Abstract
Cell adhesion molecules (CAMs) are involved in various immune-mediated diseases. This study was conducted to investigate the association of single nucleotide polymorphisms (SNPs) of CAMs with Behçet’s disease (BD) in a Chinese Han population. A two-stage association study was carried out in 1149 BD patients and 2107 normal controls. Genotyping of 43 SNPs was performed using MassARRAY System (Sequenom), polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and TaqMan SNP assays. The expression of CD6 and CD11c was examined by real-time PCR and cytokine production was measured by ELISA. A significantly higher frequency of the CT genotype, and a lower frequency of the CC genotype and C allele of CD6 rs11230563 were observed in BD as compared with controls. Analysis of CD11c rs2929 showed that patients with BD had a significantly higher frequency of the GG genotype and G allele, and a lower frequency of the AG genotype as compared with controls. Functional experiments showed an increased CD11c expression and increased production of TNF-α and IL-1beta by LPS stimulated PBMCs in GG carriers of CD11c rs2929 compared to AA/AG carriers. Our study provides evidence that CD6 and CD11c are involved in the susceptibility to BD in a Chinese Han population.
Uveitis is a vision-threatening intraocular inflammatory disease and a major cause of visual impairment and blindness. Behçet’s disease (BD) has been shown as the most common sight threatening uveitis entity in China1. BD is a recurrent systemic inflammatory disease characterized by major symptoms such as orogenital ulcers, skin lesions, and intraocular inflammation2. As yet, the pathogenesis of BD remains unclear. BD patients are currently treated with various immunosuppressive agents, but unraveling of the inflammatory pathways could lead to a tailored specific therapeutic approach that may prevent the blinding complications of the disease.
Migration of cells to the site of inflammation is a key event during uveitis and has been investigated previously in both animal models3 and in clinical uveitis4. Most of these studies have examined the role of CAMs protein expression in BD5. A thorough immunogenetic approach of CAMs in this disease has not yet been addressed and was therefore the subject of the study presented here.
Cell adhesion molecules (CAMs) are a group of proteins involved in cell binding or interaction between extracellular matrix (ECM) and cells. CAMs have been classified into four protein families: Ig (immunoglobulin) superfamily (IgSF, CAMs), the integrins (ITGA, IGTB), the cadherins (CDH), the selectins, and other uncategorized molecules. At an early stage of inflammation, leukocytes first adhere to the activated endothelium, and then infiltrate into the vessel wall, in association with an increasing expression of CAMs6. Various studies have focused on protein expression of CAMs in patients with inflammatory and autoimmune diseases7. Blocking ICAM-1 has been shown to significantly weaken the migration of Th1- and Th17-polarized cells8.
An abundance of gene association studies with CAMs, including ICAM19,10,11, ICAM311, ICAM512, ITGAV13,14,15, ITGB16, LAMB117, ALCAM18, CDH119,20, CDH2321, CDHR322, ITGAM23, selectins24,25,26, CD627,28, CD11a29, CD11c29, CD1829, CD2830, CD4431, CD4828, CD5811,30, CD8031,32,33, CD8611, CD10334, and CD22628,35 have recently been reported for several inflammatory or autoimmune diseases. Although some studies have addressed the association of selected CAMs with uveitis10,29, no reports are available concerning the association between genetic polymorphisms of the complete family of cell adhesion molecules with ocular BD. In this study we therefore investigated whether genetic variants of cell adhesion molecules may confer susceptibility to BD in a Chinese Han population. We identified two genetic loci, rs11230563 in CD6 and rs2929 in CD11c, that contribute to the risk of BD.
Results
Clinical characteristics of patients with BD
The clinical characteristics, gender and age of the enrolled BD patients and controls are displayed in Table 1. The genotype frequencies of the 43 SNPs did not deviate from the Hardy–Weinberg equilibrium in the controls.
Table 1. Clinical characteristics, gender and age of BD patients with uveitis.
| Clinical features | Total | % |
|---|---|---|
| Patients with BD | ||
| Mean age ± SD | 34.0 ± 9.1 | |
| Male | 952 | 82.9 |
| Female | 197 | 17.1 |
| Uveitis | 1149 | 100 |
| Oral ulcer | 1067 | 92.9 |
| Genital ulcer | 629 | 54.7 |
| Skin lesion | 846 | 73.6 |
| Arthritis | 205 | 17.8 |
| Pathergy reaction | 234 | 20.4 |
| Controls | ||
| Mean age ± SD | 39.6 ± 11.7 | |
| Male | 1156 | 54.9 |
| Female | 951 | 45.1 |
Allele and genotype frequencies of tested SNPs in patients and controls in the first stage study
A total of 391 BD patients and 603 healthy controls were enrolled in the first stage study. An increased frequency of the rs2929 GG genotype in CD11c was observed in patients with BD (Pc = 0.034, OR = 1.69) (Table 2). The frequency of the AG genotype of rs2929 in patients with BD was significantly lower than that in normal controls (Pc = 0.019, OR = 0.56) (Table 2). In the case of rs11230563 in CD6, an increased frequency of the CT genotype was observed in BD patients (Pc = 8.624 × 10−4, OR = 1.94), whereas a decreased frequency of the C allele and CC genotype (Pc = 1.371 × 10−3, OR = 0.59; Pc = 7.380 × 10−4, OR = 0.52, respectively) was found (Table 2). We were not able to detect a significant association between the other 41 SNPs investigated and risk of acquiring BD (Supplementary Table S1).
Table 2. Association of two SNPs with Behçet’s Disease.
| Gene | SNP | Stage | Genotype/Allele | BD |
Controls |
P Value | Pc Value | OR (95% CI) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | |||||||
| ITGAX (CD11c) | rs2929 | First | AA | 15 | 3.8 | 23 | 3.8 | 0.986 | NS | 1.01 (0.52–1.95) |
| AG | 82 | 21.0 | 193 | 32.0 | 1.453 × 10−4 | 0.019 | 0.56 (0.42–0.76) | |||
| GG | 294 | 75.2 | 387 | 64.2 | 2.606 × 10−4 | 0.034 | 1.69 (1.27–2.25) | |||
| G | 670 | 85.7 | 967 | 80.2 | 0.002 | NS | 1.48 (1.16–1.89) | |||
| Second | AA | 12 | 1.6 | 40 | 2.7 | 0.107 | NS | 0.59 (0.31–1.13) | ||
| AG | 180 | 23.7 | 470 | 31.3 | 1.974 × 10−4 | 1.184 × 10−3 | 0.69 (0.56–0.84) | |||
| GG | 566 | 74.7 | 994 | 66.1 | 3.135 × 10−5 | 1.881 × 10−4 | 1.51 (1.24–1.84) | |||
| G | 1312 | 86.5 | 2458 | 81.7 | 3.904 × 10−5 | 7.808 × 10−5 | 1.44 (1.21–1.71) | |||
| Combined | AA | 27 | 2.3 | 63 | 3.0 | 0.287 | NS | 0.78 (0.50–1.23) | ||
| AG | 262 | 22.8 | 663 | 31.5 | 1.618 × 10−7 | 2.087 × 10−5 | 0.64 (0.55–0.76) | |||
| GG | 860 | 74.8 | 1381 | 65.5 | 4.320 × 10−8 | 5.573 × 10−6 | 1.56 (1.33–1.84) | |||
| G | 1982 | 86.2 | 3425 | 81.3 | 3.252 × 10−7 | 1.398 × 10−5 | 1.45 (1.25–1.67) | |||
| CD6 | rs11230563 | First | CC | 252 | 64.5 | 468 | 77.6 | 5.721 × 10−6 | 7.380 × 10−4 | 0.52 (0.39–0.69) |
| CT | 128 | 32.7 | 121 | 20.1 | 6.685 × 10−6 | 8.624 × 10−4 | 1.94 (1.45–2.59) | |||
| TT | 11 | 2.8 | 14 | 2.3 | 0.629 | NS | 1.22 (0.55–2.71) | |||
| C | 632 | 80.8 | 1057 | 87.6 | 3.189 × 10−5 | 1.371 × 10−3 | 0.59 (0.46–0.76) | |||
| Second | CC | 501 | 66.1 | 1137 | 75.6 | 1.811 × 10−6 | 1.087 × 10−5 | 0.63 (0.52–0.76) | ||
| CT | 238 | 31.4 | 349 | 23.2 | 2.714 × 10−5 | 1.628 × 10−4 | 1.52 (1.25–1.84) | |||
| TT | 19 | 2.5 | 18 | 1.2 | 0.020 | NS | 2.12 (1.11–4.07) | |||
| C | 1240 | 81.8 | 2623 | 87.2 | 1.176 × 10−6 | 2.352 × 10−5 | 0.66 (0.56–0.78) | |||
| combined | CC | 753 | 65.5 | 1605 | 76.2 | 8.501 × 10−11 | 1.097 × 10−8 | 0.60 (0.51–0.70) | ||
| CT | 366 | 31.9 | 470 | 22.3 | 2.532 × 10−9 | 3.266 × 10−7 | 1.63 (1.39–1.91) | |||
| TT | 30 | 2.6 | 32 | 1.5 | 0.029 | NS | 1.74 (1.05–2.88) | |||
| C | 1872 | 81.5 | 3680 | 87.3 | 1.766 × 10−10 | 7.594 × 10−9 | 0.64 (0.56–0.73) | |||
Pc, Bonferroni corrected p value; NS, not significant; SNP, single nucleotide polymorphism.
Allele and genotype frequencies of tested SNPs in patients and controls in the second stage study and combined study
To further verify the observed association of CD6 and CD11c with BD, we replicated the associated SNPs rs2929 and rs11230563 using a different cohort of patients that included 758 cases and 1504 controls. The results showed that the frequencies of the rs2929/CD11c GG genotype and G allele were significantly higher in BD patients (Pc = 1.881 × 10−4, OR = 1.51; Pc = 7.808 × 10−5, OR = 1.44, respectively), and lower frequencies of the AG genotype (Pc = 1.184 × 10−3, OR = 0.69) when compared with controls (Table 2). As for rs11230563/CD6, decreased frequencies of the C allele and CC genotype (Pc = 2.352 × 10−5, OR = 0.66; Pc = 1.087 × 10−5, OR = 0.63, respectively) were found, whereas an increased frequency of the CT genotype was observed (Pc = 1.628 × 10−4, OR = 1.52) (Table 2). Combining the data from the first and second stage study showed that rs2929 in CD11c was associated with the susceptibility to BD (GG genotype: Pc = 5.573 × 10−6, OR = 1.56; AG genotype: Pc = 2.087 × 10−5, OR = 0.64; G allele: Pc = 1.398 × 10−5, OR = 1.45) (Table 2), and that rs11230563 in CD6 also conferred susceptibility to BD (CC genotype: Pc = 1.097 × 10−8, OR = 0.60; CT genotype: Pc = 3.266 × 10−7, OR = 1.63; C allele: Pc = 7.594 × 10−9, OR = 0.64) (Table 2).
Because the gender distribution of patient and control groups was different, we also calculated genotype and allele distribution of rs11230563 and rs2929 according to gender in controls and patients separately. The result showed that the genotype and allele frequencies of rs2929 showed significant differences between patients and controls in both male (G allele: Pc = 5.034 × 10−4, OR = 1.37; AG genotype: Pc = 1.289 × 10−3, OR = 0.69; GG genotype: Pc = 6.102 × 10−4, OR = 1.46) and female patients (G allele: Pc = 0.002, OR = 1.69; AG genotype: Pc = 0.012, OR = 0.57; GG genotype: Pc = 0.006, OR = 1.84). Likewise, the genotype and allele frequencies of rs11230563 also showed significant differences between patients and controls in both male (C allele: Pc = 2.290 × 10−7, OR = 0.64; CT genotype: Pc = 1.886 × 10−6, OR = 1.66; CC genotype: Pc = 2.480 × 10−7, OR = 0.59) and female patients (C allele: Pc = 0.002, OR = 0.63; CC genotype: Pc = 0.012, OR = 0.60) (Table 3). We subsequently explored whether the association with rs2929 and rs11230563 behaved as dominant or recessive using univariate and multivariate logistic regression analysis. The CD11c-rs2929 A allele association with BD behaved as a dominant model (Table 4). The CD6-rs11230563 T allele association with BD behaved as both dominant and recessive (Table 5).
Table 3. The distribution of two SNPs in patients with BD and healthy controls by gender basis.
| SNP | Allele/Genotype | Male |
P Value | Pc Value | OR (95% CI) | Female |
P Value | Pc Value | OR (95% CI) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| BD (n = 952) | Controls (n = 1156) | BD (n = 197) | Controls (n = 951) | ||||||||
| rs2929 | AA | 24 | 36 | NS | NS | 0.81 (0.48–1.36) | 3 | 27 | NS | NS | 0.53 (0.16–1.76) |
| AG | 219 | 349 | 2.149 × 10−4 | 1.289 × 10−3 | 0.69 (0.57–0.84) | 43 | 314 | 0.002 | 0.012 | 0.57 (0.39–0.82) | |
| GG | 709 | 771 | 1.017 × 10−4 | 6.102 × 10−4 | 1.46 (1.21–1.76) | 151 | 610 | 0.001 | 0.006 | 1.84 (1.29–2.62) | |
| G | 1637 | 1891 | 2.517 × 10−4 | 5.034 × 10−4 | 1.37 (1.16–1.61) | 345 | 1534 | 0.001 | 0.002 | 1.69 (1.23–2.33) | |
| rs11230563 | CC | 624 | 883 | 4.133 × 10−8 | 2.480 × 10−7 | 0.59 (0.49–0.71) | 129 | 722 | 0.002 | 0.012 | 0.60 (0.43–0.84) |
| CT | 305 | 256 | 3.144 × 10−7 | 1.886 × 10−6 | 1.66 (1.36–2.01) | 61 | 214 | 0.011 | NS | 1.55 (1.10–2.17) | |
| TT | 23 | 17 | NS | NS | 1.66 (0.88–3.12) | 7 | 15 | NS | NS | 2.30 (0.93–5.72) | |
| C | 1553 | 2022 | 1.145 × 10−7 | 2.290 × 10−7 | 0.64 (0.54–0.75) | 319 | 1658 | 0.001 | 0.002 | 0.63 (0.47–0.83) | |
NS, not significant; SNP, single nucleotide polymorphism; BD: Behçet’s Disease.
Table 4. Logistic regression analysis of the risk of BD patients with CD11c/rs2929 in additive co-dominant, dominant and recessive models.
| Model | Genotype | Control (N = 2107) | Case (N = 1149) | Univariate logistic regression |
Multivariate logistic regressiona |
||
|---|---|---|---|---|---|---|---|
| OR(95% CI) | Pb | OR(95% CI) | Pb | ||||
| Additive | 0.6892 (0.5973,0.7951) | <0.0001 | 0.7052 (0.6055,0.8213) | <0.0001 | |||
| Co-dominant | GG | 1381 (65.54%) | 860 (74.85%) | Ref. | Ref. | ||
| AG | 663 (31.47%) | 262 (22.80%) | 0.6346 (0.5373,0.7495) | <0.0001 | 0.6436 (0.5389,0.7688) | <0.0001 | |
| AA | 63 (2.99%) | 27 (2.35%) | 0.6882 (0.4350,1.0889) | 0.1104 | 0.7455 (0.4566,1.2172) | 0.2403 | |
| Dominant | GG | 1381 (65.54%) | 860 (74.85%) | Ref. | Ref. | ||
| AG+AA | 726 (34.46%) | 289 (25.15%) | 0.6393 (0.5444,0.7507) | <0.0001 | 0.6521 (0.5493,0.7742) | <0.0001 | |
| Recessive | GG+AG | 2044 (97.01%) | 1122 (97.65%) | Ref. | Ref. | ||
| AA | 63 (2.99%) | 27 (2.35%) | 0.7808 (0.4945,1.2327) | 0.2882 | 0.8422 (0.5169,1.3721) | 0.4903 | |
95% CI 95% confidence interval, OR odds ratio.
aThe age, sex were adjusted in the multivariate logistic regression model;
bThe hypothesis test was performed using Wald χ2 test.
Table 5. Logistic regression analysis of the risk of BD patients with CD6/rs11230563 in additive co-dominant, dominant and recessive models.
| Model | Genotype | Control (N = 2107) | Case (N = 1149) | Univariate logistic regression |
Multivariate logistic regressiona |
||
|---|---|---|---|---|---|---|---|
| OR(95% CI) | Pb | OR(95% CI) | Pb | ||||
| Additive | 1.5890 (1.3787,1.8313) | <0.0001 | 1.6129 (1.3839,1.8797) | <0.0001 | |||
| Co-dominant | CC | 1605 (76.17%) | 753 (65.54%) | Ref. | Ref. | ||
| CT | 470 (22.31%) | 366 (31.85%) | 1.6598 (1.4120,1.9513) | <0.0001 | 1.6494 (1.3855,1.9635) | <0.0001 | |
| TT | 32 (1.52%) | 30 (2.61%) | 1.9983 (1.2053,3.3130) | 0.0073 | 2.3023 (1.3293,3.9877) | 0.0029 | |
| Dominant | CC | 1605 (76.17%) | 753 (65.54%) | Ref. | Ref. | ||
| CT+TT | 502 (23.83%) | 396 (34.46%) | 1.6814 (1.4362,1.9685) | <0.0001 | 1.6874 (1.4237,2.0000) | <0.0001 | |
| Recessive | CC+CT | 2075 (98.48%) | 1119 (97.39%) | Ref. | Ref. | ||
| TT | 32 (1.52%) | 30 (2.61%) | 1.7385 (1.0509,2.8759) | 0.0313 | 2.0056 (1.1607,3.4656) | 0.0126 | |
95% CI 95% confidence interval, OR odds ratio.
aThe age, sex were adjusted in the multivariate logistic regression model;
bThe hypothesis test was performed using Wald χ2 test.
Stratified analysis of rs2929 and rs11230563 with main clinical features of Behcet’s disease
A stratified analysis was carried out to examine the association of rs2929 and rs11230563 with the main clinical features of BD. The main clinical features of BD included genital ulcer, skin lesions, arthritis, positive pathergy reaction and hypopyon. No significant association was found for the individual extraocular manifestations of BD with the tested SNPs (Supplementary Tables S2 and S3).
The influence of rs11230563 on CD6 and rs2929 on CD11c expression
The aforementioned results revealed that genetic polymorphisms of CD6 and CD11c are associated with susceptibility to BD. We subsequently investigated mRNA expression of CD6 and CD11c in PBMCs from 32 healthy individuals with a known genotype. No significant association was found in CD11c expression between the various genotypes of rs2929 when PBMCs were left unstimulated (Fig. 1). However, following stimulation with LPS, the mRNA expression of CD11c rs2929 in GG cases was significantly increased as compared to AA/AG carriers (P = 1.692 × 10−3) (Fig. 1). Different genotypes of rs11230563 did not affect CD6 mRNA expression levels regardless whether PBMCs had been stimulated with LPS or not (Supplementary Figure 1).
Figure 1. The influence of various rs2929 genotypes on the expression of CD11c.
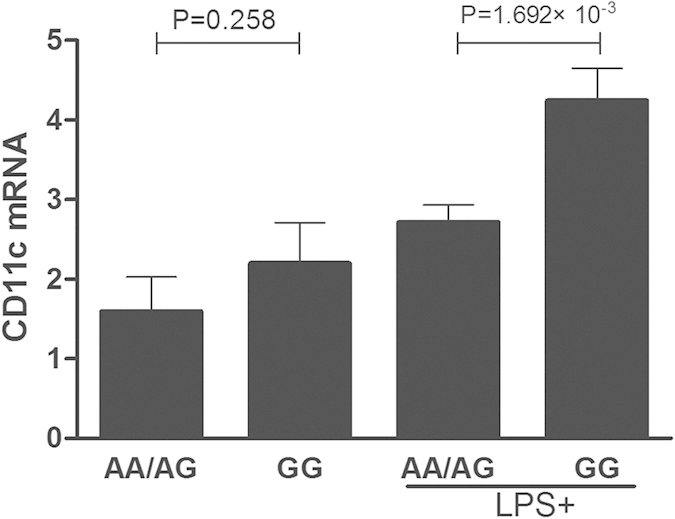
CD11c expression in non-stimulated PBMCs and LPS stimulated PBMCs from normal controls carrying different genotypes of rs2929 (AA/AG = 16, GG = 16).
The influence of rs2929 on cytokine production
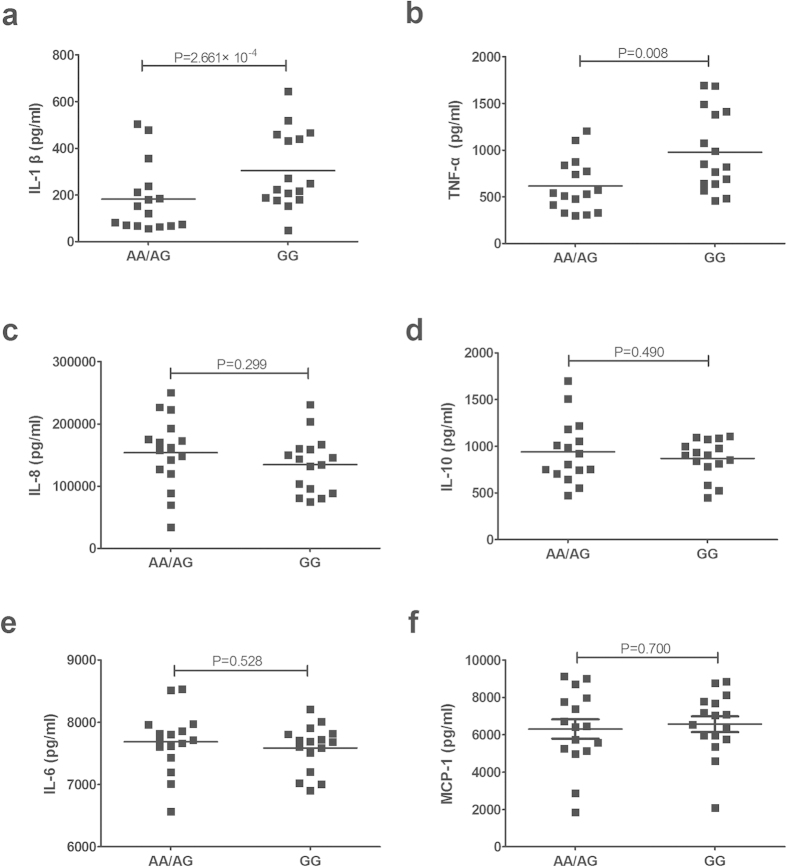
As we found that different genotypes of rs2929 could affect CD11c expression following LPS stimulation, further functional experiments were performed to investigate whether different genotypes of rs2929 could also affect the cytokine production of LPS stimulated PBMCs. This was performed using cells obtained from 32 healthy genotyped individuals. We chose to study IL-1beta, IL-6, IL-8, IL-10, TNF-α and MCP-1, cytokines that have been shown to play a crucial role in the pathogenesis of BD. IL-1beta production by stimulated PBMCs from GG carriers was higher than that seen in AA/AG (P = 2.661 × 10−4) carriers (Fig. 2a). Similarly, an increased TNF-α production by stimulated PBMCs was also observed in GG compared to AA/AG (P = 0.008) carriers (Fig. 2b). No effect of the various rs2929 genotypes on the release of the other four cytokines could be detected (Fig. 2c–f).
Figure 2. The influence of rs2929 on cytokine production.
The production of IL-1beta (a), TNF-a (b), IL-8 (c), IL-10 (d), IL-6 (e) and MCP-1 (f) by LPS stimulated PBMCs from normal controls carrying different genotypes of rs2929 (AA/AG = 16, GG = 16).
Discussion
The role of genetic polymorphisms in genes coding for cell adhesion molecules in the risk of developing BD was investigated in this study and showed that rs2929 of CD11c and rs11230563 of CD6 were significantly associated with BD. Using functional studies we found that the relative mRNA expression levels of CD11c were increased in individuals with the GG genotype of rs2929. Additionally, IL-1beta and TNF-α production by PBMCs were significantly increased in individuals with the GG genotype of rs2929 as compared with the other two genotypes.
We were not able to find evidence for an association between genetic variants of ICAM1, CD11a, CD18 and BD susceptibility. Earlier studies performed by other groups demonstrated a significant association of rs5498, rs11574944, rs2230429, rs235326 with BD risk10,29. The lack of association in our study may be due to geographical, ethnical or clinical differences.
Although earlier studies showed evidence of an association with SNPs of CD11a, CD11c, CD18 in BD patients from Korea29 and with SNPs of ICAM1 in BD patients from Tunisia10, the genetic polymorphisms of cell adhesion molecules have not yet been reported for large cohorts of BD patients in Chinese Han. Our data are similar to the Korean study29 that showed that the frequencies of the CD11c rs2929 GG genotype and G allele were significantly higher in BD patients than in controls. The odds ratio for the G allele association with BD was 1.4 in the Korean study and in our study it was 1.46. The G allele of rs2929 is a common variant in both Chinese Han and in Koreans with a frequency of 86% and 67%, respectively. In the Korean study, the patients were recruited at a department of dermatology and 61% of patients exhibited ocular symptoms. The fact that a similar association was seen with rs2929 whether patients had uveitis or not suggests that the association is not confined to those with ocular disease, which is also in agreement with our findings showing no difference between our different BD subgroups. On the other hand, the Korean study did note that the CD11c GG genotype of rs2929 was significantly more frequent in the patients with arthritis than in those without (66.3% versus 49.1%) as well as in patients with neurologic involvement compared to those without (10.0% versus 55.9%). We were not able to confirm the association found in Korean BD patients for the C allele of rs2230429 in the CD11c (OR = 1.7), which may be due to a difference in the patient population studied. An association of the CD11c rs2929 A allele with gastric ulcers has been reported for Caucasians36 whereas earlier studies from this group could not provide evidence for an association with inflammatory bowel disease37. Why gastric ulcers are associated with the A allele and BD is associated with the C allele is not clear.
The CD11 gene cluster maps on chromosome 16p11–12 and includes the integrin alpha L (CD11a), integrin alpha M (CD11b), integrin alpha X (CD11c), and integrin alpha D (CD11d) chain36. These integrin alpha chains form a heterodimeric molecule with the same integrin beta chain (CD18) and constitute a family of integral membrane glycoproteins expressed on leukocytes that play an essential role in the migration of white blood cells to the site of inflammation. Our observation that rs2929 G allele carriers show an enhanced expression of these adhesion molecules is in line with the observation that BD is a disease with an exaggerated neutrophil response leading to a vasculitis in many organs and tissues38.
The association we described between the CD6 SNP rs11230563 has not yet been reported earlier in BD, although a haplotype of CD6 containing rs11230563 and rs2074225 was shown to be associated with MS39. A recent study found that the rs12360861 in CD6 was associated with MS, although functional studies were not able to show that these CD6 SNPs affected its mRNA levels40. Studies in Caucasians demonstrated a significant association of the G allele of rs17824933 in CD6 with MS risk12, although this could not be confirmed in Korean patients24. The MS risk allele in the rs17824933 CD6 locus may result in an alternative splicing of the gene41 and this locus has been found to affect proliferation of CD4 (+) T cells42.
CD6 is a membrane glycoprotein, which is mainly expressed on T cells. It interacts with its ligand activated leukocyte cell adhesion molecule (ALCAM/CD166), which is expressed on many cell types including nervous system cells, epithelial cells, mesenchymal cells, endothelial cells and leukocytes43. The CD6 ALCAM interaction plays an important role in the immune response44 and has been implicated in autoimmune diseases such as multiple sclerosis27, rheumatoid arthritis45 and psoriasis46. A humanized monoclonal antibody directed against CD6, named Itoluzimab has recently been introduced and several clinical trials have now published preliminary data in several autoimmune diseases such as psoriasis47 and rheumatoid arthritis45.
The role of CD6 and ALCAM in ocular inflammatory disease has not received much attention until now, although expression of ALCAM has been described for retinal vascular endothelial cells48. ALCAM expression on the retinal vascular endothelium is probably involved in neovascularization and not directly on the transmigration of inflammatory T cells into the retina8. Our finding concerning an association of CD6 polymorphisms with BD may therefore be related to an effect of CD6 variants on T cell activation rather than on the egress of these cells into the retina. We could not find an effect of the rs11230563 CD6 on the mRNA expression by PBMCs, but as mentioned above, it might directly influence CD6 function39. Further experiments are needed to unravel the functional role of CD6 variants in BD pathogenesis.
It is worthwhile to point out that the odds ratios were toward different directions between the homozygotes and heterozygotes. The genotype data may be due to the dominant behavior of the A allele of rs2929 and T allele of rs11230563. In view of the relatively low frequency of the AA genotype (rs2929) and the TT genotype (rs11230563) both in patients and controls, we did not find a protective effect of the AA genotype of rs2929 (Pc > 0.05, OR = 0.78) or a positive predisposing effect of the TT genotype of rs11230563 (Pc > 0.05, OR = 1.74).
The present study has some limitations that should be noted. Firstly, though we tried to match the controls for gender, 82.9 percent of our BD cases were male, while 54.9 percent of controls were male. Analysis of our data according to gender however showed that the association between rs11230563 and rs2929 and BD was seen in both the female and male population. Secondly, our study identified rs2929 of CD11c and rs11230563 of CD6 as potential risk factors in the susceptibility for BD in a Chinese Han population, but the exact mechanism whereby these variants affect the disease pathogenesis were not clarified and deserves further study. Functional experiments showed that the CD11c risk variant influenced CD11c expression and the production of several pro-inflammatory cytokines. We only examined the effect of rs2929 in healthy genotyped controls because the BD patient population is extremely heterogeneous due to a variable inflammatory course and due to the immunosuppressive drug treatment used. Thirdly, we only chose previously reported loci in the family of adhesion molecules, which were known to be associated with autoimmune diseases and it cannot be ruled out that other SNPs in cell adhesion molecules can be associated with BD. A detailed sequence analysis of the identified risk genes should be performed to investigate whether rare variants of these factors might also be involved, thus strengthening the functional role of these factors in BD development. Last but not the least, since our BD patients were recruited from an ophthalmology department, further confirmations should be done by investigating BD patients originating from other medical fields such as the rheumatology or dermatology departments.
In summary, our study showed that CD6 rs11230563 and CD11c rs2929 polymorphisms are associated with susceptibility to BD in a Chinese Han population. Further studies are needed to reveal the mechanisms whereby CD6 and CD11c expression and function are regulated in BD and whether this knowledge can be used to develop novel therapeutic approaches.
Methods
Participants
From April 2008 to October 2015, 1149 BD patients, visiting the First Affiliated Hospital of Chongqing Medical University (Chongqing, China), were selected as the study population. BD patients were strictly diagnosed based upon the criteria of the International Study Group for BD49. A group of 2107 healthy individuals, who matched ethnically (Han Chinese) and geographically with the patients, served as the control group. A two-stage case-control association study was carried out. The first-stage study cohort consisted of 391 BD patients and 603 normal controls. In the second stage, a different set of 758 BD patients and 1504 normal controls were enrolled. The study received the approval of the First Affiliated Hospital of Chongqing Medical University Ethics Research Committee and all the investigated subjects provided informed consent before collection of blood. The tenets of the Declaration of Helsinki (2013) were adhered to during all procedures of this study. All methods were carried out in accordance with the approved guidelines.
Single nucleotide polymorphisms selection
Single nucleotide polymorphism (SNP) selection was based on previous studies on the association between cell adhesion molecules and inflammatory and autoimmune disease9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35. Linkage disequilibrium (LD) and Minor Allele Frequency (MAF) were analyzed by Haploview 4.2 software. We eliminated several SNPs which were not polymorphic in the Chinese population and finally selected 43 SNPs (the MAF at each locus was required to be >0.05 in Han Chinese in Beijing, with an r2-value of LD <0.8 between adjacent markers) in 26 molecules. The SNPs tested in this study included 4 SNPs (rs281432, rs5498, rs3093030, rs281437) of ICAM19,10,12, 1 SNP (rs2278442) of ICAM311, 1 SNP (rs2228615) of ICAM512, 3 SNPs (rs3738919, rs3768777, rs3911238) of ITGAV13,14,15, 1 SNP (rs3809865) of ITGB316, 1 SNP (rs886774) of LAMB117;1 SNP (rs6437585) of ALCAM18, 5 SNPs (rs1777241, rs1078621, rs7203337, rs10431923, rs1728785) of CDH119,20, 1 SNP (rs1417210) of CDH2321, 1 SNP (rs6967330) of CDHR322, 1 SNP (rs11150610) of ITGAM23, 1 SNP (rs10800469) of E-selectin24, 1 SNP (rs2205849) of L-selectin25, 1 SNP (rs3917657) of P-selection26, 2 SNPs (rs12288280, rs11230563) of CD627,28, 1 SNP (rs11574944) of CD11a29, 2 SNPs (rs2230429, rs2929) of CD11c29, 1 SNP (rs235326) of CD1829, 1 SNP (rs1980422) of CD2830, 2 SNPs (rs736374, rs10768122) of CD4431, 1 SNP (rs4656958) of CD4828, 2 SNPs (rs2300747, rs11586238) of CD5811,30, 4 SNPs (rs4688013, rs2222631, rs59374417, rs4330287) of CD8031,32,33, 1 SNP (rs4308217) of CD8611, 1 SNP (rs2891) of CD10334 and 2 SNPs (rs763361, rs727088) of CD22628,35.
DNA extraction and genotyping
Genomic DNA was extracted from venous blood samples of BD patients and healthy controls using the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, California, USA). All SNPs were assayed using a matrix-assisted laser desorption/ionization time of flight mass spectrometry platform (Sequenom, San Diego, CA) in the first stage, following the manufacturer’s instruction. Samples were genotyped by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) for rs11230563 in the second stage. Digestion products were separated on a 4% agarose gel and stained with GoldView TM (SBS Genetech, Beijing, China). Rs2929 (TagMan assay ID: C_9607211_10) genotypes were analyzed using the TaqMan® SNP Genotyping Assay (Applied Biosystems, Foster City, CA, USA) on the Applied Biosystems 7500 Real-Time PCR system in the second stage. The analysis was conducted using TaqMan® Genotyper Software. Five percent of study samples in a random fashion were sequenced directly to assure the validity of the SNP genotyping method used. The success rate of all SNPs genotyping ranged from 98.0% to 100%.
Cell isolation and culture
Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood samples of healthy controls by Ficoll-Hypaque density-gradient centrifugation. Isolated PBMCs (1 × 106 cells per well) were seeded in 24-well plates and cultured in RPMI medium 1640 supplemented with 10% fetal calf serum (FCS, Greiner, Wemmel, Belgium), 100 U/ml penicillin, 100ug/ml streptomycin. In order to detect the production of IL-1beta, IL-6, IL-8, IL-10, TNF-α and MCP-1, PBMCs were cultured with 100 ng/ml lipopolysaccharide (LPS, L2880, Sigma, Missouri, USA) for 24 hours.
Real-time polymerase chain reaction
Total RNA was extracted from unstimulated PBMCs and LPS stimulated PBMCs with TRIzol (Invitrogen, Carlsbad, CA) followed by reverse transcription using a transcriptase kit. The sequences of the sense and antisense primers were as follows: CD6: 5′-TGCACAATCTGTCCACTCCC-3′ and 5′-AACGATGGAGGGGATGAGGA-3′; CD11c: 5′-CCGACCATATCTGCCAGGAC-3′ and 5′-CCCGTCATTCCACACCATCA-3′. β-actin was selected as the internal reference gene and its expression was measured by the following primers: forward 5′-GGATGCAGAAGGAGATCACTG-3′ and reverse 5′-CGATCCACACGGAGTACTT-3′. The assays were performed on a 7500 real-time instrument (ABI). Relative expression levels were calculated by the 2−ΔΔCt method.
Enzyme linked immunosorbent Assay (ELISA)
The concentration of IL-1beta, IL-6, IL-8, IL-10, TNF-α and MCP-1 in PBMC culture supernatants was detected by the human Duoset ELISA development kit (R&D Systems, Minneapolis, Minnesota, USA) based upon the manufacturer’s protocols.
Statistical analysis
As to SNP analysis, Hardy-Weinberg equilibrium (HWE) was tested by the Chi-square test. Genotype frequencies were calculated by direct counting. Allele and genotype frequencies were compared between patients and controls by the chi-square test using SPSS version 19.0. P values were evaluated with the χ2 test or Fisher’s exact test. Genotype testing was performed by comparing one genotype versus the other two pooled together. P values were corrected (Pc) for multiple comparisons with the Bonferroni correction by multiplying with the number of analyses performed according to the methods of Jiang et al.50 and Fang et al.51, and Pc <0.05 was considered statistically significant. Risks were assessed by odds ratios (ORs) with 95% confidence intervals (CIs). To investigate whether associations could be elaborated by either dominant or recessive models, the data of rs2929 and rs11230563 genotype frequencies were analyzed by Univariate logistic regression and Multivariate logistic regression. The non-parametric Mann-Whitney test was used to compare CD6, CD11c expression and cytokine levels among three genotype groups.
Additional Information
How to cite this article: Zheng, M. et al. Genetic polymorphisms of cell adhesion molecules in Behcet's disease in a Chinese Han population. Sci. Rep. 6, 24974; doi: 10.1038/srep24974 (2016).
Supplementary Material
Acknowledgments
This work was supported by Natural Science Foundation Major International (Regional) Joint Research Project (81320108009), Key Project of Natural Science Foundation (81130019), National Natural Science Foundation Project (31370893), Basic Research program of Chongqing (cstc2013jcyjC10001), Chongqing Key Laboratory of Ophthalmology (CSTC, 2008CA5003), National Key Clinical Specialties Construction Program of China, Key Project of Health Bureau of Chongqing (2012-1-003), Chongqing Science & Technology Platform and Base Construction Program (cstc2014pt-sy10002). Fundamental and Advanced Research Program of Chongqing (cstc2015jcyjA10022), Science and Technology Project of Chongqing Municipal Education Commission (KJ1500236), Scientific Research Program of Science and Technology Commission of Yuzhong District of Chongqing (20150102).
Footnotes
Author Contributions P.Y. designed experiment; M.Z., L.Z. and H.Y. performed laboratory work and analyzed data; M.Z. and A.K. wrote the manuscript. J.H., Q.C., G.H., Y.H. and G.Y. collected blood of all patients and controls. All authors read and approved the final manuscript.
References
- Yang P. et al. Clinical patterns and characteristics of uveitis in a tertiary center for uveitis in China. Curr. Eye Res. 30, 943–948 (2005). [DOI] [PubMed] [Google Scholar]
- Pineton C. M., Wechsler B., Geri G., Cacoub P. & Saadoun D. New insights into the pathogenesis of Behçet’s disease. Autoimmun. Rev. 11, 687–698 (2012). [DOI] [PubMed] [Google Scholar]
- Xu H., Forrester J. V., Liversidge J. & Crane I. J. Leukocyte trafficking in experimental autoimmune uveitis: breakdown of blood-retinal barrier and upregulation ofcellular adhesion molecules. Invest. Ophthalmol. Vis. Sci. 44, 226–234 (2003). [DOI] [PubMed] [Google Scholar]
- La H. E. et al. Adhesion molecules in iris biopsy specimens from patients with uveitis. Br. J. Ophthalmol. 82, 432–437 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchio E., Matsumoto T., Tanaka S. I. & Ohno S. Soluble intercellular adhesion molecule-1 (ICAM-1), CD4, CD8 and interleukin-2 receptor in patients with Behçet’s disease and Vogt-Koyanagi-Harada’s disease. Clin. Exp. Rheumatol. 17, 179–184 (1999). [PubMed] [Google Scholar]
- Firestein G. S. Evolving concepts of rheumatoid arthritis. Nature 423, 356–361 (2003). [DOI] [PubMed] [Google Scholar]
- Sfikakis P. P. & Tsokos G. C. Clinical use of the measurement of soluble cell adhesion molecules in patients with autoimmune rheumatic diseases. Clin. Diagn. Lab Immunol. 4, 241–246 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj A. S., Schewitz-Bowers L. P., Wei L., Lee R. W. & Smith J. R. Intercellular adhesion molecule 1 mediates migration of Th1 and Th17 cells across human retinal vascular endothelium. Invest. Ophthalmol. Vis. Sci. 54, 6917–6925 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. et al. Genetic influences of the intercellular adhesion molecule 1 (ICAM-1) gene polymorphisms in development of Type 1 diabetes and diabetic nephropathy. Diabet. Med. 23, 1093–1099 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben D. I., Karray E. F., Sassi F. & Hamzaoui K. Intercellular adhesion molecule 1 K469E gene polymorphism is associated with presence of skin lesions in Tunisian Behçet’s disease patients. Tissue Antigens. 75, 74–78 (2010). [DOI] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. et al. Variation in the ICAM1-ICAM4-ICAM5 locus is associated with systemic lupus erythematosus susceptibility in multiple ancestries. Ann. Rheum. Dis. 71, 1809–1814 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis-Moffatt J. E. et al. The ITGAV rs3738919 variant and susceptibility to rheumatoid arthritis in four Caucasian sample sets. Arthritis Res. Ther. 11, R152 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koca S. S. et al. The rs3768777-G allele of ITGAV gene is associated with rheumatoid arthritis. Rheumatol. Int. 34, 693–698 (2014). [DOI] [PubMed] [Google Scholar]
- Shakiba E. et al. The ITGAV-rs3911238 polymorphism is associated with disease activity in rheumatoid arthritis. Iran J. Allergy Asthma Immunol. 13, 356–363 (2014). [PubMed] [Google Scholar]
- Zhang Y. et al. Genetic variation of ITGB3 is associated with asthma in Chinese Han children. Plos One 8, e56914 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- UK IBD Genetics Consortium et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat. Genet. 41, 1330–1334 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M. et al. ALCAM--novel multiple sclerosis locus interfering with HLA-DRB1*1501. J. Neuroimmunol. 258, 71–76 (2013). [DOI] [PubMed] [Google Scholar]
- Muise A. M. et al. Polymorphisms in E-cadherin (CDH1) result in a mis-localised cytoplasmic protein that is associated with Crohn’s disease. Gut. 58, 1121–1127 (2009). [DOI] [PubMed] [Google Scholar]
- Van S. S. et al. HNF4α and CDH1 are associated with ulcerative colitis in a Dutch cohort. Inflamm. Bowel Dis. 17, 1714–1718 (2011). [DOI] [PubMed] [Google Scholar]
- Tang X. F. et al. Association analyses identify three susceptibility Loci for vitiligo in the Chinese Han population. J. Invest. Dermatol. 133, 403–410 (2013). [DOI] [PubMed] [Google Scholar]
- Bochkov Y. A. et al. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc. Natl. Acad. Sci. USA 112, 5485–5490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos P. S. et al. A comprehensive analysis of shared loci between systemic lupus erythematosus (SLE) and sixteen autoimmune diseases reveals limited genetic overlap. Plos Genet. 7, e1002406 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. et al. The common variants of E-selectin gene in Graves’ disease. Genes Immun. 9, 182–186 (2008). [DOI] [PubMed] [Google Scholar]
- Alkhateeb A., Karasneh J., Abbadi H., Hassan A. & Thornhill M. Association of cell adhesion molecule gene polymorphisms with recurrent aphthous stomatitis. J. Oral Pathol. Med. 42, 741–746 (2013). [DOI] [PubMed] [Google Scholar]
- Morris D. L. et al. Variation in the upstream region of P-Selectin (SELP) is a risk factor for SLE. Genes Immun. 10, 404–413 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T. J. et al. Associations of CD6, TNFRSF1A and IRF8 polymorphisms with risk of inflammatory demyelinating diseases. Neuropathol. Appl. Neurobiol. 39, 519–530 (2013). [DOI] [PubMed] [Google Scholar]
- Jostins L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. R., Park K. S., Park Y. J., Bang D. & Lee E. S. CD11a, CD11c, and CD18 gene polymorphisms and susceptibility to Behçet’s disease in Koreans. Tissue Antigens. 84, 398–404 (2014). [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S. et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat. Genet. 41, 1313–1318 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y. et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat. Genet. 44, 676–680 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. Genome-wide search followed by replication reveals genetic interaction of CD80 and ALOX5AP associated with systemic lupus erythematosus in Asian populations. Ann. Rheum. Dis. Apr 10 (2015). [DOI] [PubMed] [Google Scholar]
- Hinks A. et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat. Genet. 45, 664–669 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heron M. et al. Effect of variation in ITGAE on risk of sarcoidosis, CD103 expression, and chest radiography. Clin. Immunol. 133, 117–125 (2009). [DOI] [PubMed] [Google Scholar]
- Hafler J. P. et al. CD226 Gly307Ser association with multiple autoimmune diseases. Genes Immun. 10, 5–10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmig S. et al. Haplotype analysis of the CD11 gene cluster in patients with chronic Helicobacter pylori infection and gastriculcer disease. Tissue. Antigens. 65, 271–274 (2005). [DOI] [PubMed] [Google Scholar]
- Frenzel H. et al. Mutation detection and physical mapping of the CD11 gene cluster in association with inflammatory bowel disease. Immunogenetics 53, 835–842 (2002). [DOI] [PubMed] [Google Scholar]
- Sahin S., Akoglu T., Direskeneli H., Sen L. S. & Lawrence R. Neutrophil adhesion to endothelial cells and factors affecting adhesion in patients with Behçet’s disease. Ann. Rheum. Dis. 55, 128–133 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan B. et al. Fine mapping and functional analysis of the multiple sclerosis risk gene CD6. Plos One 8, e62376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M. et al. ALCAM and CD6–multiple sclerosis risk factors. J. Neuroimmunol. 276, 98–103 (2014). [DOI] [PubMed] [Google Scholar]
- Gloria V. G. et al. T cell activation regulates CD6 alternative splicing by transcription dynamics and SRSF1. J Immunol. 193, 391–399 (2014). [DOI] [PubMed] [Google Scholar]
- Kofler D. M., Severson C. A., Mousissian N., De Jager P. L. & Hafler D. A. The CD6 multiple sclerosis susceptibility allele is associated with alterations in CD4+ T cell Proliferation. J. Immunol. 187, 3286–3291 (2011). [DOI] [PubMed] [Google Scholar]
- Swart G. W. Activated leukocyte cell adhesion molecule (CD166/ALCAM): developmental and mechanistic aspects of cell clustering and cell migration. Eur. J. Cell Biol. 81, 313–321 (2002). [DOI] [PubMed] [Google Scholar]
- Bowen M. A. et al. Cloning, mapping, and characterization of activated leukocyte-cell adhesion molecule (ALCAM), a CD6 ligand. J. Exp. Med. 181, 2213–2220 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez P. C. et al. A clinical exploratory study with itolizumab, an anti-CD6 monoclonal antibody, in patients with rheumatoid arthritis. Results Immunol. 2, 204–211 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero E. et al. CD6 molecule may be important in the pathological mechanisms of lymphocytes adhesion to human skin in psoriasis and ior t1 MAb a possible new approach to treat this disease. Autoimmunity 29, 155–156 (1999). [DOI] [PubMed] [Google Scholar]
- Aira L. E. et al. Immunological and histological evaluation of clinical samples from psoriasis patients treated with anti-CD6 itolizumab. MAbs. 6, 783–793 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. R., Chipps T. J., Ilias H., Pan Y. & Appukuttan B. Expression and regulation of activated leukocyte cell adhesion molecule in human retinal vascular endothelial cells. Exp. Eye Res. 104, 89–93 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disease ISGfBs. Criteria for diagnosis of Behcet’s disease. Lancet 335, 1078–1080 (1990). [PubMed] [Google Scholar]
- Jiang Z. et al. IL-23R gene confers susceptibility to Behcet’s disease in a Chinese Han population. Ann. Rheum. Dis. 69, 1325–1328 (2010). [DOI] [PubMed] [Google Scholar]
- Fang J. et al. Association of TLR2 gene polymorphisms with ocular Behcet’s disease in a Chinese Han population. Invest. Ophthalmol. Vis. Sci. 54, 8384–8392 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.