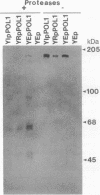
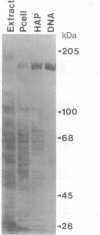
Abstract
We have overexpressed the POL1 gene of the yeast Saccharomyces cerevisiae and purified the resulting DNA polymerase alpha polypeptide in an apparently intact form. We attached the purified DNA polymerase covalently to an agarose matrix and used this matrix to chromatograph extracts prepared from yeast cells. At least six proteins bound to the yeast DNA polymerase alpha matrix that did not bind to a control matrix. We speculate that these proteins might be DNA polymerase alpha accessory proteins. Consistent with this interpretation, one of the binding proteins, which we have named POB1 (polymerase one binding), is required for normal chromosome transmission. Mutations in this gene cause increased chromosome loss and an abnormal cell morphology, phenotypes that also occur in the presence of mutations in the yeast alpha or delta polymerase genes. These results suggest that the interactions detected by polymerase affinity chromatography are biologically relevant and may help to illuminate the architecture of the eukaryotic DNA replication machinery.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B. M., Barry J., Bedinger P., Formosa T., Jongeneel C. V., Kreuzer K. N. Studies on DNA replication in the bacteriophage T4 in vitro system. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):655–668. doi: 10.1101/sqb.1983.047.01.077. [DOI] [PubMed] [Google Scholar]
- Alberts B. M. The DNA enzymology of protein machines. Cold Spring Harb Symp Quant Biol. 1984;49:1–12. doi: 10.1101/sqb.1984.049.01.003. [DOI] [PubMed] [Google Scholar]
- Bambara R. A., Jessee C. B. Properties of DNA polymerases delta and epsilon, and their roles in eukaryotic DNA replication. Biochim Biophys Acta. 1991 Jan 17;1088(1):11–24. doi: 10.1016/0167-4781(91)90147-e. [DOI] [PubMed] [Google Scholar]
- Burton Z. F., Killeen M., Sopta M., Ortolan L. G., Greenblatt J. RAP30/74: a general initiation factor that binds to RNA polymerase II. Mol Cell Biol. 1988 Apr;8(4):1602–1613. doi: 10.1128/mcb.8.4.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg M. D., Kelly T. J. Animal virus DNA replication. Annu Rev Biochem. 1989;58:671–717. doi: 10.1146/annurev.bi.58.070189.003323. [DOI] [PubMed] [Google Scholar]
- Drubin D. G. Actin and actin-binding proteins in yeast. Cell Motil Cytoskeleton. 1990;15(1):7–11. doi: 10.1002/cm.970150103. [DOI] [PubMed] [Google Scholar]
- Formosa T., Barry J., Alberts B. M., Greenblatt J. Using protein affinity chromatography to probe structure of protein machines. Methods Enzymol. 1991;208:24–45. doi: 10.1016/0076-6879(91)08005-3. [DOI] [PubMed] [Google Scholar]
- Formosa T., Burke R. L., Alberts B. M. Affinity purification of bacteriophage T4 proteins essential for DNA replication and genetic recombination. Proc Natl Acad Sci U S A. 1983 May;80(9):2442–2446. doi: 10.1073/pnas.80.9.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt J., Li J. Interaction of the sigma factor and the nusA gene protein of E. coli with RNA polymerase in the initiation-termination cycle of transcription. Cell. 1981 May;24(2):421–428. doi: 10.1016/0092-8674(81)90332-9. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Li J. The nusA gene protein of Escherichia coli. Its identification and a demonstration that it interacts with the gene N transcription anti-termination protein of bacteriophage lambda. J Mol Biol. 1981 Mar 25;147(1):11–23. doi: 10.1016/0022-2836(81)90076-0. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985 Jul;110(3):381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W. D., Rao M. R., Erdile L. F., Kelly T. J., Kolodner R. D. An essential Saccharomyces cerevisiae single-stranded DNA binding protein is homologous to the large subunit of human RP-A. EMBO J. 1990 Jul;9(7):2321–2329. doi: 10.1002/j.1460-2075.1990.tb07404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. W. Vacuolar proteases in yeast Saccharomyces cerevisiae. Methods Enzymol. 1990;185:372–386. doi: 10.1016/0076-6879(90)85033-k. [DOI] [PubMed] [Google Scholar]
- Kellogg D. R., Field C. M., Alberts B. M. Identification of microtubule-associated proteins in the centrosome, spindle, and kinetochore of the early Drosophila embryo. J Cell Biol. 1989 Dec;109(6 Pt 1):2977–2991. doi: 10.1083/jcb.109.6.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lehman I. R., Kaguni L. S. DNA polymerase alpha. J Biol Chem. 1989 Mar 15;264(8):4265–4268. [PubMed] [Google Scholar]
- Ma H., Kunes S., Schatz P. J., Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58(2-3):201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. Z., Goodman H. M., O'Farrell P. H. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977 Dec;12(4):1133–1141. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- Pizzagalli A., Valsasnini P., Plevani P., Lucchini G. DNA polymerase I gene of Saccharomyces cerevisiae: nucleotide sequence, mapping of a temperature-sensitive mutation, and protein homology with other DNA polymerases. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3772–3776. doi: 10.1073/pnas.85.11.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F., Gerring S. L., Connelly C., Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990 Feb;124(2):237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer K. F., Ingles C. J., Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990 Jun 28;345(6278):783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- Tajima S., Lauffer L., Rath V. L., Walter P. The signal recognition particle receptor is a complex that contains two distinct polypeptide chains. J Cell Biol. 1986 Oct;103(4):1167–1178. doi: 10.1083/jcb.103.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thömmes P., Hübscher U. Eukaryotic DNA replication. Enzymes and proteins acting at the fork. Eur J Biochem. 1990 Dec 27;194(3):699–712. doi: 10.1111/j.1432-1033.1990.tb19460.x. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T., Melendy T., Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990 Aug 9;346(6284):534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]