of baculovirus-based AAV production in insect cells: (1) By raising the relative amounts of VP1 in AAV5 capsids, the infectivity per genomic particle (gp/ml) was enhanced 100-fold, exceeding AAV5 infectivity of existing Sf9- or 293 cell-based AAV5 production systems. (2) Packaging of nonvector DNA, including rep/cap DNA sequences amplified during rAAV5 production, was drastically reduced by deletion of the RBE while maintaining rAAV5 vector yields and infectivity. This is the first report on a baculovirus-based AAV5 production system that combines high burst sizes with high relative infectivity and minimal copackaging of foreign DNA. These combined improvements render the OneBac system particularly suitable for scalable production of AAV5 vectors for clinical gene therapy.
The VP1 content of AAV vectors as the major determinant for infectivity
Baculovirus-based rAAV production systems were initially developed for prototype AAV2, and a few additional serotypes. Baculovirus-expressed AAV5 cap consistently failed to produce infectious rAAV5 vectors in insect cells.12,13 The previously described OneBac system also suffered from reduced AAV5 infectivity despite high burst sizes per cell. Low VP1 levels were the most likely explanation, a finding that was consistent with the results from the three- or two-Bac AAV5 production systems. The commonly accepted 1:1:10 ratio of VP1, VP2, and VP3 was first described for wild-type AAV2 capsids19 and believed to be universal for all other AAV serotypes. While VP1 is not required for the assembly of intact capsids,20 it has a high impact on AAV infectivity.14 The unique VP1 N-terminus harbors a phospholipase domain (PLA2) that is required for endosomal escape and infectivity. The PLA2 domain is highly conserved among the AAV serotypes; only the AAV5 PLA2 is more divergent and possibly less active.12,21 In the three-Bac system, fusion of the VP1 unique region of AAV2 to the VP2-VP3 ORF of AAV5 raised infectivity, but at the price of hybrid AAV5/2 capsids.12,13
The here-presented side-by-side comparison of AAV5 vectors with low and high VP1 content underlines the importance of sufficient and authentic VP1 for full AAV5 infectivity. For OneBac 2.0 AAV5 VP1 expression was enhanced by reintroduction of the original ATG start codon combined with a splicing-mediated expression strategy described previously to coexpress VP1–3 of AAV2, AAV1, AAV6, and AAV8 in a recombinant baculovirus backbone.17 The described VP1 expression strategy may be helpful for additional AAV serotypes in order to boost infectivity. High infectivity rates per particle represent a prime issue in AAV gene therapy, allowing lower AAV doses, minimizing potential adverse effects, and, last but not least, decreasing production costs.
Avoiding potential packaging signals in helper gene constructs
In gene therapy, collateral packaging of nonvector DNA represents a critical safety issue. Previously, we showed that combined rep/cap sequences as potential source of AAV wildtype were relatively low in the OneBac system.15 Further PCR analysis of helper gene subsequences described here revealed that copackaging of isolated rep or cap genes was significantly higher, exceeding that of HEK 293 cell-derived AAV preparations. Most OneBac producer cell lines were generated using the AAV2 rep expression construct. The RBE as part of the AAV ITRs also represents the AAV2 packaging signal. It is present on both the AAV rep and the AAV cap expression plasmids used for the generation of the Sf9 cell lines. Solely by deleting the RBE in either plasmid, the ratio of copackaged rep genes decreased dramatically to <0.001% and of copackaged cap genes to 0.02% per genomic particle. This virtually proves that the RBE acts as a packaging signal in the OneBac system. Rep78 has been shown to interact with the RBE and release packageable RBE-containing AAV sequences from wildtype AAV2 plasmids. Considerable copackaging of rep/cap sequences is also mediated by a cryptic RBE in the AAV p5 promoter that was described before in plasmid transfection-based AAV production.22
Recently, an AAV production system based on stable HeLa cell lines was presented that harbors the rep and cap genes and the ITR-flanked rAAV genome.23 Considerable collateral packaging of rep and cap was reported, up to 0.62% and 0.25%, respectively, of vector genome-containing capsids. In rAAV2 vector lots for a clinical trial, the reported percentage of packaged cap genes ranged between 0.016% and 0.021%.24 These numbers are comparable to those of the described RBE-deleted insect cell lines for rAAV5 production. In contrast to copackaged rep or cap sequences in mammalian AAV production systems, residual packaged AAV genes in the OneBac system will not be expressed upon transduction of human cells. Their expression is regulated by the baculovirus polyhedrin promoter that is not active in mammalian cells. Another issue of AAV production by plasmid transfection represents the fact that significant proportions of encapsidated DNA arise from “reverse” packaging at the ITRs.25 This was shown to lead to collateral packaging of prokaryotic DNA sequences, for example, ampicillin genes, derived from the plasmid backbone.26 Reverse packaging could be reduced by the use of oversized plasmid backbones that exceeded the packaging capacity of AAV vectors.24
In the OneBac system, the vector genome cassette resides on the 134 kb baculovirus genome that is too large for reverse packaging. Another source of DNA impurities can arise from the host cell genome as shown for collateral packaging of chromosomal DNA with functional RBE homologs.27 For Spodoptera frugiperda a draft genome has been published recently.28 A blast search using the Spodobase database showed that the full 16 bp RBE sequence of the AAV2 ITR or the AAVS1 region of human chromosome 19 was not present in sequencing contigs of Spodoptera frugiperda (Mietzsch et al., unpublished). It cannot be excluded that sequence motifs with small variations exist that allow binding of Rep78. The only existing method to analyze whether and to what extent chromosomal DNA from Sf9 genome is packaged into AAV capsids will be next-generation sequencing of rAAV vector pools.
Rep and cap constructs are amplified by a baculovirus-mediated replication mechanism
The RBE in the rep and cap expression constructs used for the generation of the individual Sf9 cell lines of the OneBac system was initially thought to be important for rescue and amplification. Amplification levels range between 200- and 500-fold. However, by deletion of the RBE we could show that it is not necessary for rep and cap amplification after baculovirus infection. Even in the absence of the RBE, rep and cap genes are still amplified up to 65-fold. Obviously, this is fully sufficient since the AAV yields are maintained. Our data suggest that the entire expression cassettes are replicated in this process. This can be deduced from the fact that the number of bsd genes after amplification was approximately as high as the number of rep and cap genes combined. It is therefore highly likely that the hr2-0.9 element drives the amplification. The homologous regions (hr) of baculovirus have been implicated both as transcriptional enhancers29 and as origins of DNA replication.30
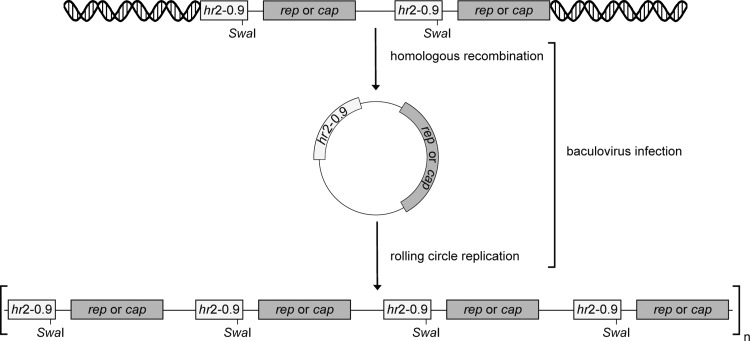
In the stable Sf9 cell lines of the OneBac system, the integrated rep and cap genes are present as two or more head-to-tail concatemers16 (Fig. 4). We show in this study that upon baculovirus infection of Sf9 cells the cap expression cassettes persist as high-molecular-weight concatemers. Baculovirus replication promotes high-frequency homologous recombinations assumedly exploiting the repeated hr elements as cis signals.31 We therefore assume that comparable recombination events occur in baculovirus-infected OneBac cell lines (Fig. 4). Baculovirus replication was described to occur by a rolling circle mechanism32 involving the hr elements.33 This is consistent with the notion that the high-molecular-weight rep/cap concatemers described here at high copy numbers represent the result of baculovirus-induced rolling circle replication.
Figure 4.
Postulated mechanism leading to high-molecular-weight concatemers of the rep and cap expression cassettes. In uninfected stable Sf9 cells, the rep and cap expression cassettes are integrated as two or more head-to-tail concatemers into the host genome. Baculovirus infection promotes high-frequency homologous recombinations assumedly exploiting the repeated hr elements as cis signals and a rolling circle replication mechanism initiated at the homologous regions (hr), leading to long continuous DNA molecules that contain multiple copies of the rep or cap expression construct.
Supplementary Material
Acknowledgments
The authors thank Catrin Stutika and Daniela Hüser of the Heilbronn lab for helpful discussions and critical reading of the article. Generous financial support was provided to R.H. and M.M. by the German academic exchange service DAAD (PROMOS and PPP).
Author Disclosure
R.H. and S.Z. are inventors of patents related to rAAV technology. S.Z. is inventor in a patent on the inducible insect cell-based system for highly efficient production of recombinant AAV vectors (US 20120100606). R.H. owns equity in a company that is commercializing AAV for gene therapy.
References
- 1.Kastelein JJ, Ross CJ, Hayden MR. From mutation identification to therapy: discovery and origins of the first approved gene therapy in the Western world. Hum Gene Ther 2013;24:472–478 [DOI] [PubMed] [Google Scholar]
- 2.Nathwani AC, Tuddenham EG, Rangarajan S, et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med 2011;365:2357–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mingozzi F, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet 2011;12:341–355 [DOI] [PubMed] [Google Scholar]
- 4.Boutin S, Monteilhet V, Veron P, et al. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther 2010;21:704–712 [DOI] [PubMed] [Google Scholar]
- 5.Zinn E, Vandenberghe LH. Adeno-associated virus: fit to serve. Curr Opin Virol 2014;8C:90–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paneda A, Lopez-Franco E, Kaeppel C, et al. Safety and liver transduction efficacy of rAAV5-cohPBGD in nonhuman primates: a potential therapy for acute intermittent porphyria. Hum Gene Ther 2013;24:1007–1017 [DOI] [PubMed] [Google Scholar]
- 7.Virella-Lowell I, Zusman B, Foust K, et al. Enhancing rAAV vector expression in the lung. J Gene Med 2005;7:842–850 [DOI] [PubMed] [Google Scholar]
- 8.Lebherz C, Maguire A, Tang W, et al. Novel AAV serotypes for improved ocular gene transfer. J Gene Med 2008;10:375–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grimm D, Kay MA, Kleinschmidt JA. Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. Mol Ther 2003;7:839–850 [DOI] [PubMed] [Google Scholar]
- 10.Urabe M, Ding C, Kotin RM. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum Gene Ther 2002;13:1935–1943 [DOI] [PubMed] [Google Scholar]
- 11.Smith RH, Levy JR, Kotin RM. A simplified baculovirus-AAV expression vector system coupled with one-step affinity purification yields high-titer rAAV stocks from insect cells. Mol Ther 2009;17:1888–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohlbrenner E, Aslanidi G, Nash K, et al. Successful production of pseudotyped rAAV vectors using a modified baculovirus expression system. Mol Ther 2005;12:1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urabe M, Nakakura T, Xin KQ, et al. Scalable generation of high-titer recombinant adeno-associated virus type 5 in insect cells. J Virol 2006;80:1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girod A, Wobus CE, Zadori Z, et al. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J Gen Virol 2002;83:973–978 [DOI] [PubMed] [Google Scholar]
- 15.Mietzsch M, Grasse S, Zurawski C, et al. OneBac: platform for scalable and high-titer production of adeno-associated virus serotype 1–12 vectors for gene therapy. Hum Gene Ther 2014;25:212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aslanidi G, Lamb K, Zolotukhin S. An inducible system for highly efficient production of recombinant adeno-associated virus (rAAV) vectors in insect Sf9 cells. Proc Natl Acad Sci USA 2009;106:5059–5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H. Intron splicing-mediated expression of AAV Rep and Cap genes and production of AAV vectors in insect cells. Mol Ther 2008;16:924–930 [DOI] [PubMed] [Google Scholar]
- 18.Berger I, Fitzgerald DJ, Richmond TJ. Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol 2004;22:1583–1587 [DOI] [PubMed] [Google Scholar]
- 19.Becerra SP, Koczot F, Fabisch P, et al. Synthesis of adeno-associated virus structural proteins requires both alternative mRNA splicing and alternative initiations from a single transcript. J Virol 1988;62:2745–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruffing M, Zentgraf H, Kleinschmidt JA. Assembly of viruslike particles by recombinant structural proteins of adeno-associated virus type 2 in insect cells. J Virol 1992;66:6922–6930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popa-Wagner R, Porwal M, Kann M, et al. Impact of VP1-specific protein sequence motifs on adeno-associated virus type 2 intracellular trafficking and nuclear entry. J Virol 2012;86:9163–9174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nony P, Chadeuf G, Tessier J, et al. Evidence for packaging of rep-cap sequences into adeno-associated virus (AAV) type 2 capsids in the absence of inverted terminal repeats: a model for generation of rep-positive AAV particles. J Virol 2003;77:776–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin J, Frederick A, Luo Y, et al. Generation and characterization of adeno-associated virus producer cell lines for research and preclinical vector production. Hum Gene Ther Methods 2013;24:253–269 [DOI] [PubMed] [Google Scholar]
- 24.Hauck B, Murphy SL, Smith PH, et al. Undetectable transcription of cap in a clinical AAV vector: implications for preformed capsid in immune responses. Mol Ther 2009;17:144–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright JF. Manufacturing and characterizing AAV-based vectors for use in clinical studies. Gene Ther 2008;15:840–848 [DOI] [PubMed] [Google Scholar]
- 26.Chadeuf G, Ciron C, Moullier P, et al. Evidence for encapsidation of prokaryotic sequences during recombinant adeno-associated virus production and their in vivo persistence after vector delivery. Mol Ther 2005;12:744–753 [DOI] [PubMed] [Google Scholar]
- 27.Hüser D, Weger S, Heilbronn R. Packaging of human chromosome 19-specific adeno-associated virus (AAV) integration sites in AAV virions during AAV wild-type and recombinant AAV vector production. J Virol 2003;77:4881–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kakumani PK, Malhotra P, Mukherjee SK, et al. A draft genome assembly of the army worm, Spodoptera frugiperda. Genomics 2014;104:134–143 [DOI] [PubMed] [Google Scholar]
- 29.Guarino LA, Gonzalez MA, Summers MD. Complete sequence and enhancer function of the homologous DNA regions of Autographa californica nuclear polyhedrosis virus. J Virol 1986;60:224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leisy DJ, Rohrmann GF. Characterization of the replication of plasmids containing hr sequences in baculovirus-infected Spodoptera frugiperda cells. Virology 1993;196:722–730 [DOI] [PubMed] [Google Scholar]
- 31.Martin DW, Weber PC. DNA replication promotes high-frequency homologous recombination during Autographa californica multiple nuclear polyhedrosis virus infection. Virology 1997;232:300–309 [DOI] [PubMed] [Google Scholar]
- 32.Oppenheimer DI, Volkman LE. Evidence for rolling circle replication of Autographa californica M nucleopolyhedrovirus genomic DNA. Arch Virol 1997;142:2107–2113 [DOI] [PubMed] [Google Scholar]
- 33.Rohrmann GF. Chapter 5 DNA replication and genome processing. In: Baculovirus Molecular Biology, 3rd edition [Internet]. (National Center for Biotechnology Information, Bethesda MD: ). 2013. Available at www.ncbi.nlm.nih.gov/books/NBK138300/ [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.