Abstract
The basal body is a highly organized structure essential for the formation of cilia. Basal bodies dock to a cellular membrane through their distal appendages (also known as transition fibers) and provide the foundation on which the microtubules of the ciliary axoneme are built. Consequently, basal body position and orientation dictates the position and orientation of its cilium. The heart of the basal body is the mother centriole, the older of the two centrioles inherited during mitosis and which is comprised of nine triplet microtubules arranged in a cylinder. Like all ciliated organisms, mice possess basal bodies, and studies of mouse basal body structure have made diverse important contributions to the understanding of how basal body structure impacts the function of cilia. The appendages and associated structures of mouse basal bodies can differ in their architecture from those of other organisms, and even between murine cell types. For example, basal bodies of immotile primary cilia are connected to daughter centrioles, whereas those of motile multiciliated cells are not. The last few years have seen the identification of many components of the basal body, and the mouse will continue to be an extremely valuable system for genetically defining their functions.
Keywords: Basal body, Mus musculus, Transition fiber, Distal appendage, Subdistal appendage, Basal foot, Rootlet, Daughter centriole, Primary cilium, Motile cilium
The organism: Mus musculus
The house mouse Mus musculus is the vertebrate most widely used in biomedical research. The long history of mouse genetics, building off of the work of Victorian mouse fanciers and amplified by the development of both forward and reverse genetic approaches, has provided a rich, tractable and powerful set of genetic tools in mouse [1]. As we share 99% of our genes with mice, they are useful for modeling many aspects of human basal body function. For example, some ciliopathies, such as primary ciliary dyskinesia (PCD) and Meckel syndrome (MKS), are well modeled by mouse mutations in orthologous genes [2–4]. However, other ciliopathies such as nephronophthisis (NPHP) and Joubert syndrome (JBTS) are imperfectly recapitulated with existing mouse mutations [5–9]. As the ability to humanize portions of the mouse genome and create tailored mutations increases, it may be possible to more accurately model complex phenotypes related to cilia and basal bodies.
Murine basal body structure
Murine basal bodies contain triplet microtubules
The nine triplet microtubules that make up the barrel of the basal body are named A, B, and C from internal to external. As with all centrioles, the plane of the triplet microtubules is tilted such that the vector from the A-tubule to the C-tubule, if viewed from the proximal end of the centriole, points counterclockwise [10, 11]. The doublet microtubules of the ciliary axoneme are contiguous with the basal body A- and B-tubules, whereas the C-tubule terminates within the distal centriole or in a region between the basal body and the cilium called the transition zone [12–16].
Different murine cell types display different basal body architectures
Distinct types of mouse cells possess structurally and functionally distinct types of cilia. The principal types of cilia are immotile primary cilia that can interpret intercellular signals, highly modified signaling cilia such as the photoreceptor connecting cilium, motile nodal cilia involved in left–right axis determination, immotile multicilia of the olfactory receptor neurons, motile multicilia that move fluid in the lung, brain ventricles and fallopian tubes, and the sperm flagellum.
While it is unclear whether basal bodies of distinct types of cilia contain sets of proteins unique to that ciliary type, the structure of the cilium itself can vary in ways that may be dependent on the basal body [17]. One example is the transition zone, a region between the basal body and cilium characterized by Y-fibers connecting the microtubules to the ciliary membrane. The transition zone can be short, such as in fibroblasts, or long, such as in photoreceptors.
Many motile cilia, such as those of tracheal and ependymal cells, have a central pair of microtubules in addition to the nine doublets of the axoneme (the so-called 9 + 2 arrangement of microtubules). Mutations in mouse Hydin, which encodes a protein associated with the central pair microtubules, causes defects in ciliary bending and beat frequency, suggesting that the central pair is critical for normal ciliary motility [18, 19].
However, not all motile cilia have a central pair. For example, most nodal cilia lack the central pair [20]. Consistent with the absence of the central pair in nodal cilia, human HYDIN mutations affect the motility of the cilia of the respiratory tract but do not cause left–right axis defects [21]. Conversely, not all 9 + 2 cilia are motile. Olfactory sensory neurons possess 9 + 2 cilia, but not the dynein arms required for ciliary motility [22].
Unlike the peripheral axonemal microtubules, the central pair microtubules are not continuous with microtubules of the basal body: they arise in the transition zone distal to the basal body. How the basal body influences whether the axoneme possesses the central pair remains unclear, but, at least in invertebrates, central pair formation depends on basal body components such as BLD10/CEP135 [17].
The mouse δ- and ε-tubulin genes
Consistent with the proposed link between δ- and ε-tubulin and the presence of triplet microtubules in centrioles, the mouse genome contains orthologs of the genes encoding δ- and ε-tubulin, Tubd1 and Tube1 [23, 24]. In Chlamydomonas, δ-tubulin is essential for the production of two and only two flagella and the production of triplet microtubules in the basal body: mutants lack the C-tubule [25]. Chlamydomonas ε-tubulin is critical for basal body formation or maintenance and is required for the formation of both basal body doublet and triplet microtubules [26]. In mammalian cells, δ-tubulin localizes to the spindle poles and co-immunoprecipitates with γ-tubulin, and ε-tubulin localizes to the subdistal appendage of the basal body [27, 28]. Answering the question of whether the function of mouse δ- and ε-tubulin is similar or distinct from that in Chlamydomonas awaits functional genetic analysis.
Accessory structures of mouse basal bodies
Murine basal bodies are accompanied by, depending on the phase of the cell cycle and cell type, either no (in the instances of sperm and multiciliated cells), one (monociliated cells in G1, G0, or early S phase), or three (monociliated cells in late S or G2 phase) centrioles [29, 30]. During G1 phase of most ciliated cells, the proximal end of the basal body is connected to the proximal end of the daughter centriole by a linkage, and the daughter centriole is oriented roughly orthogonally to the basal body [31]. In contrast, the basal bodies of motile multiciliated cells are not physically associated with daughter centrioles, although the daughter centrioles do have crucial roles in the generation of the many basal bodies possessed by these cells [32].
Basal bodies are surrounded by pericentriolar material. The pericentriolar material is comprised of proteins such as Pericentrin, appears moderately electron dense by EM, and nucleates the minus ends of many cytoplasmic microtubules [33–35]. On the periphery of the pericentriolar material exist large electron-dense protein complexes called centriolar satellites involved in ciliogenesis and centriole duplication [36–38].
Mouse basal body appendages
Murine basal bodies possess a variety of appendages, including a rootlet, distal appendages, and subdistal appendages or a basal foot. Indeed, the basal body is distinguished from daughter centrioles and procentrioles by the presence of these appendages. The relationship of subdistal appendages to the basal foot is unclear. Both project from the sides of the basal body at nearly the same position, approximately 350 nm from the proximal end of the basal body, and both are associated with microtubule nucleation [39, 40]. Basal bodies have up to nine subdistal appendages, but only one or two basal feet. The basal foot further differs from subdistal appendages in that it is larger and is more electron dense. Subdistal appendages and basal feet are mutually exclusive and have some of the same genetic requirements, suggesting that subdistal appendages may coalesce to form the basal foot [41]. During G2 phase, the subdistal appendages or basal foot are lost and do not reappear until the next G1 phase [42, 43].
The nine distal appendages project outward from the distal end of the basal body and are required for membrane docking and ciliogenesis [44–46]. Once the basal body docks to a membrane, distal appendages are often referred to as transition fibers. The distal appendages possess Cep164, Cep89, Cep83, Fbf1, and Sclt1, and all five are involved in ciliogenesis, with Cep83 being specifically important for membrane docking [45, 46].
The rootlet is a thick (80–100 nm) striated bundle of filaments that projects from the proximal end of the basal body and extends close to the nucleus [47]. Striations orthogonal to the filament axis are present at intervals of ~55–75 nm [47]. Rootlets are associated with basal bodies of both motile and immotile cilia, such as photoreceptor cells. In this cell type, the rootlet extends from the outer segment, a highly specialized modified cilium, to the synaptic terminal at the opposite end of the cell [48, 49]. One component of the rootlet is Rootletin [47, 50]. Consistent with the rootlet being dispensable for ciliary motility and signaling, a mutation in mouse Rootletin (also known as Crocc) that disrupts rootlet formation does not abrogate ciliary beating or phototransduction [51]. However, this mutation causes photoreceptor degeneration and may reduce mucociliary clearance, suggesting that mechanical support provided by the rootlet is essential for the long-term maintenance of ciliary function [51, 52].
In addition to its function in the rootlet, Rootletin, together with C-Nap1, forms fibers that connect mother and daughter centrioles and may function in centrosome cohesion [31, 53, 54]. Mutations in the human homolog of C-Nap1, called CEP250 or CEP2, are associated with Usher syndrome, a disease characterized by retinitis pigmentosa and hearing loss [55]. In cattle, mutations in C-Nap1 are associated with microcephaly, suggesting that the linker between mother and daughter centrioles plays important roles in neural development in mammals [56].
Noteworthy EM studies of mouse basal bodies
Many investigators have analyzed both rodent and other vertebrate basal bodies, thereby revealing that the basal bodies of vertebrates are highly similar. For example, Sergei Sorokin described the formation of primary cilia in rat tissue and organ cultures of chicken duodenum [57]. In addition, he and Ronald Gordon defined the ultrastructure of motile cilia in the rat lung [58, 59]. Ellen Dirksen examined the structure of basal bodies in the mouse fallopian tube [60]. Wilsman et al. [44] performed serial EM studies of primary cilia in chondrocytes. The micrographs of serial sections in chondrocytes show with remarkable clarity the relative orientation of the transition fibers, the basal foot, and the triplet microtubules. More recently, Kazuhiro et al. performed electron tomographic studies demonstrating the role of Odf2 in the formation of distal and subdistal appendages [39]. The electron tomograms nicely show the ultrastructure of the basal body appendages in three dimensions (Fig. 1).
Fig. 1.
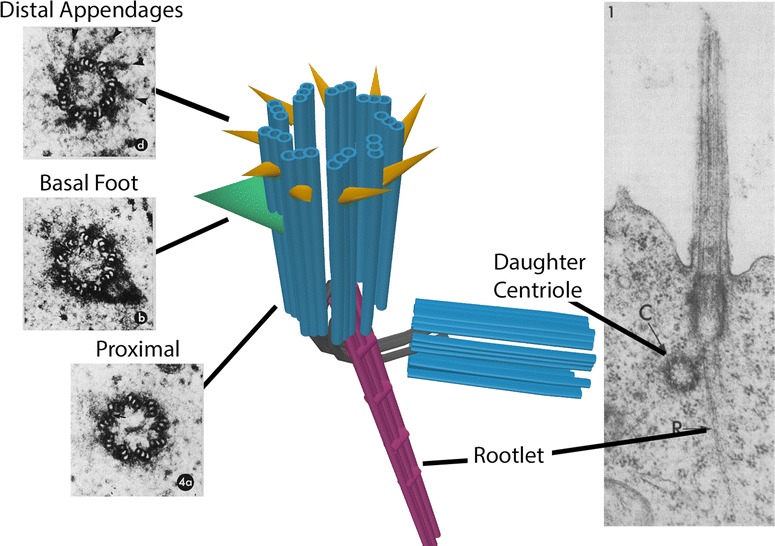
Basal body ultrastructure. At the distal end of the basal body, distal appendages or transition fibers are blades that have ninefold symmetry and radiate outward from the triplet microtubules. Proximal to the distal appendages is the basal foot, an electron-dense and cone-shaped structure projecting from one side of the basal body. Some basal bodies have multiple basal feet. The proximal end of the basal body is attached to the striated rootlet and to filaments that connect the basal body to the proximal end of the daughter centriole. Motile cilia in multiciliated cells lack associated daughter centrioles. Image credits: serial-section electron micrographs of transition fibers, the basal foot, and proximal basal body were originally published in [44]. Electron micrograph illustrating the striated rootlet and the daughter centriole was originally published in [14]
Early studies identifying basal body features
The basal foot was described in 1954 by Fawcett and Porter as “a small process” extending from the anterior face of the amphibian basal body [61]. In rodents, Rhodin and Dalhamn in 1956 identified the basal foot as a “cytoplasmic process” on basal bodies in rat tracheal epithelial cells [62]. By 1961, the term basal foot was used by Gibbons in his studies of cilia in the gills of the freshwater mussels [10]. In mouse embryonic fibroblasts, Wheatley identified the basal foot in 1969 [63].
The transition fibers were identified later than the basal foot. In 1972, Anderson identified transition fibers in mammalian fallopian tube cells, referring to them as “alar sheets” [64]. Surprisingly, the ciliary rootlet was identified before the advent of electron microscopy: Friedreich and Engelmann identified the ciliary rootlet using histological methods in the nineteenth century [61, 65, 66].
Origins of mouse basal bodies
Most basal bodies of cycling mouse cells, including many cells with primary cilia, are derived from mother centrioles inherited during mitosis [67]. Most basal bodies of non-cycling multiciliated cells are built from an electron-dense organelle called the deuterosome [32, 68]. Interestingly, it is the daughter centriole, not the mother centriole, that contributes to formation of the deuterosome [32].
In stark contrast to later phases of development, mouse cells lack centrioles during the first few cleavages following fertilization [69]. Despite the absence of centrioles, these early mouse blastomeres form and organize microtubules [70]. The pericentriolar material unassociated with centrioles may serve as the source of the MTOC activity in these cells [71, 72]. In particular, Plk4 and Cep152 can localize to an acentriolar MTOC to help organize microtubules [73].
Centrioles do not arise during development until the early blastocyst stage, indicating that these embryonic mouse cells must build centrioles de novo [74, 75]. The de novo synthesis of centrioles in human cells is error prone, suggesting that the cell’s ability to construct a structurally accurate centriole may be facilitated by the existence of a pre-existing centriole [76]. Loss of mouse Sas4 (also called Cenpj) disrupts formation of centrioles, basal bodies, and cilia, but does not halt cell cycle progression or embryonic development until midgestation, indicating that, despite their genesis early in development, they are not essential for some forms of cell division [77].
During development and in adult tissues, all ciliated cells have basal bodies
In cycling cells, the basal body becomes a mother centriole after the cilium is disassembled. This former basal body serves as a part of a spindle pole during mitosis, and thus mitosis can be considered a phase of the cell cycle during which basal bodies do not exist. Although mouse cells disassemble their cilia before entering mitosis, a heterozygous mutation in Pifo can cause mouse cells to retain a ciliary remnant into mitosis (although it may no longer be physically associated with the basal body) and, perhaps consequently, can cause mitotic defects [78]. Multiciliated cells are terminally differentiated and thus possess basal bodies during their entire lifetimes [79].
Basal body contribution to microtubule-organizing center (MTOC) activity
In addition to supporting ciliogenesis, the basal body contributes to MTOC activity although, as mentioned above, it is not essential for MTOC activity. Many microtubules are anchored in the pericentriolar material itself, but the subdistal appendages and subdistal appendage proteins, such as Ninein and the Dynactin complex, are also implicated in anchoring microtubules [35, 80–85]. How the function of microtubules originating from the pericentriolar material and those attached to the subdistal appendages or basal foot differs will be interesting to determine.
Identification of mouse basal body components
A handful of proteomics and genomics screens have identified many mouse basal body components and identified many other candidate components. A transcriptomic study of mouse tracheal epithelial cells (mTECs) during ciliogenesis has identified more than 600 genes that are upregulated during early ciliogenesis [86]. Reflecting the genesis of both basal bodies and cilia during mTEC differentiation, these upregulated genes include those that encode ciliary and basal body proteins, and thus has been a boon for the identification of basal body components.
To identify genes involved in basal body and ciliary biology, targeted and genome-wide knockdown screens have been performed in mouse cells that possess primary cilia [87, 88]. Knockdown of these genes, or orthologous genes in human cells, can result in a variety of phenotypes, including loss of cilia, short cilia, long cilia, as well as ciliary transport defects in the absence of obvious structural defects [87–89]. The cell biological origin of these phenotypes and whether the associated gene products act at the cilium, the basal body, or elsewhere remains to be elucidated in most cases.
The proteome of the mouse photoreceptor sensory cilium complex, an isolated preparation containing the axonome, the basal body, and the ciliary rootlet of the photoreceptor outer segment, has identified over 1000 candidate ciliary proteins [90]. In addition to studies in mouse cells, proteomic and genomic screens have identified novel basal body and ciliary components in human and rat cells [91–93]. For example, mass spectrometry-based proteomics analysis of centrosomes and centrosomal protein interactors has identified novel centrosomal proteins, many of which have functions relevant to basal bodies [94–97]. Given the close evolutionary relationship between mice and other mammals, the mouse orthologs of the proteins identified in such screens are likely to inform mouse basal body biology.
Two independent comparative genomics studies focused on identifying genes involved in ciliary biology. Comparison of the gene complement of unciliated organisms with those of ciliated organisms identified genes involved in ciliogenesis and ciliary function [91, 98]. Although these computational approaches do not discriminate between genes encoding basal body and ciliary proteins, a subset is likely to encode components of the basal body. For example, both studies implicated the basal body component Sas4 as being specific to ciliated organisms.
Notable basal body findings made using mice
Genetic studies in mice have been especially useful in determining the physiological functions of basal bodies. In most instances, a single mutant allele exists, providing important but limited insight into basal body function. In select cases, an allelic series provides more nuanced insight into the full range of basal body functions. For example, a hypomorphic mutation indicates that Odf2 mediates the orientation of basal feet, and proper polarization of basal feet is required for polarity of the ciliary beat in tracheal epithelial cells [99]. A stronger allele reveals that Odf2 is also essential for formation of the transition fibers and basal feet [39].
Like Odf2, Chibby homolog 1 (Cby1), a protein that localizes to the distal centriole, aids in docking of the basal body to the plasma membrane and is essential for mucociliary clearance in the airway epithelium [100–102]. One important function of Cby1 is in the recruitment of Ahi1 to the transition zone [101]. Understanding how Cby1, Odf2, and other basal body proteins orient the basal body to provide effective mucociliary clearance will provide insights into how ciliary orientation and motive force are achieved.
Other studies have helped illuminate how the distal centriole functions in ciliogenesis. For example, genetic and cell biological studies on mouse C2cd3 have demonstrated that it localizes to centriolar satellites, as well as to the distal end of centrioles, and that C2cd3 is required for formation of the distal appendages and for ciliary vesicle docking to the mother centriole [103, 104]. Loss of C2cd3 blocks removal of Cp110 from, and recruitment of Ttbk2, to the mother centriole, early steps in the initiation of ciliogenesis [104, 105]. Loss of C2cd3 also blocks recruitment of Ift88 and Cep164 to the distal appendage of the mother centriole [104]. Mouse C2cd3 mutants display phenotypes reminiscent of human ciliopathies, including severe polydactyly, situs defects, and disruption of the dorsal–ventral patterning of the neural tube [106]. Hedgehog signaling is disrupted in these mice, reflecting the essential function for cilia in transducing vertebrate Hedgehog signals [106]. Ofd1, a protein mutated in oral-facial-digital syndrome, co-localizes with C2cd3 at the distal centriole [103]. Whereas depletion of C2cd3 leads to shorter centrioles and the loss of the distal appendages, mutation of Ofd1 leads to centriole hyperelongation [103].
Another protein that localizes to the distal end of centrioles, Talpid3, interacts with Cp110 and regulates ciliogenesis [107–109]. Talpid3 mutant mice lack primary cilia, and have situs, neural tube, and facial defects [107, 110]. Taken together, these results indicate that the distal centriole appears to be an important locale where a complex of proteins coordinates with Cp110 to initiate ciliogenesis.
EHD1 and EHD3 are yet additional distal centriole proteins that are required for ciliary genesis. EHD1 is involved in ciliary vesicle formation and the removal of Cp110 [111, 112]. Future investigation of potential centriole “capping” proteins, as well as other proteins that regulate centriole length, will help reveal how architecture varies in different cell types to promote the diverse functions of the basal body.
Genetic studies have the advantage of having the capacity to identify regulatory inputs that do not directly involve basal body components, or even protein-coding genes. For example, the microRNAs miR-34/449 may promote the biogenesis of motile cilia by repressing Cp110 [113]. Consequently, mutant mice lacking these microRNAs are infertile and display defective mucociliary clearance [113].
Strengths and future of basal body research in Mus musculus
Due to the organism’s genetic tractability, the mouse is the most commonly used experimental organism to study vertebrate development and to model human disease. One of the strengths of the mouse as an experimental organism is the ability to tailor the genome, a strength that is growing as a new generation of genetic tools becomes widely used. Genes required for basal body formation or function can be mutated, and phenotypes can be analyzed in a wide range of cell types with a wide variety of ciliary types, illuminating the function of basal bodies in development, physiology, and disease.
Another strength of the mouse as a model organism is the breadth of research tools available. For example, there are many antibodies available for the detection of basal body and ciliary proteins. A weakness of the mouse is the difficulty in acquiring sufficient material for some approaches, such as the proteomics of basal bodies in specific cell types.
Future prospects for research on basal bodies in the mouse are diverse. How is duplication of the basal body controlled in primary ciliated and multiciliated cells? What role do basal bodies have in transducing developmental cues, such as Hedgehog signals? How do basal bodies interact with the planar cell polarity pathway to control the orientation of the motile cilia that move external fluids? Genetic modeling of basal body-associated diseases, such as ciliopathies, in mice will continue to help identify the cell biological origins of human disease, but also will illuminate the diverse functions of basal bodies in fundamental cellular processes such as ciliogenesis, ciliary motility, centriole duplication, and microtubule organization.
Authors’ contributions
GG and JFR conceived of and wrote the manuscript. Both authors read and approved the final manuscript.
Acknowledgements
We thank Andrew Kodani for critical comments. Our work was supported by grants from the NIH (AR054396 and GM095941) to JFR and (1F32GM109714) to GG. Work was supported by an award from the Burroughs Wellcome Fund (1014012) to GG.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- MTOC
Microtubule organizing center
- EM
electron microscopy
Contributor Information
Galo Garcia, III, Email: galo.garcia@ucsf.edu.
Jeremy F. Reiter, Email: jeremy.reiter@ucsf.edu
References
- 1.Paigen K. One hundred years of mouse genetics: an intellectual history. II. the molecular revolution (1981–2002) Genetics. 2003;163:1227–1235. doi: 10.1093/genetics/163.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee L, Campagna DR, Pinkus JL, Mulhern H, Wyatt TA, Sisson JH, Pavlik JA, Pinkus GS, Fleming MD. Primary ciliary dyskinesia in mice lacking the novel ciliary protein Pcdp1. Mol Cell Biol. 2008;28:949–957. doi: 10.1128/MCB.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chih B, Liu P, Chinn Y, Chalouni C, Komuves LG, Hass PE, Sandoval W, Peterson AS. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol. 2011;14:61–72. doi: 10.1038/ncb2410. [DOI] [PubMed] [Google Scholar]
- 4.Roberson EC, Dowdle WE, Ozanturk A, Garcia-Gonzalo FR, Li C, Halbritter J, Elkhartoufi N, Porath JD, Cope H, Ashley-Koch A, Gregory S, Thomas S, Sayer JA, Saunier S, Otto EA, Katsanis N, Davis EE, Attié-Bitach T, Hildebrandt F, Leroux MR, Reiter JF. TMEM231, mutated in orofaciodigital and Meckel syndromes, organizes the ciliary transition zone. J Cell Biol. 2015;209:129–142. doi: 10.1083/jcb.201411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Gonzalo FR, Corbit KC, Sirerol-Piquer MS, Ramaswami G, Otto EA, Noriega TR, Seol AD, Robinson JF, Bennett CL, Josifova DJ, Garcia-Verdugo JM, Katsanis N, Hildebrandt F, Reiter JF. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43:776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damerla RR, Cui C, Gabriel GC, Liu X, Craige B, Gibbs BC, Francis R, Li Y, Chatterjee B, San Agustin JT, Eguether T, Subramanian R, Witman GB, Michaud JL, Pazour GJ, Lo CW. Novel Jbts17 mutant mouse model of Joubert syndrome with cilia transition zone defects and cerebellar and other ciliopathy related anomalies. Hum Mol Genet. 2015;24:3994–4005. doi: 10.1093/hmg/ddv137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang S-T, Chiou Y-Y, Wang E, Lin H-K, Lee S-P, Lu H-Y, Wang C-KL, Tang M-J, Li H. Targeted disruption of Nphp1 causes male infertility due to defects in the later steps of sperm morphogenesis in mice. Hum Mol Genet. 2008;17:3368–3379. doi: 10.1093/hmg/ddn231. [DOI] [PubMed] [Google Scholar]
- 8.Jiang S-T, Chiou Y-Y, Wang E, Chien Y-L, Ho H-H, Tsai F-J, Lin C-Y, Tsai S-P, Li H. Essential role of nephrocystin in photoreceptor intraflagellar transport in mouse. Hum Mol Genet. 2009;18:1566–1577. doi: 10.1093/hmg/ddp068. [DOI] [PubMed] [Google Scholar]
- 9.Won J, Marín de Evsikova C, Smith RS, Hicks WL, Edwards MM, Longo-Guess C, Li T, Naggert JK, Nishina PM. NPHP4 is necessary for normal photoreceptor ribbon synapse maintenance and outer segment formation, and for sperm development. Hum Mol Genet. 2011;20:482–496. doi: 10.1093/hmg/ddq494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibbons IR. The relationship between the fine structure and direction of beat in gill cilia of a lamellibranch mollusc. J Biophys Biochem Cytol. 1961;11:179–205. doi: 10.1083/jcb.11.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Hara PT. Spiral tilt of triplet fibers in human leukocyte centrioles. J Ultrastruct Res. 1970;31:195–198. doi: 10.1016/S0022-5320(70)90154-1. [DOI] [PubMed] [Google Scholar]
- 12.Sorokin SP. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci. 1968;3:207–230. doi: 10.1242/jcs.3.2.207. [DOI] [PubMed] [Google Scholar]
- 13.Cordier AC. Ultrastructure of the cilia of thymic cysts in “nude” mice. Anat Rec. 1975;181:227–249. doi: 10.1002/ar.1091810206. [DOI] [PubMed] [Google Scholar]
- 14.Webber WA, Lee J. Fine structure of mammalian renal cilia. Anat Rec. 1975;182:339–343. doi: 10.1002/ar.1091820307. [DOI] [PubMed] [Google Scholar]
- 15.Wilsman NJ, Farnum CE. Arrangement of C-tubule protofilaments in mammalian basal bodies. J Ultrastruct Res. 1983;84:205–212. doi: 10.1016/S0022-5320(83)80001-X. [DOI] [PubMed] [Google Scholar]
- 16.Paintrand M, Moudjou M, Delacroix H, Bornens M. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J Struct Biol. 1992;108:107–128. doi: 10.1016/1047-8477(92)90011-X. [DOI] [PubMed] [Google Scholar]
- 17.Carvalho-Santos Z, Machado P, Alvarez-Martins I, Gouveia SM, Jana SC, Duarte P, Amado T, Branco P, Freitas MC, Silva STN, Antony C, Bandeiras TM, Bettencourt-Dias M. BLD10/CEP135 is a microtubule-associated protein that controls the formation of the flagellum central microtubule pair. Dev Cell. 2012;23:412–424. doi: 10.1016/j.devcel.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Davy BE, Robinson ML. Congenital hydrocephalus in hy3 mice is caused by a frameshift mutation in hydin, a large novel gene. Hum Mol Genet. 2003;12:1163–1170. doi: 10.1093/hmg/ddg122. [DOI] [PubMed] [Google Scholar]
- 19.Lechtreck K-F, Delmotte P, Robinson ML, Sanderson MJ, Witman GB. Mutations in hydin impair ciliary motility in mice. J Cell Biol. 2008;180:633–643. doi: 10.1083/jcb.200710162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Odate T, Takeda S, Narita K, Kawahara T: 9 + 0 and 9 + 2 cilia are randomly dispersed in the mouse node. Microscopy (Oxf). 2015: dfv352. [DOI] [PubMed]
- 21.Olbrich H, Schmidts M, Werner C, Onoufriadis A, Loges NT, Raidt J, Banki NF, Shoemark A, Burgoyne T, TurkiAl S, Hurles ME, Köhler G, Schroeder J, Nürnberg G, Nürnberg P, Chung EMK, Reinhardt R, Marthin JK, Nielsen KG, Mitchison HM, Omran H. Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left-right body asymmetry. Am J Hum Genet. 2012;91:672–684. doi: 10.1016/j.ajhg.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menco BP. Ciliated and microvillous structures of rat olfactory and nasal respiratory epithelia. A study using ultra-rapid cryo-fixation followed by freeze-substitution or freeze-etching. Cell Tissue Res. 1984;235:225–241. doi: 10.1007/BF00217846. [DOI] [PubMed] [Google Scholar]
- 23.Ross I, Clarissa C, Giddings TH, Winey M. ε-tubulin is essential in Tetrahymena thermophila for the assembly and stability of basal bodies. J Cell Sci. 2013;126:3441–3451. doi: 10.1242/jcs.128694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winey M, O’Toole E. Centriole structure. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130457–20130457. doi: 10.1098/rstb.2013.0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dutcher SK, Trabuco EC. The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes delta-tubulin, a new member of the tubulin superfamily. Mol Biol Cell. 1998;9:1293–1308. doi: 10.1091/mbc.9.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dutcher SK, Morrissette NS, Preble AM, Rackley C, Stanga J. ε-Tubulin is an essential component of the centriole. Mol Biol Cell. 2002;13:3859–3869. doi: 10.1091/mbc.E02-04-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang P, Stearns T. δ-Tubulin and ε-tubulin: two new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nat Cell Biol. 2000;2:30–35. doi: 10.1038/71350. [DOI] [PubMed] [Google Scholar]
- 28.Chang P, Giddings TH, Winey M, Stearns T. ε-tubulin is required for centriole duplication and microtubule organization. Nat Cell Biol. 2003;5:71–76. doi: 10.1038/ncb900. [DOI] [PubMed] [Google Scholar]
- 29.Conduit PT, Wainman A, Raff JW. Centrosome function and assembly in animal cells. Nat Rev Mol Cell Biol. 2015;16:611–624. doi: 10.1038/nrm4062. [DOI] [PubMed] [Google Scholar]
- 30.Sorokin SP. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci. 1968;3:207–230. doi: 10.1242/jcs.3.2.207. [DOI] [PubMed] [Google Scholar]
- 31.Bahe S, Stierhof Y-D, Wilkinson CJ, Leiss F, Nigg EA. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J Cell Biol. 2005;171:27–33. doi: 10.1083/jcb.200504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jord Al A, Lemaître AI, Delgehyr N, Faucourt M, Spassky N, Meunier A. Centriole amplification by mother and daughter centrioles differs in multiciliated cells. Nature. 2014;516:104–107. doi: 10.1038/nature13770. [DOI] [PubMed] [Google Scholar]
- 33.Woodruff JB, Wueseke O, Hyman AA. Pericentriolar material structure and dynamics. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130459. doi: 10.1098/rstb.2013.0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- 35.Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA. Microtubule nucleation by γ-Tubulin-containing rings in the centrosome. Nature. 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- 36.Tollenaere MAX, Mailand N, Bekker-Jensen S. Centriolar satellites: key mediators of centrosome functions. Cell Mol Life Sci. 2015;72:11–23. doi: 10.1007/s00018-014-1711-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubo A, Sasaki H, Yuba-Kubo A, Tsukita S, Shiina N. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J Cell Biol. 1999;147:969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kodani A, Yu TW, Johnson JR, Jayaraman D, Johnson TL, AlGazali, Sztriha L, Partlow JN, Kim H, Krup AL, Dammermann A, Krogan NJ, Walsh CA, Reiter JF. Centriolar satellites assemble centrosomal microcephaly proteins to recruit CDK2 and promote centriole duplication. eLife. 2015;4:555. doi: 10.7554/eLife.07519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tateishi K, Yamazaki Y, Nishida T, Watanabe S, Kunimoto K, Ishikawa H, Tsukita S. Two appendages homologous between basal bodies and centrioles are formed using distinct Odf2 domains. J Cell Biol. 2013;203:417–425. doi: 10.1083/jcb.201303071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/S0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- 41.Kodani A, Salomé Sirerol-Piquer M, Seol A, Manuel Garcia-Verdugo J, Reiter JF. Kif3a interacts with Dynactin subunit p150Glued to organize centriole subdistal appendages. EMBO J. 2013;32:597–607. doi: 10.1038/emboj.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robbins E, Jentzsch G, Micali A. The centriole cycle in synchronized HeLa cells. J Cell Biol. 1968;36:329–339. doi: 10.1083/jcb.36.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vorobjev IA, Chentsov Yu S. Centrioles in the cell cycle I. Epithelial cells. J Cell Biol. 1982;93:938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilsman NJ, Farnum CE, Reed-Aksamit DK. Incidence and morphology of equine and murine chondrocytic cilia. Anat Rec. 1980;197:355–361. doi: 10.1002/ar.1091970309. [DOI] [PubMed] [Google Scholar]
- 45.Graser S, Stierhof Y-D, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanos BE, Yang H-J, Soni R, Wang W-J, Macaluso FP, Asara JM, Tsou M-FB. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 2013;27:163–168. doi: 10.1101/gad.207043.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, Liu X, Yue G, Adamian M, Bulgakov O, Li T. Rootletin, a novel coiled-coil protein, is a structural component of the ciliary rootlet. J Cell Biol. 2002;159:431–440. doi: 10.1083/jcb.200207153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sjostrand FS. The ultrastructure of the inner segments of the retinal rods of the guinea pig eye as revealed by electron microscopy. J Cell Physiol. 1953;42:45–70. doi: 10.1002/jcp.1030420104. [DOI] [PubMed] [Google Scholar]
- 49.Spira AW, Milman GE. The structure and distribution of the cross-striated fibril and associated membranes in guinea pig photoreceptors. Am J Anat. 1979;155:319–337. doi: 10.1002/aja.1001550304. [DOI] [PubMed] [Google Scholar]
- 50.Klotz C, Bordes N, Laine MC, Sandoz D, Bornens M. A protein of 175,000 daltons associated with striated rootlets in ciliated epithelia, as revealed by a monoclonal antibody. Cell Motil Cytoskeleton. 1986;6:56–67. doi: 10.1002/cm.970060108. [DOI] [PubMed] [Google Scholar]
- 51.Yang J, Gao J, Adamian M, Wen X-H, Pawlyk B, Zhang L, Sanderson MJ, Zuo J, Makino CL, Li T. The ciliary rootlet maintains long-term stability of sensory cilia. Mol Cell Biol. 2005;25:4129–4137. doi: 10.1128/MCB.25.10.4129-4137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gilliam JC, Chang JT, Sandoval IM, Zhang Y, Li T, Pittler SJ, Chiu W, Wensel TG. Three-dimensional architecture of the rod sensory cilium and its disruption in retinal neurodegeneration. Cell. 2012;151:1029–1041. doi: 10.1016/j.cell.2012.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J, Adamian M, Li T. Rootletin interacts with C-Nap1 and may function as a physical linker between the pair of centrioles/basal bodies in cells. Mol Biol Cell. 2006;17:1033–1040. doi: 10.1091/mbc.E05-10-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J Cell Biol. 2000;151:837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khateb S, Zelinger L, Mizrahi-Meissonnier L, Ayuso C, Koenekoop RK, Laxer U, Gross M, Banin E, Sharon D. A homozygous nonsense CEP250 mutation combined with a heterozygous nonsense C2orf71 mutation is associated with atypical Usher syndrome. J Med Genet. 2014;51:460–469. doi: 10.1136/jmedgenet-2014-102287. [DOI] [PubMed] [Google Scholar]
- 56.Floriot S, Vesque C, Rodriguez S, Bourgain-Guglielmetti F, Karaiskou A, Gautier M, Duchesne A, Barbey S, Fritz S, Vasilescu A, Bertaud M, Moudjou M, Halliez S, Cormier-Daire V, EL Hokayem J, Nigg EA, Manciaux L, Guatteo R, Cesbron N, Toutirais G, Eggen A, Schneider-Maunoury S, Boichard D, Sobczak-Thépot J, Schibler L. C-Nap1 mutation affects centriole cohesion and is associated with a Seckel-like syndrome in cattle. Nat Commun. 2015;6:6894. doi: 10.1038/ncomms7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sorokin S. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J Cell Biol. 1962;15:363–377. doi: 10.1083/jcb.15.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sorokin SP. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci. 1968;3:207–230. doi: 10.1242/jcs.3.2.207. [DOI] [PubMed] [Google Scholar]
- 59.Gordon RE. Three-dimensional organization of microtubules and microfilaments of the basal body apparatus of ciliated respiratory epithelium. Cell Motil. 1982;2:385–391. doi: 10.1002/cm.970020407. [DOI] [PubMed] [Google Scholar]
- 60.Dirksen ER. Centriole morphogenesis in developing ciliated epithelium of the mouse oviduct. J Cell Biol. 1971;51:286–302. doi: 10.1083/jcb.51.1.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fawcett DW, Porter KR. A study of the fine structure of ciliated epithelia. J Morphol. 1954;94:221–281. doi: 10.1002/jmor.1050940202. [DOI] [Google Scholar]
- 62.Rhodin J, Dalhamn T. Electron microscopy of the tracheal ciliated mucosa in rat. Z Zellforsch Mikrosk Anat. 1956;44:345–412. doi: 10.1007/BF00345847. [DOI] [PubMed] [Google Scholar]
- 63.Wheatley DN. Cilia in cell-cultured fibroblasts. I. On their occurrence and relative frequencies in primary cultures and established cell lines. J Anat. 1969;105:351–362. [PMC free article] [PubMed] [Google Scholar]
- 64.Anderson RG. The three-dimensional structure of the basal body from the rhesus monkey oviduct. J Cell Biol. 1972;54:246–265. doi: 10.1083/jcb.54.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Engelmann TW. Zur Anatomie und Physiologie der Flimmerzellen. Pflugers Arch. 1880;23:505–535. doi: 10.1007/BF01637532. [DOI] [Google Scholar]
- 66.Friedreich PDN. Einiges über die Structur der Cylinder- und Flimmerepithelien. Archiv f pathol Anat. 1858;15:535–539. doi: 10.1007/BF01914889. [DOI] [Google Scholar]
- 67.Kobayashi T, Dynlacht BD. Regulating the transition from centriole to basal body. J Cell Biol. 2011;193:435–444. doi: 10.1083/jcb.201101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klos Dehring DA, Vladar EK, Werner ME, Mitchell JW, Hwang P, Mitchell BJ. Deuterosome-mediated centriole biogenesis. Dev Cell. 2013;27:103–112. doi: 10.1016/j.devcel.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Szollosi D, Calarco P, Donahue RP. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J Cell Sci. 1972;11:521–541. doi: 10.1242/jcs.11.2.521. [DOI] [PubMed] [Google Scholar]
- 70.Houliston E, Pickering SJ, Maro B. Redistribution of microtubules and pericentriolar material during the development of polarity in mouse blastomeres. J Cell Biol. 1987;104:1299–1308. doi: 10.1083/jcb.104.5.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Telzer BR, Rosenbaum JL. Cell cycle-dependent, in vitro assembly of microtubules onto pericentriolar material of HeLa cells. J Cell Biol. 1979;81:484–497. doi: 10.1083/jcb.81.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gould RR, Borisy GG. The pericentriolar material in Chinese hamster ovary cells nucleates microtubule formation. J Cell Biol. 1977;73:601–615. doi: 10.1083/jcb.73.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coelho PA, Bury L, Sharif B, Riparbelli MG, Fu J, Callaini G, Glover DM, Zernicka-Goetz M. Spindle formation in the mouse embryo requires Plk4 in the absence of centrioles. Dev Cell. 2013;27:586–597. doi: 10.1016/j.devcel.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abumuslimov SS, Nadezhdina ES, Chentsov IS. An electron microscopic study of centriole and centrosome morphogenesis in the early development of the mouse. Tsitologiia. 1994;36:1054–1061. [PubMed] [Google Scholar]
- 75.Calarco-Gillam PD, Siebert MC, Hubble R, Mitchison T, Kirschner M. Centrosome development in early mouse embryos as defined by an autoantibody against pericentriolar material. Cell. 1983;35:621–629. doi: 10.1016/0092-8674(83)90094-6. [DOI] [PubMed] [Google Scholar]
- 76.Wang WJ, Acehan D, Kao C-H, Jane WN, Uryu K, Tsou M-FB. De novo centriole formation in human cells is error-prone and does not require SAS-6 self-assembly. eLife. 2015;4:e10586. doi: 10.7554/eLife.10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bazzi H, Anderson KV. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc Natl Acad Sci USA. 2014;111:E1491–E1500. doi: 10.1073/pnas.1400568111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kinzel D, Boldt K, Davis EE, Burtscher I, Trümbach D, Diplas B, Attié-Bitach T, Wurst W, Katsanis N, Ueffing M, Lickert H. Pitchfork regulates primary cilia disassembly and left-right asymmetry. Dev Cell. 2010;19:66–77. doi: 10.1016/j.devcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brooks ER, Wallingford JB. Multiciliated cells. Curr Biol. 2014;24:R973–R982. doi: 10.1016/j.cub.2014.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mogensen MM, Malik A, Piel M, Bouckson-Castaing V, Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci. 2000;113(Pt 17):3013–3023. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- 81.Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abal M, Piel M, Bouckson-Castaing V, Mogensen M, Sibarita J-B, Bornens M. Microtubule release from the centrosome in migrating cells. J Cell Biol. 2002;159:731–737. doi: 10.1083/jcb.200207076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Delgehyr N, Sillibourne J, Bornens M. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci. 2005;118:1565–1575. doi: 10.1242/jcs.02302. [DOI] [PubMed] [Google Scholar]
- 84.Quintyne NJ, Gill SR, Eckley DM, Crego CL, Compton DA, Schroer TA. Dynactin is required for microtubule anchoring at centrosomes. J Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quintyne NJ, Schroer TA. Distinct cell cycle-dependent roles for dynactin and dynein at centrosomes. J Cell Biol. 2002;159:245–254. doi: 10.1083/jcb.200203089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hoh RA, Stowe TR, Turk E, Stearns T. Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PLoS ONE. 2012;7:e52166. doi: 10.1371/journal.pone.0052166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lai CK, Gupta N, Wen X, Rangell L, Chih B, Peterson AS, Bazan JF, Li L, Scales SJ. Functional characterization of putative cilia genes by high-content analysis. Mol Biol Cell. 2011;22:1104–1119. doi: 10.1091/mbc.E10-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wheway G, Schmidts M, Mans DA, Szymanska K, Nguyen TMT, Racher H, Phelps IG, Toedt G, Kennedy J, Wunderlich KA, Sorusch N, Abdelhamed ZA, Natarajan S, Herridge W, van Reeuwijk J, Horn N, Boldt K, Parry DA, Letteboer SJF, Roosing S, Adams M, Bell SM, Bond J, Higgins J, Morrison EE, Tomlinson DC, Slaats GG, van Dam TJP, Huang L, Kessler K, et al. An siRNA-based functional genomics screen for the identification of regulators of ciliogenesis and ciliopathy genes. Nat Cell Biol. 2015;17:1074–1087. doi: 10.1038/ncb3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim J, Lee JE, Heynen-Genel S, Suyama E, Ono K, Lee K, Ideker T, Aza-Blanc P, Gleeson JG. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464:1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu Q, Tan G, Levenkova N, Li T, Pugh EN, Rux JJ, Speicher DW, Pierce EA. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6:1299–1317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/S0092-8674(04)00412-X. [DOI] [PubMed] [Google Scholar]
- 92.Mayer U, Küller A, Daiber PC, Neudorf I, Warnken U, Schnölzer M, Frings S, Möhrlen F. The proteome of rat olfactory sensory cilia. Proteomics. 2009;9:322–334. doi: 10.1002/pmic.200800149. [DOI] [PubMed] [Google Scholar]
- 93.Ostrowski LE, Blackburn K, Radde KM, Moyer MB, Schlatzer DM, Moseley A, Boucher RC. A proteomic analysis of human cilia: identification of novel components. Mol Cell Proteomics. 2002;1:451–465. doi: 10.1074/mcp.M200037-MCP200. [DOI] [PubMed] [Google Scholar]
- 94.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 95.Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I, Falkenby LG, Bennetzen M, Westendorf J, Nigg EA, Uhlen M, Hyman AA, Andersen JS. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 2011;30:1520–1535. doi: 10.1038/emboj.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gupta GD, Coyaud E, Gonçalves J, Mojarad BA, Liu Y, Wu Q, Gheiratmand L, Comartin D, Tkach JM, Cheung SWT, Bashkurov M, Hasegan M, Knight JD, Lin Z-Y, Schueler M, Hildebrandt F, Moffat J, Gingras A-C, Raught B, Pelletier L. A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell. 2015;163:1484–1499. doi: 10.1016/j.cell.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, Hyman AA, Mann M. A Human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 98.Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N, Dutcher SK. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/S0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 99.Kunimoto K, Yamazaki Y, Nishida T, Shinohara K, Ishikawa H, Hasegawa T, Okanoue T, Hamada H, Noda T, Tamura A, Tsukita S, Tsukita S. Coordinated ciliary beating requires Odf2-mediated polarization of basal bodies via basal feet. Cell. 2012;148:189–200. doi: 10.1016/j.cell.2011.10.052. [DOI] [PubMed] [Google Scholar]
- 100.Voronina VA, Takemaru K-I, Treuting P, Love D, Grubb BR, Hajjar AM, Adams A, Li F-Q, Moon RT. Inactivation of Chibby affects function of motile airway cilia. J Cell Biol. 2009;185:225–233. doi: 10.1083/jcb.200809144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee YL, Santé J, Comerci CJ, Cyge B, Menezes LF, Li F-Q, Germino GG, Moerner WE, Takemaru K-I, Stearns T. Cby1 promotes Ahi1 recruitment to a ring-shaped domain at the centriole-cilium interface and facilitates proper cilium formation and function. Mol Biol Cell. 2014;25:2919–2933. doi: 10.1091/mbc.E14-02-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Burke MC, Li F-Q, Cyge B, Arashiro T, Brechbuhl HM, Chen X, Siller SS, Weiss MA, O’Connell CB, Love D, Westlake CJ, Reynolds SD, Kuriyama R, Takemaru K-I. Chibby promotes ciliary vesicle formation and basal body docking during airway cell differentiation. J Cell Biol. 2014;207:123–137. doi: 10.1083/jcb.201406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thauvin-Robinet C, Lee JS, Lopez E, Herranz-Pérez V, Shida T, Franco B, Jego L, Ye F, Pasquier L, Loget P, Gigot N, Aral B, Lopes CAM, St-Onge J, Bruel A-L, Thevenon J, González-Granero S, Alby C, Munnich A, Vekemans M, Huet F, Fry AM, Saunier S, Rivière J-B, Attié-Bitach T, Garcia-Verdugo JM, Faivre L, Mégarbané A, Nachury MV. The oral-facial-digital syndrome gene C2CD3 encodes a positive regulator of centriole elongation. Nat Genet. 2014;46:905–911. doi: 10.1038/ng.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ye X, Zeng H, Ning G, Reiter JF, Liu A. C2cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc Natl Acad Sci USA. 2014;111:2164–2169. doi: 10.1073/pnas.1318737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tsang WY, Dynlacht BD. CP110 and its network of partners coordinately regulate cilia assembly. Cilia. 2013;2:9. doi: 10.1186/2046-2530-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hoover AN, Wynkoop A, Zeng H, Jia J, Niswander LA, Liu A. C2cd3 is required for cilia formation and Hedgehog signaling in mouse. Development. 2008;135:4049–4058. doi: 10.1242/dev.029835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bangs F, Antonio N, Thongnuek P, Welten M, Davey MG, Briscoe J, Tickle C. Generation of mice with functional inactivation of talpid3, a gene first identified in chicken. Development. 2011;138:3261–3272. doi: 10.1242/dev.063602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yin Y, Bangs F, Paton IR, Prescott A, James J, Davey MG, Whitley P, Genikhovich G, Technau U, Burt DW, Tickle C. The Talpid3 gene (KIAA0586) encodes a centrosomal protein that is essential for primary cilia formation. Development. 2009;136:655–664. doi: 10.1242/dev.028464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kobayashi T, Kim S, Lin Y-C, Inoue T, Dynlacht BD. The CP110-interacting proteins Talpid3 and Cep290 play overlapping and distinct roles in cilia assembly. J Cell Biol. 2014;204:215–229. doi: 10.1083/jcb.201304153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ede DA, Kelly WA. DEVELOPMENTAL ABNORMALITIES IN THE TRUNK AND LIMBS OF THE TALPID3 MUTANT OF THE FOWL. J Embryol Exp Morphol. 1964;12:339–356. [PubMed] [Google Scholar]
- 111.Lu Q, Insinna C, Ott C, Stauffer J, Pintado PA, Rahajeng J, Baxa U, Walia V, Cuenca A, Hwang Y-S, Daar IO, Lopes S, Lippincott-Schwartz J, Jackson PK, Caplan S, Westlake CJ. Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat Cell Biol. 2015;17:531–531. doi: 10.1038/ncb3155. [DOI] [PubMed] [Google Scholar]
- 112.Bhattacharyya S, Rainey MA, Arya P, Dutta S, George M, Storck MD, McComb RD, Muirhead D, Todd GL, Gould K, Datta K, Waes JG-V, Band V, Band H. Endocytic recycling protein EHD1 regulates primary cilia morphogenesis and SHH signaling during neural tube development. Sci Rep. 2016;6:20727. doi: 10.1038/srep20727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Song R, Walentek P, Sponer N, Klimke A, Lee JS, Dixon G, Harland R, Wan Y, Lishko P, Lize M, Kessel M, He L. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature. 2014;510:115–120. doi: 10.1038/nature13413. [DOI] [PMC free article] [PubMed] [Google Scholar]


