Abstract
The contractile forces in individual cells drive the tissue processes, such as morphogenesis and wound healing, and maintain tissue integrity. In these processes, α-catenin molecule acts as a tension sensor at cadherin-based adherens junctions (AJs), accelerating the positive feedback of intercellular tension. Under tension, α-catenin is activated to recruit vinculin, which recruits actin filaments to AJs. In this study, we revealed how α-catenin retains its activated state while avoiding unfolding under tension. Using single-molecule force spectroscopy employing atomic force microscopy (AFM), we found that mechanically activated α-catenin fragment had higher mechanical stability than a non-activated fragment. The results of our experiments using mutated and segmented fragments showed that the key intramolecular interactions acted as a conformational switch. We also found that the conformation of α-catenin was reinforced by vinculin binding. We demonstrate that α-catenin adaptively changes its conformation under tension to a stable intermediate state, binds to vinculin, and finally settles into a more stable state reinforced by vinculin binding. Our data suggest that the plastic characteristics of α-catenin, revealed in response to both mechanical and biochemical cues, enable the functional-structural dynamics at the cellular and tissue levels.
A combination of contractile forces in individual cells drives tissue dynamics such as morphogenesis1,2,3 and wound healing4. The cadherin-based adherens junctions (AJs) function as direct links between the contractile actomyosin cytoskeletons of different cells5,6. AJs balance the intercellular tensions by the adaptive assembly of the cytoskeletal actin filaments7,8,9,10. In this mechano-adaptive mechanism, α-catenin, a tension-sensing component of AJs, is critical regulator of vinculin binding11,12,13, which recruits another actin filament to AJ14,15.
The molecular mechanisms of the regulation of vinculin binding by α-catenin have been investigated using various approaches. The molecular and cellular study of Yonemura et al.16 has revealed that α-catenin under intercellular tension exposes the cryptic vinculin binding site (VBS). The process is triggered by the release of the autoinhibited conformation caused by an intramolecular interaction between its MI domain (residues 275–391) containing VBS (residues 325–360) and the vinculin-inhibitory MII–MIII domain (residues 510–697). The autoinhibited conformation has been determined in the crystallized α-catenin [PDB code: 4K1N]17, where the vinculin-binding surface of VBS is buried in the helix bundle of MI domain that is structurally stabilized by MII–MIII helix bundles. Single-molecule experiments using magnetic tweezers have suggested that the disruption of MI/MII–MIII interaction requires only approximately 5-pN tension18, which is close to the range of forces generated by a single myosin molecule19. Thereby, the MI/MII–MIII interaction holds the key to the mechanical activation of α-catenin, recruiting vinculin under tension.
However, α-catenin mechanically activated under tension faces a critical problem common to all tension-sensing proteins. Usually, an external force acts as a biomolecule denaturant20,21. A study22 using atomic force microscopy (AFM) has reported a complete unfolding of a protein in approximately 30 s under low tension of approximately 13 pN. Furthermore, α-helical proteins such as α-catenin have lower mechanical stabilities than other β-sheet proteins23. During the tension-sensing process, the helical conformation of VBS should be conserved to associate with vinculin24. Thus, the most significant question is how does α-catenin, one of the tension-sensing proteins, retain its activated state while avoiding the successive unfolding under denaturing tension.
Here, we revealed that α-catenin under tension changed its conformation to a stable intermediate state. We investigated the mechanical behaviors of mutated, segmented, and vinculin-bound α-catenin using single-molecule force spectroscopy (SMFS)25,26,27 employing AFM. To reveal the conformational changes in α-catenin under tension, we adopted the following two types of loading conditions: (a) direct loading without mechanical activation and (b) loading with a holding period to wait for mechanical activation.
Results
Mechano-adaptive conformational change in MI-MIII
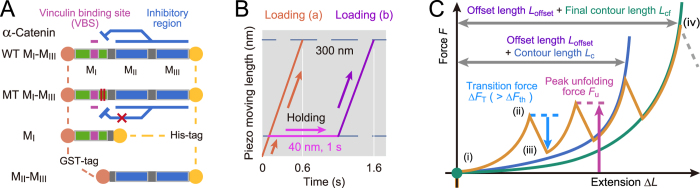
We used four types of α-catenin fragments (Fig. 1A): wild type (WT) MI-MIII (residues 276–634), mutant (MT) MI-MIII (M319G and R326E), MI (residues 276–393), and MII–MIII (residues 385–634). The mutant fragment, in which the autoinhibiting MI/MII–MIII interaction is disrupted, behaved as an activated α-catenin that interacts with vinculin under no tension (Supplementary Figure 1). The weakened MI/MII–MIII interaction causes a change in the angle between helix bundles MII and MIII17 and the destabilization of helix bundle MI28. α-Catenin fragments bound to the glass substrate at the C-terminus exploiting NTA-Ni2+-His6 affinity were loaded at the N-terminus using GST-GSH affinity. The autoinhibitory WT MI-MIII fragment was examined in the following two types of loading conditions (Fig. 1B): (a) direct loading (orange line) and (b) loading with a holding period (magenta line) to wait for mechanical activation. The force curves corresponding to the single α-catenin molecules were extracted and their characteristics were analyzed (Fig. 1C); we calculated contour length Lc to identify intermediate states during unfolding and the measured peak unfolding force Fu to estimate the mechanical stabilities of the abovementioned α-catenin fragments.
Figure 1. SMFS using AFM.
(A) α-Catenin fragments: WT MI-MIII (residues 276–634), mutant (MT) MI-MIII (M319G and R326E), MI (residues 276–393), and MII–MIII (residues 385–634). (B) Loading conditions (a) and (b). Single α-catenin molecules were loaded by NanoWizard 3 BioAFM (JPK Instruments, Berlin, Germany). The piezo-actuator was moved upward by 300 nm at a constant speed of 500 nm/s (“Loading (a)” in this study, orange line). To analyze the mechanical behavior of WT MI-MIII after mechanical activation, we introduced a holding time of 1 s at 40 nm of constant piezo-moving length (“Loading (b),” orange (initial loading), magenta (holding), and purple (further loading)). (C) Force curve analysis. First, the force curves with saw-tooth patterns (state (i) to (iv), orange curve) caused by single-molecule behaviors were identified, with the rupture force and final stiffness in the last peak (state (iv)). Next, the offset length Loffset was determined by WLC-model fitting to the last peak (green curve) with the fixed final contour length Lcf (143.2 nm (WT/MT MI-MIII), 46.8 nm (MI), and 99.6 nm (MII–MIII)). Finally, we measured the contour length Lc and peak unfolding force Fu at each force peak with transition force ΔFT (state (ii) to (iii)) greater than the threshold ΔFth (Methods: “Force curve analysis”).
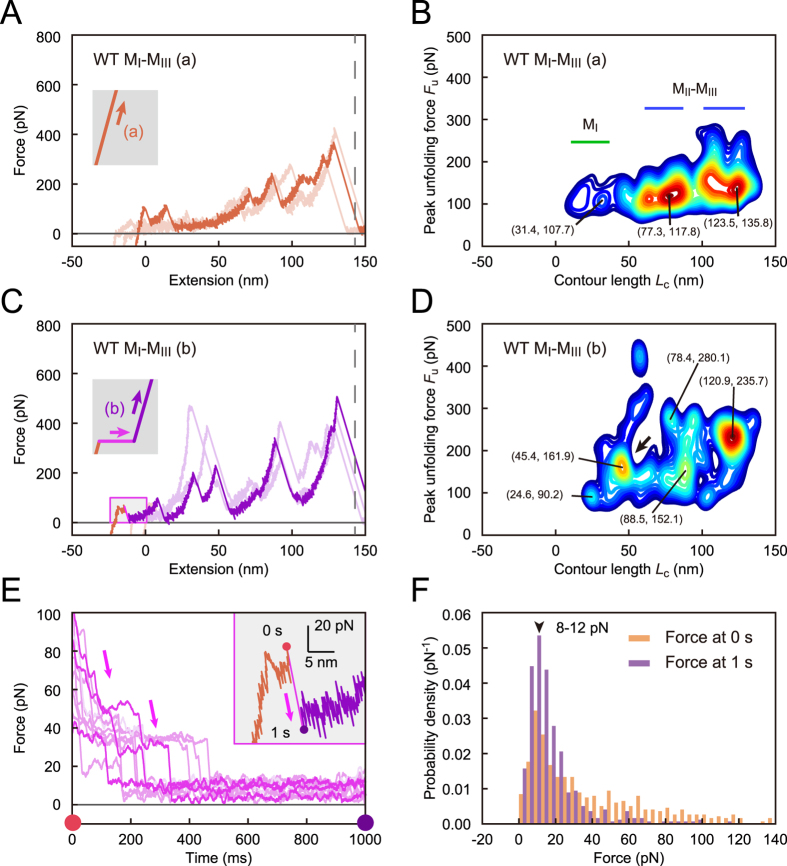
WT MI-MIII fragment in loading (a) unfolded each domain under tension. To analyze the stochastic unfolding trajectories from obtained force curves (Fig. 2A), we evaluated the number density of force peaks, nd (Lc, Fu) on the basis of scatter plots of Fu versus Lc. The contour map of the number density nd showed three wide horizontal regions with similar intervals (Fig. 2B; the contour map with regard to net extension is shown in Supplementary Figure 2A). The low nd in the initial region (green line) indicated that the weak helix bundle of MI unfolded easily, with infrequent force peaks. However, the last two regions (blue lines) with high nd, i.e., with frequent force peaks, corresponded to two stable helix bundles of MII–MIII domain. These results indicated that MI-MIII domain unfolding depended on the mechanical stability of their helix bundles under direct loading.
Figure 2. Conformational change in MI-MIII domain under tension.
(A) Force curves for WT MI-MIII fragment in loading (a). The force curves are shifted to the left by the offset length Loffset. (B) Contour map of number density nd of force peaks in loading (a) based on 797 force curves. Color contours are set from the maximum value of nd (red) to 0.4 × the value (blue). (C) Force curves in loading (b). (D) Contour map in loading (b) based on 514 force curves. (E) Force relaxation curve during the holding time. (F) The probability density of force at 0 s (orange bars) and 1 s (purple bars) during the holding time.
However, WT MI-MIII in loading (b) showed force peaks within a rather greater force range (Fig. 2C), and the contour map of nd showed greater peak unfolding force Fu (Fig. 2D) than those in loading (a) (Fig. 2B). This result indicated that the conformation of MI-MIII domain changed to another intermediate state that required higher tension for further unfolding. As the peak unfolding force Fu increased along the entire contour length Lc (Fig. 2D), the conformational change must have occurred at all three MI-MIII domains. The contour map showed a specific peak at Lc of approximately 45.4 nm (arrow, Fig. 2D); the net extension was approximately 19.0 nm (Supplementary Figure 2B), <40% of completely extended length of MI domain. This observation suggested that VBS in MI domain was conserved in this region. During the holding time, the force relaxation curve showed stepwise relaxations (Fig. 2E), implying dynamic transitions in the MI-MIII conformation. The force distribution at the beginning (0 s) of the holding time settled into a narrow distribution approximately 10 pN at the end of this period (1 s, Fig. 2F), showing that the MI-MIII conformation finally equilibrated under low tension. We suggest that MI/MII–MIII interaction was partly diminished during the holding time; the average extension of approximately 2.8 nm in the holding period was longer than the reported extension28 that was approximately 1.0 nm for the mechanical activation of MI-MIII domain. The probability density of the force peaks in loading (b) in the initial extension was lower than that in loading (a), which supported this suggestion (Supplementary Figure 2C). Thus, we revealed that MI-MIII domain of α-catenin adaptively changes the conformation to another stable state under low tension, with weakened MI/MII–MIII interaction.
MI/MII–MIII interaction as a conformational switch
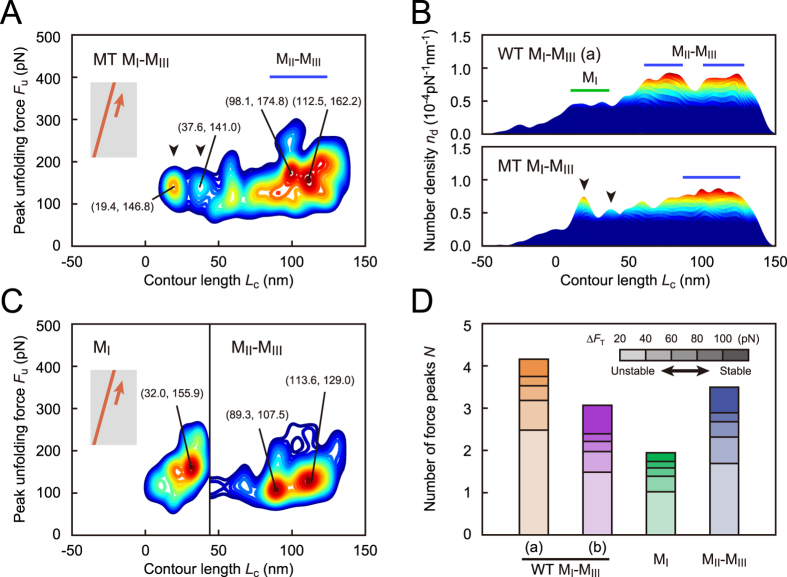
To elucidate the conformational changes in the MI-MIII domain with weakened MI/MII–MIII interaction, we examined the MT MI-MIII fragment (force curves are shown in Supplementary Figure 3A). The contour map for MT MI-MIII showed two specific regions (Lc = 19.4 nm and 37.6 nm, arrowheads in Fig. 3A; the contour map with regard to net extension is shown in Supplementary Figure 3B). The regions had greater peak unfolding forces Fu (146.8 pN and 141.0 pN) than those for WT in loading (a) (Fig. 2B). Moreover, the number densities nd in these regions were higher than those in WT (arrowheads, the side view of nd distribution in Fig. 3B). Furthermore, in the later region (50 nm < Lc < 150 nm), two separate regions observed in WT MI-MIII (blue lines, upper part of Fig. 3B) fused into one broad region in MT (lower part of Fig. 3B). This observation indicated that MII–MIII domain changed its conformation because of weakened MI/MII–MIII interaction. The result for MT MI-MIII indicated that weakened MI/MII–MIII interaction triggers the changes in MI-MIII conformation.
Figure 3. Mechanical behaviors of mutated and segmented α-catenin.
(A) Contour map for MT MI-MIII fragment based on 576 force curves. (B) Comparison of the number density nd in contour maps from a side view in WT and MT MI-MIII fragments. (C) Contour maps for MI and MII–MIII fragments based on 181 and 785 force curves, respectively. The contour map for MII–MIII fragment is shifted to the right by 46.8 nm, which corresponds to the fully-extended length of MI domain. (D) The average number of force peaks N per curve. The stacked bar graphs display N values, with color intensity illustrating the range of transition force ΔFT. The substructure stability increases with the increasing ΔFT values.
Individual MI and MII–MIII fragments showed one and two regions, respectively (Fig. 3C; force curves are shown in Supplementary Figure 3C,D) with higher nd values (Supplementary Figure 3E) than those for MI-MIII fragments, indicating that MI and MII–MIII domains settled into an innate stable state without interacting with each other. In particular, the peak unfolding force for the MI domain (155.9 pN, Fig. 3C) was approximately 50% greater than that for the corresponding region of WT MI-MIII in loading (a) (107.7 pN, Fig. 2B). This result indicated that the weak helix bundle of MI changed the conformation to another state with higher mechanical stability without the interaction with MII–MIII domain.
To compare the mechanical stabilities of α-catenin fragments, we analyzed the number of force peaks N per curve, as shown in Fig. 3D. The stacked bar graphs (Fig. 3D) display N values, with color intensity illustrating the range of transition force ΔFT for force peaks (Fig. 1C). The greater ΔFT values correspond to more stable substructures. For WT MI-MIII fragment (orange and purple bars, Fig. 3D), N values at large ΔFT (>100 pN, associated with a stable substructure) increased in loading (b) compared with that in loading (a). On the other hand, the value of N at small ΔFT (20 pN to 40 pN, associated with unstable substructure) decreased in loading (b). This result supported our idea that the conformation of WT MI-MIII changed to a more stable state with weakened MI/MII–MIII interaction under tension. In addition, the sum of the N values for MI (green bar, Fig. 3D) and MII–MIII (blue bar) was greater than that for WT MI-MIII in loading (a). This result suggested that the MI/MII–MIII interaction destabilized the MI and MII–MIII conformations under no force. On the basis of the results for mutated and segmented α-catenin fragments, we determined that the MI/MII–MIII interaction acted as an intramolecular switch to induce the mechano-adaptive conformational change of MI-MIII domain.
Reinforcing vinculin binding to MI-MIII
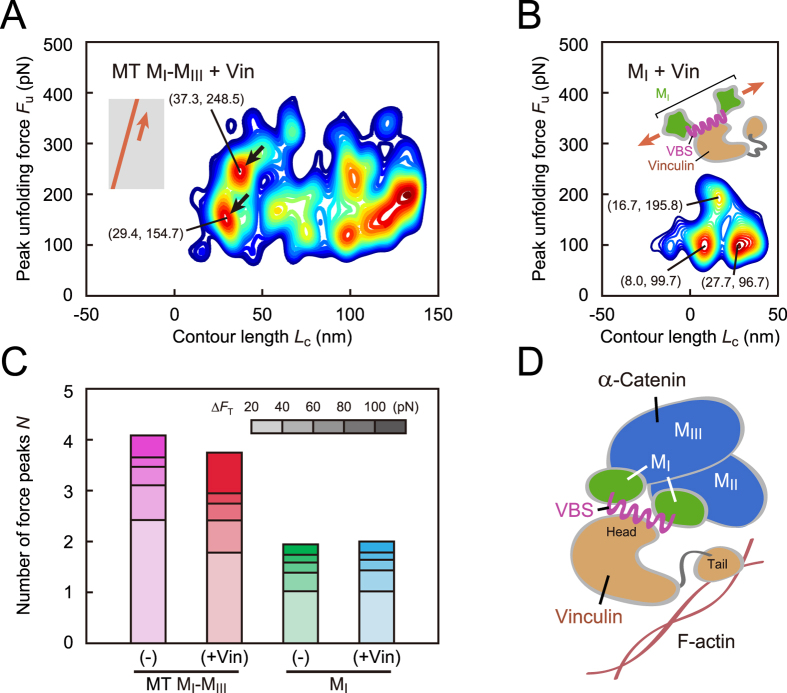
To elucidate how the vinculin binding affects the mechanical behavior of α-catenin, we examined the α-catenin fragments after vinculin treatment. Vinculin binds to an α-catenin molecule with the head domain, and the tail domain associates with another actin filament. The contour map of nd for vinculin-bound MT MI-MIII showed characteristic regions (Lc = 29.4 nm and 37.3 nm; arrows in Fig. 4A; force curves are shown in Supplementary Figure 4A) at a greater peak unfolding force Fu (154.7 pN and 248.5 pN) than that for MT MI-MIII without vinculin (Fig. 3A). This result indicated that MI-MIII domain was reinforced by vinculin binding. In contrast to MT MI-MIII, vinculin-bound MI (Fig. 4B; force curves are shown in Supplementary Figure 4B) showed two regions at smaller Fu (99.7 pN and 96.7 pN) than MI without vinculin (155.9 pN, Fig. 3C). Vinculin-bound MI showed one region at a rather greater Fu (195.8 pN, Fig. 4B). This result indicated that the conformation of vinculin-bound MI comprised a stable helix of VBS and two unstable substructures remaining in this domain.
Figure 4. The effect of vinculin binding on mechanical behavior of α-catenin.
(A) Contour map for vinculin-bound MT MI-MIII fragment based on 571 force curves. (B) Contour map for vinculin-bound MI fragment based on 111 force curves. The conformation of vinculin-bound MI domain comprised a stable helix of VBS with high Fu and two unstable substructures with low Fu. (C) Changes in the average number of force peaks N caused by vinculin binding. (D) Schematic of the molecular complex consisting of α-catenin, vinculin, and actin filament (F-actin). MI-MIII domain was reinforced by vinculin binding at the head domain, in which the MII–MIII domain stabilized the conformation of vinculin-bound MI domain. The tail domain of vinculin associates with another actin filament.
Comparing the results for MT MI-MIII and MI fragments, we concluded that the MII–MIII domain stabilized the unstable vinculin-bound MI domain. Thus, the number of force peaks N at large ΔFT (>100 pN) for MT MI-MIII was increased by vinculin binding (red bar, Fig. 4C), with a decrease in N at small ΔFT (20 pN–40 pN). The number of force peaks for MI did not change significantly (cyan bar, Fig. 4C). The stabilizing role of MII–MIII domain was further confirmed in the analysis of total energy Etot for completely unfolding (Supplementary Figure 5). Etot for MI decreased after vinculin binding (cyan bar) that was caused by greater decrease in unfolding energy for two unstable substructures than the increase in unfolding energy for stable VBS helix. However, MT MI-MIII increased after vinculin binding (red bar). The results for vinculin-bound α-catenin fragments revealed that MI-MIII domain was reinforced by vinculin binding at the head domain (Fig. 4D), with MII–MIII domain stabilizing the conformation of the vinculin-bound MI.
Changes in the persistence length of MI-MIII as a polymer chain
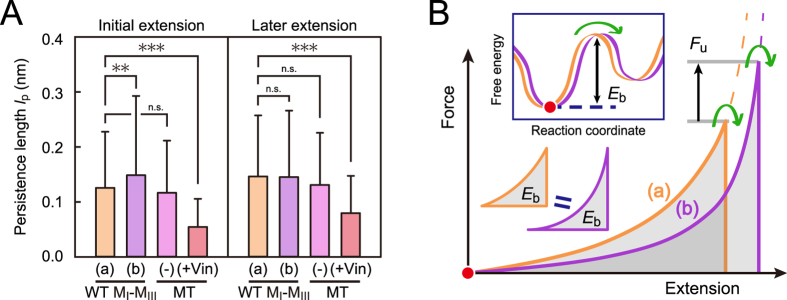
To examine the changes in the α-catenin molecule as a polymer chain, we analyzed the persistence length lp (Fig. 5A). In the initial extension (0 nm < Lc ≤ 72 nm, the left plot in Fig. 5A), the persistence length lp for WT MI-MIII in loading (b) (purple bar) was greater than in loading (a) (orange bar). This result indicated that the persistence length was increased by mechanical activation, resulting in the decreased tensile force required to a certain amount of extension (Fig. 5B). No significant differences were observed in the later extension (72 nm < Lc < 144 nm, the right plot in Fig. 5A). After vinculin binding, lp settled to a much smaller value in the entire extension (red bars in Fig. 5A), indicating that the MI-MIII domain was immobilized because of the conformational reinforcement caused by vinculin.
Figure 5. Changes in the persistence length of α-catenin as a polymer chain.
(A) Comparison of persistence lengths lp in MI-MIII fragments. Statistical significance of the differences was analyzed using t-test (**p < 0.01 and ***p < 0.005). The error bars show standard deviations (S.D.). Persistence length lp was analyzed in the initial extension (0 nm < Lc ≤ 72 nm) and the later extension (72 nm < Lc < 144 nm). (B) The effect of persistence length on the peak unfolding force. If we postulate the same energy barrier Eb for the next intermediate state in MI-MIII before and after the activation (orange (a) and purple (b) lines), greater peak unfolding force Fu will be required to overcome the Eb in the activated MI-MIII (b), with greater lp, than in MI-MIII (a).
Here, the increase in lp of MI-MIII by mechanical activation was consistent with the increase in the peak unfolding force Fu. We can postulate the same energy barrier Eb for the next intermediate state in MI-MIII before and after the activation (Fig. 5B) on the basis of the total unfolding energy Etot (Supplementary Figure 5). Further, the activated MI-MIII (b), with greater lp, should be extended more to overcome the energy barrier Eb than MI-MIII (a), with smaller lp, resulting in a greater peak unfolding force Fu. Thus, the MI-MIII domain of α-catenin under tension changes the conformation to an intermediate state with a larger persistence length, and finally settles into the immobilized state caused by the reinforcement with vinculin.
Discussion
On the basis of our single-molecule experiments, we revealed the mechano-adaptive sensory mechanism of α-catenin. Under physiologically possible low tension, α-catenin adaptively changed the conformation to a stable intermediate state. Such mechano-adaptive conformational changes enable α-catenin at AJs to retain the activated state under tension, without successive unfolding, to function as a robust tension sensor. Our findings could be one solution to a paradox. The tension-sensing proteins require mechanical forces for their activation; however, they have to be stable under such conditions because the mechanical forces function as basic protein denaturants. Furthermore, we revealed that vinculin-binding reinforces α-catenin conformation; the stable α-catenin-vinculin complex contributes to the tight anchoring of adhesive molecules at AJs29 by recruiting another actin filament. Therefore, we suggest that the mechano-adaptive sensory ability of α-catenin arises from its molecular plasticity in response to both mechanical and biochemical cues.
Our results for mutated (MT MI-MIII) and segmented (MI, MII–MIII) α-catenin reveal that the MI/MII–MIII interaction acts as a conformational switch to the intermediate state, where MI and MII–MIII domains change to the conformations with increased stability. Our results are consistent with the previous reports that the helix bundle of MI requires the structural stabilization by MII–MIII28 and that helix bundles MII–MIII are approximately 180°-rotated in conformation without MI domain30,31. However, there were some discrepancies between the results for MT MI-MIII without mechanical activation and those for WT MI-MIII in loading (b) with mechanical activation. Thus, we should consider both the importance of tension for the drastic conformational changes and the conformational switch caused by MI/MII–MIII interaction.
In our experiments, the full-length vinculin firmly bound to MT MI-MIII fragment, with an increase in peak unfolding force Fu and a decrease in the persistence length lp. The full-length vinculin assumes an autoinhibited conformation for α-catenin-binding32. Our results suggested that the VBS exposed in the mechanically-activated α-catenin opens the autoinhibited conformation of vinculin to make a stable α-catenin–vinculin complex. Our data supported the previous reports24,33 that α-catenin and vinculin are “co-activated” for interacting with each other under tension. This type of a force-induced vinculin-activation mechanism of α-catenin could be conserved in talin and α-actinin that are similar adhesive proteins constituting a molecular complex with vinculin and actin filament34.
The loading rate used here was appropriate for the qualitative analysis of the mechanical behavior of α-catenin. Our results for WT MI-MIII in loading (a) were in agreement with the results of the experiments (performed using magnetic tweezers) using a low loading rate (approximately 4 pN/s)18. Moreover, the “holding” method utilized in our study allowed us to analyze the conformational changes under physiologically possible low tension of approximately 10 pN. Thus, by introducing the holding time into fast loadings, we succeeded in analyzing the conformational changes of α-catenin molecules under low tension while efficiently exploring their unfolding trajectories.
The persistence length Lp appeared in this study was smaller than that reported (approximately 0.4 nm) for titin molecules35. This discrepancy may be related to the secondary structures of molecules. A previous study36 employing molecular dynamics simulation revealed that a protein with high φ dihedral potential shows a small persistence length of 0.19 nm. This suggests that α-helical proteins such as α-catenin, of which φ dihedral angle is more constrained than β-sheet proteins such as titin, exhibit smaller persistence length. In addition, subcomponents in α-catenin molecules, such as helix bundles, could decrease the persistence length Lp.
Based on our study, we propose a novel concept of “mechano-adaptive” molecules that fulfill their innate functions by adapting to the cellular forces. Under these forces, biomolecules such as proteins and nucleic acids may change their conformations, mechanical behaviors, and chemical properties. Such molecular-scale changes under mechanical forces could affect force-induced phenomena at cellular and tissue level. We believe that our study may be the basis for future studies investigating the concept of mechano-adaptive sensory mechanism.
Methods
Protein purification
DNA fragments of mouse WT αE-catenin MI-MIII, MT MI-MIII, MI, and MII–MIII were amplified by PCR and cloned into the pGEX6P-3 vector (GE Healthcare). All plasmids were verified by DNA sequencing and transformed into Escherichia coli strain BL21Star (DE3) (Invitrogen) cells for protein expression. Protein expression was performed at 20 °C in Luria–Bertani medium containing 0.1 mM isopropyl-β-D-thiogalactopyranoside. Cells expressing αE-catenin were suspended in 20 mM Tris-HCl buffer (pH 8.0) containing 150 mM NaCl and disrupted by sonication. After ultracentrifugation, the supernatant was applied onto a Glutathione Sepharose 4B column (GE Healthcare). Eluted proteins were further purified by anion-exchange (HiTrap Q HP, GE Healthcare) and gel filtration (Superdex 200 pg, GE Healthcare) chromatography.
Chemical modification
For SMFS, a glass substrate and AFM tip were treated using a chemical modification process21. The glass substrate was modified with α-catenin at its C-terminus and AFM tip was modified with glutathione, which interacts with N-terminal GST-tag of α-catenin. The glass substrate was oxidized and treated with 2% MPTMS/ethanol for 15 min. The substrate was then treated with 2 mM maleimide-C3-NTA (Mal-C3-NTA; DOJINDO Lab.)/PBS for 30 min, with 10 mM NiCl2 (Wako Pure Chemical Industries)/Milli-Q for 30 min, and washed with PBS. α-Catenin fragments (10 μM for each fragment) were modified by NTA-Ni2+-His6 affinity binding for 1 h and finally washed with working buffer (10 mM HEPES, 150 mM NaCl, pH 7.2). For the SMFS of vinculin-bound α-catenin fragments, the α-catenin-modified substrate was further incubated with 1 μM full-length vinculin/PBS for 30 min and washed with working buffer. Silicon nitride AFM tip (OMCL-TR400PSA-1; spring constant, 0.02 N/m, Olympus Co.) was first oxidized using ozone cleaner and treated with 2% APTES/ethanol for 15 min. The tip was then treated with 1.5 mM Mal-PEG-NHS ester/PBS for 30 min and with 10 mM glutathione/PBS for 1 h. The remaining maleimide was quenched with 50 mM 2-mercaptoethanol/HEPES and finally washed with working buffer.
Force curve analysis
Force curves with saw-tooth peaks, caused by conformational transitions of α-catenin, were analyzed using the in-house software (Fig. 1C). First, we extracted the force curves for completely extended single α-catenin molecules, based on the thresholds of force and stiffness at the rupture event, assuming an 85%-extended worm-like chain model37,
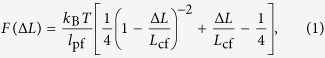 |
where kB is the Boltzmann constant, T is temperature [300 K], lpf is the final persistence length [0.4 nm] based on a previous report35. The final contour length Lcf was estimated as 143.2 nm (WT/MT MI-MIII), 46.8 nm (MI), and 99.6 nm (MII–MIII) based on the number of residues. The threshold of the stiffness excluded the curves with low stiffness for aggregated molecules. For the extracted curves, we determined the length of offset Loffset, corresponding to PEG length (approximately 45 nm) and the tip curvature radius, by fitting WLC model allowing baseline offset as
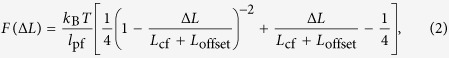 |
to the final peak at the rupture event (green line, Fig. 1C), where Loffset was a fitting parameter. Finally, we identified the intermediate force peaks with the transition force ΔFT (cyan arrow, Fig. 1C) based on the threshold ΔFth (40 pN) and calculated the contour length Lc and the persistence length lp by fitting WLC model with the determined Loffset to the force peaks as
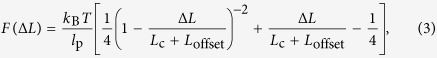 |
by considering that the PEG linker, without any substructures, can be fully unfolded at each intermediate state. To analyze the mechanical stabilities for intermediate states, we measured peak unfolding force Fu (magenta arrow, Fig. 1C). We adopted lower ΔFth (20 pN) to detect the force peaks caused by unfolding of unstable substructures.
Number density of force peaks
Based on scatter plots of peak unfolding force Fu versus contour length Lc, we evaluated the number density nd (Lc, Fu) of force peaks per curve by taking the product of the average number of force peaks per one curve N and the probability density P (Lc, Fu) calculated using two-variable Gaussian distribution function as
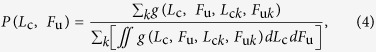 |
where
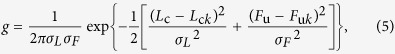 |
in which the summation for k meant the summation for all scattered data points.
Additional Information
How to cite this article: Maki, K. et al. Mechano-adaptive sensory mechanism of α-catenin under tension. Sci. Rep. 6, 24878; doi: 10.1038/srep24878 (2016).
Supplementary Material
Acknowledgments
This work was partly supported by CREST from JST, Japan, and by the Grant-in-Aid for Challenging Exploratory Research (15K13832) from the JSPS, Japan, and by the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Dynamic Approaches to Living System) from the Ministry of Education, Culture, Sports, Science (MEXT) and Japan Agency for Medical Research and development (AMED), Japan. Koichiro Maki was supported by the JSPS Research Fellowships for Young Scientists.
Footnotes
Author Contributions K.M., S.W.H. and T.A. designed and performed SMFS experiments. S.Y., Y.H. and T.H. constructed the samples and performed the biochemical characterizations. All authors contributed to data interpretation. K.M., S.Y., T.H. and T.A. contributed to manuscript preparation.
References
- Martin A. C., Gelbart M., Fernandez-Gonzalez R., Kaschube M. & Wieschaus E. F. Integration of contractile forces during tissue invagination. J. Cell Biol. 188, 735–749 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M. et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56 (2011). [DOI] [PubMed] [Google Scholar]
- Lecuit T., Lenne P.-F. & Munro E. Force generation, transmission, and integration during cell and tissue morphogenesis. Annu. Rev. Cell Dev. Biol. 27, 157–184 (2011). [DOI] [PubMed] [Google Scholar]
- Tamada M., Perez T. D., Nelson W. J. & Sheetz M. P. Two distinct modes of myosin assembly and dynamics during epithelial wound closure. J. Cell Biol. 176, 27–33 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. J. C. & Tepass U. Adherens junctions: from molecules to morphogenesis. Nat. Rev. Mol. Cell Biol. 11, 502–514 (2010). [DOI] [PubMed] [Google Scholar]
- Meng W. & Takeichi M. Adherens junction: molecular architecture and regulation. Cold Spring Harb. Perspect. Biol. 1, a002899; 10.1101/cshperspect.a002899 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y. et al. Actomyosin tension is required for correct recruitment of adherens junction components and zonula occludens formation. Exp. Cell Res. 312, 1637–1650 (2006). [DOI] [PubMed] [Google Scholar]
- Levayer R. & Lecuit T. Oscillation and polarity of E-cadherin asymmetries control actomyosin flow patterns during morphogenesis. Dev. Cell 26, 162–175 (2013). [DOI] [PubMed] [Google Scholar]
- Engl W., Arasi B., Yap L. L., Thiery J. P. & Viasnoff V. Actin dynamics modulate mechanosensitive immobilization of E-cadherin at adherens junctions. Nat. Cell Biol. 16, 584–591 (2014). [DOI] [PubMed] [Google Scholar]
- Sato K., Ogawa Y., Ito S., Fujisawa S. & Minami K. Strain magnitude dependent intracellular calcium signaling response to uniaxial stretch in osteoblastic cells. J. Biomech. Sci. Eng. 10, 15–00242; 10.1299/jbse.15-00242 (2015). [DOI] [Google Scholar]
- Lecuit T. α-Catenin mechanosensing for adherens junctions. Nat. Cell Biol. 12, 522–524 (2010). [DOI] [PubMed] [Google Scholar]
- Yonemura S. A mechanism of mechanotransduction at the cell-cell interface: emergence of α-catenin as the center of a force-balancing mechanism for morphogenesis in multicellular organisms. Bioessays 33, 732–736 (2011). [DOI] [PubMed] [Google Scholar]
- Kim T.-J. et al. Dynamic visualization of α-catenin reveals rapid, reversible conformation switching between tension states. Curr. Biol. 25, 218–224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grashoff C. et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466, 263–266 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huveneers S. et al. Vinculin associates with endothelial VE-cadherin junctions to control force-dependent remodeling. J. Cell Biol. 196, 641–652 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S., Wada Y., Watanabe T., Nagafuchi A. & Shibata M. α-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12, 533–542 (2010). [DOI] [PubMed] [Google Scholar]
- Ishiyama N. et al. An autoinhibited structure of α-catenin and its implications for vinculin recruitment to adherens junctions. J. Biol. Chem. 288, 15913–15925 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M. et al. Force-dependent conformational switch of α-catenin controls vinculin binding. Nat. Commun. 5, 4525; 10.1038/ncomms5525 (2014). [DOI] [PubMed] [Google Scholar]
- Finer J. T., Simmons R. M. & Spudich J. A. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature 368, 113–119 (1994). [DOI] [PubMed] [Google Scholar]
- Oberhauser A. F., Hansma P. K., Carrion-Vazquez M. & Fernández J. M. Stepwise unfolding of titin under force-clamp atomic force microscopy. Proc. Natl. Acad. Sci. USA 98, 468–472 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa I., Kosuri P., Alegre-Cebollada J., Garcia-Manyes S. & Fernández J. M. Force dependency of biochemical reactions measured by single-molecule force-clamp spectroscopy. Nat. Protoc. 8, 1261–1276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Garcia-Manyes S., Sarkar A., Badilla C. L. & Fernández J. M. Mechanical characterization of protein L in the low-force regime by electromagnetic tweezers/evanescent nanometry. Biophys. J. 96, 3810–3821 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann T. & Dougan L. Single molecule force spectroscopy using polyproteins. Chem. Soc. Rev. 41, 4781–4796 (2012). [DOI] [PubMed] [Google Scholar]
- Choi H.-J. et al. αE-catenin is an autoinhibited molecule that coactivates vinculin. Proc. Natl. Acad. Sci. USA 109, 8576–8581 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki K., Han S.-W. & Adachi T. β-Catenin as a tension transmitter revealed by AFM nanomechanical testing. Cell. Mol. Bioeng. 8, 14–21 (2015). [Google Scholar]
- Lv C. et al. Single-molecule force spectroscopy reveals force-enhanced binding of calcium ions by gelsolin. Nat. Commun. 5, 4623; 10.1038/ncomms5623 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D. H. et al. Mechanical force releases nascent chain–mediated ribosome arrest in vitro and in vivo. Science 348, 457–460 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. Structural determinants of the mechanical stability of α-catenin. J. Biol. Chem. 290, 18890–18903 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas W. A. et al. α-Catenin and vinculin cooperate to promote high E-cadherin-based adhesion strength. J. Biol. Chem. 288, 4957–4969 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta S., Drees F., Takai Y., Nelson W. J. & Weis W. I. Biochemical and structural definition of the l-afadin- and actin-binding sites of α-catenin. J. Biol. Chem. 277, 18868–18874 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Dokurno P., Tonks N. K. & Barford D. Crystal structure of the M-fragment of α-catenin: implications for modulation of cell adhesion. EMBO J. 20, 3645–3656 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D. M., Chen H., Johnson R. P., Choudhury B. & Craig S. W. Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. J. Biol. Chem. 280, 17109–17117 (2005). [DOI] [PubMed] [Google Scholar]
- Peng X., Maiers J. L., Choudhury D., Craig S. W. & DeMali K. A. α-Catenin uses a novel mechanism to activate vinculin. J. Biol. Chem. 287, 7728–7737 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bois P. R., O’Hara B. P., Nietlispach D., Kirkpatrick J. & Izard T. The vinculin binding sites of talin and α-actinin are sufficient to activate vinculin. J. Biol. Chem. 281, 7228–7236 (2006). [DOI] [PubMed] [Google Scholar]
- Carrion-Vazquez M. et al. Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. USA 96, 3694–3699 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnemann G., Giganti D., Fernández J. M. & Berne B. J. Elasticity, structure, and relaxation of extended proteins under force. Proc. Natl. Acad. Sci. USA 110, 3847–3852 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C., Marco J. F., Siggia E. D. & Smith S. Entropic elasticity of lambda-phage DNA. Science 265, 1599–1600 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.