Abstract
Heterogeneity in rates of survival, growth and reproduction among viruses is related to virus particle (i.e. virion) size, but we have little understanding of the factors that govern the four to five orders of magnitude in virus size variation. Here, we analyse variation in virion size in 67 double-stranded DNA viruses (i.e. dsDNA) that span all major biomes, and infect organisms ranging from single-celled prokaryotes to multicellular eukaryotes. We find that two metrics of virion size (i.e. virion volume and genome length) decrease by about 55-fold as the temperature of occurrence increases from 0 to 40°C. We also find that gene overlap increases exponentially with temperature, such that smaller viruses have proportionally greater gene overlap at higher temperatures. These results indicate dsDNA virus size increases with environmental temperature in much the same way as the cell or genome size of many host species.
Keywords: latitudinal gradient, thermal gradient, phage, cell size, viral ecology
1. Introduction
Viruses play a major role in governing the diversity and abundance of species through their effects on the evolution and ecology of their hosts [1–3]. They may strongly impact the biological systems they inhabit, from effects on human health to effects on population dynamics and biogeochemical cycling [4–7]. The strength of these impacts depends on the individual-level attributes of virions (i.e. virus particles) that affect rates of survival, growth and reproduction [8–9], and influence population-level dynamics [8–9]. However, little is known about how these basic ‘life-history’ features of viruses vary across species and environments given the tremendous structural and functional diversity present in viruses [7,10].
Many key features of viruses that affect virility (e.g. mutation rate, burst size, multiplication rate, decay rate, etc.) appear related to virus genome length and/or virion volume [11–13]. Similar to living organisms, smaller-sized viruses tend to exhibit higher rates of growth, decay and mutation than larger-sized viruses (i.e. ‘r-selected’) [7]. Yet, we have little understanding of the factors that govern virus size variation in viruses at broad scales. From the small polyomaviruses to the recently discovered megaviruses, virus size varies by at least four orders of magnitude (2571 to 75 × 106 nm3) across all virus types (ssDNA, dsDNA, RNA) [14].
Here, we focus on understanding the size variation of double-stranded DNA (i.e. dsDNA) viruses. These viruses are found in all biomes, and infect all major groups of organisms, from single-celled prokaryotes to multicellular eukaryotes. The 28 families of dsDNA viruses currently recognized vary considerably in size (approx. two orders of magnitude), shape (e.g. spherical to rod-like) and basic genetics (e.g. linear and circular genomes) [3,14].
We evaluate our hypothesis that dsDNA virus size decreases exponentially with increasing temperature, as has been observed for cell volumes and genome sizes of some single-celled prokaryotes and eukaryotes [15,16]. We first examine if either of two independent, but highly correlated measures of virus size [14], virion volume and virus genome length, decline with increasing temperature. We do so by analysing data from 67 dsDNA viruses that vary in their taxonomic affiliations (18+ families), host types (prokaryotes, single-celled eukaryotes and multicellular eukaryotes) and environments (terrestrial, freshwater, marine). The temperature of occurrence in these viruses ranges from near zero for those inhabiting polar environments to over 40°C for those inhabiting endothermic vertebrates. We then examine whether gene overlap increases exponentially with temperature among dsDNA viruses, as has been observed among single-celled organisms [17]. These analyses provide a first step towards better understanding how and why dsDNA viruses vary in size across species and environments.
2. Material and methods
(a). Data collection
Data were collected from the literature (electronic supplementary material, appendix 1) for dsDNA viruses from locations ranging from the Siberian permafrost at 60°N [18] to the Chilean Sea at 33°S [19]. Data include viruses that inhabit diverse host species (nine prokaryotes, seven single-celled eukaryotes, 16 vertebrate endotherms (10 mammals, six birds), 17 vertebrate ectotherms (three reptiles, three amphibians and 11 fishes) and four invertebrate ectotherms; see electronic supplementary material).
(b). Virion volumes and genome lengths
Volumes of both enveloped and non-enveloped virions were estimated from linear dimensions. For enveloped virions or non-enveloped virions with capsids, we used length or diameter measures of the capsid, defined as the innermost protein shell of the virion. For non-enveloped virions without capsids, we used length or diameter measures of the outermost layer, not including fibrils. Virion volumes were categorized into one of four shape categories following previous work (icosahedral, spherical, ovoid and rod; [14]), and standard geometric formulae were used to calculate volumes [14]. Genome lengths of viruses were taken from the literature.
(c). Temperature estimates
Temperatures of occurrence were estimated based on one of three measures of temperature: the internal temperature of the host inhabited by the virus (n = 30), the temperature at which the virus was isolated (n = 7) or the temperature at which the host was most infected by the virus (n = 30). The use of these three temperature measures, while not strictly equivalent, offers the best available approximation of virus temperatures of occurrence for purposes of analyses. Differences in the error associated with any single measure of temperature are likely small relative to the range of temperatures considered here (1–42.1°C).
(d). Virus gene overlap
The proportion of gene overlap in dsDNA viruses was estimated at the family level for nine of the 18+ virus families given available data (see electronic supplementary material) [20]. Overlap estimates did not include regulatory regions or overlap within the same reading frame [20]. Temperatures of occurrence at the family level were estimated as the mean temperature of occurrence of species considered here from those families.
(e). Analysis
To evaluate relationships with temperature, we first performed ordinary least-squares regression on natural log-transformed data using R v. 3.0.1 [21]. Analyses were performed on the complete set of dsDNA viruses, and on two subsets of data representing species occupying single-celled or multicellular hosts. Accounting for the possible effects of evolutionary relatedness among dsDNA viruses was not deemed feasible, in part because viruses are polyphyletic and do not share a common evolutionary history [22]. Additionally, we performed a bootstrapping procedure of the regressions to account for possible pseudo-replication in the observed relationships. We also performed partial correlation analyses on the relationships between virion volume and genome length with temperature (see electronic supplementary material).
3. Results
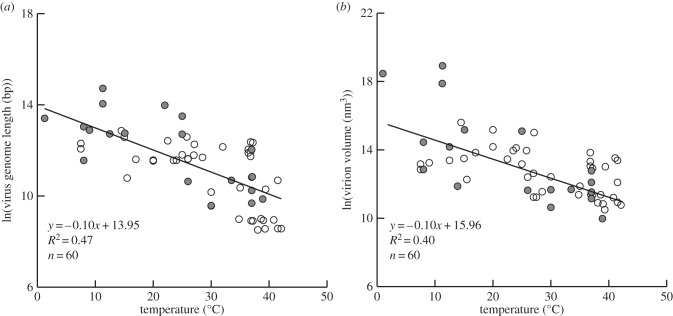
Both dsDNA genome length (bp) and virion volume (nm3) decreased exponentially with increasing temperature (figure 1a,b). Both relationships were highly significant (p < 0.001), with temperature explaining 47% and 40% of the variation in genome length and volume, respectively. The slopes of the fitted lines in both relationships (−0.10) show that on average virus genome length and virion volume decrease about 55-fold from 0 to 40°C (figure 1a,b). Among only species with single-celled hosts, similar relationships with temperature were observed for both genome length (y = −0.10x + 14.12; R² = 0.52; n = 21) and virion volume (y = −0.16x + 17.09; R² = 0.57; n = 19). Among only those with multicellular hosts, similar relationships with temperature were also observed for genome length (y = −0.09x + 13.67; R² = 0.38; n = 39) and virion volume (y = −0.065x + 14.56; R² = 0.27, n = 41). Bootstrapping analyses and partial correlation analyses support the results described for figures 1 and 2 (see electronic supplementary material).
Figure 1.
(a,b) Effect of temperature of occurrence on (a) virus genome length and (b) virion volume for dsDNA viruses. Species associated with single-celled or multicellular hosts are indicated by shaded or open symbols, respectively.
Figure 2.
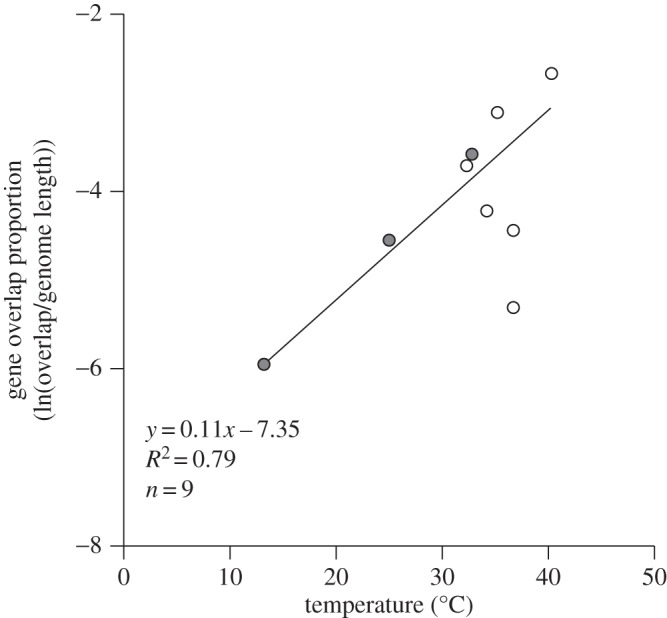
Effect of temperature of occurrence on the proportion of gene overlap across nine families of dsDNA viruses. Species associated with single-celled or multicellular hosts are indicated by shaded or open symbols, respectively.
In contrast, the proportion of gene overlap among nine families of dsDNA viruses increased exponentially with increasing temperature (figure 2; y = 0.11x − 7.35; R² = 0.79; n = 9; p < 0.001). The slope of this relationship (0.11, figure 2) is nearly inverse to the relationships with temperature shown for virus genome length and virion volume (−0.10, figure 1). Based on this slope, gene overlap increases about 81-fold from 0 to 40°C. Both virus families with single-celled hosts (n = 3) and virus families with multicellular hosts (n = 6) show this pattern (figure 2).
4. Discussion
Results show virion volume and genome length decrease exponentially with increasing temperature in dsDNA viruses, whereas the proportion of gene overlap shows the inverse relationship with temperature (figures 1 and 2). Still, many factors in addition to temperature may influence these two metrics of virus size (e.g. host-specific differences), as reflected by the unexplained variation in figure 1a,b. Surprisingly, little is known on the topic of virus size variation [14].
The relationships of virion volume, genome length and gene overlap with temperature are qualitatively similar to those previously observed in some single-celled and multicellular organisms, including host species [15–17,23,24]. This suggests ‘genome streamlining’ (e.g. reduction in proportion of noncoding DNA) in these viruses may compensate for any functional loss that accompanies a reduction in genome length at higher temperatures—as in single-celled organisms [17,20]. It also suggests that the size of dsDNA viruses has evolved with the sizes of the cells or genomes of their hosts across gradients in temperature.
Finally, the macroscale patterns shown here may point to trade-offs in virus life history across gradients in temperature. At the individual level, smaller viruses have shorter generation times and higher rates of replication; our results suggest that this trade-off may vary systematically across gradients in temperature [7,13]. At the ecosystem level, dsDNA viral abundance in marine environments is positively correlated with host cell abundance, and increases with temperature [25,26]. Our results suggest that virus size and abundance may be inversely related, such that total viral biomass varies relatively little across temperature gradients. These individual-level and ecosystem-level trade-offs have previously been shown to occur in a diversity of living organisms (e.g. plants, animals, microbes) and have been attributed to energetic trade-offs related to metabolism. At the individual level, Pearl's rate of living hypothesis postulates that the rate at which living species operate is inversely related to their lifespan based on a finite lifetime energy budget [27]. At the ecosystem level, Damuth's energetic equivalence rule postulates that the population density of living organisms is inversely related to individual size given finite resource availability [28]. On the surface, these explanations would not appear to explain the aforementioned trade-offs in viruses, because viruses have no intrinsic metabolism. Perhaps, however, these trade-offs in viruses are a consequence of the energetic constraints imposed by their hosts.
Supplementary Material
Acknowledgements
We acknowledge Juan Pablo Gomez for advice on statistical analyses.
Data accessibility
All data used in analyses are presented in the electronic supplementary material.
Authors' contributions
R.L.N. and J.F.G. designed, analysed, drafted and approved the final version of the manuscript. Both authors will be held accountable for its accuracy.
Competing interests
We declare we have no competing interests.
Funding
R.L.N. was supported by USEPA STAR Fellowship 91745201.
References
- 1.Rohwer F, Thurber RV. 2009. Viruses manipulate the marine environment. Nature 459, 207–212. ( 10.1038/nature08060) [DOI] [PubMed] [Google Scholar]
- 2.Forterre P, Prangishvili D. 2009. The great billion-year war between ribosome- and capsid-encoding organisms (cells and viruses) as the major source of evolutionary novelties. Ann. NY Acad. Sci. 1178, 65–77. ( 10.1111/j.1749-6632.2009.04993.x) [DOI] [PubMed] [Google Scholar]
- 3.Krupovic M, Bamford DH. 2011. Double-stranded DNA viruses: 20 families and only five different architectural principles for virion assembly. Curr. Opin. Virol. 1, 118–124. ( 10.1016/j.coviro.2011.06.001) [DOI] [PubMed] [Google Scholar]
- 4.Fuhrman JA. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548. ( 10.1038/21119) [DOI] [PubMed] [Google Scholar]
- 5.Bamford DH, Grimes JM, Stuart DI. 2005. What does structure tell us about virus evolution?. Curr. Opin. Struct. Biol. 15, 655–663. ( 10.1016/j.sbi.2005.10.012) [DOI] [PubMed] [Google Scholar]
- 6.Suttle CA. 2005. Viruses in the sea. Nature 437, 356–361. ( 10.1038/nature04160) [DOI] [PubMed] [Google Scholar]
- 7.Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812. ( 10.1038/nrmicro1750) [DOI] [PubMed] [Google Scholar]
- 8.Hurst CJ. (ed.). 2000. Viral ecology. San Diego, CA: Academic Press. [Google Scholar]
- 9.Nowak M, May RM. 2000. Virus dynamics: mathematical principles of immunology and virology. Oxford, UK: Oxford University Press. [Google Scholar]
- 10.Breitbart M, Rohwer F. 2005. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13, 278–284. ( 10.1016/j.tim.2005.04.003) [DOI] [PubMed] [Google Scholar]
- 11.Brown CM, Lawrence JE, Campbell DA. 2006. Are phytoplankton population density maxima predictable through analysis of host and viral genomic DNA content? J. Mar. Biol. Assoc. UK 86, 491–498. ( 10.1017/S0025315406013397) [DOI] [Google Scholar]
- 12.Sanjuan R, Nebot MR, Chirico N, Mansky LM, Belshaw R. 2010. Viral mutation rates. J. Virol. 84, 9733–9748. ( 10.1128/JVI.00694-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Paepe M, Taddei F. 2006. Viruses’ life history: towards a mechanistic basis of a trade-off between survival and reproduction among phages. PLoS Biol. 4, e193 ( 10.1371/journal.pbio.0040193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui J, Schlub TE, Holmes CE. 2014. An allometric relationship between the genome length and virion volume of viruses. J. Virol. 88, 6403–6410. ( 10.1128/JVI.00362-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morán XAG, López-Urrutia A, Calvo-Díaz A, Li WKW. 2010. Increasing importance of small phytoplankton in a warmer ocean. Glob. Change Biol. 16, 1137–1144. ( 10.1111/j.1365-2486.2009.01960.x) [DOI] [Google Scholar]
- 16.Daufresne M, Lengfellner K, Sommer U. 2009. Global warming benefits the small in aquatic ecosystems. Proc. Natl Acad. Sci. USA 106, 12 788–12 793. ( 10.1073/pnas.0902080106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saha D, Panda A, Podder S, Ghosh TC. 2015. Overlapping genes: a new strategy of thermophilic stress tolerance in prokaryotes. Extremophiles 19, 345–353. ( 10.1007/s00792-014-0720-3) [DOI] [PubMed] [Google Scholar]
- 18.Legendre M, et al. 2014. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl Acad. Sci. USA 111, 4274–4279. ( 10.1073/pnas.1320670111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillipe N, et al. 2013. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341, 281–286. ( 10.1126/science.1239181) [DOI] [PubMed] [Google Scholar]
- 20.Chirico N, Vianelli A, Belshaw R. 2010. Why genes overlap in viruses. Proc. R. Soc. B 277, 3809–3817. ( 10.1098/rspb.2010.1052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R Development Core Team. 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 22.Moreira D, López-García P. 2009. Ten reasons to exclude viruses from the tree of life. Nat. Rev. Microbiol. 7, 306–311. ( 10.1038/nrmicro2108) [DOI] [PubMed] [Google Scholar]
- 23.Mousing EA, Ellegaard M, Richardson K. 2014. Global patterns in phytoplankton community size structure—evidence for a direct temperature effect. Mar. Ecol. Prog. Ser. 497, 25–38. ( 10.3354/meps10583) [DOI] [Google Scholar]
- 24.López-Urrutia Á, Morán XAG. 2015. Temperature affects the size-structure of phytoplankton communities in the ocean. Limnol. Oceanogr. 60, 733–738. ( 10.1002/lno.10049) [DOI] [Google Scholar]
- 25.Danovaro R, Corinaldesi C, Dell'Anno A, Fuhrman JA, Middelburg JJ, Noble RT, Suttle CA. 2011. Marine viruses and global climate change. FEMS Microbiol. Rev. 35, 993–1034. ( 10.1111/j.1574-6976.2010.00258.x) [DOI] [PubMed] [Google Scholar]
- 26.Jiang SC, Paul JH. 1994. Seasonal and diel abundance of viruses and occurrence of lysogeny/bacteriocinogeny in the marine environment. Mar. Ecol. Prog. Ser. 104, 163–172. ( 10.3354/meps104163) [DOI] [Google Scholar]
- 27.Pearl R. 1928. The rate of living. Being an account of some experimental studies on the biology of life duration. New York, NY: Knopf. [Google Scholar]
- 28.Damuth J. 1987. Interspecific allometry of population density in mammals and other animals: the independence of body mass and population energy-use. Biol. J. Linn. Soc. 31, 193–246. ( 10.1111/j.1095-8312.1987.tb01990.x) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in analyses are presented in the electronic supplementary material.