Abstract
Centromere protein B (CENP-B) is one of the major proteins involved in centromere formation, binding to centromeric repetitive DNA by recognizing a 17 bp motif called the CENP-B box. Hominids (humans and great apes) carry large numbers of CENP-B boxes in alpha satellite DNA (AS, the major centromeric repetitive DNA of simian primates). Only negative results have been reported regarding the presence of the CENP-B box in other primate taxa. Consequently, it is widely believed that the CENP-B box is confined, within primates, to the hominids. We report here that the common marmoset, a New World monkey, contains an abundance of CENP-B boxes in its AS. First, in a long contig sequence we constructed and analysed, we identified the motif in 17 of the 38 alpha satellite repeat units. We then sequenced terminal regions of additional clones and found the motif in many of them. Immunostaining of marmoset cells demonstrated that CENP-B binds to DNA in the centromeric regions of chromosomes. Therefore, functional CENP-B boxes are not confined to hominids. Our results indicate that the efficiency of identification of the CENP-B box may depend largely on the sequencing methods used, and that the CENP-B box in centromeric repetitive DNA may be more common than researchers previously thought.
Keywords: primates, marmoset, centromeric repetitive DNA, molecular evolution, alpha satellite DNA
1. Introduction
The amino acid sequence of CENP-B is highly conserved in a wide range of mammals (amino acid sequence identity of 92% between human and mouse) [1–3]. This protein plays a significant role in the assembly of the centromere by binding to centromeric repetitive DNA [4–7]. However, the CENP-B is considered non-essential to the survival of its host organism because CENP-B gene knockout mice are viable [8]. CENP-B binds to centromeric repetitive DNA by recognizing a 17 bp motif called the CENP-B box [4]. The CENP-B box is also considered non-essential because the CENP-B protein is non-essential. The term ‘non-essential’ can be expressed as s ≠ 1, where s is the selection coefficient used in population genetics [9] and the contribution of box-carrying and box-free ‘alleles’ to the fitness of the host is given by 1 and 1 − s, respectively. However, ‘non-essential’ simply implies that the host can survive without a CENP-B box, and does not exclude the possibility that the possession of a CENP-B box leads to a higher survival rate of the host. A fact to be considered in this context is that sequence blocks similar to the CENP-B box have been found in some mammalian species, including horses, dogs and elephants [10], in addition to humans and mice, in which functional CENP-B boxes are present [4,5]. Moreover, CENP-B boxes are densely distributed in the functionally active region but rarely found in pericentric regions of human and mouse repetitive DNAs [4]. These observations suggest that s takes a small positive value. If s is positive, the CENP-B box may be found in more species than have been documented when suitable detection methods are used.
Simian primates constitute the infraorder Simiiformes, the phylogenetic structure of which is shown in the electronic supplementary material, figure S1, and carry alpha satellite DNA (AS) as their major centromeric repetitive DNA [11]. The CENP-B box was described as a 17 bp motif (YTTCGTTGGAARCGGGA), based on sequence information derived from humans [4], in which the underlined nucleotides form the core recognition sequence [5]. In this report, we use the term CENP-B box in its broad sense: a 17 bp nucleotide block containing the core recognition sequence (NTTCGNNNNANNCGGGN). Humans carry CENP-B boxes in all autosomes and the X chromosome [4]. In a study of a wide range of primates [12], great apes (chimpanzee, gorilla and orangutan) were shown to carry CENP-B boxes in all autosomes and the X chromosome, but the results were negative for two gibbon species (white-handed gibbon and siamang), five Old World monkeys (African green monkey, Japanese macaque, mantled guereza, silvery lutung and red shanked douc langur), a New World monkey (howler monkey) and five non-simian primates (Philippine tarsier, three galagos and ring-tailed lemur). Based on these results, it was proposed that the CENP-B box carried by modern humans emerged in the lineage leading to hominids (humans and great apes) after its divergence from the small ape lineage [12]. For the approximately 20 years that ensued, to our knowledge, there has been no report of the presence of the CENP-box in non-hominid primates, although genome-sequencing projects have been completed, or are in progress, for many primate species. We have recently identified multiple copies of the CENP-B box sequence in the common marmoset (Callithrix jacchus), a New World monkey. We describe features of the nucleotide sequences and distribution pattern, as well as evidence for the binding of CENP-B to DNA in the centromeric regions of marmoset chromosomes.
2. Material and methods
The common marmoset (individual identification number 186) used as source of DNA and culture cell samples was the same as that used in our previous study [13]. The genomic library prepared for the previous study (vector, fosmid pCC1FOS; insert, 40 to 44 kb fragments produced by mechanical shearing) was screened for clones containing AS using the same method described therein. Cell culturing and immunofluorescence staining were performed basically as described previously [14]. Details of these methods, as well as the antibodies used for CENP-A and CENP-B, are described in the electronic supplementary material, Supplementary methods.
3. Results
AS of the common marmoset comprises repeat units of a median size of 345 bp [13]. In our previous study [13], we identified a higher order repeat structure (in which a block of multiple repeat units forms a larger repeat unit and larger repeat units are repeated in tandem; electronic supplementary material, figure S2) in a contig sequence of marmoset AS (GenBank accession number LC030305). Because CENP-B box motifs are often associated with higher order repeat structures in human AS, we surveyed the 13.1 kb marmoset sequence for a CENP-B box. The sequence contained 38 AS repeat units, and 17 of them were found to contain the CENP-B box sequence (figure 1a). These repeat units were distributed in a pattern of specific intervals (electronic supplementary material, figure S2). The repetition of this specific pattern coincided with the repetition of the larger repeat units (electronic supplementary material, figure S2).
Figure 1.
Distribution of CENP-B box sequences. (a) Distribution along a single contig sequence. The 38 repeat units contained in the 13.1 kb AS sequence were numbered from 1 to 38. The alignment of the sequences of the CENP-B box and its flanking regions is shown. ‘Con’ indicates the consensus sequence. Nucleotides that match those in the CENP-B box sequence are shown in magenta. An asterisk indicates a complete matching with the CENP-B box sequence. (b) Distribution among random samples of repeat units. The names of the fosmid clones are listed to the left of the alignment.
To estimate the frequency of AS repeat units carrying the CENP-B box sequence, we isolated 24 AS-carrying fosmids from the marmoset genomic library using methods previously described [13]. We sequenced one end of the insert fragments of the 24 clones using a universal primer (GenBank LC064994–LC065017). Each sequence read contained one or two repeat units of the full size and partial units in its head and tail regions (electronic supplementary material, figure S3), showing the presence or absence of the CENP-B box sequence in two or three consecutive repeat units. It is likely from the electronic supplementary material, figure S3, that the CENP-B box sequence often appears in two or three, or possibly more, repeat units. As our screening method does not use a specific clone as a probe, the 24 clones we isolated can be regarded as random samples of marmoset AS. If multiple units are used, however, a deviation from randomness is introduced into the frequency estimation. For this reason, we used only the first intact unit in each sequence read. Of the 24 such repeat units, seven contained the CENP-B box sequence (figure 1b; electronic supplementary material, figure S4). Thus, the frequency of AS repeat units carrying the CENP-B box sequence was estimated to be approximately 1 in 3.
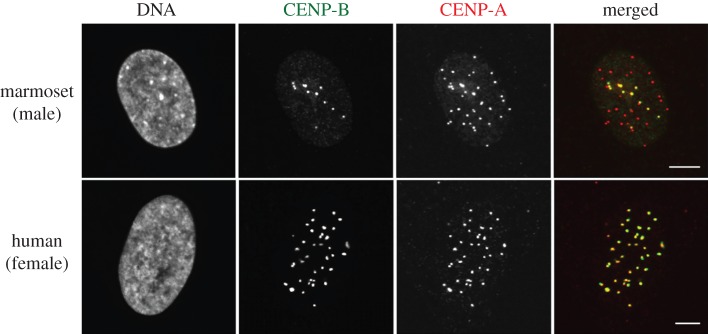
We then conducted an immunofluorescence staining analysis of marmoset cells in order to determine if the CENP-B protein binds to centromeric DNA (figure 2). CENP-A is a highly conserved protein (amino acid sequence identity of 70% between human and mouse) [15] absolutely required for centromere function, and a signal for CENP-A accumulation on the chromosome indicates location of the centromere [16]. On the other hand, a signal for CENP-B accumulation indicates that CENP-B binds to DNA at the position of the signal. Therefore, the overlap of a CENP-A signal and a CENP-B signal indicates that CENP-B binds to DNA at the centromere. In human HeLa cells, all red signals (CENP-A) and green signals (CENP-B) overlapped with each other, as seen in the merged photograph. This is consistent with the existence of CENP-B boxes in AS of all normal human chromosomes. In marmoset cells, 39.3 ± 3.43 (s.d.) signals/cell for CENP-A and 11.7 ± 1.76 signals/cell for CENP-B were observed (n = 57 cells). Interestingly, the few CENP-B signals detected were found to overlap completely with CENP-A signals, resulting in 29.7% ± 2.84% of CENP-A signals perfectly colocalizing with CENP-B signals. These results indicate that CENP-B binds to centromeric DNA of approximately 30% of the marmoset chromosomes.
Figure 2.
Results of immunofluorescence staining experiments. Cells were co-stained with antibodies against CENP-B (green) and CENP-A (red). DNA was visualized with DAPI. Scale bar represents 5 µm.
4. Discussion
The CENP-B protein carries a DNA-binding domain at its N-terminus and binds to DNA by recognizing the CENP-B box [4]. Our previous study using synthetic DNAs demonstrated that the CENP-B box is required for the CENP-B function [6]. In this study, we have identified the CENP-B box sequence that is present in marmoset AS at approximately a frequency of 1 in 3 repeat units. We have also confirmed that CENP-B binds to centromeric DNA of approximately 30% of the marmoset chromosomes. These results indicate that the CENP-B box sequences we identified in marmoset AS serve as functional CENP-B boxes. A strict confirmation requires molecular-level analyses of marmoset chromosomes, including ChIP-based analyses, but these require significantly more time to carry out. Preliminary assays have shown positive results and we will publish fully confirmed results in another report.
This is the first report to our knowledge to confirm the presence of functional CENP-B boxes in non-hominid primates. Whether the marmoset CENP-B boxes emerged independently of hominid CENP-B boxes is unclear. If the two CENP-B boxes were located at different positions of the AS repeat units in hominids and the marmoset, this would be regarded as evidence for independent origins. We attempted to make this comparison but failed because the sequence identities of the AS repeat units were not high enough to establish an accurate alignment.
CENP-B binds to centromeres of all chromosomes, with the exception of the Y chromosome, in hominids. In marmosets, however, CENP-B binding was observed in approximately 30% of the chromosomes. Considering that the CENP-B box is non-essential, this may reflect the possibility that the number of chromosomes carrying the CENP-B box fluctuates by random genetic drift after its emergence. If natural selection favouring AS repeat units that harbour the CENP-B box is strong enough (s is large enough) to balance elimination by random genetic drift and mutational decay, the CENP-B box currently residing in marmoset AS might attain fixation in all marmoset chromosomes after many generations.
CENP-B boxes are embedded in centromeric repetitive DNA. In genome sequence databases constructed through next-generation sequencing, repetitive DNA is generally underrepresented and susceptible to artificial alterations because of the difficulty in assembling contigs [17,18]. The centromere regions are still left as large gaps even in the human sequence databases. In this study, we used our original method for obtaining long and accurate contig sequences, of which the strategy was fully described in our previous reports [13,19], and its essence is shown in the electronic supplementary material, Supplementary methods. This approach was vital to the successful identification of higher order repeat structures, and subsequently CENP-B box sequences associated with these structures, in marmoset AS. It is thus likely that the efficiency of identification of CENP-B boxes depends largely on the sequencing methods used. This raises the possibility that the presence of the CENP-B box in centromeric repetitive DNA may be more common than researchers previously thought.
Supplementary Material
Acknowledgements
We are grateful to Koichiro Otake for helpful discussion, and Yuki Enomoto for technical assistance. We thank four reviewers for constructive comments to improve the manuscript.
Ethics
All animal experiments were approved by the Animal Care and Use Committee of Kyoto University Primate Research Institute (project number 2015-088-02).
Data accessibility
Sequence data are available from GenBank with the accession numbers stated in the text.
Authors' contributions
H.H. established cell lines of the marmoset. K.K. and H.M. conducted molecular biology experiments. A.K. performed sequencing experiments. A.S., K.S. and K.O. analysed sequence data. All authors contributed to writing and revising the manuscript and agree to take responsibility for the content therein.
Competing interests
The authors declare they have no competing interests.
Funding
This work was supported by Grants-in-Aid from the MEXT of Japan (23114005, 26251040 and 15H04427 to A.K., and 23114008 to H.M.) and the Kazusa DNA Research Institute Foundation (to H.M.).
References
- 1.Earnshaw WC, et al. 1987. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J. Cell Biol. 104, 817–829. ( 10.1083/jcb.104.4.817) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schueler MG, Swanson W, Thomas PJ, Green ED. 2010. Adaptive evolution of foundation kinetochore proteins in primates. Mol. Biol. Evol. 27, 1585–1597. ( 10.1093/molbev/msq043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan KF, Glass CA. 1991. CENP-B is a highly conserved mammalian centromere protein with homology to the helix-loop-helix family of proteins. Chromosoma 100, 360–370. ( 10.1007/BF00337514) [DOI] [PubMed] [Google Scholar]
- 4.Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. 1989. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 109, 1963–1973. ( 10.1083/jcb.109.5.1963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masumoto H, Nakano M, Ohzeki J. 2004. The role of CENP-B and alpha-satellite DNA: de novo assembly and epigenetic maintenance of human centromeres. Chromosome Res. 12, 543–556. ( 10.1023/B:CHRO.0000036593.72788.99) [DOI] [PubMed] [Google Scholar]
- 6.Ohzeki J, Nakano M, Okada T, Masumoto H. 2002. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 159, 765–775. ( 10.1083/jcb.200207112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H. 2007. CENP-B controls centromere formation depending on the chromatin context. Cell 131, 1287–1300. ( 10.1016/j.cell.2007.10.045) [DOI] [PubMed] [Google Scholar]
- 8.Hudson DF, et al. 1998. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J. Cell Biol. 141, 309–319. ( 10.1083/jcb.141.2.309) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright S. 1931. Evolution in Mendelian populations. Genetics 16, 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alkan C, et al. 2011. Genome-wide characterization of centromeric satellites from multiple mammalian genomes. Genome Res. 21, 137–145. ( 10.1101/gr.111278.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cellamare A, et al. 2009. New insights into centromere organization and evolution from the white-cheeked gibbon and marmoset. Mol. Biol. Evol. 26, 1889–1900. ( 10.1093/molbev/msp101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haaf T, Mater AG, Wienberg J, Ward DC. 1995. Presence and abundance of CENP-B box sequences in great ape subsets of primate-specific alpha-satellite DNA. J. Mol. Evol. 41, 487–491. ( 10.1007/BF00160320) [DOI] [PubMed] [Google Scholar]
- 13.Sujiwattanarat P, Thapana W, Srikulnath K, Hirai Y, Hirai H, Koga A. 2015. Higher-order repeat structure in alpha satellite DNA occurs in New World monkeys and is not confined to hominoids. Sci. Rep. 5, 10315 ( 10.1038/srep10315) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohzeki J, et al. 2012. Breaking the HAC barrier: histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J. 31, 2391–2402. ( 10.1038/emboj.2012.82) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalitsis P, MacDonald AC, Newson AJ, Hudson DF, Choo KH. 1998. Gene structure and sequence analysis of mouse centromere proteins A and C. Genomics 47, 108–114. ( 10.1006/geno.1997.5109) [DOI] [PubMed] [Google Scholar]
- 16.Fukagawa T, Earnshaw WC. 2014. The centromere: chromatin foundation for the kinetochore machinery. Dev. Cell 30, 496–508. ( 10.1016/j.devcel.2014.08.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koga A. 2012. Under-representation of repetitive sequences in whole-genome shotgun sequence databases: an illustration using a recently acquired transposable element. Genome 55, 172–175. ( 10.1139/g11-088) [DOI] [PubMed] [Google Scholar]
- 18.Thapana W, Sujiwattanarat P, Srikulnath K, Hirai H, Koga A. 2014. Reduction in the structural instability of cloned eukaryotic tandem-repeat DNA by low-temperature culturing of host bacteria. Genet. Res. 96, e13 ( 10.1017/S0016672314000172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koga A, Hirai Y, Terada S, Jahan I, Baicharoen S, Arsaithamkul V, Hirai H. 2014. Evolutionary origin of higher-order repeat structure in alpha-satellite DNA of primate centromeres. DNA Res. 21, 407–415. ( 10.1093/dnares/dsu005) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data are available from GenBank with the accession numbers stated in the text.