Here, we describe the dramatic response of a patient with an ETV6-NTRK3-driven mammary analogue secretory carcinoma to treatment with a pan-Trk inhibitor, and the development of acquired resistance linked to a novel NTRK3 mutation that interferes with drug binding. This case emphasizes how molecular profiling can identify therapies for rare diseases and dissect mechanisms of drug resistance.
Keywords: ETV6-NTRK3, TrkC, mammary analogue secretory carcinoma, entrectinib
Abstract
Background
Mammary analogue secretory carcinoma (MASC) is a recently described pathologic entity. We report the case of a patient with an initial diagnosis of salivary acinic cell carcinoma later reclassified as MASC after next-generation sequencing revealed an ETV6-NTRK3 fusion.
Patients and methods
This alteration was targeted with the pan-Trk inhibitor entrectinib (Ignyta), which possesses potent in vitro activity against cell lines containing various NTRK1/2/3 fusions.
Results
A dramatic and durable response was achieved with entrectinib in this patient, followed by acquired resistance that correlated with the appearance of a novel NTRK3 G623R mutation. Structural modeling predicts that this alteration sterically interferes with drug binding, correlating to decreased sensitivity to drug inhibition observed in cell-based assays.
Conclusions
This first report of clinical activity with TrkC inhibition and the development of acquired resistance in an NTRK3-rearranged cancer emphasize the utility of comprehensive molecular profiling and targeted therapy for rare malignancies (NCT02097810).
introduction
Mammary analogue secretory carcinoma (MASC) of the salivary gland is a recently identified salivary cancer subtype that commonly originates in the parotid gland [1]. Before its designation as a separate entity, MASCs were predominantly grouped with other low-grade salivary cancer histologies [most commonly acinic cell carcinoma (AciCC)]. In 2010, identification of the ETV6-NTRK3 translocation t(12:15)(p13;q25) confirmed these tumors to be a molecularly distinct disease; the histologic resemblance to secretory carcinoma of the breast (which also harbors this fusion gene) inspired the designation ‘mammary analogue secretory carcinoma’ [2, 3]. Of note, while ETV6-NTRK3 represents the most common fusion in MASCs, some cases have been found to harbor rearrangements involving ETV6 and a non-NTRK3 partner [4].
Recurrent gene rearrangements such as ETV6-NTRK3 are a critical mechanism of oncogenic activation for the neurotrophic tyrosine receptor kinase genes, NTRK1, NTRK2, and NTRK3, in human malignancies [5]. These genes encode a family of tropomyosin receptor kinase proteins (TrkA, TrkB, and TrkC, respectively) that are involved in nervous system development. Apart from salivary gland tumors, these rearrangements have been identified in lung, thyroid, and colon cancers, as well as sarcomas, spitzoid neoplasms, and primary brain tumors [6]. Fusion of the intact tyrosine kinase domain of NTRK1, NTRK2, or NTRK3 with a variety of upstream partners results in dysregulated activation of several biochemical signaling pathways that promote oncogenic initiation and growth [7].
Here, we provide the first report of a dramatic clinical response to TrkC inhibition in an NTRK3-rearranged malignancy and the development of acquired resistance linked to a novel genomic mechanism.
materials and methods
genomic profiling
Broad, hybrid-capture-based next-generation sequencing was carried out using the Integrated Mutational Profiling of Actionable Cancer Targets (MSK-IMPACT) assay [8] and sequenced on a HiSeq 2500 (Illumina, San Diego, CA). Four hundred ten cancer-related genes were interrogated, capturing base substitutions, small INDELS, copy number alterations, and select rearrangements. To detect somatic structural aberrations via MSK-IMPACT, a framework was developed that first aligns raw reads to the reference human genome (hg19) using the Burrows-Wheeler Alignment tool. Duplicates are then filtered using the Picard-tools java package (samtools) and searched for candidate structural rearrangements using DELLY. All candidate somatic structural aberrations were filtered, annotated using in-house tools, and manually reviewed using the Integrative Genomics Viewer.
The FACETS algorithm was used to estimate tumor purity, ploidy, and allele-specific copy number [9]. Cancer cell fraction was estimated from mutant allele frequency, and corrected for tumor purity and the copy number state of the region of the gene containing the mutation. RNA sequencing was carried out using the Archer™ FusionPlex™ assay (ArcherDx, Boulder, CO); libraries were sequenced on an Illumina MiSeqDx. NTRK3 break-apart was assessed by a fluorescence in situ hybridization (FISH). For TrkC immunohistochemistry (IHC), 4-µm tissue sections on positively charged slides were deparaffinized and rehydrated, and antigens were retrieved for 30 min in a Tris–EDTA (pH 9) at 98°C. Both a TrkC-specific antibody [TrkC Antibody (798): sc-117, Santa Cruz Biotechnology] (Santa Cruz Biotechnology, Dallas, TX) and a primary antibody mixture that consisted of a cocktail of anti-pan-Trk (C17F1 Rabbit mAb, 1:25 dilution, Cell Signaling), anti-ROS1 (D4D6 Rabbit mAb, 1:500 dilution, Cell Signaling), and anti-ALK (C17F1 Rabbit mAb, 1:500 dilution, Cell Signaling) were used to amplify signal development.
targeted therapy administration
Crizotinib was obtained from a commercial supply. Computed tomography (CT) imaging was carried out at baseline and at various time points on therapy (3, 10, and 18 weeks) to monitor response. The TrkA, TrkB, TrkC, ROS1, and ALK tyrosine kinase inhibitor entrectinib (RXDX-101) was administered on a phase I clinical trial (NCT02097810). The primary end point of the dose escalation phase of this trial was to determine the maximum tolerated and recommended phase II doses of entrectinib. A conventional 3 + 3 dose escalation design was used. The patient was enrolled in the 400 mg/m2 cohort of the dose escalation phase, and a fixed daily dose of entrectinib was calculated. Response was assessed via the Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 at protocol-defined intervals [10]. Treatment was administered until disease progression or unacceptable toxicity.
functional and structural studies
cDNAs encoding various NTRK1-3 gene rearrangements (TPM3-NTRK1, LMNA-NTRK1, ETV6-NTRK1, VCL-NTRK2, AFAP1-NTRK2, ETV6-NTRK2, ETV6-NTRK3, and ETV6-NTRK3 G623R) were inserted into the lentiviral vector pVL-EF1a-MCS-IRES-Puro (BioSettia, San Diego, CA) and introduced into the murine IL-3 dependent pro-B-cell Ba/F3. Proliferation assays were conducted with various concentrations of vehicle, entrectinib, TSR-011, and LOXO-101. LOXO-101 [11] and TSR-011 [12] were synthesized by Bioduro (Beijing, China) based on publicly disclosed structures. Plates were incubated at 37°C in 5% CO2 for 72 h, after which cell viability was assessed by measuring ATP content using Cell Titer-Glo® Luminescent Cell Viability assay (Promega). IC50s were determined by 4-parameter curve fit with variable slope.
To assess the impact of entrectinib upon signaling pathways, Ba/F3-ETV6-NTRK3 cells were treated with 300 nM entrectinib, a concentration that is approximately 50% of the clinically achievable minimal concentration (Cmin). Cell lysates were prepared 4 h post treatment and probed for phosphorylated/total Trk, PLCγ1, PI3K (p85), MAPK, and Stat3. All primary antibodies were purchased from Cell Signaling Technology except for anti-β-actin (Millipore). Structural assessments of entrectinib binding were obtained using Glide docking implemented in Maestro [13]. Receptor coordinates were obtained from PDB (code: 4YMJ).
results
ETV6-NTRK3 identification and tumor reclassification
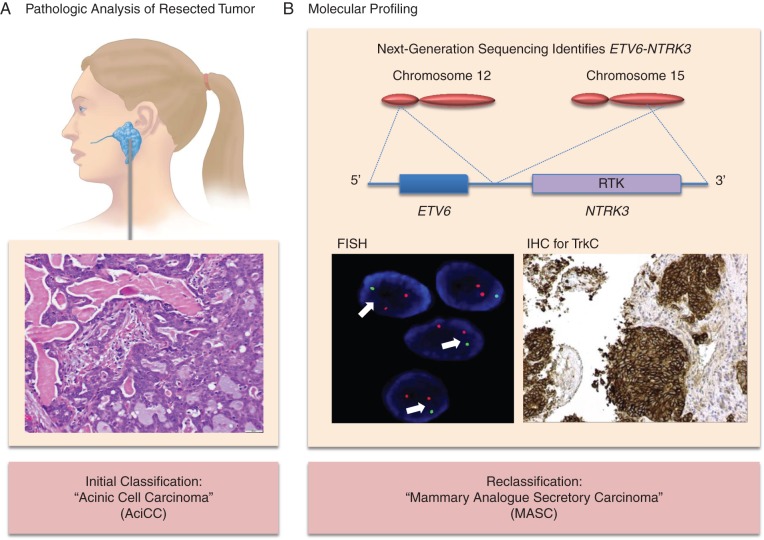
A 34-year-old female presented in January 2006 with a growing left parotid mass. A left superficial parotidectomy was carried out, revealing a stage III (pT3N0M0) parotid cancer initially classified as AciCC (Figure 1A). Surgical margins were microscopically involved with tumor, and postoperative intensity-modulated radiation therapy was administered to the left parotid and ipsilateral lower neck.
Figure 1.
ETV6-NTRK3 identification results in tumor reclassification. In panel A, pathology from a superficial parotidectomy revealed an infiltrative carcinoma with predominantly tubular and microcystic/macrocystic growth patterns and intraluminal secretions. Tumor cells exhibited granular eosinophilic cytoplasm with occasional vacuolation and had relatively bland cytology with mild atypia. A diagnosis of acinic cell carcinoma (AciCC) was made. In panel B, the ETV6-NTRK3 fusion detected via broad, hybrid-capture-based next-generation sequencing is depicted. Reciprocal translocation between chromosome 12 and chromosome 15 resulted in fusion of ETV6 exons 1–5 to NTRK3 exons 15–20 containing the receptor tyrosine kinase (RTK) domain. On the lower left, a positive break-apart fluorescence in situ hybridization (FISH) NTRK3 assay is shown with split signals (arrows). On the lower right, immunohistochemistry for TrkC revealed strong staining signifying TrkC overexpression. The patient's diagnosis was reclassified as mammary analogue secretory carcinoma (MASC).
In August 2011 the patient was diagnosed with biopsy-confirmed metastatic disease involving the lung, pleura, mediastinum, and chest wall. She was initially asymptomatic and managed with active surveillance. She eventually developed pleuritic right-sided chest pain caused by enlarging right-sided pleural metastases which were treated with two palliative surgical resections and three different lines of cytotoxic chemotherapy (vinorelbine, carboplatin and paclitaxel, and doxorubicin). A more detailed history is outlined in supplementary Table S1, available at Annals of Oncology online.
Pathologic review of a right lower lobe paraesophageal mass resected in February 2013 revealed a carcinoma morphologically similar to the patient's initial salivary tumor (supplementary Figure S1A, available at Annals of Oncology online). Broad, hybrid-capture-based next-generation sequencing of the mass identified an ETV6-NTRK3 t(12;15)(p13.2;q25.3) rearrangement (Figure 1B). Additional testing by FISH using an NTRK3 break-apart probe was positive (Figure 1B). IHC carried out with a TrkC-specific antibody (Figure 1B) and a Trk antibody cocktail that also detects TrkC (supplementary Figure S1B, available at Annals of Oncology online) revealed strong staining, confirming TrkC expression. RNA sequencing confirmed the presence of ETV6-NTRK3 (supplementary Figure S1C, available at Annals of Oncology online). The patient's malignancy was thus reclassified as MASC.
crizotinib therapy
Based on in vitro data indicating that the multitargeted kinase inhibitor crizotinib may modestly inhibit TrkC kinase in the context of the ETV6-NTRK3 rearrangement [8], the patient was treated with crizotinib. With the initiation of therapy and the performance of intercostal neurolysis, she quickly experienced resolution of her pleuritic pain. Repeat CT imaging at 3 and 10 weeks revealed stable disease (2% and 19% reduction in disease burden, respectively). A CT scan at 18 weeks unfortunately revealed disease progression accompanied by recurrent, tumor-related, chest wall pain. Additionally, the patient developed acute, painful swelling of the hands, legs, and feet bilaterally, determined to be consistent with a paraneoplastic hypertrophic osteoarthropathy in the setting of progressive malignancy. Nonsteroidal anti-inflammatory therapy was initiated with minimal improvement in symptoms.
response and acquired resistance to entrectinib
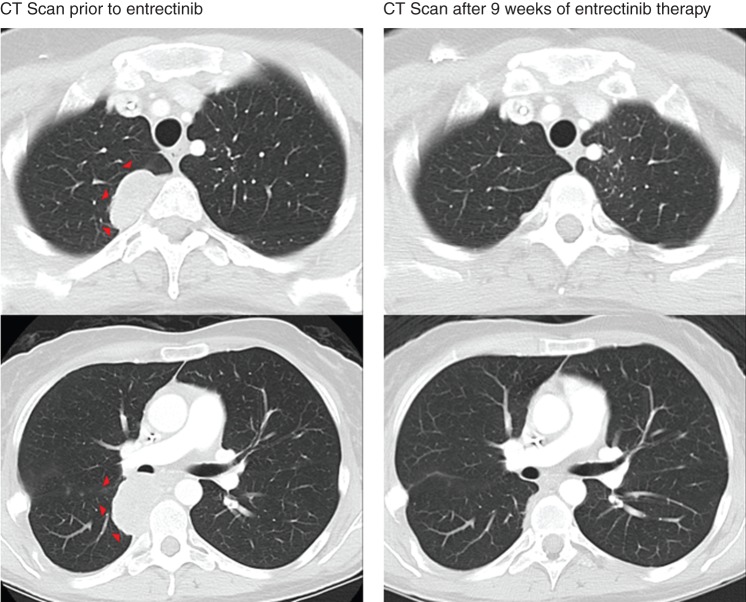
We hypothesized that TrkC activity remained the central oncogenic driver of the patient's disease despite progression on crizotinib and that inhibition with a potent Trk inhibitor would produce tumor regression. She was thus enrolled on to a phase I clinical trial of the pan-Trk inhibitor, entrectinib (NCT02097810). The patient noted a substantial decrease in hypertrophic osteoarthropathy-related limb edema within the first 4 weeks of treatment. These symptoms resolved by week 8 of therapy. CT imaging at 9 weeks (Figure 2) revealed a dramatic partial response with decreased pleural-based metastases. Response was confirmed at 13 weeks, and further disease shrinkage was noted at 21 weeks (89% reduction in tumor burden).
Figure 2.
A durable partial response is achieved with entrectinib therapy in an ETV6-NTRK3-rearranged mammary analogue secretory carcinoma. Computed tomography (CT) imaging of the patient after progression on crizotinib and before entrectinib therapy is shown on the left. Repeat CT imaging at 9 weeks revealed a dramatic partial response to therapy (RECIST v1.1) with an interval decrease and resolution of pleural-based metastases in the right hemithorax (arrows). This response was confirmed at 13 weeks and further shrinkage was noted at 21 weeks. A best radiologic response of 89% reduction in tumor burden from baseline was achieved.
After 7 months of therapy, imaging revealed asymptomatic disease progression in a solitary tumor site in the right lower lobe of the lung. Although this met RECIST criteria for progressive disease, given stable response at all other metastatic sites and the absence of tumor-related symptoms, she continued entrectinib at an increased dose (supplementary Table S1, available at Annals of Oncology online). Serial imaging at 8 and 9 months showed more modest growth of the same mass and continued stable response at other sites. Given ongoing clinical benefit and acceptable tolerability, she remained on entrectinib. Assessment 10 months into treatment unfortunately revealed further disease progression in not only the right lower lobe tumor, but also several other adjacent pulmonary nodules, leading to discontinuation of entrectinib.
NTRK3 G623R identification
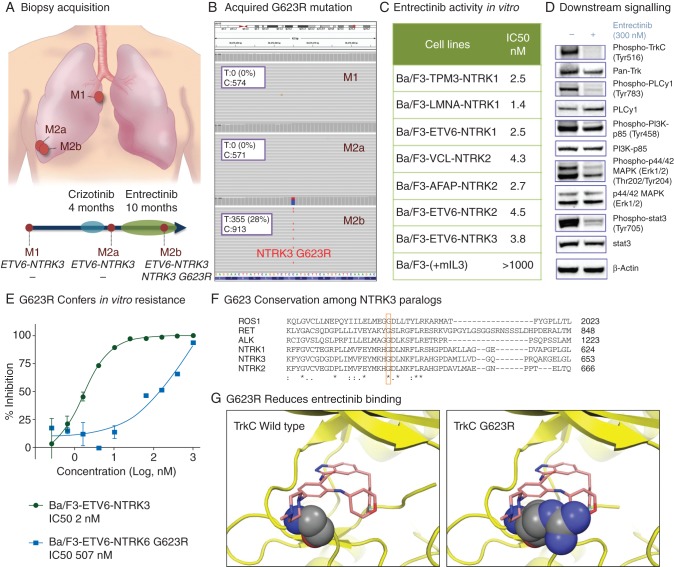
Next-generation sequencing was carried out on three tumor biopsies: M1 (paraesophageal right lower lobe mass acquired before crizotinib), M2a (separate pleural-based right lower lobe mass acquired before entrectinib), and M2b (pleural-based right lower lobe mass immediately adjacent to M2a that progressively enlarged on entrectinib therapy) (Figure 3A). While ETV6-NTRK3 was detected in all three tumors (supplementary Figure S2, available at Annals of Oncology online), a novel NTRK3 exon 16 G623R (c.1867G>A) mutation was identified in the M2b tumor, correlating to the development of entrectinib resistance (Figure 3B). The cancer cell fraction estimate for NTRK3 G623R was 90%, indicating its presence at a clonal level. Concurrent mutations in RB1 and MYC identified in M2a were maintained in M2b (supplementary Figure S3, available at Annals of Oncology online).
Figure 3.
The development of clinical entrectinib resistance is mediated by the appearance of a novel NTRK3 G623R mutation. In panel A, areas of tumor acquisition via serial biopsies are depicted: before crizotinib (M1, paraesophageal right lower lobe mass), after progression on crizotinib and before entrectinib (M2a, pleural-based right lower lobe mass), and after progression on entrectinib (M2b, pleural-based right lower lobe mass immediately adjacent to M2a). In panel B, broad, hybrid-capture-based next-generation sequencing confirmed the appearance of an NTRK3 G623R mutation after progression on entrectinib (M2b) that was not present in pre-entrectinib tumor samples (M1 and M2a). Panel C depicts the antiproliferative activity of entrectinib in engineered Ba/F3 cells expressing a variety of Trk fusion proteins with IC50s ranging from 1.4 to 4.5 nM. Entrectinib was found to inhibit phospho-TrkC and phospho-PLCy1, with less inhibition of PI3K, MAPK, and Stat3 as depicted in panel D. In panel E, introduction of the NTRK3 G623R mutation into the ETV6-NTRK3 construct (Ba/F3-ETV6-NTRK3 G623R) conferred reduced sensitivity to entrectinib, increasing the IC50 value in the proliferation assays by more than 250-fold relative to the Ba/F3-ETV6-NTRK3 cells. Homology alignment in panel F suggests that the native glycine at position 623 of TrkC is highly conserved among TrkC paralogs. A comparison to glycine residues at position 1202 of ALK, position 2032 of ROS1, and position 595 of TrkA, in addition to other paralogs, is shown. Panel G depicts the binding of entrectinib to both wild-type TrkC and NTRK3 G623-mutant TrkC. Extensive hydrogen bonding and hydrophobic interactions between wild-type TrkC and entrectinib occur in the ATP binding pocket where the G623 residue is located (left). The substitution of arginine for glycine at position 623 results in steric hindrance that decreases the binding of entrectinib to mutant TrkC (right).
A review of histology did not identify morphologic changes associated with the development of acquired resistance to therapy. While the patient's primary tumor resected in 2006 showed a lower grade histology with a mitotic count of 2/10 high-power fields and no tumor necrosis, pathologic features of all the metastatic tumors (M1, M2a, and M2b) were similar and more aggressive, each with a mitotic count of 6/10 high-power fields and notable tumor necrosis.
functional and structural evaluation
To evaluate the hypothesis that the NTRK3 G623R mutation mediates resistance to entrectinib, we investigated Trk inhibitor susceptibility in Ba/F3 cell lines overexpressing NTRK constructs. Entrectinib demonstrated potent antiproliferative activity in cell lines overexpressing various NTRK family rearrangements (TPM3-NTRK1, LMNA-NTRK1, ETV6-NTRK1, VCL-NTRK2, AFAP1-NTRK2, ETV6-NTRK2, ETV6-NTRK3) with IC50 values ranging between 1 and 5 nM (Figure 3C). This effect was Trk fusion-specific as entrectinib had no effect on parental Ba/F3 cells (IC50 >1000 nM) or Ba/F3 cells transfected with empty lentiviral vector.
For the ETV6-NTRK3 fusion, entrectinib was more potent than other Trk inhibitors: TSR-011 (Tesaro), LOXO-101 (LOXO), and crizotinib (IC50 values of 2 nM for entrectinib, 14 nM for LOXO-101, 59 nM for TSR-011, and 88 nM for crizotinib, supplementary Figures S4A and S4B, available at Annals of Oncology online). Western blots confirmed Trk targeting with entrectinib as phosphorylation of both TrkC and PLCγ1 was substantially reduced with drug exposure. Other signaling proteins downstream of TrkC also had reduced phosphorylation with entrectinib, including PI3K (p85), MAPK, and Stat3 (Figure 3D).
Introduction of the NTRK3 G623R mutation to the ETV6-NTRK3 construct (Ba/F3-ETV6-NTRK3 G623R) conferred reduced sensitivity to entrectinib, increasing the IC50 value in the proliferation assays more than 250-fold (2 to 507 nM) relative to the Ba/F3-ETV6-NTRK3 cells (Figure 3E). The NTRK3 G623R mutation conferred even greater loss of sensitivity to the other tested Trk inhibitors, TSR-011 (Tesaro) and LOXO-101 (LOXO), eliciting IC50 proliferation values of >1000 nM (supplementary Figure S4C, available at Annals of Oncology online).
Investigation into the structural impact of the G623R point mutation revealed a potential mechanism of relative resistance to entrectinib and other Trk inhibitors. The glycine at position 623 lies in the kinase domain of TrkC, a codon that homology alignment suggested is highly conserved in other kinases, including ALK (position 1202), ROS1 (position 2032), and NTRK1 (position 595, Figure 3F). Structural analysis (Maestro) revealed extensive hydrogen bonding and hydrophobic interactions between entrectinib and the ATP binding pocket of wild-type TrkC where the G623 residue is located. The NTRK3 G623R mutation creates steric hindrance that functionally reduces the binding of entrectinib with mutant TrkC (Figure 3G).
discussion
In this article, we describe the first clinical response to TrkC inhibition in a patient with an NTRK3-rearranged malignancy, underscoring the role of Trk fusion proteins as targetable drivers of oncogenesis. This observation has widespread implications across a number of adult and pediatric hematologic/nonhematologic cancers where NTRK3 rearrangements have been identified [7]. We demonstrate that entrectinib (RXDX-101), a multikinase inhibitor with activity against TrkA, TrkB, TrkC, ROS1, and ALK, has potent in vitro activity against a variety of NTRK rearrangements, including TPM3-NTRK1, LMNA-NTRK1, ETV6-NTRK1, VCL-NTRK2, AFAP1-NTRK2, ETV6-NTRK2, and ETV6-NTRK3.
While crizotinib has modest in vitro activity against Trk [14], it is significantly less potent against ETV6-NTRK3 in comparison to entrectinib, correlating to the modest benefit achieved clinically in this patient. This observation suggests that the degree of Trk inhibition may be a critical determinant of response, emphasizing the need for potent Trk inhibitors such as entrectinib to achieve meaningful clinical outcomes in patients with Trk-driven tumors. Drug development strategies for these agents will need to take into account that NTRK rearrangements occur both frequently in rare cancers, and at a low incidence in more common tumors.
This experience also represents the first reported case of resistance to TrkC inhibition mediated by the appearance of an NTRK3 G623R mutation. Structural and cellular studies suggest that this alteration reduces entrectinib (and other tested Trk inhibitors) inhibition of TrkC by sterically disrupting drug binding to the kinase domain. Interestingly, solvent front mutations resulting in amino acid substitution at paralogous positions of ALK and ROS1 (ALK G1202R [15, 16] and ROS1 G2032R [17]), have previously been identified as mechanisms of acquired resistance to ALK and ROS1 inhibition by crizotinib in ALK- and ROS1-rearranged tumors, respectively. More importantly, the TrkC G623 and TrkA G595 residues are paralogous, confirming the work carried out by Russo et al. who identified NTRK1 G595R as a mechanism of acquired resistance to entrectinib in a patient with LMNA-NTRK1-rearranged colorectal cancer [18]. We recommend that molecular profiling be carried out at the onset of acquired resistance to Trk inhibition in NTRK-rearranged tumors to identify NTRK3 G623R, NTRK1 G595R, and the analogous mutation NTRK2 G639R. Ongoing drug discovery efforts should focus on the development of targeted therapies with activity against these mutations.
Finally, this report emphasizes the tremendous potential of comprehensive molecular profiling to impact oncologic diagnostics and therapeutics for cancer patients [19], particularly those with rare malignancies for whom effective or standard therapies are lacking [11, 20]. In this case, profiling prompted pathologic reclassification of the patient's diagnosis from AciCC to MASC and identified a class of novel drugs that would not have otherwise been considered for her. In a series of nonparotid AciCC cases, 79% of tumors harbored ETV6-NTRK3 rearrangements, resulting in a change in diagnosis to MASC [21]. While salient histologic features may suggest a diagnosis of MASC [22], we recommend that all suspected MASC neoplasms, including zymogen-poor AciCC and other morphologically similar low-grade salivary tumors, undergo molecular profiling for ETV6-NTRK. For those with recurrent or metastatic disease, such an approach could potentially translate to clinically meaningful therapeutic options.
funding
This study was funded in part by NIH/NCI Cancer Center Support (grant no. P30 CA008748).
disclosure
AD has received honoraria or consulting fees for Ignyta, Exelixis, Roche, and Genentech; GL, GW, NRC, RS, JL, ZH, JC, DL, ECM, and PM are employees or stockholders of Ignyta; AF has consulted for Intervention Insights; SVL has consulted for Genentech and Boehringer Ingelheim; AS has received honoraria or consulting fees from Ignyta, Pfizer, Novartis, Roche, Genentech, Ariad, Daiichi-Sankyo, Blueprint Medicines, and EMD Serono. All remaining authors have declared no conflicts of interest.
Supplementary Material
acknowledgements
We acknowledge Andrew Drilon for creating body diagrams for Figures 1 and 3, and Ryma Benayed of Memorial Sloan Kettering Cancer Center for performing RNA sequencing for ETV6-NTRK3 to confirm RNA sequencing results carried out at Ignyta.
references
- 1.Skalova A, Vanecek T, Sima R et al. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol 2010; 34: 599–608. [DOI] [PubMed] [Google Scholar]
- 2.Tognon C, Knezevich SR, Huntsman D et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell 2002; 2: 367–3376. [DOI] [PubMed] [Google Scholar]
- 3.Ito Y, Ishibashi K, Masaki A et al. Mammary analogue secretory carcinoma of salivary glands: a clinicopathologic and molecular study including 2 cases harboring ETV6-X fusion. Am J Surg Pathol 2015; 39: 602–610. [DOI] [PubMed] [Google Scholar]
- 4.Skálová A, Vanecek T, Simpson RH et al. Mammary analogue secretory carcinoma of salivary glands: molecular analysis of 25 ETV6 gene rearranged tumors with lack of detection of classical ETV6-NTRK3 fusion transcript by standard RT-PCR: report of 4 cases harboring ETV6-X gene fusion. Am J Surg Pathol 2016; 40(1): 3–13. [DOI] [PubMed] [Google Scholar]
- 5.Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun 2014; 5: 4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaishnavi A, Capelletti M, Le AT et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med 2013; 19: 1469–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaishnavi A, Le AT, Doebele RC. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov 2015; 5: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng DT, Mitchell T, Zehir A et al. MSK-IMPACT: a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015; 17: 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen R, Seshan V. FACETS: fraction and allele-specific copy number estimates from tumor sequencing. Department of Epidemiology and Biostatistics Working Paper Series. Working Paper 29. 2015.
- 10.Eisenhauer EA, Therasse P, Bogaerts J et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45: 228–247. [DOI] [PubMed] [Google Scholar]
- 11.Doebele RC, Davis LE, Vaishnavi A et al. An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer Discov 2015; 5: 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilcoxen KM. Modulating certain tyrosine kinases. Google Patents WO2013074518 A1, 2013.
- 13.Maestro Release 2015-1. New York, NYC: Schrodinger, Inc. 2015. [Google Scholar]
- 14.Taipale M, Krykbaeva I, Whitesell L et al. Chaperones as thermodynamic sensors of drug-target interactions reveal kinase inhibitor specificities in living cells. Nat Biotechnol 2013; 31: 630–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katayama R, Shaw AT, Khan TM et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung cancers. Sci Transl Med 2012; 4: 120ra17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Awad MM, Engelman JA, Shaw AT. Acquired resistance to crizotinib from a mutation in CD74-ROS1. N Engl J Med 2013; 369: 1173. [DOI] [PubMed] [Google Scholar]
- 17.Ignatius Ou SH, Azada M, Hsiang DJ et al. Next-generation sequencing reveals a novel NSCLC ALK F1174V mutation and confirms ALK G1202R mutation confers high-level resistance to alectinib (CH5424802/RO5424802) in ALK-rearranged NSCLC patients who progressed on crizotinib. J Thorac Oncol 2014; 9: 549–553. [DOI] [PubMed] [Google Scholar]
- 18.Russo M, Misale S, Wei G et al. Acquired resistance to the TRK inhibitor entrectinib in colorectal cancer. Cancer Discov 2016; 6: 36–44. [DOI] [PubMed] [Google Scholar]
- 19.Drilon A, Wang L, Arcila ME et al. Broad, hybrid capture-based next-generation sequencing identifies actionable genomic alterations in lung adenocarcinomas otherwise negative for such alterations by other genomic testing approaches. Clin Cancer Res 2015; 21: 3631–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bishop JA. Unmasking MASC: bringing to light the unique morphologic, immunohistochemical and genetic features of the newly recognized mammary analogue secretory carcinoma of salivary glands. Head Neck Pathol. 2013; 7: 35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bishop JA, Yonescu R, Batista D, Eisele DW, Westra WH. Most nonparotid “acinic cell carcinomas” represent mammary analog secretory carcinomas. Am J Surg Pathol 2013; 37: 1053–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Connor A, Perez-Ordonez B, Shago M, Skalova A, Weinreb I. Mammary analog secretory carcinoma of salivary gland origin with the ETV6 gene rearrangement by FISH: expanded morphologic and immunohistochemical spectrum of a recently described entity. Am J Surg Pathol 2012; 36: 27–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.