Abstract
Social living poses challenges for individual fitness because of the increased risk of disease transmission among conspecifics. Despite this challenge, sociality is an evolutionarily successful lifestyle, occurring in the most abundant and diverse group of organisms on earth—the social insects. Two contrasting hypotheses predict the evolutionary consequences of sociality on immune systems. The social group hypothesis posits that sociality leads to stronger individual immune systems because of the higher risk of disease transmission in social species. By contrast, the relaxed selection hypothesis proposes that social species have evolved behavioural immune defences that lower disease risk within the group, resulting in lower immunity at the individual level. We tested these hypotheses by measuring the encapsulation response in 11 eusocial and non-eusocial insect lineages. We built phylogenetic mixed linear models to investigate the effect of behaviour, colony size and body size on cellular immune response. We found a significantly negative effect of colony size on encapsulation response (Markov chain Monte Carlo generalized linear mixed model (mcmcGLMM) p < 0.05; phylogenetic generalized least squares (PGLS) p < 0.05). Our findings suggest that insects living in large societies may rely more on behavioural mechanisms, such as hygienic behaviours, than on immune function to reduce the risk of disease transmission among nest-mates.
Keywords: disease transmission, sociality, encapsulation, phylogenetic correction
1. Introduction
A key challenge of living in large societies is dealing with pathogens and parasites, which is as true for primates, including humans, as it is for naked mole rats or social insects. Because of crowded living conditions and frequent physical interactions, social living makes individuals potentially more vulnerable to socially transmitted pathogens and the diseases they cause [1]. For example, since the origin of agriculture and large settlements, diseases—from the bubonic plague to HIV—have threatened human populations [2]. However, humans have controlled the risk and spread of some diseases with the development of public health, antibiotics and vaccination [3]. Insect societies also confront and respond to strong selective pressures from diseases [4], worsened by the high genetic relatedness among nest-mates that enables pathogens and parasites to easily sweep through a host insect colony [5]. Even though social insects have been remarkably successful ecologically and evolutionarily [6], the mechanisms behind their ability to resist disease remain enigmatic.
Heightened individual immunity could combat disease risk in social insects, as purported in hominid evolution [7]. However, increased immune function can be costly whether because of metabolic costs or, as is the case in humans, because of the risk of immune overreaction to host tissue [8,9]. These costs lead to potential evolutionary trade-offs between immune function and other functionally important traits—such as metabolism or reproduction—that could reduce individual fitness and colony productivity [10–12]. In the light of these trade-offs, selection might favour less metabolically expensive ‘behavioural immunity’ in social insects who construct nests with antimicrobial properties, exhibit auto- and allogrooming, and many other behaviours that reduce disease transmission and the likelihood of infection [13]. Such behaviours of ‘social immunity’ may be a key adaptive response to compensate for the increasing risk of disease associated with more frequent interactions among nest-mates as colony size increases [14].
Two hypotheses predict how individual immune systems should respond to the evolution of sociality and larger colony sizes. The social group hypothesis (SGH) argues that social insects might evolve more heightened immune responses than solitary organisms, despite their behavioural immunity [14]. Alternatively, the relaxed selection hypothesis (RSH) predicts that behavioural immunity reduces disease risk such that costly internal immune functions are maintained at lower levels [14–16]. A direct test of these hypotheses would measure the investment in physiological and behavioural immunity as a function of sociality or colony size.
Here, we tested the SGH and RSH by contrasting cellular immune function across multiple insect lineages. We quantified encapsulation response, an important part of the innate immune system in insects, in which haemocytes bind to large parasites after they have entered the host body to immobilize and kill them. Finding higher encapsulation in social insect lineages would support the SGH, whereas higher encapsulation response in solitary species would support the RSH. We used phylogenetic generalized linear mixed models to evaluate the effects of behaviour (eusocial or not), maximum colony size, incubation temperature and body size on the cellular immune response of diverse species of insects.
2. Material and methods
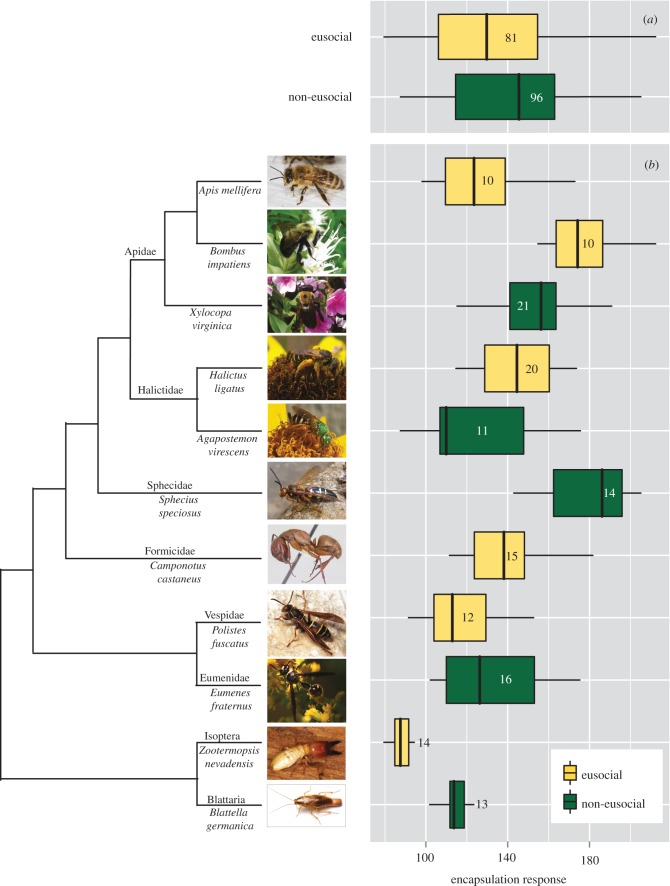
We collected specimens from 11 insect lineages, including bees, wasps, ants, termites and cockroaches, for the immune assays (figure 1). Live individuals were immunologically challenged using 3 mm nylon microfilaments coated with lipopolysaccharide, to induce an immune response, and inserted in the specimen abdomen using sterile technique [17,18]. Individuals were anaesthetized on ice to immobilize them and minimize stress during the procedure. After the probes were introduced in the insect bodies, individuals were incubated for a 4 h period in two temperature treatments (25°C and 35°C). Then, we removed the probes and photographed them under a scope Nikon SMZ 800 using consistent light and camera settings. Encapsulation density was quantified from the melanin deposition on the probe as the mean greyness of the nylon microfilament through image analyses in the software ImageJ [19] (electronic supplementary material, appendix S1).
Figure 1.
Levels of encapsulation response quantified in eusocial and non-eusocial insect lineages. (a) Boxplot of the encapsulation response in eusocial (yellow) and non-eusocial (green) lineages. (b) Boxplot of the encapsulation response across the sampled insect lineages. Numbers in boxplots show sample size. Colony size, not eusociality, had a significantly negative effect on encapsulation response. See main text for details. Photo credits: Apis mellifera (Wikipedia), Bombus impatiens (Elsa Youngsteadt), Xylocopa virginica (Margarita M. López-Uribe), Halictus ligatus (Elsa Youngsteadt), Agapostemon virescens (Wikipedia), Sphecius speciosus (Elsa Youngsteadt), Camponotus castaneus (Wikipedia), Polistes fuscatus (Wikipedia), Eumenes fraternus (Wikipedia), Zootermopsis nevadensis (Will Chatfield-Taylor) and Blattella germanica (Matt Bertone).
Lineages were coded as ‘eusocial’ if they exhibited a reproductive division of labour and overlapping generations (electronic supplementary material, appendix S2). However, this binary classification of eusociality is a simplification of the variability in social complexity between the focal species. Therefore, we included colony size as a secondary predictor variable of immune response in our analysis with the assumption that species with larger colonies tend to be at increased risk from socially transmitted pathogens. To control for differences in body size across the different lineages, we measured body length of each taxa and included this continuous variable in our model. Colony size was log transformed to normalize the data and improve their fit to our model.
To quantify the phylogenetic signal in our dataset, we calculated lambda for the two continuous variables in our dataset: colony size and body size. To account for the non-independent evolutionary history among the 11 lineages in our dataset, we calculated the inverse phylogenetic covariance matrix with the function inverseA in ‘ape’ [20], after building a maximum likelihood phylogeny using seven genes (electronic supplementary material, appendix S3). This pairwise phylogenetic matrix was incorporated as a random effect in a multivariate generalized linear mixed model using the R package ‘MCMCglmm’ [21] (electronic supplementary material, appendix S3). For this analysis, we estimated the effect of sociality, colony size, temperature and body size on encapsulation response. The Heidelberg and Welch diagnostic was used to corroborate convergence of the MCMC runs. We used Deviance Information Criterion (DIC) to determine the models that best explain the variability in immune function across the focal lineages. Interaction terms between factors were also assessed. We used an ordinary generalized least-square (GLS) model and a phylogenetic GLS with Pagel's ‘lambda’ correlation structure using the R package ‘ape’ [20] to quantify the effect of the predictor variables on encapsulation response (electronic supplementary material, appendix S3).
3. Results
We detected that encapsulation response was lower in insect species with larger colony size using the MCMCglmm (posterior mean = −4.93; 95% CI = [−10.20, −0.05]) and PGLS approaches (−2.55, p = 0.037). This corresponds to a 2% change in encapsulation response (mean greyness) [−4.93; CI = −10.20, −0.049] with every 10-fold increase in colony size. The high lambda values detected for the continuous variables (colony size λ = 0.78; body size λ = 0.77) and the lower Akaike Information Criterion (AIC) values for phylogenetically corrected models suggest that the best models included phylogeny fitted as a random effect (table 1). Encapsulation response was not significantly lower in eusocial than in non-eusocial lineages (posterior mean = 17.84; 95% CI = [−5.03, 40.04]; figure 1). However, DIC values indicated that colony size, eusociality and body size similarly contributed to the variability in encapsulation response. We found a significant effect of body size in the PGLS model (2.89; p = 0.043) but no significant interaction was detected between body size and other variables (table 1). No significant effect of temperature (posterior mean = −3.52; 95% CI = [−10.21, 3.40]) was found on encapsulation response (table 1).
Table 1.
Statistical models showing coefficients and significance (in parenthesis) of main effects and interactions from the MCMCglmm and PGLS approaches. Significant terms are shown in bold. DIC and AICc values are reported for MCMCglmm and PGLS models, respectively.
| sociality | colony size | body size | temperature | |
|---|---|---|---|---|
| MCMC generalized linear mixed model | ||||
| main effect | 17.84 (−5.03, 40.04) | −4.93 (−10.20, −0.049) | 14.16 (3.27, 25.58) | −3.52 (−10.21, 3.40) |
| interaction with body size | −6.57 (−29.21, 15.17) | 0.81 (−4.99, 6.83) | — | — |
| interaction with colony size | −92.34 (−471.46, 233.13) | — | — | — |
| phylogenetic generalized least squares | ||||
| main effect | 15.27 (0.273) | −2.55 (0.037) | 2.89 (0.043) | — |
| interaction with body size | 1.19 (0.675) | 0.053 (0.934) | — | — |
| interaction with colony size | −3.51980 (0.6850) | — | — | — |
| models of trait evolution | ||||
| DIC (independent) | 1409.102 | 1409.286 | 1409.608 | 1410.107 |
| DIC (Pagel) | 1409.102 | 1409.005 | 1409.608 | 1410.354 |
| AIC (independent) | 107.379 | 107.434 | 111.164 | — |
| AIC (Pagel) | 108.481 | 103.829 | 105.377 | — |
| Pagel's lambda | — | 0.787 | 0.771 | — |
4. Discussion
Our results suggest that encapsulation response of social insects decreases with increasing colony size, in support of RSH. Even though the effect size we found was small [−4.93; CI = −10.20, −0.049], we detected reduced encapsulation in species with larger colonies after correcting for phylogeny. The significantly negative association between colony size and encapsulation response suggests that species with larger colonies rely more heavily on behavioural immune mechanisms to decrease disease transmission among group members [22]. Other aspects of ‘social behaviour’ that do not relate to colony size may be more subtle than we could capture in this categorical metric [23].
The lower number of immune-related genes in the genome of Apis mellifera, compared with dipterans and other hymenopterans, support predictions from RSH [15]. However, new evidence suggests that the reduced repertoire of immune genes in A. mellifera precedes the evolution of sociality in bees [24], a complexity not yet well accounted for. Nonetheless, the accelerated rates of evolution in immune genes of honeybees and ants also suggest that immune systems are under relaxed selection in social insect lineages, as supported by Harpur & Zayed [15]. By contrast, evidence from external immunity—immune defence mediated by external non-immunological antimicrobial compounds—shows the opposite trend. A study comparing the strength of external antimicrobial secretions between solitary, semisocial and social bees found evidence of progressively stronger antimicrobial activity in species with higher levels of sociality [25]. Stronger antimicrobial activity is a form of heightened immune response [26], as expected under SGH, and suggests that selection is strongly acting on external immune defences in social insects. Internal and external immune responses appear to be under different selective pressures.
In our analyses, variation within species in encapsulation response accounted for a large proportion of the variation present in our study (figure 1). Large heterogeneity in immune response within taxa may result from background differences in immune levels of wild individuals, as a result of age or previous immune challenges. However, we still found significant differences among taxa suggesting that species-specific selective pressures play key roles in immune systems. We did not detect a significant effect of temperature on encapsulation. Suboptimal environmental temperature is often predicted to decrease immune function in insects [27] but temperature can interact with other environmental factors, such as nutrition or population density, resulting in nonlinear responses [28].
Overall, our results suggest that encapsulation response is reduced in social lineages with larger colonies, supporting the RSH hypothesis that social insects rely on behavioural immunity to reduce disease transmission. These findings help elucidate how sociality and colony-size drive selection on different aspects of immune systems in insects.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We would like to thank Holden Appler, Karly Dugan and Jennifer Fulp for field and laboratory assistance; Sarah Diamond and Carlos Botero for advice on comparative phylogenetic methods; and members of the Tarpy Lab for comments on previous versions of the manuscript.
Ethics
During field collections and encapsulation experiments, we minimized handling time and stress of individuals as much as possible. Procedures used in this study were based on standard protocols for this type of immune assays.
Data accessibility
Files for raw encapsulation data, seven-gene alignment for phylogenetic reconstruction and maximum-likelihood estimated phylogeny are available from Dryad: http://dx.doi.org/10.5061/dryad.sj066.
Authors' contributions
M.M.L.-U., S.D.F., R.R.D and D.R.T. designed the experiment; M.M.L.-U. and W.B.S. conducted the experiment; M.M.L.-U. performed the data analysis and wrote the manuscript; all authors revised, approved the manuscript and are accountable for the work herein.
Competing interests
We declare no conflict of interest.
Funding
This study was funded by the CALS Dean's Enrichment Grant from North Carolina State University.
References
- 1.Cremer S, Armitage SA, Schmid-Hempel P. 2007. Social immunity. Curr. Biol. 17, R693–R702. ( 10.1016/j.cub.2007.06.008) [DOI] [PubMed] [Google Scholar]
- 2.Wolfe ND, Dunavan CP, Diamond J. 2007. Origins of major human infectious diseases. Nature 447, 279–283. ( 10.1038/nature05775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dobson AP, Carper ER. 1996. Infectious diseases and human population history. Bioscience 46, 115–126. ( 10.2307/1312814) [DOI] [Google Scholar]
- 4.Cremer S, Sixt M. 2009. Analogies in the evolution of individual and social immunity. Phil. Trans. R. Soc. B 364, 129–142. ( 10.1098/rstb.2008.0166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmid-Hempel P. 1998. Parasites in social insects. Princeton, NJ: Princeton University Press. [Google Scholar]
- 6.Wilson EO. 1971. The insect societies. Cambridge, MA: Belknap Press of Harvard University Press. [Google Scholar]
- 7.Schmid-Hempel P. 2003. Variation in immune defence as a question of evolutionary ecology. Proc. R. Soc. Lond. B 270, 357–366. ( 10.1098/rspb.2002.2265) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parker BJ, Barribeau SM, Laughton AM, de Roode JC, Gerardo NM. 2011. Non-immunological defense in an evolutionary framework. Trends Ecol. Evol. 26, 242–248. ( 10.1016/j.tree.2011.02.005) [DOI] [PubMed] [Google Scholar]
- 9.Schwarz F, Fong JJ, Varki A. 2015. Human-specific evolutionary changes in the biology of siglecs. Adv. Exp. Med. Biol. 842, 1–16. ( 10.1007/978-3-319-11280-0_1) [DOI] [PubMed] [Google Scholar]
- 10.Evans JD, Pettis JS. 2005. Colony-level impacts of immune responsiveness in honey bees, Apis mellifera. Evolution 59, 2270–2274. ( 10.1111/j.0014-3820.2005.tb00935.x) [DOI] [PubMed] [Google Scholar]
- 11.Lazzaro BP, Galac MR. 2006. Disease pathology: wasting energy fighting infection. Curr. Biol. 16, R964–R965. ( 10.1016/j.cub.2006.10.015) [DOI] [PubMed] [Google Scholar]
- 12.Lazzaro BP, Little TJ. 2009. Immunity in a variable world. Phil. Trans. R. Soc. B 364, 15–26. ( 10.1098/rstb.2008.0141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baer B, Schmid-Hempel P. 2001. Unexpected consequences of polyandry for parasitism and fitness in the bumblebee, Bombus terrestris. Evolution 55, 1639–1643. ( 10.1111/j.0014-3820.2001.tb00683.x) [DOI] [PubMed] [Google Scholar]
- 14.Evans J, et al. 2006. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 15, 645–656. ( 10.1111/j.1365-2583.2006.00682.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harpur BA, Zayed A. 2013. Accelerated evolution of innate immunity proteins in social insects: adaptive evolution or relaxed constraint? Mol. Biol. Evol. 30, 1665–1674. ( 10.1093/molbev/mst061) [DOI] [PubMed] [Google Scholar]
- 16.Viljakainen L, Evans JD, Hasselmann M, Rueppell O, Tingek S, Pamilo P. 2009. Rapid evolution of immune proteins in social insects. Mol. Biol. Evol. 26, 1791–1801. ( 10.1093/molbev/msp086) [DOI] [PubMed] [Google Scholar]
- 17.Wilson-Rich N, Dres ST, Starks PT. 2008. The ontogeny of immunity: development of innate immune strength in the honey bee (Apis mellifera). 54, 1392–1399. [DOI] [PubMed] [Google Scholar]
- 18.Appler RH, Frank SD, Tarpy DR. 2015. Within-colony variation in the immunocompetency of managed and feral honey bees (Apis mellifera L.) in different urban landscapes. Insects 6, 912–925. ( 10.3390/insects6040912) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. ( 10.1038/nmeth.2089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. ( 10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 21.Hadfield JD. 2010. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Stat. Softw. 33, 1–22. ( 10.18637/jss.v033.i02)20808728 [DOI] [Google Scholar]
- 22.Evans JD, Spivak M. 2010. Socialized medicine: individual and communal disease barriers in honey bees. J. Invertebr. Pathol. 103, S62–S72. ( 10.1016/j.jip.2009.06.019) [DOI] [PubMed] [Google Scholar]
- 23.Schwarz MP, Richards MH, Danforth BN. 2007. Changing paradigms in insect social evolution: insights from halictine and allodapine bees. Annu. Rev. Entomol. 52, 127–150. ( 10.1146/annurev.ento.51.110104.150950) [DOI] [PubMed] [Google Scholar]
- 24.Barribeau SM, et al. 2015. A depauperate immune repertoire precedes evolution of sociality in bees. Genome Biol. 16, 83 ( 10.1186/s13059-015-0628-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stow A, Briscoe D, Gillings M, Holley M, Smith S, Leys R, Silberbauer T, Turnbull C, Beattie A. 2007. Antimicrobial defences increase with sociality in bees. Biol. Lett. 3, 422–424. ( 10.1098/rsbl.2007.0178) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otti O, Tragust S, Feldhaar H. 2014. Unifying external and internal immune defences. Trends Ecol. Evol. 29, 625–634. ( 10.1016/j.tree.2014.09.002) [DOI] [PubMed] [Google Scholar]
- 27.Blanford S, Thomas MB. 1999. Host thermal biology: the key to understanding host–pathogen interactions and microbial pest control? Agric. For. Entomol. 1, 195–202. ( 10.1046/j.1461-9563.1999.00027.x) [DOI] [Google Scholar]
- 28.Triggs A, Knell RJ. 2012. Interactions between environmental variables determine immunity in the Indian meal moth Plodia interpunctella. J. Anim. Ecol. 81, 386–394. ( 10.1111/j.1365-2656.2011.01920.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Files for raw encapsulation data, seven-gene alignment for phylogenetic reconstruction and maximum-likelihood estimated phylogeny are available from Dryad: http://dx.doi.org/10.5061/dryad.sj066.