Abstract
Genotypic variability based genome-wide association studies (vGWASs) can identify potentially interacting loci without prior knowledge of the interacting factors. We report a two-stage approach to make vGWAS applicable to diseases: firstly using a mixed model approach to partition dichotomous phenotypes into additive risk and non-additive environmental residuals on the liability scale and secondly using the Levene’s (Brown-Forsythe) test to assess equality of the residual variances across genotype groups per marker. We found widespread significant (P < 2.5e-05) vGWAS signals within the major histocompatibility complex (MHC) across all three study cohorts of rheumatoid arthritis. We further identified 10 epistatic interactions between the vGWAS signals independent of the MHC additive effects, each with a weak effect but jointly explained 1.9% of phenotypic variance. PTPN22 was also identified in the discovery cohort but replicated in only one independent cohort. Combining the three cohorts boosted power of vGWAS and additionally identified TYK2 and ANKRD55. Both PTPN22 and TYK2 had evidence of interactions reported elsewhere. We conclude that vGWAS can help discover interacting loci for complex diseases but require large samples to find additional signals.
Genome-wide association studies (GWASs) have been extremely successful in highlighting loci associated with complex diseases such as rheumatoid arthritis (RA)1,2,3. While GWASs may continue to reveal novel loci based on the additive assumption, analyzing non-additive effects, such as gene-gene (GxG) and/or gene-environment (GxE) interactions, could provide novel insights into the underlying regulatory mechanisms1,4,5,6. Exploring GxG and/or GxE interactions in GWAS data has been attempted but, despite intensive efforts, the results so far have been relatively disappointing mainly due to low power of detection7,8,9. On the other hand, these results implicate that gene interactions could be prevalent but unlikely carry a big effect each and thus require a large sample to detect6,9. Currently available methods for detecting GxG interactions suffer from unmet computational and statistical challenges in supporting powerful meta-analysis of multiple cohorts9, and further potentially interacting environmental factors are not necessarily measured in study cohorts. Therefore, innovative approaches are needed to improve studying non-additive effects10.
One promising approach is genotypic variability based genome-wide association study (vGWAS) to search for loci with substantial genotypic variability as potential interaction signatures, i.e. differences of phenotypic variation across three SNP genotypes which tend to be small when only additive effects are important but large when non-additive effects such as interactions are important11,12. Such variability could be heritable to some extent and thus give rise to a new avenue of mapping genetic variants13,14. Hence, vGWAS has a great advantage in prioritizing potentially interacting loci without requiring prior knowledge of interaction types and interacting factors15,16. Additional explicit tests of GxE and/or GxG interactions are needed but only for the identified vGWAS loci, leading to a power increase attributed to a much reduced number of multiple tests17,18. Furthermore, studying biology of the identified vGWAS loci may inform what environmental factors should be further investigated. Therefore, vGWAS could be a cost effective approach to boost studies of GxG and GxE interactions based on existing GWAS data. However, vGWAS loci may not necessarily interact because other factors such as overdominance and scaling (i.e. various transformations) could also generate apparent genotypic variability19,20,21. Therefore, explicit tests of interactions and careful interpretation are essential to claim interacting signals.
There have been several successful applications of vGWAS in quantitative traits using either GWAS18,19,22 or gene expression data17,23,24. For example, Brown et al.17 reported abundant vGWAS signals within the major histocompatibility complex (MHC) region using RNA sequence data from lymphoblastoid cell lines derived from the TwinsUK cohort and subsequently confirmed GxE and GxG interactions underlying the signals. Yang et al.19 identified a well-known locus FTO in a vGWAS of body mass index in 170,000 samples from multiple GWAS cohorts, which is believed to interact with physical activity and/or lifestyle25. Intriguingly, the majority of the reported vGWAS loci are known major GWAS loci (i.e. with strong additive effects based on allelic means): FTO in body mass index19, LEPR in C-reactive protein levels and ICAM1 in soluble ICAM1 levels18, SLC2A9 in serum urate22, MHC in autoimmune diseases5. These observations led to a concern whether vGWAS might simply rediscover GWAS signals due to the mean-variance correlation, i.e. a distinct difference in allelic means likely accompanied by obvious genotypic variability in a locus19,20,21. Nevertheless, increasing evidence suggest that these major GWAS loci tend to have multiple roles and are indeed involved in interactions26,27,28,29, therefore it is biologically plausible to see them having signals in both GWAS and vGWAS analyses.
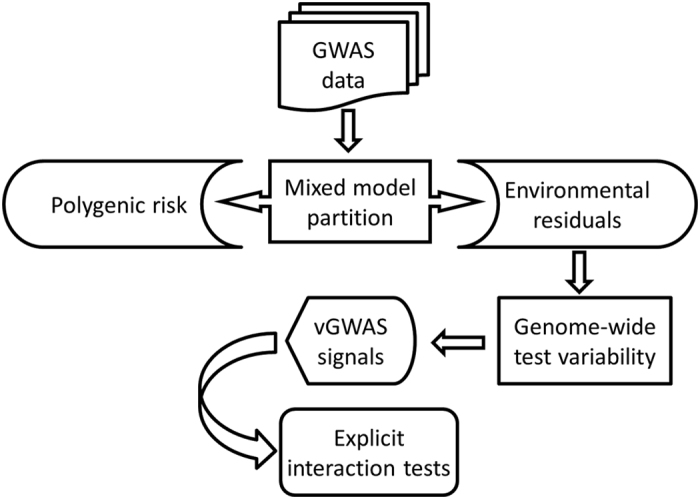
So far vGWAS has been little exploited in complex diseases probably because phenotypes classified either as a disease case or a healthy control provide little continuous variation. This problem can be remedied by applying a two-stage vGWAS approach: firstly using a mixed model method implemented in software such as GCTA30 to partition polygenic additive risk and non-additive environmental residuals for each sample on the liability scale31,32,33 and secondly using the Levene’s (Brown-Forsythe) test to assess equality of the residual variances across genotype groups per marker. It is therefore of interest to find out if vGWAS can also identify interacting loci in complex diseases. Here we report a pilot vGWAS of RA using a large GWAS cohort for discovery and two independent cohorts for replication. We aim to demonstrate that vGWAS is also useful to study disease traits.
Materials and Methods
Study cohorts and quality control (QC)
We used the Rheumatoid Arthritis Consortium International for Immunochip samples recruited in the UK (RACI-UK), US (RACI-US) and Sweden (RACI-SE, i.e. Swedish Epidemiological Investigation of Rheumatoid Arthritis) as described previously2. Each cohort was genotyped with the high density Immunochip34 in accordance with Illumina protocols and has been described in detail elsewhere2. Briefly, all RA patients fulfilled the 1987 criteria of the American College of Rheumatology and were tested for anti-citrullinated peptide antibody (ACPA) with a status of either seropositive (ACPA+), or seronegative (ACPA-) or unassigned. All participants provided written informed consent for participation. Further details of the sample collections in the three study cohorts can be found in the Supplementary Note by Eyre et al.2. This study was approved by the North West Research Ethics Committee (MREC 99/8/84). All experiments (e.g. genotyping) were performed in accordance with relevant guidelines and regulations.
We excluded SNPs on sex chromosomes and samples of non-European origin. RA cases that were either ACPA- or ACPA unassigned were also excluded in each cohort to avoid disease heterogeneity. A rigorous QC was conducted using PLINK35 for each cohort as described previously2 followed by additional QC based on the criteria suggested for accurate GCTA prediction of genetic risk31,32: minor allele frequency >0.01, SNP call rate >0.95, sample call rate >0.99, deviation from Hardy–Weinberg Equilibrium P < 1.0e-04.
Two-stage vGWAS and Statistical analysis
Figure 1 illustrates the workflow of the proposed two-stage vGWAS approach with an extension of explicit interaction tests. In this study GCTA was used to compute the genetic relationship matrix (GRM) and subsequently the first ten principal components (PCs) and then polygenic liability risk for each unrelated individual was predicted by imposing a GRM relatedness threshold of 0.15 recommended for Immunochip33 while setting the disease prevalence as 0.01 and fitting gender and the first 10 PCs as covariates in the mixed model. The resultant environmental residuals were tested for variance heterogeneity across three SNP genotypes using an R package VariABEL36 implemented the Levene’s (Brown-Forsythe) test that requires no assumption of normally distributed phenotypes:
Figure 1. Workflow of the two-stage vGWAS approach with an extension of explicit interaction tests.
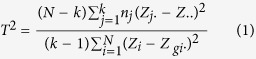 |
where the environmental residuals were the trait y; Zi = |yi - ỹgi| is the deviation of y of the ith sample (yi) and the median of y in samples with genotype g (ỹgi); N is the sample size and k is the total possible genotypes; nj is the number of samples with genotype j; Zj. is the mean deviation from the median for genotype j and Z. is the mean deviation from the overall median. When N is large, T2 is an approximate χ2 test taking two degrees of freedom.
To be extra cautious about the impact of normality, we also compared the vGWAS results based on the raw environmental residuals with those based on fully normalized environmental residuals using rank transformation available in the GenABEL package37. We found the normalization led to overly conservative vGWAS results in RACI-US and RACI-SE (Supplementary Figure S1) and thus reported only the results based on the raw environmental residuals.
We derived the significance threshold as 2.5e-05 based on permutation with 1000 iterations where a vGWAS was carried out using the randomly permutated environmental residuals and the lowest P value was recorded in each iteration (Supplementary Figure S2). We performed vGWAS in each of the three cohorts and used RACI-UK as the discovery cohort and examined any vGWAS significant SNPs for direct replication in RACI-US and RACI-SE, i.e. the same SNP with a vGWAS P value less than 5.0e-02.
For comparison we also performed a conventional GWAS for each cohort using PLINK and following the previous study2 that used a subset of independent common SNPs (i.e. minor allele frequency >0.05 and minimum inter-SNP linkage disequilibrium (LD)) to compute GRM and subsequently the first 10 PCs. A forward selection approach was applied to the genome-wide significant (P < 5.0e-08) GWAS SNPs identified in RACI-UK iteratively to identify a set of independent signals to represent the additive background. In each iteration the logistic regression model fitting the covariates and any pre-selected SNPs was used to test all remaining SNPs and select the most associated SNP for next round until none had a P value less than 5.0e-02.
We tested interactions between the identified vGWAS SNPs in RACI-UK as follows: a) PLINK was used to compute pairwise LD (in r2) for all the identified SNPs and then only pairs of SNPs in low LD (i.e. r2 < 0.1)9 were selected for interaction tests; b) the logistic regression model was used to test interactions for each of the k selected pairs of SNPs by fitting a pair of SNPs and their interaction terms as well as covariates of gender and the first 10 PCs derived from the GRM based on the subset of independent SNPs, with a significance of threshold defined as 0.05/(k*(k-1)/2); c) significant pairwise interactions were assessed while conditioning on the additive background of independent GWAS signals and unique pairs of interactions were identified using the forward selection approach above. Variance explained by the selected epistatic SNP pairs was calculated using a full logistic regression model fitting all the covariates, the additive background and the independent pairs of SNPs and their interactions. The identified epistatic SNP pairs were tested for replication individually in RACI-US and RACI-SE and the total variance explained was calculated following the approach above.
We further merged the three study cohorts into one and then performing the two-stage vGWAS as well as GWAS adjusted for cohort and the covariates described above to explore the impact of increased sample size. After the QC and GCTA analysis the combined cohort had 18,405 unrelated samples (5954 cases and 12,451 controls) and 107,144 SNPs in the vGWAS and GWAS. We quoted SNP genomic locations in the GRCh38/hg38 version throughout.
Results
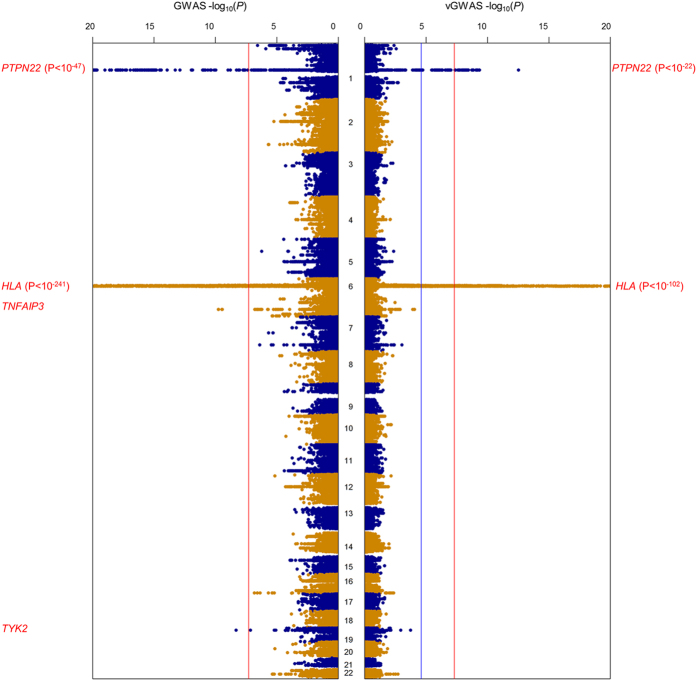
The vGWAS analysis identified 1639 significant SNPs (P < 2.5e-05) in the RACI-UK cohort where the Immunochip-wide heritability of ACPA+ RA was estimated as 24.7% (Table 1). About 92% of the identified vGWAS SNPs mapped to MHC and the remaining mapped to PTPN22 – both were major RA associated loci in the conventional GWAS that in addition identified TNFAIP3 and TYK2 at the genome-wide significance level (P < 5.0e-08) (Fig. 2). Forward selection of the significant GWAS SNPs led to an additive background composed of 20 independent SNPs (Supplementary Table S1). The quantile-quantile (QQ) plots of both the vGWAS (Supplementary Figure S1) and GWAS (Supplementary Figure S3) analyses in RACI-UK showed no sign of inflation.
Table 1. Study cohorts and vGWAS results*.
| cohort | SNPs | cases | controls | heritability (se) | vGWAS |
|---|---|---|---|---|---|
| RACI-UK | 124 363 | 2402 | 8411 | 0.247 (0.013) | 1639 |
| RACI-US | 118 550 | 1800 | 2125 | 0.246 (0.017) | 154 |
| RACI-SE | 123 146 | 1755 | 1931 | 0.185 (0.018) | 343 |
*only ACPA+ cases were included in each cohort; heritability (se): polygenic heritability estimate and standard error in brackets.
Figure 2. Aligned Manhattan plots of GWAS (left) and vGWAS (right) analyses in RACI-UK.
P values are at the -log10 scale; red line represents GWAS genome-wide significance threshold; blue line represents vGWAS significance threshold derived from permutation; significant loci are annotated to genes in red (or blue if reached only the vGWAS significance threshold).
In contrast, the vGWAS analyses detected only 154 SNPs in RACI-US (heritability estimated as 24.6%) and 343 in RACI-SE (heritability estimated as 18.5%) under the same significance threshold, suggesting the two independent cohorts were underpowered probably due to the relatively small samples used (Table 1). The QQ plots for RACI-US and RACI-SE indicated slight deflation in the vGWAS analyses (Supplementary Figure S1) but not in the GWAS analyses (Supplementary Figure S3). Nearly all the vGWAS SNPs identified in both RACI-US and RACI-SE also mapped to MHC, except for rs2322659 (P = 7.7e-06) mapped to LCT in RACI-US (Supplementary Figures S4 and S5).
We found that a quarter (i.e. 413 out of 1639) of the identified vGWAS SNPs in RACI-UK had direct replication in both the RACI-US and RACI-SE cohorts (Supplementary Table S2) and they were all within the MHC region spanning the Class III and Class II. In addition, the two top vGWAS SNPs rs2476601 (P = 6.7e-22) and rs6679677 (P = 1.4e-21) within PTPN22 in RACI-UK each had a direct replication only in RACI-SE with a P value of 3.3e-02 and 3.0e-02 respectively, suggesting genotypic variability within the locus was detectable only with a large sample size. The top vGWAS SNP rs2322659 within LCT in RACI-US had no replication in either RACI-UK or RACI-SE.
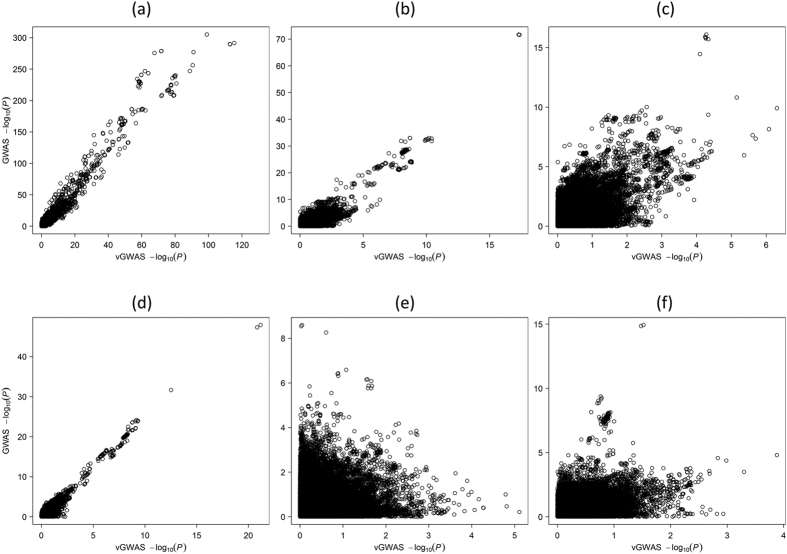
Interestingly, in RACI-UK the most strongly associated vGWAS SNP rs1964995 (intergenic between HLA-DRA and HLA-DRB5) was about 134 kb away from the most strongly associated GWAS SNP rs6931277 (near HLA-DRB1) (Fig. 3a), and the two SNPs were also the top vGWAS and GWAS signals respectively in both RACI-US and RACI-SE (Supplementary Table S2). These results (re)confirm that the MHC region harbors both strong additive and non-additive effects29,38 and suggest that the top non-additive sites could differ from the top additive loci. Nevertheless, the top GWAS SNP rs6931277 could also be involved in interactions as it had a strong vGWAS signal (i.e. P = 1.2e-96, 2.9e-07 and 7.1e-11 in RACI-UK, RACI-US and RACI-SE respectively).
Figure 3. Concordance plots of GWAS P values against vGWAS P values.
(a) the Combined cohort using all SNPs; (b) the Combined cohort excluding SNPs within the MHC region; (c) the Combined cohort excluding SNPs within the MHC and PTPN22 regions; (d) the RACI-UK cohort excluding SNPs within the MHC region; (e) the RACI-US cohort excluding SNPs within the MHC region; (f) the RACI-SE cohort excluding SNPs within the MHC region.
We tested interactions for 11,871 pairs of the RACI-UK significant vGWAS SNPs with r2 < 0.1. Based on a significance threshold of 4.2e-06 derived from Bonferroni correction, we found a large number of significant interactions all within MHC, of which 2,962 pairs of SNPs had a conditional interaction P value between 3.4e-07 and 5.0e-02 independent of the additive background. Applying forward selection to the 2,962 pairs and requiring a conditional interaction P < 1.0e-03 in each step, we identified 10 independent epistatic pairs of SNPs that spanned the entire MHC region and were replicated in both RACI-US and RACI-SE except for the rs9366778 - rs2239707 pair in RACI-US (Table 2, Supplementary Figure S6). In contrast to the additive background that explained 17.8% of the phenotypic variance in RACI-UK, the 10 epistatic pairs jointly explained about 2.5% of the phenotypic variance mainly (i.e. 1.9%) by interactions, suggesting the existence of widespread GxG interactions within MHC that are individually moderate/weak (on average explaining <0.2% of phenotypic variance) but jointly appreciable. The additive background and the 10 epistatic pairs explained 17.1% and 1.7% (0.8% by interactions) of the phenotypic variance in RACI-US, and 14.5% and 1.7% (0.7% by interactions) in RACI-SE.
Table 2. The ten independent epistatic pairs of SNPs within MHC and their interaction P values conditioning on the additive background in RACI-UK and without conditioning on the additive background in each of the three study cohorts*.
| SNP1 | Position1 | SNP2 | Position2 | Conditional_P | Unconditional_P_UK | Unconditional_P_US | Unconditional_P_SE |
|---|---|---|---|---|---|---|---|
| rs805286 | 31 711 530 | rs532098 | 32 610 275 | 2.5e-07 | 1.3e-16 | 6.9e-04 | 4.6e-04 |
| rs9266629 | 31 379 045 | rs3830076 | 32 128 467 | 1.0e-04 | 2.2e-14 | 4.4e-04 | 1.5e-04 |
| rs9267833 | 32 210 123 | rs2858312 | 32 699 453 | 2.5e-03 | 2.3e-14 | 2.2e-18 | 9.9e-12 |
| rs3177928 | 32 444 658 | rs9368737 | 32 690 652 | 5.6e-04 | 7.7e-16 | 2.9e-02 | 1.0e-02 |
| rs3997849 | 32 714 625 | rs3892710 | 32 715 085 | 3.2e-05 | 1.9e-38 | 5.0e-18 | 3.2e-15 |
| rs9366778 | 31 301 396 | rs2239707 | 31 557 542 | 7.1e-04 | 1.6e-14 | 8.7e-02 | 2.6e-03 |
| rs2517448 | 31 094 890 | rs2256594 | 32 219 095 | 3.2e-04 | 3.3e-17 | 1.3e-05 | 2.8e-02 |
| rs4947342 | 32 685 293 | rs2857126 | 32 809 363 | 1.0e-03 | 6.5e-13 | 1.0e-03 | 2.2e-02 |
| rs241433 | 32 831 018 | rs2239701 | 32 837 272 | 7.9e-04 | 9.5e-24 | 5.1e-07 | 6.2e-03 |
| rs2736426 | 31 777 507 | rs11244 | 32 812 946 | 6.3e-04 | 2.0e-16 | 4.6e-04 | 2.3e-12 |
*SNP1(2): the first (second) epistatic SNP; Position1(2): the genomic postion of the SNP1(2); Conditional_P: the interaction P value in the final model fitting covariates, the additive effects of the 20 independent GWAS SNPs and the 10 epistatic pairs of SNPs and their interactions; Unconditional_P_UK (US or SE): the interaction P value corrected for covariates but not the additive background in RACI-UK (US or SE); P values greater than 0.05 underlined.
Combining the three study cohorts together boosted power for both the GWAS and vGWAS analyses as expected (Supplementary Figure S7), with no signs of inflation or deflation (Supplementary Figure S8). The vGWAS analysis identified two new significant loci ANKRD55 and TYK2 in addition to MHC and PTPN22, which were all genome-wide significant in the GWAS analysis. The top vGWAS SNPs rs71624119 for ANKRD55 (P = 4.9e-07) and rs34536443 for TYK2 (P = 7.0e-06) both had only weak or no signals in individual member cohorts, i.e. the P values were 4.4e-03, 2.5e-02 and 7.9e-02 in RACI-UK, RACI-US and RACI-SE respectively for rs71624119; and 1.8e-04, 3.4e-01 and 2.1e-01 respectively for rs34536443. In contrast, the GWAS analysis identified additional 12 genome-wide significant loci that were insignificant in the vGWAS (Supplementary Figure S7).
We plotted the vGWAS P values against the GWAS counterparts for the Combined cohort as well as each individual cohort to assess the impact of the mean-variance correlation (Fig. 3). In the Combined cohort, high concordance between GWAS and vGWAS was observed when the MHC and/or PTPN22 regions were included (Fig. 3a,b) but greatly diminished if excluding both MHC and PTPN22 (Fig. 3c). In individual cohorts, GWAS and vGWAS concordance was observed only in RACI-UK even when including PTPN22 (Fig. 3d,e,f). These results suggested that the mean-variance correlation might influence vGWAS directly mainly at the major GWAS loci. In addition, we reran the vGWAS analyses while fitting the additive effects of the pre-identified GWAS SNPs as additional covariates at either stage in RACI-UK and found little differences in the results, indicating the vGWAS signals were unlikely driven by the differences of allelic means or scaling factors.
Discussion
We showed that the two-stage vGWAS approach successfully identified MHC that harbored widespread genotypic variability of non-additive risk of RA across the three study cohorts. Such non-additive genotypic variability could be explained in part by GxG interactions across the entire MHC region that were independent of the strong additive effects, and had a moderate/weak effect individually but an appreciable joint effect. We also identified PTPN22 as a significant vGWAS locus in the discovery cohort that however was replicated in only one independent cohort. Combining the three study cohorts boosted the power of vGWAS and detected two additional loci ANKRD55 and TYK2 that are yet to be replicated. These results suggest that the two-stage vGWAS indeed works for complex diseases.
Our results of multiple GxG interactions within MHC are in line with the recent reports that focused on a few preselected MHC loci (e.g. HLA-DRB1) and used high resolution imputation and a much increased sample size to fine map within-locus interactions29,39. Similar fine mapping efforts are needed to accurately compute the additive background and subsequently refine GxG interactions across the entire MHC region. There is not information of environmental factors other than gender in the study cohorts to test GxE interactions. However, there have been several reports of GxE interactions (e.g. with smoking and alcohol) in RA involving MHC and/or PTPN2240,41,42,43,44. Besides, there is also evidence of non-additive effects and heterozygous advantage in MHC variants from other autoimmune diseases29,38,39, all supporting the notion that MHC is highly diverse owing to strong natural selection pressure in order to cope with ever changing environmental pathogens45.
One major concern about vGWAS applications is potential conflation caused by the mean-variance correlation that is hardly disentangled, i.e. variance can be mathematically modelled as some function of the mean20,46. Here we showed that the impact of such a correlation was strong only in the major RA associated loci (i.e. MHC and PTPN22) that tended to be involved in GxG and/or GxE interactions. We argue that co-existence of strong additive and non-additive effects in the major associated loci may be biologically important9 rendering these loci the key regulators of the study phenotype and even other related phenotypes (e.g. MHC regulates a number of autoimmune diseases). From this perspective, detection of the major GWAS loci should not be regarded as a rediscovery but in fact an assurance that vGWAS can at least pick up the key interacting loci for dichotomous disease phenotypes.
vGWAS may be more fruitful in meta-analysis of multiple cohorts as demonstrated in the Combined cohort analysis. We showed that the newly discovered vGWAS loci, like those in GWAS meta-analysis, had weaker association signals than MHC or PTPN22, suggesting that they are unlikely involved in strong interactions and thus require more samples to detect9. ANKRD55 is associated with multiple diseases including RA and multiple sclerosis3,47 but its biological functions and involvement in statistical interaction are not yet clear. TYK2 is also a pleiotropic gene associated with multiple autoimmune diseases and was found interacting with IRF5 in lupus but the interactions seemed weak and inconsistent48,49,50. Further work is required to validate their non-additive roles in RA. We anticipate that future enlarged vGWAS meta-analyses may detect additional, especially novel loci that may be driven more by the variance rather than the mean.
vGWAS is also useful to increase power of detection of interactions via an effective reduction of multiple tests concerning only the prioritized loci. For example, testing GxG interactions benefited from a much relaxed significance threshold as 4.2e-06 after filtering on vGWAS significant SNPs and then their pairwise LD in this study. Caution is recommended when using such filtering because vGWAS could miss true interacting loci with limited genotypic variability9,11,12. Nevertheless, such a filtering is practically useful to prioritize the main non-additive loci particularly when a big sample size is available.
This pilot study has a few limitations. Firstly, the low power of detection in individual cohorts (particularly in RACI-US and RACI-SE). In addition to sample size and SNP density, power of a vGWAS is also determined by the effect sizes of the interactions involved and the unobserved interacting factors11,36. The MHC signals appeared significantly in every cohort probably because they were involved in multiple interactions as suggested previously17. Secondly, uncertainties associated with the estimated environmental residuals due to hidden or unaccounted factors. Factors influencing the accuracy of polygenic risk prediction (e.g. sample size, case control ratio, SNP density, population structure, sample relatedness and ascertainment)31,32,33 will also influence the vGWAS power. Population histories and/or special environments may also cause biases if not unaccounted for properly. For example, RACI-SE was the only cohort using matched cases and controls2, which might explain in part the relatively low estimate of heritability (Table 1) and the slight deflation observed in the vGWAS QQ plot (Supplementary Figure S1). Thirdly, this study concerned only the Immunochip platform for an autoimmune disease. Further investigations using GWAS arrays and other types of diseases would be useful to provide a complete view of this approach. A carefully designed simulation study is also recommended to assess the factors above as well as to quantify the impact of the mean-variance correlation comprehensively. Such a simulation study may also help develop a consensus protocol to facilitate future vGWAS meta-analysis of diseases.
Additional Information
How to cite this article: Wei, W.-H. et al. Major histocompatibility complex harbors widespread genotypic variability of non-additive risk of rheumatoid arthritis including epistasis. Sci. Rep. 6, 25014; doi: 10.1038/srep25014 (2016).
Supplementary Material
Acknowledgments
We thank the anonymous referees for their valuable comments. We are grateful to Drs Chris Haley, Soumya Raychaudhuri, Heather Cordell, Toby Johnson and Frank Dudbridge for valuable discussions about the vGWAS method. We thank Drs Sang Hong Lee, Jian Yang and Guo-Bo Chen for suggestions on the GCTA software and Dr Maksim Struchalin for discussions about the VariABEL package. WHW is funded by the Higher Education Funding Council for England (HEFCE) and currently by Cure Kids NZ. AY is funded by the innovative medicines initiative joint undertaking (IMI JU) funded project BeTheCure (contract number 115142-2). The authors acknowledge the assistance given by IT Services and the use of the Computational Shared Facility at The University of Manchester. The authors thank the Wellcome Trust Sanger Institute Genotyping Facility for genotyping of the UK Rheumatoid Arthritis Genetics samples funded by the Arthritis Research UK grant reference number 17552 and the Manchester Biomedical Research Centre. This work was made possible by funds from the Arthritis Foundation (S.R., principal investigator) and the US National Institutes Health (K08AR055688 to S.R. and 1R01AR062886-01 to P.I.W.d.B). P. Gilbert prepared the UK samples. We acknowledge use of DNA from the UK Blood Services collection of Common Controls (UKBS-CC collection), which is funded by Wellcome Trust grant 076113/C/04/Z and by the US National Institutes for Health research program grant to the National Health Service Blood and Transplant (RP-PG-0310-1002). We acknowledge the use of DNA from the British 1958 Birth Cohort collection, which is funded by the UK Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. The North American Rheumatoid Arthritis Consortium and analysis of other US patient and control collections at the Feinstein Institute were supported by the US National Institutes of Health RO1-AR-4-4422, NO1-AR-2-2263; NO1-AR1-2256, RO1 AI068759 and RC2AR059092-01, with additional support from the Eileen Ludwig Greenland Center for Rheumatoid Arthritis and the family of Robert S. Boas.
Footnotes
Author Contributions W.H.W., S.E. and J.W. designed and directed the study. W.H.W. performed data analyses and wrote the manuscript. J.B. and J.M. contributed to data analyses. All authors contributed to the manuscript discussion, writing and reviewing.
References
- Diogo D., Okada Y. & Plenge R. M. Genome-wide association studies to advance our understanding of critical cell types and pathways in rheumatoid arthritis: recent findings and challenges. Curr Opin Rheumatol 26, 85–92 (2014). [DOI] [PubMed] [Google Scholar]
- Eyre S. et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet 44, 1336–1340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viatte S., Plant D. & Raychaudhuri S. Genetics and epigenetics of rheumatoid arthritis. Nat Rev Rheumatol 9, 141–153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes M., Cortes A., van Heel D. A. & Brown M. A. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet 14, 661–673 (2013). [DOI] [PubMed] [Google Scholar]
- Zuk O., Hechter E., Sunyaev S. R. & Lander E. S. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci USA 109, 1193–1198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. Gene–environment-wide association studies: emerging approaches. Nat Rev Genet 11, 259–272 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B. Genotype to phenotype: lessons from model organisms for human genetics. Nat Rev Genet 14, 168–178 (2013). [DOI] [PubMed] [Google Scholar]
- Wei W.-H., Hemani G. & Haley C. S. Detecting epistasis in human complex traits. Nat Rev Genet 15, 722–733 (2014). [DOI] [PubMed] [Google Scholar]
- Robinson M. R., Wray N. R. & Visscher P. M. Explaining additional genetic variation in complex traits. Trends Genet 30, 124–132 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struchalin M. V., Dehghan A., Witteman J. C., van Duijn C. & Aulchenko Y. S. Variance heterogeneity analysis for detection of potentially interacting genetic loci: method and its limitations. BMC Genet 11, 92 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnegard L. & Valdar W. Recent developments in statistical methods for detecting genetic loci affecting phenotypic variability. BMC Genet 13, 63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Pettersson M., Rönnegård L. & Carlborg Ö. Inheritance Beyond Plain Heritability: Variance-Controlling Genes in Arabidopsis thaliana. PLos Genet 8, e1002839 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles J. F. et al. Behavioral idiosyncrasy reveals genetic control of phenotypic variability. Proc Natl Acad Sci USA 112, 6706–6711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer D. S. Selection in different environments: effects on environmental sensitivity (reaction norm) and on mean performance. Genet Res (Camb) 56, 57–70 (1990). [Google Scholar]
- Visscher P. M. & Posthuma D. Statistical power to detect genetic Loci affecting environmental sensitivity. Behav Genet 40, 728–733 (2010). [DOI] [PubMed] [Google Scholar]
- Brown A. A. et al. Genetic interactions affecting human gene expression identified by variance association mapping. Elife 3, e01381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare G., Cook N. R., Ridker P. M. & Chasman D. I. On the use of variance per genotype as a tool to identify quantitative trait interaction effects: a report from the Women’s Genome Health Study. PLos Genet 6, e1000981 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. et al. FTO genotype is associated with phenotypic variability of body mass index. Nature 490, 267–272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Elston R., Morris N. & Zhu X. What Is the Significance of Difference in Phenotypic Variability across SNP Genotypes? Am J Hum Genet 93, 390–397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggart C. J. et al. Novel Approach Identifies SNPs in SLC2A10 and KCNK9 with Evidence for Parent-of-Origin Effect on Body Mass Index. PLos Genet 10, e1004508 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topless R. et al. Association of SLC2A9 genotype with phenotypic variability of serum urate in pre-menopausal women. Front Genet 6, 313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse A. M. & Cai J. J. Genetic variants contribute to gene expression variability in humans. Genetics 193, 95–108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Yang E., Brinkmeyer-Langford C. L. & Cai J. J. Additive, epistatic, and environmental effects through the lens of expression variability QTL in a twin cohort. Genetics 196, 413–425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks P. W., Pearson E. & Florez J. C. Gene-environment and gene-treatment interactions in type 2 diabetes: progress, pitfalls, and prospects. Diabetes Care 36, 1413–1421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M. et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med 373, 895–907 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Reyes T. et al. Interaction of dietary fat intake with APOA2, APOA5 and LEPR polymorphisms and its relationship with obesity and dyslipidemia in young subjects. Lipids Health Dis 14, 106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W. H. et al. Abundant local interactions in the 4p16.1 region suggest functional mechanisms underlying SLC2A9 associations with human serum uric acid. Hum Mol Genet 23, 5061–5068 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz T. L. et al. Widespread non-additive and interaction effects within HLA loci modulate the risk of autoimmune diseases. Nat Genet 47, 1085–1090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S. H., Goddard M. E. & Visscher P. M. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H. et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet 44, 247–250 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Wray N. R., Goddard M. E. & Visscher P. M. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet 88, 294–305 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. B. et al. Estimation and partitioning of (co)heritability of inflammatory bowel disease from GWAS and immunochip data. Hum Mol Genet 23, 4710–4720 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes A. & Brown M. A. Promise and pitfalls of the Immunochip. Arthritis Res Ther 13, 101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struchalin M. V., Amin N., Eilers P. H., van Duijn C. M. & Aulchenko Y. S. An R package “VariABEL” for genome-wide searching of potentially interacting loci by testing genotypic variance heterogeneity. BMC Genet 13, 4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko Y. S., Ripke S., Isaacs A. & van Duijn C. M. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23, 1294–1296 (2007). [DOI] [PubMed] [Google Scholar]
- Goyette P. et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat Genet 47, 172–179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X. et al. Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat Genet 47, 898–905 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallberg H. et al. Gene-gene and gene-environment interactions involving HLA-DRB1, PTPN22, and smoking in two subsets of rheumatoid arthritis. Am J Hum Genet 80, 867–875 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlson E. W. et al. Gene-environment interaction between HLA-DRB1 shared epitope and heavy cigarette smoking in predicting incident rheumatoid arthritis. Ann Rheum Dis 69, 54–60 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B. et al. Associations of smoking and alcohol consumption with disease activity and functional status in rheumatoid arthritis. J Rheumatol 41, 24–30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. W. et al. Reevaluation of the interaction between HLA-DRB1 shared epitope alleles, PTPN22, and smoking in determining susceptibility to autoantibody-positive and autoantibody-negative rheumatoid arthritis in a large UK Caucasian population. Arthritis Rheum 60, 2565–2576 (2009). [DOI] [PubMed] [Google Scholar]
- Kim K. et al. Interactions between amino-acid-defined MHC class II variants and smoking for seropositive rheumatoid arthritis. Arthritis Rheumatol 67, 2611–2623 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R. et al. Gene map of the extended human MHC. Nat Rev Genet 5, 889–899 (2004). [DOI] [PubMed] [Google Scholar]
- Shen X. & Ronnegard L. Issues with data transformation in genome-wide association studies for phenotypic variability. F1000Res 2, 200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alloza I. et al. ANKRD55 and DHCR7 are novel multiple sclerosis risk loci. Genes Immun 13, 253–257 (2012). [DOI] [PubMed] [Google Scholar]
- Hellquist A. et al. Evidence for genetic association and interaction between the TYK2 and IRF5 genes in systemic lupus erythematosus. J Rheumatol 36, 1631–1638 (2009). [DOI] [PubMed] [Google Scholar]
- Suarez-Gestal M., Calaza M. & Gonzalez A. Lack of interaction between systemic lupus erythematosus-associated polymorphisms in TYK2 and IRF5. J Rheumatol 37, 676–677; author reply 678 (2010). [DOI] [PubMed] [Google Scholar]
- Tang L. et al. Genetic association and interaction between the IRF5 and TYK2 genes and systemic lupus erythematosus in the Han Chinese population. Inflamm Res 64, 817–824 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.