Abstract
Accurate and reliable gene expression data from qPCR depends on stable reference gene expression for potential gene functional analyses. In this study, 15 reference genes were selected and analyzed in various sample sets including abiotic stress treatments (salt, cold, water stress, heat, and abscisic acid) and tissues (leaves, roots, seedlings, panicle, and mature seeds). Statistical tools, including geNorm, NormFinder and RefFinder, were utilized to assess the suitability of reference genes based on their stability rankings for various sample groups. For abiotic stress, PP2A and CYP were identified as the most stable genes. In contrast, EIF4α was the most stable in the tissue sample set, followed by PP2A; PP2A was the most stable in all the sample set, followed by EIF4α. GAPDH, and UBC1 were the least stably expressed in the tissue and all the sample sets. These results also indicated that the use of two candidate reference genes would be sufficient for the optimization of normalization studies. To further verify the suitability of these genes for use as reference genes, SbHSF5 and SbHSF13 gene expression levels were normalized using the most and least stable sorghum reference genes in root and water stressed-leaf tissues of five sorghum varieties. This is the first systematic study of the selection of the most stable reference genes for qPCR-related assays in Sorghum bicolor that will potentially benefit future gene expression studies in sorghum and other closely related species.
Keywords: qPCR, RefFinder, Sorghum bicolor, gene expression stability, reference gene, normalization
Introduction
Sorghum is the fifth most important cereal crop that is widespread in the semi-arid regions of the world with an annual production of 65.5 Mt (FAO, 2011), possesses strong drought-tolerance traits and a high forage value. Sorghum seeds are utilized for both human and animal feed, and cultivars are processed for syrup, sugar, and alcohol. Additionally, sorghum cultivars have great economic potential as they can be valorized for second-generation biofuels to produce environment-friendly energy (Vermerris, 2011; Calviño and Messing, 2012). Sorghum is related to many C4 plants including maize, sugarcane, foxtail millet and switchgrass, which tend to have larger polyploid genomes containing repetitive sequences (Price et al., 2005). In contrast, sorghum is a diploid with a small genome (750 Mbp) and possesses an extraordinary germplasm diversity that greatly aids gene discovery and analysis through comparative and functional genomics, making it highly useful as model cereal for structural and functional genomic studies aimed at improving agronomically important traits (Paterson et al., 2009).
Coupled with species phenotypes, functional genomics can provide important insights into complex biological processes including abiotic stress responses. Functional genomic studies are now being used to identify the roles of regulatory and structural genes. The importance of sorghum functional genomics has increased following the sequencing of its genome (Paterson et al., 2009). Despite a wide range of available experimental approaches for exploring gene expression at the transcriptional level, e.g., northern blotting, ribonuclease protection assays, reverse transcription PCR (RT-PCR), quantitative real-time PCR (qPCR) and DNA microarrays (Valasek and Repa, 2005). Among them, qPCR is the most efficient for quantification of gene expression levels due to its simplicity, sensitivity, accuracy and cost (Wong and Medrano, 2005); however, despite these advantages, the utility of this method is often limited due to the absence of reliable reference genes for qPCR data normalization.
Classically, most of the reference genes fall in the category of housekeeping genes, which have major roles in the maintenance of basic cellular metabolism irrespective of physiological conditions (Bustin, 2002). These reference genes have been widely used in qPCR assays in molecular and functional genomic studies as internal calibrators (Vandesompele et al., 2002; Eisenberg and Levanon, 2013); however, recent studies have shown that these reference genes are often not stably expressed under various experimental conditions (Thellin et al., 1999; Zhu et al., 2013; Gimeno et al., 2014). Consequently, the selection and validation of suitable reference genes that are uniformly and stably expressed across experimental conditions have become imperative (Jian et al., 2008; Li et al., 2012). Various statistical tools, e.g., geNorm, NormFinder, BestKeeper and RefFinder, have been developed to determine reference gene suitability for qPCR data normalization (Vandesompele et al., 2002; Andersen et al., 2004; Pfaffl et al., 2004). Stable reference gene validation has been performed in many plant species, including model species and crop plants such as Arabidopsis thaliana (Czechowski et al., 2005; Remans et al., 2008), Brassica napus (Yang et al., 2014a; Machado et al., 2015), Hordium vulgare (Zmienko et al., 2015), Fagopyrum tataricum (Demidenko et al., 2011), Cajanus cajan (Sinha et al., 2015), Oryza sativum (Jain et al., 2006; Li et al., 2010; Ji et al., 2014), Arachis hypogaea (Reddy et al., 2013), Pennisetum glaucum (Saha and Blumwald, 2014; Reddy et al., 2015b), Solanum tuberosum (Nicot et al., 2005; Mariot et al., 2015), Glycine max (Jian et al., 2008; Nakayama et al., 2014), Saccharum officinarum (Iskandar et al., 2004; Ling et al., 2015), Lycopersicon esculentum (Expósito-Rodríguez et al., 2008), Triticum aestivum (Paolacci et al., 2009), and Zea mays (Manoli et al., 2012; Lin et al., 2014).
Until now, sorghum qPCR assays were performed using a single reference gene to quantify target gene expression in response to various experimental conditions. Traditional reference genes including 18S RNA, Actin, EIF4α, Tubulin, and Ubiquitin have been used as calibrators for quantifying sorghum gene expression in response to abiotic stresses and during plant growth and development (Yang et al., 2004; Jain et al., 2008; Cook et al., 2010; Shen et al., 2010; Wang et al., 2010; Dugas et al., 2011; Ishikawa et al., 2011; Koegel et al., 2013; Li et al., 2013; Gelli et al., 2014; Shakoor et al., 2014; Yang et al., 2014b; Yin et al., 2014; Kebrom and Mullet, 2015; Li et al., 2015; Walder et al., 2015). Nevertheless, the reference genes used in these studies were randomly selected from various sources without any experimental validation. Moreover, single gene quantification qPCR assays are well known to frequently exhibit variability in gene expression under various experimental conditions (Dheda et al., 2005). The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines developed for the proper selection (Bustin et al., 2009) and validation of stable candidate reference genes for qPCR experiments highly recommend averaging data from more than 2 reference genes (Bustin et al., 2010). To date, only one study has evaluated reference genes in virus-infected Sorghum bicolor tissues (Zhang et al., 2013), and to the best of our knowledge, there are no systematic studies regarding the selection of suitable stable sorghum reference genes in various tissues and abiotic stress conditions.
This study tested 15 potential candidate reference genes, Acyl Carrier Protein 2 (ACP2), ADP-Ribosylation Factor (ADPRF), alpha Tubulin (α-TUB), beta Tubulin (β-TUB), Eukaryotic Initiation Factor 4A-1 (EIF4a), Elongation Factor P (EF-P), Glyceraldehyde 3-phosphate Dehydrogenase (GAPDH), Malonyl CoA-Acyl Carrier Protein (MACP), Malate Dehydrogenase (MDH), Cyclophilin/Peptidylprolyl Isomerase (CYP), Protein Phosphatase 2C (PP2C), 6-Phosphogluconate Dehydrogenase (6PGDH), S-adenosyl methionine Decarboxylase (SAMDC), Serine/threonine-Protein Phosphatase (PP2A), and Ubiquitin Protein 1, Isoform c (UBC1). The stabilities of these reference genes were analyzed in sets of samples from treatments with various abiotic stresses (salt, cold, heat, water stress, and ABA stress) and tissues (seedlings, leaves, panicles, roots and mature seeds). Additionally, to verify and support the identification of the best-ranked candidate reference genes, SbHSF5 and SbHSF13 gene expression levels were assayed. The stable reference genes identified in this study will be helpful in future qPCR-based molecular and functional studies in sorghum and related species.
Materials and methods
Plant materials and abiotic stress treatments
Sorghum bicolor genotypes (Parbani Moti, Phule Vasudha (PVS), S35, M35, and BTx623) seeds were collected from the sorghum breeding unit of the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) in Patancheru, India. Seeds were sown in pots filled with soil mixture (3:2:1 clay:sand:manure) in a glasshouse. Various abiotic stress treatments (salt, cold, heat, water stress, and ABA stresses) were performed and various tissues (seedlings, leaves, panicles, roots, and mature seeds) were sampled according to (Reddy et al., 2015a). Leaf tissues were collected from sorghum cultivars including Parbani Moti, Phule Vasudha, S35, M35, and BTx623 were grown under control conditions and under progressive drought where at normalized transpiration ratio (NTR) of reached at 0.1 (10% of soil moisture remaining in the pot) (Vadez and Sinclair, 2001) then leaf samples were collected. Tissue samples were collected from three individual seedlings for three independent biological replicates, immediately snap frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Total RNA isolation and cDNA synthesis
Total RNA was extracted from tissues using the Plant RNA mini spin kit (MACHEREY-NAGEL GmbH and Co. KG, Neumann-Neander-Straße 6-8, Duren, Germany) with the in-column DNase I treatment according to the manufacturer's instructions. Sample quantity and purity were determined using a NanoVue Plus Spectrophotometer (GE Healthcare). RNA samples with an OD260/OD280 absorbance ratio between 1.9 and 2.2 were used for further analysis, and RNA integrity was assessed on 1% denaturing formaldehyde agarose gel. For each sample, 1 μg of total RNA was reverse transcribed using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) and oligo (dT) primers according to manufacturer's instructions. The reverse transcribed cDNAs were then diluted 1:12 with nuclease-free water and used for qPCR analysis.
Candidate reference gene selection and qPCR primer design
Fifteen candidate reference genes were identified from the literature and the sequences of their corresponding homologs were extracted from the NCBI and the Phytozome databases (Table 1). The selected candidate reference genes included GAPDH, UBC1, MACP, ACP2, α-TUB, EF-P, 6PGDH, SAMDC, CYP, MDH, ADPRF, β-TUB, PP2A, EIF4α, and PP2C that were earlier reported to be stably expressed in monocots such as rice, pearl millet, barley, foxtail millet, wheat, and maize (Jain et al., 2006; Paolacci et al., 2009; Manoli et al., 2012; Kumar et al., 2013; Reddy et al., 2015b; Zmienko et al., 2015). Primers were designed using the Primer3 software (Untergasser et al., 2012) with the following parameters: 59–62°C annealing temperature, 20–22 bp primer length, 45–55% GC contents, and 90–150 bp amplicon length (Table 1). Pooled and diluted cDNA samples were used for qPCR and 2% agarose gel electrophoresis was used to check primer pair specificity prior to sequencing. The sequences amplified by each primer combination were compared with GenBank sequences using the BLASTN and BLASTX algorithms to verify amplicon specificity.
Table 1.
Comprehensive details of the reference genes used for the normalization in Sorghum.
| S.No. | Gene symbol | Gene name | Accession number | Cellular function | Primers (F/R) (5′'–3′') | Amplicon length (bp) | Tm (°C) | PCR efficiency | Regression coefficient (R2) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | GAPDH | Glyceraldehyde 3-phosphate-Dehydrogenase | XM_002439118 | Glycolysis and gluconeogenesis | AAGGCCGGCATTGCTTTGAAT ACATGTGGCAGATCAGGTCGA |
107 | 84.1 | 1.00 | 0.998 |
| 2 | UBC1 | Ubiquitin Protein 1 Isoform C | XM_002452660 | Protein degradation | AGAGGCTCATCTTCGCTGGG CTCAGTTAACACGGCCACCAC |
120 | 88.1 | 0.98 | 0.994 |
| 3 | MACP | Malonyl CoA-Acyl Carrier protein | XM_002465363 | Fatty acid biosynthesis | GCATTGAGAACATCGGGGCTT ATGAGTGGAAACTTCGTTCCA |
139 | 82.8 | 0.98 | 0.997 |
| 4 | ACP2 | Acyl Carrier Protein-2 | XM_002462703 | Fatty acid and Polyketides biosynthesis | ACGAACTTGTTGCGGCAGAAG GAACAAGAAGGGATGCGCTGG |
110 | 84.3 | 1.00 | 0.996 |
| 5 | α-TUB | Alpha –Tubulin | XM_002466490 | Cytoskeleton structure protein | GAGGGTGAGTTCTCTGAGGCC CTCCCTCATCTCCATCCTCGC |
103 | 85.2 | 1.00 | 0.997 |
| 6 | EF-P | Elongation Factor P | XM_002463651 | Peptide bond synthesis | TGAAGCGGGTGAGAAGATTGT AGCCAAATCATACTCGCCCA |
114 | 80.3 | 1.00 | 0.999 |
| 7 | 6PGDH | 6-Phosphogluconate dehydrogenase | XM_002449451 | Involved in the pentose phosphate pathway | CACACGGAATGGACCAAGCTG AGAGAAATCACCCAGAGCAGCA |
91 | 85.3 | 0.97 | 0.997 |
| 8 | SAMDC | S-adenosylmethionine decarboxylase | XM_002446695 | Synthesis of polyamines | GTGGCGGACTCCTCATCTACC CCAGTTTGCGGTCCTTCACAG |
132 | 85.1 | 0.98 | 0.996 |
| 9 | CYP | Peptidylprolyl Isomerase | XM_002453800 | Cis-trans isomerization of prolineimidic peptide bonds | GTATCTGTGCTCGCCGTCTCT TTCACCCAACTCCTCAACCCC |
108 | 81.1 | 1.01 | 0.989 |
| 10 | MDH | Malate dehydrogenase | XM_002467034 | Citric acid cycle and gluconeogenesis | TGCAGTGGTGGTGAATGGAA GCGTCTTCTCTTCCGACAGC |
103 | 82.5 | 1.01 | 0.972 |
| 11 | ADP-RF | ADP-Ribosylation Factor | XM_002441244 | Regulators of vesicular traffic and actin remodeling | GTCTGTCGGATGTGGGGATGT CACAGCACACAGTCGGACATG |
136 | 84.5 | 0.96 | 0.998 |
| 12 | β-TUB | Beta Tubulin | XM_002439758 | Cytoskeleton structure protein | GCGTGTGAGTCATCCGTTCAC CCGCCTCAGTGAACTCCATCT |
98 | 84.3 | 1.03 | 0.999 |
| 13 | PP2A | Serine/threonine-Protein Phosphatase | XM_002453490 | Control the specific dephosphorylation | AACCCGCAAAACCCCAGACTA TACAGGTCGGGCTCATGGAAC |
138 | 82.7 | 0.97 | 0.991 |
| 14 | EIF4α | Eukaryotic Initiation Factor 4A | XM_002451491 | Eukaryotic translation | CAACTTTGTCACCCGCGATGA TCCAGAAACCTTAGCAGCCCA |
144 | 84.8 | 1.02 | 0.993 |
| 15 | PP2C | Protein Phosphatase 2C | XM_002468507 | Signal transduction | GTAACCCTTCCCGCGAAATCC GCGCTACACTTTGCTGCTTTT |
140 | 85.4 | 0.94 | 0.992 |
Details of candidate reference genes, NCBI accession number, their putative functions, primer sequences, product size and amplicon characteristics, i.e., melting temperature, PCR efficiency and regression coefficient values.
Quantitative real-time RT-PCR (qPCR)
The qPCR reactions were performed using a Realplex Real-Time PCR system (Eppendorf, Germany) and SYBR Green mix (Bioline) in 96 well optical reaction plates (Axygen, USA) sealed with ultra-clear sealing film (Platemax). The reactions were performed in a 10 μl total volume containing 5 μl of 2x SensiMix SYBR No ROX mix (Bioline), 400 nM of each primer, 1.0 μl of diluted cDNA and nuclease-free water. The reaction conditions were 95°C for 2 min, followed by 40 cycles of 15 s at 95°C and 30 s at 62°C with fluorescent signal recording. After amplification, melt curves were generated for each reaction to ensure specific amplification. All qPCR reactions, including the non-template control, were performed in biological and technical triplicates. The mean values obtained from the nine values (triplicates of each biological triplicate) were used to calculate the final quantification cycle values (Cq).
Data analysis
The quantitative cycle (Cq) values were recorded using the RT-PCR system default settings in which the baseline was automatically corrected and threshold values were estimated using the noise band mode. Statistical analysis (mean and CV) of the Cq values was performed using a Microsoft Excel 2010 spreadsheet. PCR efficiencies (E) for candidate genes were evaluated using the dilution series method and pooled cDNA samples. The 12-fold diluted pooled cDNA sample was used for 2-fold serial dilutions. Five serially diluted cDNA samples were used as templates for the construction of standard curves for each primer pair using the above PCR composition and conditions. Standard curves were generated using linear regression based on the quantitative cycle (Cq) values for the dilution series. The correlation coefficients (R2) and slope values were obtained from the standard curves, and the PCR amplification efficiencies (E) were calculated according to the following equation: E = (10−1∕slope-1).
Statistical tools for normalization
The genEX Professional software geNorm and NormFinder algorithms (MultiD Analyses AB, Sweden) were used to identify and analyze the stably expressed gene (s) in diverse experimental samples. The raw Cq values for each gene were corrected according to their PCR efficiencies and then converted into relative quantities. The mean values for the biological replicates were used as the input data for the geNorm and NormFinder analyses. The geNorm program calculates the candidate gene expression stability (M value) using pairwise comparisons and stepwise exclusion (Vandesompele et al., 2002). Pairwise variation analysis was performed to identify the optimal number of reference genes required for normalization in each sample set using geNorm and qBase plus software (v: 2.4; Biogazelle, Belgium) (Hellemans et al., 2007). NormFinder also measures candidate reference gene expression stability using stepwise exclusion by estimating the intra- and inter-group variation. A low stability value indicates low combined variation and high expression stability (Andersen et al., 2004). RefFinder is a web-based tool (http://www.leonxie.com/referencegene.php) that integrates the four major computational programs currently available [geNorm (Vandesompele et al., 2002), NormFinder (Andersen et al., 2004), BestKeeper (Pfaffl et al., 2004), and the comparative ΔCt method (Silver et al., 2006)], and calculates the geometric mean for comprehensive ranking.
SbHSF5 and SbHSF13 genes expression
Sorghum heat shock transcription factors SbHSF5 and SbHSF13 were selected as target genes for validating stabilities of the most and least stable reference genes by quantifying the gene expression levels in various experimental samples. Sample collections and experiments were performed as described above. SbHSF5 and SbHSF13 gene expression levels were normalized using the two most stable candidate reference genes, PP2A and EIF4α, as well as the two least stable reference genes in the all sample group, UBC1 and GAPDH, individually and in combination. For reference gene validations, root tissues of variety Parbani Moti, and leaf tissues of five sorghum varieties under water stress conditions were used for estimating relative expression levels of SbHSF5 and SbHSF13 using REST software (Pfaffl et al., 2002).
Results
Primer specificity and PCR efficiency calculations
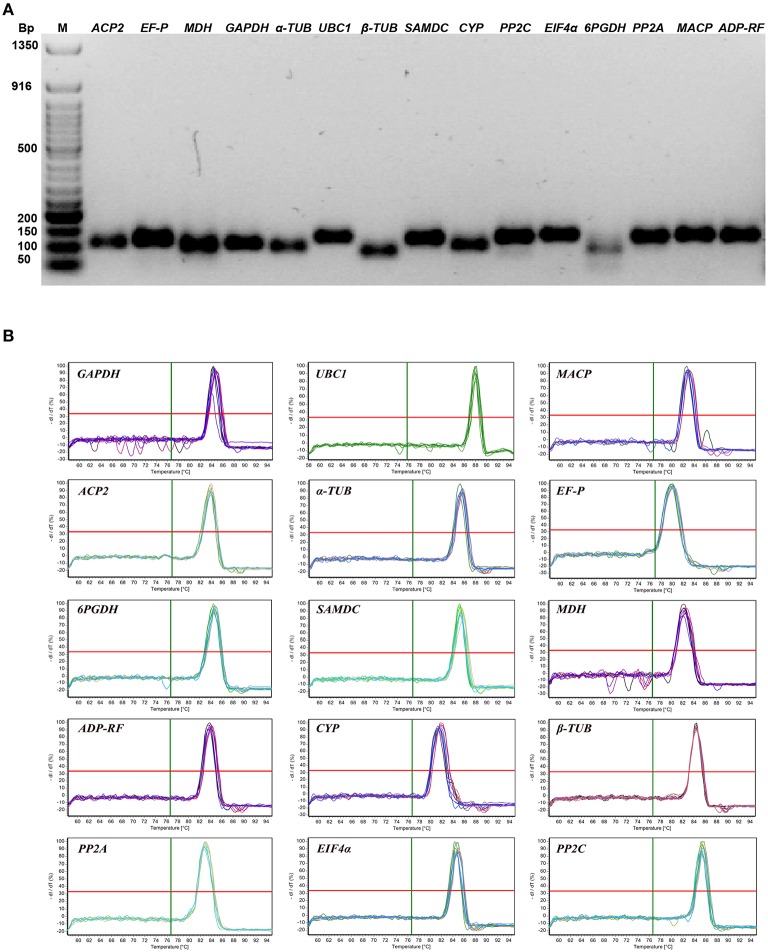
The candidate reference genes selected for this study represent various functional classes and gene families. Primer pairs for 15 candidate reference genes were used for qPCR amplification of sorghum cDNA and yielded single PCR products of the expected sizes (Figure 1A), as well as single melting curve peaks (Figure 1B). The amplification products were sequenced, and verification using the NCBI database BLASTN algorithm revealed 100% matches, demonstrating the gene specificity of the qPCR primer pairs. While the amplification efficiencies (E) of the candidate reference genes ranged from 0.94 (PP2C) to 1.03 (β-TUB), the regression coefficient values (R2) ranged from 0.972 (UBC1) to 0.999 (FE-P and β-TUB) (Table 1).
Figure 1.
Specificity of primer pairs for qPCR amplification. (A) Agarose gel (2.4%) electrophoresis showing PCR products of the expected sizes for 15 candidate genes. M: 50 bp DNA marker (NEB). (B) Dissociation curves of 15 candidate reference genes under various experimental conditions, each showing a single peak.
Sorghum reference gene expression analysis
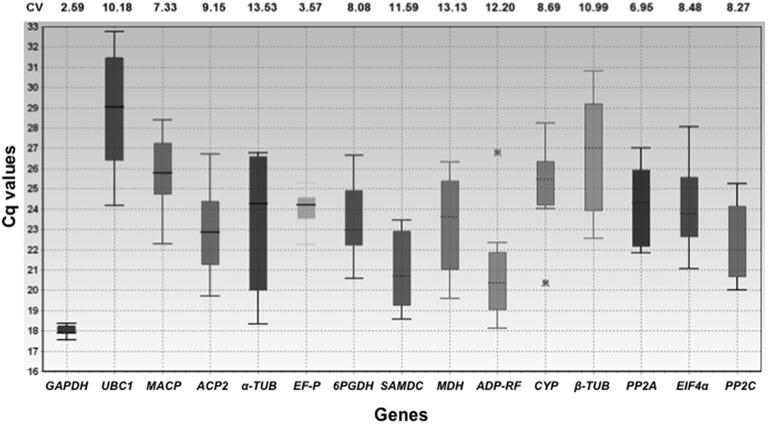
A total of 15 candidate reference genes were selected for qPCR normalization. A SYBR Green-based qPCR assay was used for transcriptional profiling of the 15 candidate reference genes in 10 samples, including five from abiotic stress conditions (water stress, salt, ABA, cold and heat shock) and five different tissues (leaves, roots, panicles, mature seeds and seedlings). Candidate reference gene transcript levels were determined using the quantification cycle (Cq) values, and these genes varied in their abundance (Figure 2). The mean candidate gene Cq values ranged from 17.70 to 28.92, with most falling between 22 and 25. GAPDH exhibited the lowest mean Cq value (17.70), indicating that GAPDH was the most highly expressed, while PP2C, ACP2, α-TUB, 6PGDH, MDH, ELFP, EIF4α, and PP2A were moderately expressed (mean Cq 22.31–24.14), whereas β-TUB (mean Cq 26.69) and UBC1 (mean Cq 28.91) were expressed at low levels. Across the tested samples, the GAPDH gene showed the least variation in expression levels (coefficient of variation 2.59%), while α-TUB was the most variable (13.53%). The degree of variation in reference gene expression across the tested samples is shown in Figure 2. These results indicated that none of the reference gene expression levels were constant and that they varied from one assay to another.
Figure 2.
Expression levels of 15 candidate reference genes in all experimental samples displaying the Ct distribution for each candidate reference gene in all tested samples. Whiskers represent the maximum and minimum Ct values, and the line across the box indicates the median value, while the asterisks marks outliers. The coefficient of variance (CV) for each gene is given as a percentage. The x-axis represents the genes and the y-axis represents the Cq values.
Reference gene ranking and expression stability analysis
The stability values for each reference gene or combination of reference genes may vary from one experimental set to another. The orders of candidate gene stability ranking for the three sample sets were determined separately to identify the most stable reference genes using three different statistical tools, geNorm, NormFinder and RefFinder.
GeNorm analysis
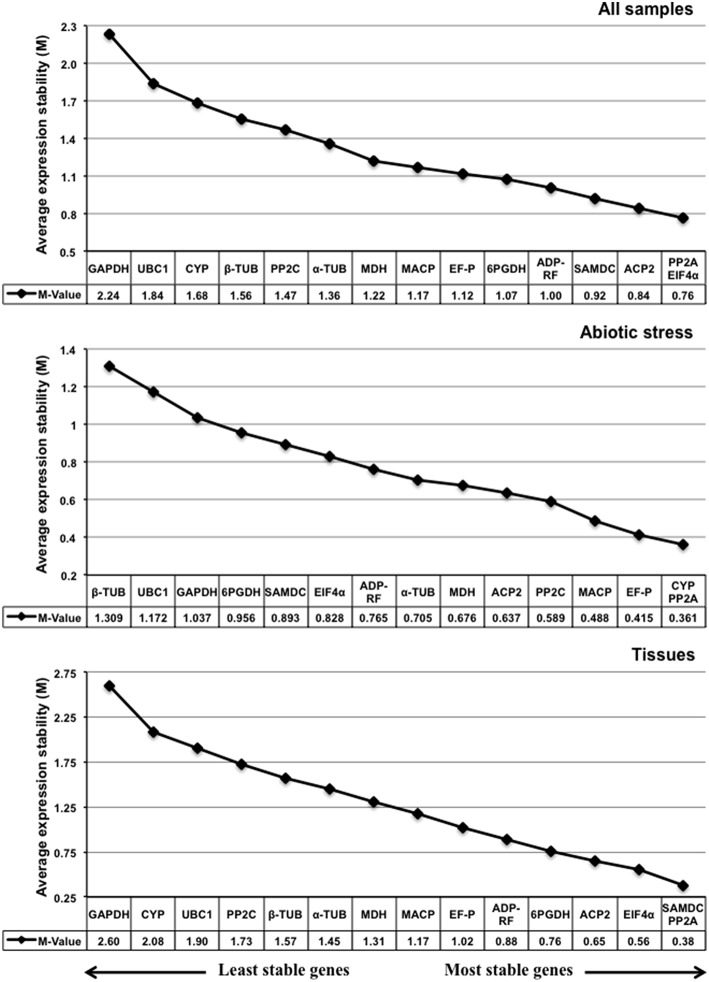
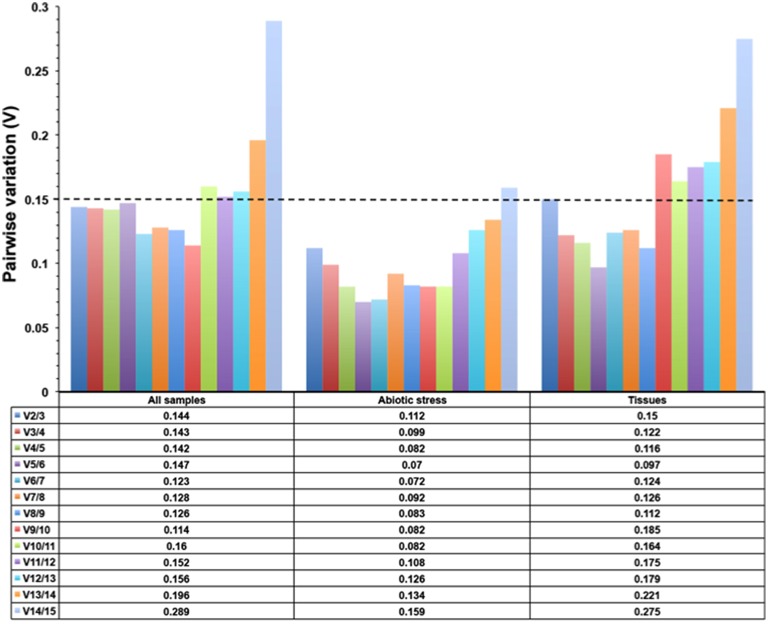
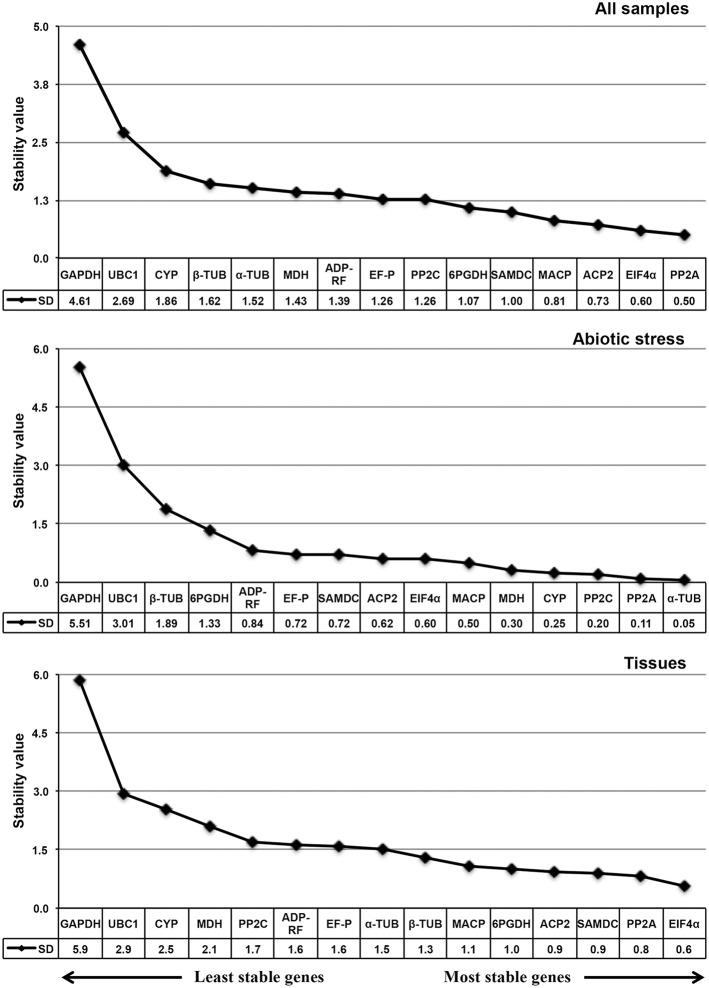
GeNorm software was used to determine the expression stability of the 15 candidate sorghum reference genes; for this analysis, lower M values indicate greater gene expression stability. The software recommends a cutoff M value of 1.5 to identify sets of reference genes that are stably expressed. Figure 3 displays the expression stability rankings for the tested candidate genes; the lowest M value was calculated for the PP2A and EIF4α pair (M = 0.76) and corresponded to the most stable expression in the all sample group, whereas the M values for GAPDH and UBC1 were considerably higher than for the remaining candidate genes (Figure 3). For the abiotic stress treatments, PP2A and CYP were the most stable reference genes, while UBC1 and β-TUB were the least stable (Figure 3). In the tissue group, SAMDC and PP2A were the most stable candidate genes while CYP and GAPDH were the least stable (Figure 3). The optimal numbers of reference genes required for gene expression normalization for the various sample groups were determined using geNorm. As shown in Figure 4, the V2/3 values of all three-sample sets were below the threshold value of 0.15 (0.144 for all pooled samples, 0.112 for the abiotic treatments, and 0.14 for the various tissue sample sets), indicating that two reference genes were sufficient for sorghum gene expression data normalization.
Figure 3.
geNorm expression stability and ranking of the 15 candidate reference genes in various sample sets. The cutoff M value was set at 1.5; a lower M value indicates greater stability and the largest value indicates the least stable reference gene. The direction of the arrow indicates the most and least stable reference genes. The most stable genes are listed on the right and the least stable genes are listed on the left.
Figure 4.
Optimal number of reference genes required for accurate normalization in all three experimental groups using the geNorm tool. The pairwise variation (Vn/Vn+1) was calculated by geNorm tool to determine the minimum number of reference genes for accurate normalization in each experimental set. The cutoff value was 0.15, below which additional reference genes are not necessary for gene expression normalization. The dotted line indicates the optimal number of reference genes.
NormFinder analysis
NormFinder rankings differed slightly from the geNorm rankings (Figure 5). Both tools ranked PP2A and EIF4α as being the most stable genes in all the sample sets, while GAPDH and UBC1 were the least stable genes (Figure 5); in contrast, α-TUB and PP2A emerged as the most stably expressed genes in the abiotic stress sample set, while geNorm ranked α-TUB eighth (Figure 5). For the tissue sample set, EIF4α and PP2A reference genes occupied the top positions, while geNorm ranked EIF4α third (Figure 5). GAPDH and UBC1 were also the least stable reference genes in the abiotic stress and tissue sample sets (Figure 5).
Figure 5.
NormFinder expression stability values and candidate reference gene ranking in Sorghum bicolor samples. Lower values indicate greater stability and larger values indicate the least stable reference genes. The direction of the arrow indicates the most and least stable reference genes. The most stable genes are listed on the right and the least stable are listed on the left.
RefFinder analysis
The comprehensive ranking of the candidate reference genes for all three experimental sample sets using RefFinder was highly consistent with the findings of geNorm and NormFinder. RefFinder analysis revealed that PP2A and EIF4α were the most stable in all the sample set (Table 2). Similarly, PP2A and CYP were highly stable under abiotic stress conditions, while EIF4α and PP2A were the two most stable genes for the tissue sample set (Table 2). Comprehensive ranking revealed that GAPDH was the least stable gene in the all sample and tissue sample sets, while β-TUB was the least stable in the abiotic sample set. UBC1 was the second most stable gene in all three-sample sets (Table 2). Comprehensive ranking for the individual abiotic stress treatments revealed a pattern similar to that of the abiotic stress sample set in which PP2A was second for heat, salt and water stresses. Similarly, EF-P remained in the third position for the ABA and clod stresses (Supplementary Table 1).
Table 2.
The stability ranking of 15 candidate Sorghum bicolor reference genes in three experimental sample sets calculated by delta Ct (DCT), geNorm (GN), NormFinder (NF), BestKeeper (BK) and RefFinder (RF) algorithms.
| Rank | All Samples | Abiotic stress | Tissues | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DCT | GN | NF | BK | RF | DCT | GN | NF | BK | RF | DCT | GN | NF | BK | RF | |
| 1 | PP2A | PP2A | PP2A | EF-P | PP2A | PP2A | CYP | PP2A | GAPDH | PP2A | EIF4α | SAMDC | EIF4A | EF-P | EIF4α |
| 2 | ACP2 | EIF4α | EIF4α | MACP | EIF4α | CYP | PP2A | CYP | EF-P | CYP | SAMDC | PP2A | PP2A | PP2C | PP2A |
| 3 | EIF4α | ACP2 | ACP2 | PP2C | ACP2 | PP2C | EF-P | α-TUB | PP2A | EF-P | PP2A | EIF4α | SAMP | CYP | SAMDC |
| 4 | MACP | SAMDC | MACP | 6PGDH | MACP | EF-P | MACP | MDH | CYP | PP2C | ACP2 | ACP2 | ACP2 | 6PGDH | ACP2 |
| 5 | SAMDC | ADP-RF | SAMDC | CYP | EF-P | MDH | PP2C | PP2C | EIF4α | α-TUB | 6PGDH | 6PGDH | 6PGDH | MACP | 6PGDH |
| 6 | 6PGDH | 6PGDH | 6PGDH | EIF4α | 6PGDH | α-TUB | ACP2 | EF-P | MACP | MACP | MACP | ADP-RF | MACP | ACP2 | EF-P |
| 7 | EF-P | EF-P | PP2C | PP2A | SAMDC | MACP | MDH | MACP | α-TUB | MDH | ADP-RF | EF-P | β-TUB | EIF4α | MACP |
| 8 | ADP-RF | MACP | EF-P | ACP2 | PP2C | ACP2 | α-TUB | ACP2 | ACP2 | GAPDH | EF-P | MACP | A-TUB | UBC1 | PP2C |
| 9 | MDH | MDH | ADP-RF | ADP-RF | ADP-RF | EIF4α | ADP-RF | EIF4α | MDH | ACP2 | β-TUB | MDH | ELFP | PP2A | ADP-RF |
| 10 | PP2C | α-TUB | MDH | SAMDC | MDH | ADP-RF | EIF4α | SAMDC | ADP-RF | EIF4α | α-TUB | α-TUB | ADPRF | SAMDC | CYP |
| 11 | α-TUB | PP2C | α-TUB | MDH | CYP | SAMDC | SAMDC | ADP-RF | PP2C | ADP-RF | MDH | B-TUB | PP2C | β-TUB | β-TUB |
| 12 | β-TUB | β-TUB | β-TUB | UBC1 | α-TUB | 6PGDH | 6PGDH | 6PGDH | SAMDC | SAMDC | PP2C | PP2C | MDH | ADP-RF | α-TUB |
| 13 | CYP | CYP | CYP | α-TUB | β-TUB | GAPDH | GAPDH | GAPDH | 6PGDH | 6PGDH | CYP | UBC1 | CYP | MDH | MDH |
| 14 | UBC1 | UBC1 | UBC1 | β-TUB | UBC1 | UBC1 | UBC1 | UBC1 | UBC1 | UBC1 | UBC1 | CYP | UBC1 | α-TUB | UBC1 |
| 15 | GAPDH | GAPDH | GAPDH | GAPDH | GAPDH | β-TUB | β-TUB | β-TUB | β-TUB | β-TUB | GAPDH | GAPDH | GAPDH | GAPDH | GAPDH |
Validation of the best and least ranked sorghum reference genes
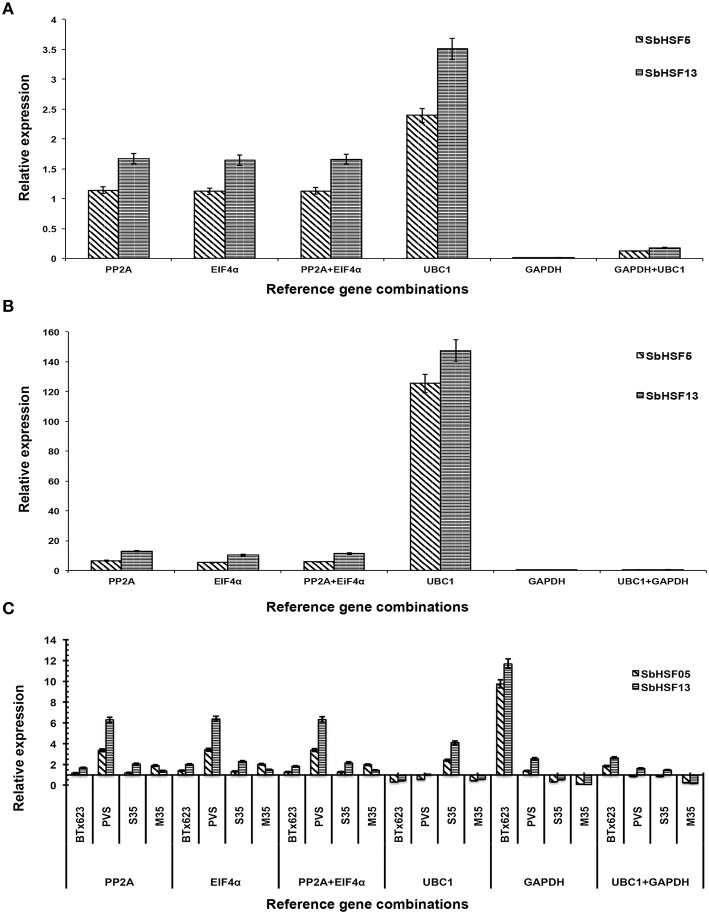
To validate the selected candidate reference genes, the expression patterns of SbHSF5 and SbHSF13 were assayed in root tissues, water-stressed samples and different varieties (Figures 6A–C). In sorghum, SbHSF5 and SbHSF13 genes have been shown to express in different tissues and induced by abiotic stress treatments (Nagaraju et al., 2015). In this study, the two most stable reference genes from all the sample set, PP2A and EIF4α, were used for normalization: in roots under normal growth conditions, SbHSF5 and SbHSF13 expression were not significantly different. In contrast, the SbHSF5 and SbHSF13 expression patterns were very different when GAPDH and UBC1, the least stable genes from all the sample set, were used for normalization. Normalization with UBC1 indicated a 2- to 3-fold increase in expression, while normalization with GAPDH and GAPDH+UBC1 indicated downregulation of both genes in root tissues under normal conditions (Figure 6A). To further validate the selected reference genes, the transcript levels were quantified in five sorghum varieties under water stress conditions (Figure 6C). For water stress samples, SbHSF5 and SbHSF13 expression levels increased1 to 12-fold under water stress conditions when normalized with the two most stable reference genes from all the sample set (PP2A and EIF4α; Figures 6B,C). As expected, normalization with the least stable genes (GAPDH and UBC1), either alone or in combination with UBC1 resulted in discrepancies for expression of both SbHSF5 and SbHSF13 gene expression (Figures 6B,C).
Figure 6.
Relative expression of SbHSF5 and SbHSF13 genes in root tissues (A) leaves during water stress treatment (B) selected varieties (C) using different reference genes selected by the RefFinder tool. SbHSF5 and SbHSF13 expression levels were normalized in a single and combined manner with either a stable or unstable reference genes. All samples were analyzed in triplicate, in three independent experiments.
Discussion
Accurate gene expression primarily relies on the stable expression of reference genes under diverse experimental conditions; however, thus far, no single reference gene has been shown to be stably expressed across experimental conditions (Olsvik et al., 2008; Cruz et al., 2009). The stability value of the most stable reference gene or the best combination of genes may vary from one experimental setup to another, meaning that the reference genes used for normalization should be validated under certain experimental conditions by statistically and experimentally. In the present study, 15 candidate reference genes were evaluated for expression stability among diverse experimental samples including various tissues and abiotic stress conditions. Expression analysis revealed that all 15 candidate reference genes varied significantly across the 10 tested samples; therefore, reference genes for optimal normalization were chosen from a set of candidate genes for each experiment either alone or in combination. To determine the comprehensive ranking of candidate reference genes in a sample set, we also used RefFinder, which considers the results of the four algorithms (geNorm, NormFinder, BestKeeper and the ΔCq method) together. Based on their stability rankings, various candidate genes have been proposed for their suitability as reference genes depending on the experimental conditions, implying that the reference genes used for abiotic stress studies in sorghum might not be suitable for gene expression analysis across genotypes and species.
In all the sample set, all three algorithms identified two reference genes, PP2A and EIF4α, as the two most stable genes. The pairwise variation calculated by geNorm also indicated that the use of two genes would be sufficient for normalization (Figure 4). Based on these results, it is inferred that PP2A and EIF4α would be appropriate for qPCR data normalization for the sorghum all sample set. For the abiotic stress treatment sample set, the pairwise variation indicated that the top ranking two genes (PP2A and CYP) could be used for normalization (Figure 4). Similarly, PP2A and EIF4α would be sufficient for the normalization of the tissue sample set data (Figure 4). PP2A was identified as one of the two most stable genes in all three studied sample sets (Table 2). This result is in agreement with previous studies of maturing B. napus embryos (Chen et al., 2010), multiple stress conditions (Wang et al., 2014), development and under various environmental conditions in A. thaliana (Czechowski et al., 2005) and Caragana intermedia (Zhu et al., 2013), as well as for virus-infected Nicotiana benthamiana (Liu et al., 2012) and Z. mays (Zhang et al., 2013). Furthermore, PP2A was the most stably expressed gene among various Striga life stages (Fernández-Aparicio et al., 2013). EIF4α was second most stably expressed gene in the all sample and tissue sample sets (Table 2). Similar results were observed in Carica papaya, where EIF4α expression was stable under most of the experimental conditions tested (Zhu et al., 2012). EIF4α was also reported as being a stably expressed gene in sexual and apomictic Brachiaria brizantha (Silveira et al., 2009), and in sexual and apomictic accessions of Cenchrus ciliaris (Simon et al., 2013). In the present study, CYP was the third most stable gene; it was stably expressed under abiotic stress treatments and in the tissue sample set (Table 2). The results obtained in the present study are in agreement with our previous studies in peanuts, where CYP was the most stably expressed gene during vegetative stages and under abiotic stress conditions (Reddy et al., 2013) and was also stably expressed in various tissues of G. max (Jian et al., 2008), Vicia faba (Gutierrez et al., 2011) and salt-stressed S. tuberosusm (Nicot et al., 2005). The CYP gene was moderately stable in developing and germinating G. max seeds (Li et al., 2012) and in bananas under various experimental conditions (Chen et al., 2011). In contrast, our recent study in Cicer arietinum indicated that CYP was the least stably expressed gene under various experimental conditions (Reddy et al., 2016), consistent with a previous study of grapevine berry development (Reid et al., 2006). In conclusion, PP2A, EIF4α, and CYP are recommended as suitable reference genes for the qPCR-based normalization of gene expression in sorghum under abiotic stress and in various tissues.
To further validate the candidate reference genes selected from the RefFinder comprehensive ranking, the expression levels of two sorghum heat shock transcription factors, SbHSF5 and SbHSF13, were assessed. Plant heat shock transcription factors play major roles in higher plants under various abiotic stresses. In sorghum, SbHSF5 and SbHSF13 are expressed under abiotic stress conditions: SbHSF5 is moderately expressed under cold stress and upregulated under drought stress, and SbHSF13 is moderately expressed under cold and heat stress and upregulated under drought and high salinity stress (Nagaraju et al., 2015). In the present study, SbHSF5 and SbHSF13 expression levels were upregulated under water stress conditions and no significant changes were observed in root tissue gene expression levels when normalized to the most stable reference genes, PP2A and EIF4α (Figures 6A,B). While, SbHSF5 and SbHSF13 genes significantly upregulated under water stress conditions in leaf tissues of all five sorghum genotypes with relatively higher expression when normalized with the sable reference genes (PP2A and EIF4α), the normalization was obscured when least stable reference gene (s) (UBC1 and GAPDH) were used (Figures 6B,C). The upregulation of SbHSF5 and SbHSF13 under this study is in agreement with another recent study in sorghum (Nagaraju et al., 2015) confirming the stability validation of the selected candidate reference genes and demonstrated the disadvantages of using unstable reference genes for normalization.
Conclusion
This study reports on the selection and validation of stable reference genes for qPCR-based gene expression studies in sorghum in response to various abiotic stresses and in different tissues. Three major statistical tools, geNorm, NormFinder and RefFinder, were used to analyze the suitability of reference genes. Some slight differences were observed between the statistical tools with regards to the selected stable reference genes. Overall, PP2A and EIF4α are the most stable candidate reference genes, and UBC1 and GAPDH were found to be least stable. The use of two reference genes would be optimal for the normalization of sorghum gene expression levels in various tissues and abiotic stress conditions. The most and least stable reference genes were further validated for their suitability as reference genes by normalizing SbHSF5 and SbHSF13 expression levels under various experimental conditions across five genotypes. This work will benefit future studies of gene expression in related experiments using different tissues and abiotic stress conditions in S. bicolor and related crops.
Author contributions
PS, KS, and VV designed the experiments. PS, DS, SK, and PB performed the experiments and data analysis. PS, DS and KS drafted the manuscript.
Funding
Financial support to PS through the INSPIRE Faculty Award (IFA11-LSPA-06) and the Young Scientist Scheme SB/YS/LS-12/2013 provided by the Department of Science and Technology, Government of India, New Delhi is gratefully acknowledged. This work was performed as part of the CGIAR Research Program on Dryland Cereals.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
PS acknowledges INSPIRE Faculty Award (IFA11-LSPA-06) and the Young Scientist Scheme (SB/YS/LS-12/2013) from the Department of Science and Technology, Government of India, New Delhi. This work was performed as part of the CGIAR Research Program on Dryland Cereals. ICRISAT is a member of the CGIAR Consortium.
Glossary
Abbreviations
- MACP
Malonyl CoA-Acyl Carrier protein
- 6PGDH
6-Phosphogluconate dehydrogenase
- SAMDC
S-adenosylmethionine decarboxylase
- CYP
Cyclophilin/Peptidylprolyl Isomerase
- PP2A
Serine/threonine-Protein Phosphatase 2A
- qPCR
Quantitative Real-Time Polymerase Chain Reaction.
References
- Andersen C. L., Jensen J. L., Ørntoft T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- Bustin S. A. (2002). Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29, 23–39. 10.1677/jme.0.0290023 [DOI] [PubMed] [Google Scholar]
- Bustin S. A., Beaulieu J. F., Huggett J., Jaggi R., Kibenge F. S. B., Olsvik P. A., et al. (2010). MIQE precis: practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol. Biol. 11:74. 10.1186/1471-2199-11-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- Calviño M., Messing J. (2012). Sweet sorghum as a model system for bioenergy crops. Curr. Opin. Biotechnol. 23, 323–329. 10.1016/j.copbio.2011.12.002 [DOI] [PubMed] [Google Scholar]
- Chen L., Zhong H. Y., Kuang J. F., Li J. G., Lu W. J., Chen J. Y. (2011). Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 234, 377–390. 10.1007/s00425-011-1410-3 [DOI] [PubMed] [Google Scholar]
- Chen X., Truksa M., Shah S., Weselake R. J. (2010). A survey of quantitative real-time polymerase chain reaction internal reference genes for expression studies in Brassica napus. Anal. Biochem. 405, 138–140. 10.1016/j.ab.2010.05.032 [DOI] [PubMed] [Google Scholar]
- Cook D., Rimando A. M., Clemente T. E., Schröder J., Dayan F. E., Nanayakkara N. P., et al. (2010). Alkylresorcinol synthases expressed in Sorghum bicolor root hairs play an essential role in the biosynthesis of the allelopathic benzoquinone sorgoleone. Plant Cell 22, 867–887. 10.1105/tpc.109.072397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz F., Kalaoun S., Nobile P., Colombo C., Almeida J., Barros L. M. G., et al. (2009). Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Mol. Breed. 23, 607–616. 10.1007/s11032-009-9259-x [DOI] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M. K., Scheible W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. 10.1104/pp.105.063743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidenko N. V., Logacheva M. D., Penin A. A. (2011). Selection and validation of reference genes for quantitative real-time PCR in buckwheat (Fagopyrum esculentum) based on transcriptome sequence data. PLoS ONE 6:e19434. 10.1371/journal.pone.0019434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheda K., Huggett J. F., Chang J. S., Kim L. U., Bustin S. A., Johnson M. A., et al. (2005). The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 344, 141–143. 10.1016/j.ab.2005.05.022 [DOI] [PubMed] [Google Scholar]
- Dugas D. V., Monaco M. K., Olsen A., Klein R. R., Kumari S., Ware D., et al. (2011). Functional annotation of the transcriptome of Sorghum bicolor in response to osmotic stress and abscisic acid. BMC Genomics 12:514. 10.1186/1471-2164-12-514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E., Levanon E. Y. (2013). Human housekeeping genes, revisited. Trends Genet. 29, 569–574. 10.1016/j.tig.2013.05.010 [DOI] [PubMed] [Google Scholar]
- Expósito-Rodríguez M., Borges A. A., Borges-Perez A., Pérez J. A. (2008). Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 8:131. 10.1186/1471-2229-8-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2011). FAOSTAT Database. Available online at: http://faostat.fao.org
- Fernández-Aparicio M., Huang K., Wafula E. K., Honaas L. A., Wickett N. J., Timko M. P., et al. (2013). Application of qRT-PCR and RNA-Seq analysis for the identification of housekeeping genes useful for normalization of gene expression values during Striga hermonthica development. Mol. Biol. Rep. 40, 3395–3407. 10.1007/s11033-012-2417-y [DOI] [PubMed] [Google Scholar]
- Gelli M., Duo Y. C., Konda A. R., Zhang C., Holding D., Dweikat I. (2014). Identification of differentially expressed genes between sorghum genotypes with contrasting nitrogen stress tolerance by genome-wide transcriptional profiling. BMC Genomics 15:179. 10.1186/1471-2164-15-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno J., Eattock N., Van Deynze A., Blumwald E. (2014). Selection and validation of reference genes for gene expression analysis in switchgrass (Panicum virgatum) using quantitative real-time RT-PCR. PLoS ONE 9:e91474. 10.1371/journal.pone.0091474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez N., Gimenez M. J., Palomino C., Avila C. M. (2011). Assessment of candidate reference genes for expression studies in Vicia faba L. by real-time quantitative PCR. Mol. Breed. 28, 13–24. 10.1007/s11032-010-9456-7 [DOI] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:r19. 10.1186/gb-2007-8-2-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R., Aoki M., Kurotani K., Yokoi S., Shinomura T., Takano M., et al. (2011). Phytochrome B regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol. Genet. Genomics 285, 461–470. 10.1007/s00438-011-0621-4 [DOI] [PubMed] [Google Scholar]
- Iskandar H. M., Simpson R. S., Casu R. E., Bonnett G. D., Maclean D. J., Manners J. M. (2004). Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene expression. Plant Mol. Biol. Rep. 22, 325–337. 10.1007/BF02772676 [DOI] [Google Scholar]
- Jain M., Chourey P. S., Li Q. B., Pring D. R. (2008). Expression of cell wall invertase and several other genes of sugar metabolism in relation to seed development in sorghum (Sorghum bicolor). J. Plant Physiol. 165, 331–344. 10.1016/j.jplph.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Jain M., Nijhawan A., Tyagi A. K., Khurana J. P. (2006). Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 345, 646–651. 10.1016/j.bbrc.2006.04.140 [DOI] [PubMed] [Google Scholar]
- Ji Y., Tu P., Wang K., Gao F., Yang W., Zhu Y., et al. (2014). Defining reference genes for quantitative real-time PCR analysis of anther development in rice. Acta Biochim. Biophys. Sin. (Shanghai). 46, 305–312. 10.1093/abbs/gmu002 [DOI] [PubMed] [Google Scholar]
- Jian B., Liu B., Bi Y. R., Hou W. S., Wu C. X., Han T. F. (2008). Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol. Biol. 9:59. 10.1186/1471-2199-9-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom T. H., Mullet J. E. (2015). Photosynthetic leaf area modulates tiller bud outgrowth in sorghum. Plant Cell Environ. 38, 1471–1478. 10.1111/pce.12500 [DOI] [PubMed] [Google Scholar]
- Koegel S., Ait Lahmidi N., Arnould C., Chatagnier O., Walder F., Ineichen K., et al. (2013). The family of ammonium transporters (AMT) in Sorghum bicolor: two AMT members are induced locally, but not systemically in roots colonized by arbuscular mycorrhizal fungi. New Phytol. 198, 853–865. 10.1111/nph.12199 [DOI] [PubMed] [Google Scholar]
- Kumar K., Muthamilarasan M., Prasad M. (2013). Reference genes for quantitative real-time PCR analysis in the model plant foxtail millet (Setaria italica L.) subjected to abiotic stress conditions. Plant Cell Tissue Organ. Cult. 115, 13–22. 10.1007/s11240-013-0335-x [DOI] [Google Scholar]
- Li J. Q., Wang L. H., Zhan Q. W., Liu Y. L., Fu B. S., Wang C. M. (2013). Sorghum bmr6 mutant analysis demonstrates that a shared MYB1 transcription factor binding site in the promoter links the expression of genes in related pathways. Funct. Integr. Genomics 13, 445–453. 10.1007/s10142-013-0335-2 [DOI] [PubMed] [Google Scholar]
- Li J., Wang L., Zhang Q., Liu Y. (2015). Map-based cloning and expression analysis of BMR-6 in sorghum. J. Genet. 94, 445–452. 10.1007/s12041-015-0550-9 [DOI] [PubMed] [Google Scholar]
- Li Q., Fan C. M., Zhang X. M., Fu Y. F. (2012). Validation of reference genes for real-time quantitative PCR normalization in soybean developmental and germinating seeds. Plant Cell Rep. 31, 1789–1798. 10.1007/s00299-012-1282-4 [DOI] [PubMed] [Google Scholar]
- Li Q. F., Sun S. S. M., Yuan D. Y., Yu H. X., Gu M. H., Liu Q. Q. (2010). Validation of candidate reference genes for the accurate normalization of real-time quantitative rT-PCR data in rice during seed development. Plant Mol. Biol. Rep. 28, 49–57. 10.1007/s11105-009-0124-1 [DOI] [Google Scholar]
- Lin F., Jiang L., Liu Y. H., Lv Y. D., Dai H. X., Zhao H. (2014). Genome-wide identification of housekeeping genes in maize. Plant Mol. Biol. 86, 543–554. 10.1007/s11103-014-0246-1 [DOI] [PubMed] [Google Scholar]
- Ling H., Wu Q., Guo J., Xu L., Que Y. (2015). Comprehensive selection of reference genes for gene expression normalization in sugarcane by real time quantitative RT-PCR. PLoS ONE 10:e97469. 10.1371/journal.pone.0097469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. S., Shi L. D., Han C. G., Yu J. L., Li D. W., Zhang Y. L. (2012). Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PLoS ONE 7:e46451. 10.1371/journal.pone.0046451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado R. D., Christoff A. P., Loss-Morais G., Margis-Pinheiro M., Margis R., Körbes A. P. (2015). Comprehensive selection of reference genes for quantitative gene expression analysis during seed development in Brassica napus. Plant Cell Rep. 34, 1139–1149. 10.1007/s00299-015-1773-1 [DOI] [PubMed] [Google Scholar]
- Manoli A., Sturaro A., Trevisan S., Quaggiotti S., Nonis A. (2012). Evaluation of candidate reference genes for qPCR in maize. J. Plant Physiol. 169, 807–815. 10.1016/j.jplph.2012.01.019 [DOI] [PubMed] [Google Scholar]
- Mariot R. F., De Oliveira L. A., Voorhuijzen M. M., Staats M., Hutten R. C., Van Dijk J. P., et al. (2015). Selection of reference genes for transcriptional analysis of edible tubers of potato (Solanum tuberosum L.). PLoS ONE 10:e0120854. 10.1371/journal.pone.0120854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraju M., Reddy P. S., Kumar S. A., Srivastava R. K., Kavi Kishor P. B., Rao D. M. (2015). Genome wide scanning and characterization of Sorghum bicolor L. heat shock transcription factors. Curr. Genom. 16, 279–291. 10.2174/1389202916666150313230812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T. J., Rodrigues F. A., Neumaier N., Marcelino-Guimarães F. C., Farias J. R., De Oliveira M. C., et al. (2014). Reference genes for quantitative real-time polymerase chain reaction studies in soybean plants under hypoxic conditions. Genet. Mol. Res. 13, 860–871. 10.4238/2014.February.13.4 [DOI] [PubMed] [Google Scholar]
- Nicot N., Hausman J. F., Hoffmann L., Evers D. (2005). Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 56, 2907–2914. 10.1093/jxb/eri285 [DOI] [PubMed] [Google Scholar]
- Olsvik P. A., Softeland L., Lie K. K. (2008). Selection of reference genes for qRT-PCR examination of wild populations of Atlantic cod Gadus morhua. BMC Res. Notes 1:47. 10.1186/1756-0500-1-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolacci A. R., Tanzarella O. A., Porceddu E., Ciaffi M. (2009). Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 10:11. 10.1186/1471-2199-10-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Bowers J. E., Bruggmann R., Dubchak I., Grimwood J., Gundlach H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556. 10.1038/nature07723 [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W., Horgan G. W., Dempfle L. (2002). Relative expression software tool (REST (c)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. 10.1093/nar/30.9.e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. 10.1023/B:BILE.0000019559.84305.47 [DOI] [PubMed] [Google Scholar]
- Price H. J., Dillon S. L., Hodnett G., Rooney W. L., Ross L., Johnston J. S. (2005). Genome evolution in the genus Sorghum (Poaceae). Ann. Bot. 95, 219–227. 10.1093/aob/mci015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy D. S., Bhatnagar-Mathur P., Cindhuri K. S., Sharma K. K. (2013). Evaluation and validation of reference genes for normalization of quantitative real-time PCR based gene expression studies in peanut. PLoS ONE 8:e78555. 10.1371/journal.pone.0078555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy D. S., Bhatnagar-Mathur P., Reddy P. S., Sri Cindhuri K., Sivaji Ganesh A., Sharma K. K. (2016). Identification and validation of reference genes and their impact on normalized gene expression studies across cultivated and wild cicer species. PLoS ONE 11:e0148451. 10.1371/journal.pone.0148451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P. S., Rao T. S. R., Sharma K. K., Vadez V. (2015a). Genome-wide identification and characterization of the aquaporin gene family in Sorghum bicolor (L.). Plant Gene 1, 18–28. 10.1016/j.plgene.2014.12.002 [DOI] [Google Scholar]
- Reddy P. S., Reddy D. S., Bhatnagar-Mathur P., Sharma K. K., Vadez V. (2015b). Cloning and validation of reference genes for normalization of gene expression studies in pearl millet [Pennisetum glaucum (L.) R. Br.] by quantitative real-time PCR. Plant Gene 1, 35–42. 10.1016/j.plgene.2015.02.001 [DOI] [Google Scholar]
- Reid K. E., Olsson N., Schlosser J., Peng F., Lund S. T. (2006). An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 6:27. 10.1186/1471-2229-6-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T., Smeets K., Opdenakker K., Mathijsen D., Vangronsveld J., Cuypers A. (2008). Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227, 1343–1349. 10.1007/s00425-008-0706-4 [DOI] [PubMed] [Google Scholar]
- Saha P., Blumwald E. (2014). Assessing reference genes for accurate transcript normalization using quantitative real-time PCR in pearl millet [Pennisetum glaucum (L.) R. Br.]. PLoS ONE 9:e106308. 10.1371/journal.pone.0106308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakoor N., Nair R., Crasta O., Morris G., Feltus A., Kresovich S. (2014). A Sorghum bicolor expression atlas reveals dynamic genotype-specific expression profiles for vegetative tissues of grain, sweet and bioenergy sorghums. BMC Plant Biol. 14:35. 10.1186/1471-2229-14-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Bai Y., Wang S., Zhang S., Wu Y., Chen M., et al. (2010). Expression profile of PIN, AUX/LAX and PGP auxin transporter gene families in Sorghum bicolor under phytohormone and abiotic stress. FEBS J. 277, 2954–2969. 10.1111/j.1742-4658.2010.07706.x [DOI] [PubMed] [Google Scholar]
- Silveira E. D., Alves-Ferreira M., Guimaraes L. A., Da Silva F. R., Carneiro V. T. (2009). Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biol. 9:84. 10.1186/1471-2229-9-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver N., Best S., Jiang J., Thein S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7:33. 10.1186/1471-2199-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon B., Conner J. A., Ozias-Akins P. (2013). Selection and validation of reference genes for gene expression analysis in apomictic and sexual Cenchrus ciliaris. BMC Res. Notes 6:397. 10.1186/1756-0500-6-397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha P., Singh V. K., Suryanarayana V., Krishnamurthy L., Saxena R. K., Varshney R. K. (2015). Evaluation and validation of housekeeping genes as reference for gene expression studies in pigeonpea (Cajanus cajan) under drought stress conditions. PLoS ONE 10:e0122847. 10.1371/journal.pone.0122847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thellin O., Zorzi W., Lakaye B., De Borman B., Coumans B., Hennen G., et al. (1999). Housekeeping genes as internal standards: use and limits. J. Biotechnol. 75, 291–295. 10.1016/S0168-1656(99)00163-7 [DOI] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B. C., Remm M., et al. (2012). Primer3-new capabilities and interfaces. Nucleic Acids Res. 40, e115. 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadez V., Sinclair T. R. (2001). Leaf ureide degradation and N(2) fixation tolerance to water deficit in soybean. J. Exp. Bot. 52, 153–159. 10.1093/jexbot/52.354.153 [DOI] [PubMed] [Google Scholar]
- Valasek M. A., Repa J. J. (2005). The power of real-time PCR. Adv. Physiol. Educ. 29, 151–159. 10.1152/advan.00019.2005 [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermerris W. (2011). Survey of genomics approaches to improve bioenergy traits in maize, sorghum and sugarcane. J. Integr. Plant Biol. 53, 105–119. 10.1111/j.1744-7909.2010.01020.x [DOI] [PubMed] [Google Scholar]
- Walder F., Brulé D., Koegel S., Wiemken A., Boller T., Courty P. E. (2015). Plant phosphorus acquisition in a common mycorrhizal network: regulation of phosphate transporter genes of the Pht1 family in sorghum and flax. New Phytol. 205, 1632–1645. 10.1111/nph.13292 [DOI] [PubMed] [Google Scholar]
- Wang S. K., Bai Y. H., Shen C. J., Wu Y. R., Zhang S. N., Jiang D. A., et al. (2010). Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct. Integr. Genomics 10, 533–546. 10.1007/s10142-010-0174-3 [DOI] [PubMed] [Google Scholar]
- Wang Z., Chen Y., Fang H. D., Shi H. F., Chen K. P., Zhang Z. Y., et al. (2014). Selection of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in Brassica napus under various stress conditions. Mol. Genet. Genomics 289, 1023–1035. 10.1007/s00438-014-0853-1 [DOI] [PubMed] [Google Scholar]
- Wong M. L., Medrano J. F. (2005). Real-time PCR for mRNA quantitation. BioTechniques 39, 75–85. 10.2144/05391RV01 [DOI] [PubMed] [Google Scholar]
- Yang H. L., Liu J., Huang S. M., Guo T. T., Deng L. B., Hua W. (2014a). Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene 538, 113–122. 10.1016/j.gene.2013.12.057 [DOI] [PubMed] [Google Scholar]
- Yang S., Murphy R. L., Morishige D. T., Klein P. E., Rooney W. L., Mullet J. E. (2014b). Sorghum phytochrome B inhibits flowering in long days by activating expression of SbPRR37 and SbGHD7, repressors of SbEHD1, SbCN8 and SbCN12. PLoS ONE 9:e105352. 10.1371/journal.pone.0105352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Scheffler B. E., Weston L. A. (2004). SOR1, a gene associated with bioherbicide production in sorghum root hairs. J. Exp. Bot. 55, 2251–2259. 10.1093/jxb/erh252 [DOI] [PubMed] [Google Scholar]
- Yin L., Wang S., Liu P., Wang W., Cao D., Deng X., et al. (2014). Silicon-mediated changes in polyamine and 1-aminocyclopropane-1-carboxylic acid are involved in silicon-induced drought resistance in Sorghum bicolor L. Plant Physiol. Biochem. 80, 268–277. 10.1016/j.plaphy.2014.04.014 [DOI] [PubMed] [Google Scholar]
- Zhang K., Niu S. F., Di D. P., Shi L. D., Liu D. S., Cao X. L., et al. (2013). Selection of reference genes for gene expression studies in virus-infected monocots using quantitative real-time PCR. J. Biotechnol. 168, 7–14. 10.1016/j.jbiotec.2013.08.008 [DOI] [PubMed] [Google Scholar]
- Zhu J. F., Zhang L. F., Li W. F., Han S. Y., Yang W. H., Qi L. W. (2013). Reference gene selection for quantitative real-time PCR normalization in Caragana intermedia under different abiotic stress conditions. PLoS ONE 8:e53196. 10.1371/journal.pone.0053196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. Y., Li X. P., Chen W. X., Chen J. Y., Lu W. J., Chen L., et al. (2012). Evaluation of new reference genes in papaya for accurate transcript normalization under different experimental conditions. PLoS ONE 7:e44405. 10.1371/journal.pone.0044405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmienko A., Samelak-Czajka A., Goralski M., Sobieszczuk-Nowicka E., Kozlowski P., Figlerowicz M. (2015). Selection of reference genes for qPCR- and ddPCR-based analyses of gene expression in Senescing Barley leaves. PLoS ONE 10:e0118226. 10.1371/journal.pone.0118226 [DOI] [PMC free article] [PubMed] [Google Scholar]