Significance
The human endogenous retrovirus (HERV) group HERV-K contains nearly intact and insertionally polymorphic integrations among humans, many of which code for viral proteins. Expression of such HERV-K proviruses occurs in tissues associated with cancers and autoimmune diseases, and in HIV-infected individuals, suggesting possible pathogenic effects. Proper characterization of these elements necessitates the discrimination of individual HERV-K loci; such studies are hampered by our incomplete catalog of HERV-K insertions, motivating the identification of additional HERV-K copies in humans. By examining >2,500 sequenced genomes, we have discovered 19 previously unidentified HERV-K insertions, including an intact provirus without apparent substitutions that would alter viral function, only the second such provirus described. Our results provide a basis for future studies of HERV evolution and implication for disease.
Keywords: HERV-K, HML-2, human endogenous retrovirus, 1000 Genomes Project, Human Genome Diversity Project
Abstract
Endogenous retroviruses (ERVs) have contributed to more than 8% of the human genome. The majority of these elements lack function due to accumulated mutations or internal recombination resulting in a solitary (solo) LTR, although members of one group of human ERVs (HERVs), HERV-K, were recently active with members that remain nearly intact, a subset of which is present as insertionally polymorphic loci that include approximately full-length (2-LTR) and solo-LTR alleles in addition to the unoccupied site. Several 2-LTR insertions have intact reading frames in some or all genes that are expressed as functional proteins. These properties reflect the activity of HERV-K and suggest the existence of additional unique loci within humans. We sought to determine the extent to which other polymorphic insertions are present in humans, using sequenced genomes from the 1000 Genomes Project and a subset of the Human Genome Diversity Project panel. We report analysis of a total of 36 nonreference polymorphic HERV-K proviruses, including 19 newly reported loci, with insertion frequencies ranging from <0.0005 to >0.75 that varied by population. Targeted screening of individual loci identified three new unfixed 2-LTR proviruses within our set, including an intact provirus present at Xq21.33 in some individuals, with the potential for retained infectivity.
During a retrovirus infection, a DNA copy of the viral RNA genome is permanently integrated into the nuclear DNA of the host cell as a provirus. The provirus is flanked by short target site duplications (TSDs), and consists of an internal region encoding the genes for replication that is flanked by identical LTRs. Infection of cells contributing to the germ line may result in a provirus that is transmitted to progeny as an endogenous retrovirus (ERV), and may reach population fixation (1). Indeed, more than 8% of the human genome is recognizably of retroviral origin (2). The majority of human ERVs (HERVs) represent ancient events and lack function due to accumulated mutations or deletions, or from recombination leading to the formation of a solitary (solo) LTR; however, several HERVs have been coopted for physiological functions to the host (3).
The HERV-K (HML-2) proviruses (4–9), so-named for their use of a Lys tRNA primer and similarity to the mouse mammary tumor virus (human MMTV like) (10), represent an exception to the antiquity of most HERVs. HML-2 has contributed to at least 120 human-specific insertions, and population-based surveys indicate as many as 15 unfixed sites, including 11 loci with more or less full-length proviruses (5, 6, 8, 9). To distinguish the latter from recombinant solo-LTRs, we refer to these elements as “2-LTR” insertions throughout this study. The majority of these insertions are estimated to have occurred within the past ∼2 My, the youngest after the appearance of anatomically modern humans (4, 8, 11). Population modeling has implied a relatively constant rate of HML-2 accumulation since the Homo-Pan divergence (5, 12, 13). All known insertionally polymorphic HML-2 proviruses have signatures of purifying selection, implying ongoing exogenous replication, and retain one or more ORFs (8, 13–15). HML-2 expression has been observed in tumor-derived tissues as well as normal placenta in the form of RNAs, proteins, and noninfectious retrovirus-like particles (3, 16–19). These unique properties raise the possibility that some HML-2 group members are still capable of replication by exogenous transmission from rare intact proviruses, from the generation of infectious recombinants via copackaged viral RNAs, or from rare viruses still in circulation in some populations. A naturally occurring infectious provirus has yet to be observed, although the well-studied “K113” provirus, which is not in the GRCh37 (hg19) reference genome but maps to chr19:21,841,544, has intact ORFs (9) and engineered recombinant HML-2 proviruses are infectious in cell types, including human cells (20, 21). The goal of this study was to enhance our understanding of such elements by identifying and characterizing additional polymorphic HML-2 insertions in the population.
The wealth of available human whole-genome sequence (WGS) data should, in principle, provide the information needed to identify transposable elements (TEs), including proviruses, in the sequenced population. However, algorithms for routine analysis of short-read (e.g., Illumina) paired-end sequence data exclude reads that do not match the reference genome. Based on read signatures stemming from such read pairs, specialized algorithms have been developed to detect TEs present within sequenced whole genomes. These methods seek to identify read pairs for which one read is mapped to a reference genome and the mate is aligned to the TE of interest (22). Additional criteria (e.g., read support, depth, presence of reads that cross the insertion junction) are then assessed to identify a confident call set. Recent applications of this general method to Illumina WGS data have indicated the presence of additional nonreference HML-2 insertions (12, 23), although validation and further characterization of these sites have been limited. Also, given the comparably short fragment lengths of typical Illumina libraries, it is not possible to distinguish between solo-LTR insertions and the presence of a 2-LTR provirus using these data alone, and experimentation is required to exclude sequencing artifacts.
To date, the number of human genomes analyzed for unfixed HML-2 proviruses is fairly small, limiting discovery of elements not present in the human reference genome, or “nonreference” elements, to those elements that are present in a relatively high proportion of individuals. Here, we build on existing detection methods to improve the efficiency of nonreference HML-2 identification from WGS data and assess the alleles present at each site. From analysis of more than 2,500 sequenced genomes, we have identified and characterized 36 nonreference insertions. We detected unique HML-2 insertions that were present in <0.05% to >75% of all samples and displayed variable presence across populations. Validation by locus-specific PCR confirmed three newly unreported 2-LTR proviruses within our dataset; one of these proviruses contains full ORFs for the viral gag, pro, pol, and env genes and lacks any obvious substitutions that would alter conserved sequence motifs, implying a potential for infectivity.
Materials and Methods
Data Analyzed.
Illumina WGS data were obtained from 1000 Genomes Project (1KGP) samples, including a total of 2,484 individuals from 26 populations (24), and 53 individuals in seven populations from the Human Genome Diversity Project (HGDP) (25, 26). The 1KGP data were downloaded in aligned Binary Alignment/Map (BAM) format (ftp://ftp.ncbi.nlm.nih.gov/1000genomes/ftp/data/). HGDP data were processed as described (26), and are available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive under accession SRP036155. Individual BAMs were merged using the Genome Analysis Toolkit (27) by population (1KGP) or dataset (HGDP). The 1KGP populations ranged from 66 to 113 individuals and had an effective coverage of ∼1,067x ± 207.4x per pooled BAM; 53 HGDP samples were pooled to a single BAM of ∼429x.
HML-2 Discovery from Read Pair Data.
Candidate nonreference HML-2 LTRs were identified using RetroSeq (28). LTR-supporting read pairs were identified by running “discover” on individual BAM files, with read alignment to the HML-2 LTR5Hs consensus elements from RepBase (29) and previous reports (20, 21). RepeatMasker (30) HERV coordinates from the GRCh37/hg19 reference were used for exclusion of previously annotated sites. RetroSeq “call” was applied to the merged BAMs (above), requiring a read support of ≥2 for a call. A maximum read depth per call of 10,000 was applied for the increased coverage of the BAMs. To capture only novel insertions, calls within 500 bp of an annotated HML-2 LTR were excluded. Other RetroSeq options were kept at default values.
Reconstruction of Viral-Genome Junctions.
For each RetroSeq candidate call, supporting read pairs and split reads within 200 bp of the assigned break were extracted from each sample and subjected to de novo assembly using CAP3 (31, 32). Assembled contigs were subjected to RepeatMasker analysis to confirm the LTR presence and type (i.e., LTR5Hs) (30), and then filtered to identify the most likely candidates, requiring separate contigs that contained the respective 5′ and 3′ HML-2 LTR edges, and the presence of ≥30 bp of both the LTR-derived and genomic sequence at each breakpoint. We examined contig pairs for the presence of 4-bp to 6-bp putative TSDs, but did not require their presence for a call. Output assemblies were aligned to the hg19 reference to confirm the position of the preintegration, or empty, site per call.
Analysis of Unmapped Reads for LTR Junction Discovery.
Unmapped reads were retrieved from BAM files with Samtools (samtools.sourceforge.net/) from all 53 HGDP samples and 825 1KGP samples (≥10 samples per 1KGP population) and searched for a sequence that matched the 5′ HML-2 LTR edge (TGTGGGGAAAAGCAAGAGA), 3′ LTR edge (GGGGCAACCCACCCATACA), or 3′ LTR variant (GGGGCAACCCACCCATTCA) that is observed in a subset of human-specific elements, requiring ≥10 bp of non-LTR sequence per read. Reads matching reference HML-2 junctions were removed. Candidate reads were then aligned to the hg19 reference to identify genomic position. Sequences with no match to hg19, with <90% identity, or that aligned to gaps or multiple genomic positions were searched against the chimpanzee (panTro4) and gorilla (gorGor3) references, and available human WGS data from the NCBI Trace Archive to identify insertions in structurally variable regions.
Validation and Sequencing.
DNA from samples yielding positive reads was obtained from Coriell or the Foundation Jean Dausset-Centre d’Étude du Polymorphisme Humain. Coordinates for each insertion were based on mapping of assembled contigs or read-captured flanking sequence to the hg19 reference. PCR was performed with 100 ng of genomic DNA using primers flanking each site to detect either the empty site or solo-LTR alleles. A separate PCR was run to infer a 2-LTR allele with a primer situated in the HML-2 5′ UTR paired with a flanking primer (6, 8, 33). Capillary sequencing was performed on at least one positive sample. The 2-LTR alleles were amplified in overlapping fragments from a single sample and sequenced to ≥4x (8, 9), and a consensus then constructed with the read traces from each site. Complete sequences are available in the NCBI GenBank under accession nos. KU054242–KU054309.
Phylogenetic Analysis.
Full-length sequences representing either solo-LTRs or proviral 5′ and 3′ LTRs were aligned to the consensus HML-2 using ClustalX (34); the alignment was then edited, and truncated LTRs were removed (8). A single neighbor-joining tree was generated from the remaining 68 insertions (90 total LTRs) using MEGA6 (35). The Kimura 2 parameter model was used for branch length estimation, with α of 2.5 and deletions treated pairwise. Support for the tree was assessed using 1,000 bootstrap replicates.
Age Estimations.
LTR divergence was used to infer the time since insertion, normalized to a neutral mutation rate of 0.24–0.45% per My as measured by calculating the divergence between orthologous human and chimpanzee HML-2 proviruses (6, 8). Alternatively, using mutation rates directly obtained from pedigrees (36) results in estimated times of insertion fourfold to ninefold as old and implausible integration times for many proviruses. Briefly, the nucleotide differences were totaled between proviral 5′ and 3′ LTRs, and the total was divided by the LTR nucleotide length. The percentage of divergence was then divided by the upper and lower bound mutation rates for age range estimation (million years) (6, 8).
In Silico Genotyping.
Genotyping was performed for both reference (hg19) and nonreference unfixed insertions using a read-based method that has been used for genotyping Alu TEs in humans (32, 37). Briefly, discrete reference (e.g., the empty state) and alternate (e.g., the insertion state) alleles were recreated for each locus, including ±600 bp upstream and downstream of the insertion point based on hg19 coordinates. Within those coordinates, Illumina read pairs that had at least one read mapped to the empty allele were extracted for each site (requiring a mapping quality score >20). Treating the reconstructed alleles as the target genome, genotype likelihoods were then determined based on remapping of those reads to either allele, with error probabilities based on read mapping quality as described previously (32, 38). Samples without reads aligning to the reconstructed reference and alternate alleles for a particular site were not genotyped at that site. Insertion allele frequencies were estimated per site for all genotyped samples as the total number of insertion alleles divided by the total number of alleles. Detection frequencies (the proportion of individuals carrying the insertion) were calculated as the number of individuals with the insertion divided by the total number of individuals genotyped at each locus. We note that the reference insertion at 7p22.1, which is present as a tandem duplication of two proviruses that share a central LTR (6, 15), was treated as a single insertion (chr7:4,622,057–4,640,031). Nine of the 36 nonreference loci could not be aligned to the hg19 reference and were excluded from genotyping: insertions within duplicated segments (we refer to these as dup1 through dup4), insertions of unusual assembled structure (10q24.2b and 15q13.1), or insertions that could not be mapped to the hg19 assembly (10q26.3, 12q24.32, and 22q11.23b).
Proportion of Provirus Carriers.
Unique 30-mers were identified from a set of 51 reference and nonreference elements from the HML2 subgroup using Jellyfish (39). Candidate 30-mers were further mapped against the GRCh37 genome reference using mrsFAST and mrFAST (40), and k-mers with >100 matches within an edit distance of two were omitted, resulting in a set of 83,343 k-mers. The position of each 30-mer in each HML2 element was determined, and 1,445 k-mers that crossed LTR-internal proviral junctions were omitted, leaving 5,698 k-mers that were unique to an LTR and 76,200 k-mers that were unique to the internal sequence. Total observed counts for each k-mer were determined in WGS sequence data from 53 HGDP and 2,453 1KP samples. The median k-mer depth for each element in each sample was determined. Median depths were normalized per sample by dividing by the maximum median depth observed for a proviral sequence. Elements with a normalized median k-mer depth ≥0.25 were considered to be present in a sample. The proportion of individuals for which an element was present was then determined for each population.
Results
HERV-K (HML-2) Insertions Discovered from WGS Data.
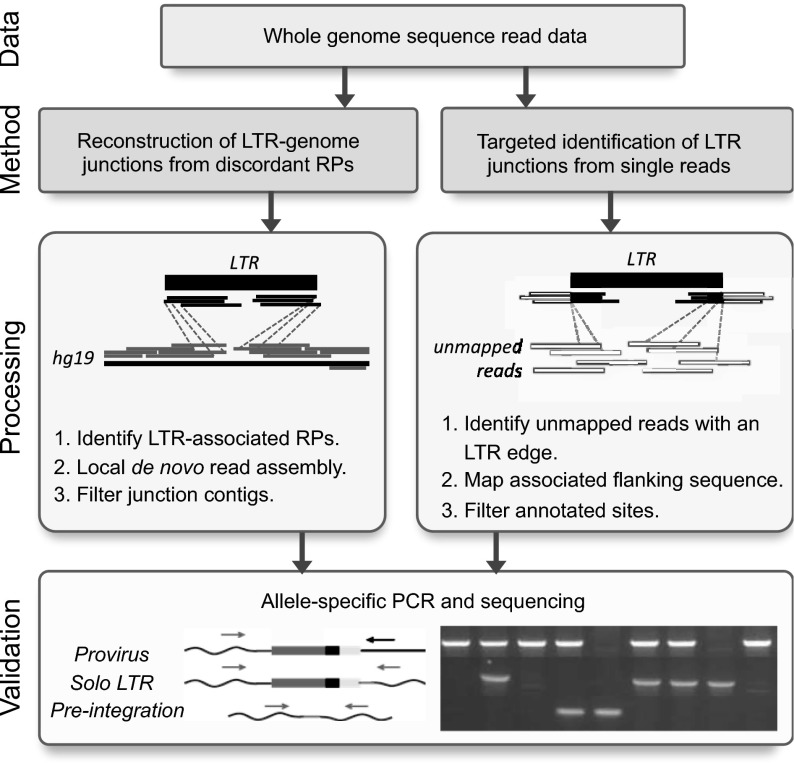
The goal of this study was to use the extensive available WGS data in the 1KGP and HGDP collections to identify relatively rare polymorphic nonreference HML-2 insertions. To make the fullest use of all sequence information available within these data, we applied two approaches to identify candidate nonreference HML-2 insertions in the raw reads for these collections (Fig. 1).
Fig. 1.
Approaches for the detection of nonreference HML-2 insertions from WGS read data. Illumina short reads were processed by one of two methods. (Left) Read pairs (RPs) were identified that have one read mapped to the genome (gray) and mate to reads that map to the sequence matching the HML-2 LTR consensus (black). Supporting reads from each site were extracted and subjected to local assembly, and the resulting contigs were analyzed for the presence of LTR–genome junctions. (Right) Unmapped reads from each sample were identified that contained a sequence corresponding to the LTR edge, and the cognate sequence was then used to determine candidate integration positions from genomic data. (Bottom) PCR and capillary sequencing were used to validate candidate insertions in reactions that used flanking primers (gray arrows) to detect the presence of a solo-LTR or empty site, or a flanking primer paired with an internal proviral primer (black arrow) to infer the presence of a full-length allele. Representative products are shown in a genotyping gel to the right.
First, we identified insertions based on read pair signatures using the program RetroSeq (28) (Fig. 1, Left). To improve the detection of insertions present in multiple samples, we combined reads within a population (1KGP) or study (HGDP) (32). Excluding calls within ±500 bp of a reference HML-2 sequence, we obtained 140.3 ± 56.1 candidate calls per pool. Next, we applied a de novo assembly approach to insertion-supporting reads to reconstruct the LTR–genome junction for as many sites as possible (32). Given the size of HML-2 LTRs (∼968 bp per LTR), we inferred the presence of an insertion based on the presence of separately assembled 5′ and 3′ breakpoints. This requirement reduced false-positive calls, for example, as caused by SVA elements (SINE-VNTR-Alu), which have high identity to bases 1–329 of the HML-2 LTR. A total of 29 candidate HML-2 insertions with a flanking sequence were assembled, including K113 (19p12b; also see Fig. S1 A and B and Table 1).
Fig. S1.
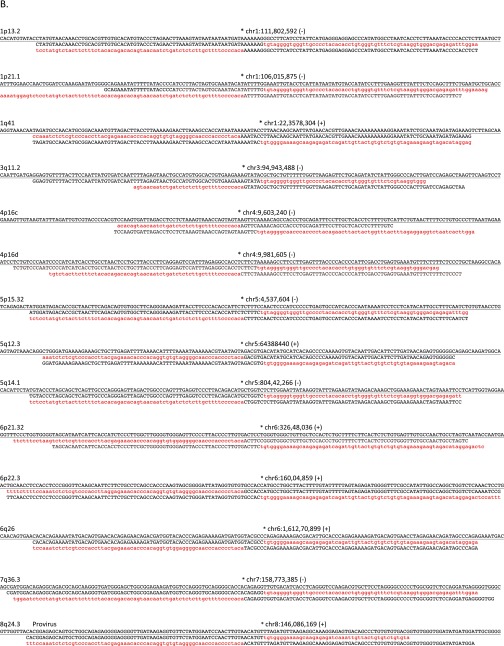
Assembly-based identification and validation of nonreference HML-2 insertions. (A, Upper) Integrative Genomics Viewer (IGV) screen shot showing Illumina paired short reads from four HGDP samples mapped to a candidate nonreference HML-2 insertion. The K113 locus at chr19:21,841,544 is illustrated as an example. Read pairs that have both reads mapped to the reference are shown in gray. Anchored reads that have discordant mated pairs (i.e., mapped to HML-2 LTRs that are present in the hg19 reference) are flagged and shown in color, and split reads (representing putative captured viral-genome junctions) are multicolored. Nearby SNPs are indicated by colored vertical lines within individual reads. HML-2 LTR-supporting read pairs, as reported in RetroSeq outputs, were then subjected to a local de novo assembly to generate contigs (boxed) corresponding to the 5′ and 3′ K113 proviral junctions. The sequence corresponding to the HML-2 LTR is shown in red; the sequence that maps to the reference is shown in black; and the candidate TSD is in shown dark red and underlined. (A, Middle) Alignment confirming overlap of the assembled contigs and presence of the TSD to the hg19 reference. Coloring is as above; the sequence corresponding to the reference is underlined. (A, Lower) Example validation screening is shown for integrations at 1p13.2 and 15q22.2 across a subset of 12 samples from the 1KGP. Each PCR contained three primers: two were designed to flank the insertion site, and a third primer was specific for the 5′ edge of the HML-2 LTR. Potential amplicons are interpreted to represent the preintegration site, solo-LTR (∼968 bp larger than the empty site), or an LTR-specific band. 2-LTR proviruses will not be amplified, but should produce an LTR-specific band. (B) Three-way alignments show overlap of HML-2–genome junctions against the hg19 reference (empty allele) for 27 validated nonreference insertions identified by breakpoint assembly. In each alignment, the reference allele is underlined and the sequence corresponding to the edges of either the 5′ or 3′ HML-2–LTR junction is shaded in red (LTR) or black (flanking). The hg19 insertion coordinates and locus are provided above each alignment; the asterisk indicates the first base of the LTR, respective of orientation (as indicated by “+” or “−” symbol). (C) Alignments corresponding to insertions identified in mining of unmapped reads. The junctions for five of seven insertions were validated in this study; the remaining two loci have been validated elsewhere (refer to main text). Sequences are shaded as above.
Table 1.
Nonreference HML-2 insertions in human genomes
| Locus | Coordinate GRCh37/hg19 | Alias* | Alleles† | Flanking region and other properties | First report in humans (source) |
| 1p13.2‡ | chr1:111,802,592 | De5;K1 | LTR, pre | L1 (L1PA6) | (41) |
| 1p21.1‡ | chr1:106,015,875 | LTR, pre | (12) | ||
| 1p31.1c | chr1:79,792,629 | LTR, pre | AluSz | This study | |
| 1q41 | chr1:223,578,304 | K2 | LTR, pre | L1 (L1MDa) | (12) |
| 3q11.2 | chr3:94,943,488 | LTR, pre | L1 (L1PA10) | This study | |
| 4p16c | chr4:9,603,240 | K6 | LTR, pre | ERV1 (HERVS71) | (12) |
| 4p16d | chr4:9,981,605 | LTR, pre | L2 L2b; SLCA29 intron 5/6 | This study | |
| 5p15.32 | chr5:4,537,604 | LTR, pre | ERV1 (LTR1C) | This study | |
| 5q12.3 | chr5:64,388,440 | Ne7;K12 | LTR, pre | L1 L1M6 | (12) |
| 5q14.1 | chr5:80,442,266 | De6/Ne1;K10 | LTR, pre | RASGRF2 intron 17 | (12) |
| 6p21.32 | chr6:32,648,036 | LTR, pre | L1 (L1PA10) | (12) | |
| 6p22.3 | chr6:16,004,859 | LTR, pre | AluSx | This study | |
| 6q26 | chr6:161,270,899 | De2;K12 | LTR, pre | (12) | |
| 7q36.3 | chr7:158,773,385 | LTR, pre | This study | ||
| 8q24.3c | chr8:146,086,169 | pro, pre | ERV1 (LTR46); COMMD5 intron 9 of transcript variant 2; gag and pro ORFs | This study | |
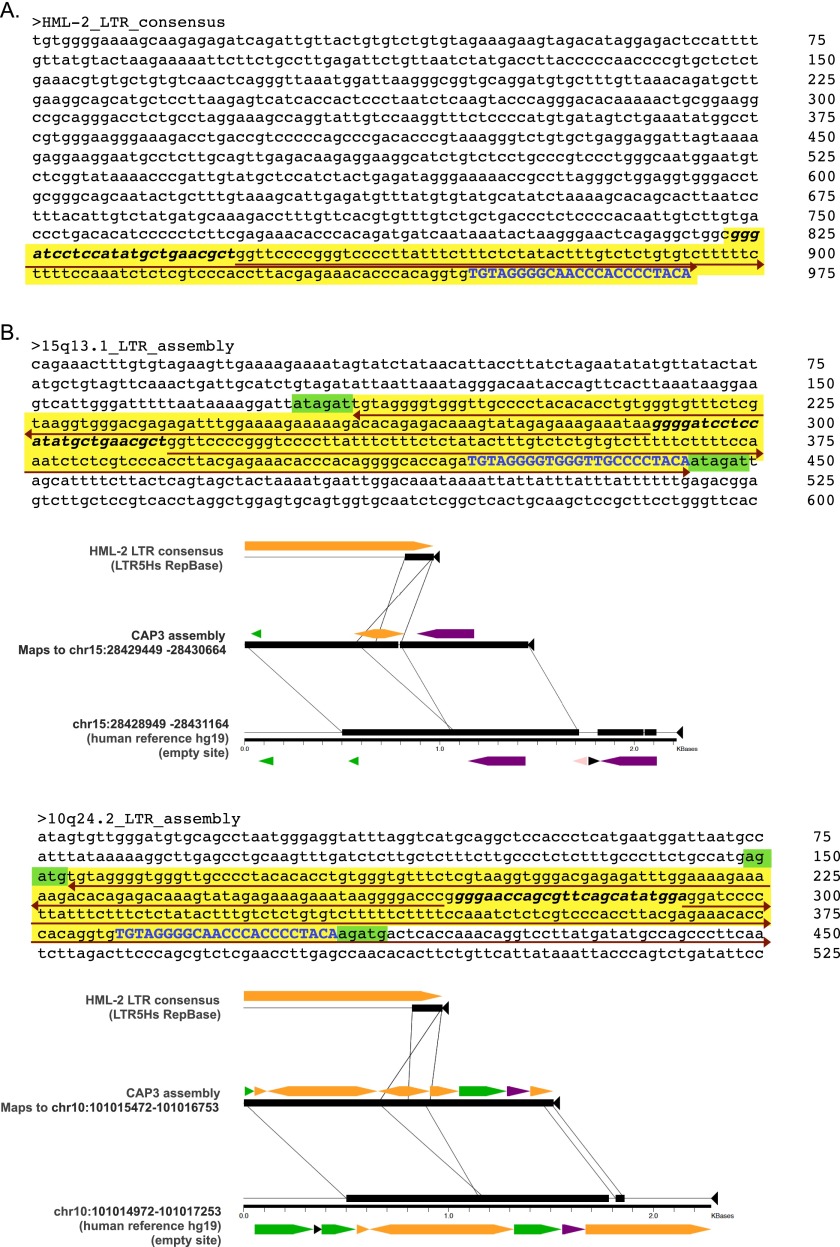
| 10q24.2b | chr10:101,016,122 | De12 | ERL MalR (MSTD); unexpected structure | This study | |
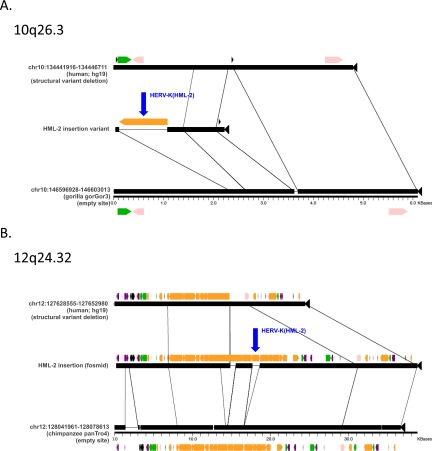
| 10q26.3§ | chr10:134,444,012 | LTR, pre | INPP5A intron 2 | This study | |
| 11q12.2 | chr11:60,449,890 | De4;K18 | LTR, pre | L1 (L1M4); LINC00301 intron 6 | (12) |
| 12q12 | chr12:44,313,657 | Ne6;K20 | LTR, pre | L1 (L1MB1); TMEM117 intron 2 | (12) |
| 12q24.31 | chr12:124,066,477 | K21 | LTR, pre | AluSx1; LOC101927415 exon 3 | (12) |
| 12q24.32§ | chr12:127,638,080–127,639,871 | LTR, pre | ERV1 (MER57); deleted in hg19; from fosmid CloneDB: AC195745.1 bases 17648–18615 | (43) | |
| 13q31.3 | chr13:90,743,183 | Ne2;K22 | LTR, pre | SINE (FLAM_A); LINC0559 intron 3 | (12) |
| 15q13.1 | chr15:28,430,088 | HERC2 intron 56; unexpected structure | This study | ||
| 15q22.2 | chr15:63,374,594 | K24 | LTR, pre | (12) | |
| 19p12b | chr19:21,841,536 | De1;K113 | pro, pre | (9) | |
| 19p12d | chr19:22,414,379 | pro, pre | Deletion in 5′ LTR; pro ORF; insertion within fosmid clone accession AC245253.1 | (41) | |
| 19p12e | chr19:22,457,244 | De11 | pro, pre | AluSq | This study |
| 19q12§ | chr19:29,855,781 | De3;K28 | LTR, pre | LOC284395 intron 9 | (12) |
| 19q13.43 | chr19:57,996,939 | Ne5 | LTR, pre | 2 kb upstream of ZNF419 | This study |
| 20p12.1 | chr20:12,402,387 | De14*;K30 | LTR, pre | (12) | |
| 22q11.23b§ | chr22:23852639–23852640 | De7;K16 | LTR, pre | ERVL-MalR (MLT1C); maps to Hg38 alt locus scaffold 22_KI270878v1_alt:156355–180653 | This study |
| Xq21.33 | chrX:93,606,603 | De9 | pro, pre | L1 (L1MD1); gag, prol, pol, env ORFs | This study |
| Dup 1§ | Not determined | LTR | Flank maps to centromere associated duplications on multiple chromosomes | This study | |
| Dup 2§ | Not determined | LTR, pre | Flank maps to duplicated regions within predicted FAM86 and ALG1L2 exonic variants | This study | |
| Dup 3§ | Not determined | LTR, pre | Flank maps to 3 segmental duplications on chr1 | This study | |
| Dup 4§ | Not determined | LTR, pre | Deletion in hg19 reference; putative empty site on chr19 within fosmid CloneDB: AC232224.2 | This study |
Reported originally in the sequenced Neandertal (Ne) or Denisovan (De) by Agoni et al. (42) or Lee et al. (51), or in modern humans (K) by Marchi et al. (12) or Lee et al. (41).
Alleles detected. LTR, solo LTR; pre, preinsertion site; pro, 2-LTR provirus.
Previously PCR validated as solo-LTR by Lee et al. (41).
Insertion is located within an encompassing structural variant not present in the hg19 reference.
As a second approach, we mined unmapped reads for evidence of LTR–genome junctions captured in reads that could not be placed on the human reference (Fig. 1, Right) and would therefore be missed using current read-based detection methods, such as RetroSeq. Using this approach permitted the identification of insertions in regions absent from the human reference. Excluding reads that could be aligned to annotated HML-2 junctions, we obtained overlap for the 29 candidate sites identified above, as well for as seven loci not found in assembled RetroSeq calls (Fig. S1C and Table 1). Our final call set includes 17 insertions identified in recent reports from Marchi et al. (12) and Lee et al. (41). The nomenclature for all sites is as maintained in those studies and in other previous reports (8, 33).
Validation and Sequencing.
We validated the presence of 34 of the 36 candidate insertions in at least one individual predicted to have the insertion (Table 1 and Dataset S1). The remaining two sites (at 10q24.2 and 15q13.1) were predicted to have an unusual inverted repeat structure based on assemblies of supporting reads at either site (Fig. S2), and could not be conclusively confirmed by sequencing, possibly due to hairpin formation. For the 34 validated nonreference sites, we confirmed 29 sites as having solo-LTRs and five sites with 2-LTR proviruses (at 8q24.3c, 19p12d, 19p12e, Xq21.33, and the published K113 provirus at 19p12b; also see Table 1). Four of the solo-LTRs were situated within duplicated segments and could not be mapped to unique positions in the hg19 reference (dup 1–dup 4), and two insertions, at 12q24.32 and 10q26.3, were located within structurally variable regions that are absent from the hg19 reference (Fig. S3). One insertion was initially mapped to the reported 9q34.11 locus (12, 41); however, comparison of the Sanger reads from its validated LTR–genome junctions revealed unexpectedly low identity in the extended flanking sequence. Our reexamination of this site indicates it maps instead to a region that is not in hg19 but is present in an alternate scaffold in the GRCh38 assembly at 22q11.23 (Table 1). This discrepancy may explain why this particular site has only been previously inferred by reads supporting only the 5′ breakpoint of the integration (12, 41, 42).
Fig. S2.
Assembled loci with unusual structure. Detailed nucleotide structure of the assembled contigs at 15q13.1 and 10q24.2. (A) Consensus LTR from the most recent HERV-K HML-2 insertions corresponds to the 968-bp LTR5_Hs consensus (RepBase release 11.03). The last 146 bp (yellow shading) are detected in two of our assembled structures; specific regions are labeled to indicate structural similarities with the two assembled elements as follows. (B) Nucleotide sequences and Miropeats alignments derived in local read assemblies at the 15q13.1 (Upper) and 10q24.2 (Lower) loci. Shading is used to discriminate LTR-derived portions from flanking genomic sequence. The yellow shading indicates an LTR5_Hs-matching sequence that assembles as an inverted repeat (red arrows) with a central unique portion (bolded and italicized in all three sequences) at those sites. The presence of a short stretch of the LTR-derived portion that is present in the hg19 reference is shown in blue, and the putative TSDs of 6 bp and 5 bp, respectively, are shaded in green. Block arrows indicate RepeatMasker annotated repeats within each aligned segment. DNA repeats are shown in pink, LTRs are shown in orange, long interspersed elements (LINEs) are shown in green, short interspersed elements (SINEs) are shown in purple.
Fig. S3.
Insertions located within genomic structural variants. Aligned sequence segments were generated using the program Miropeats (58) for HML-2 insertions in encompassing deletions that exist as structural variants in humans, located within 10q26.3 (A) and 12q24.32 (B) relative to the hg19 reference. Junction sequences corresponding to these insertions were recovered in BLAST searches of the NCBI Trace Archive; the adjacent flanking sequence maps to putative preinsertion sites within nonhuman primate reference genomes (gorilla or chimpanzee indicated as appropriate, including positions mapping to the preinsertion alleles in those references). The alignments corresponding to either insertion are arranged to indicate the absence of the insertion in the hg19 segment (Top), the presence of the insertion within the validated variant region (Middle), and the homologous sequence from the reference to which the preinsertion site could be mapped (Bottom). Lines are used to indicate segments of homology between the aligned segments. The validated LTR at 10q26.3 was sequenced in this study (HG00449), and the LTR at 12q24.32 was taken from a finished fosmid clone from Kidd et al. (43) (NIH CloneDB: AC1945745.1). Repeats are colored as in Fig. S2. The LTRs corresponding to the identified HML-2 insertions at 10q26.3 and 12q23.32 are labeled in blue with arrows.
We obtained full sequences for 30 of the 36 candidate insertions in at least one individual predicted to have the insertion (Dataset S1); these sequences included the full-length insertion at Xq21.33 that was found to have intact viral ORFs (NCBI GenBank accession no. KU054272). The remaining six insertions were extracted or reconstructed from public sequence databases for subsequent analysis as follows. The full sequence from one locus identified within a duplicated segment was reconstructed from Sanger reads corresponding to that site from the NCBI Trace Archive (dup1). The sequence flanking the insertion at 12q24.32 could be mapped to a previously sequenced fosmid clone in a region corresponding to an encompassing deletion of ∼14.3 kb in the hg19 reference (43) (Fig. S3A). Another insertion, corresponding to a 2-LTR provirus, was also from a sequenced fosmid clone (19p12d) as reported (41, 44). The complete sequence of the K113 provirus (19p12b) was from the GenBank (accession no. AY037928). One solo-LTR, 1p31.1c, was detected and validated as a solo-LTR in a single individual of the 1KGP Yoruba. We searched for, but did not find evidence of, this site in subsequent PCR screens of other samples.
Estimated Frequencies of Unfixed HML-2 Loci.
We performed in silico read-based genotyping to obtain estimations of the allele frequencies of 27 nonreference insertions with clear integration coordinates, and extended the analysis to include 13 annotated polymorphic HML-2 loci from the hg19 human reference (5, 8) (Dataset S2). Briefly, reference and alternate alleles representing each HML-2 locus were recreated, and individual genotypes were then inferred based on the remapping of proximal Illumina reads to the reconstructed alleles per site per sample (Materials and Methods). Given the larger size of the HML-2 LTR (∼968 bp) and relatively short reads in these data, 2-LTR and solo-LTR insertions are indistinguishable in read-based genotyping alone, such that genotypes were based on the presence or absence of the HML-2 insertion at each locus. Values reported below correspond to allele frequencies unless otherwise noted.
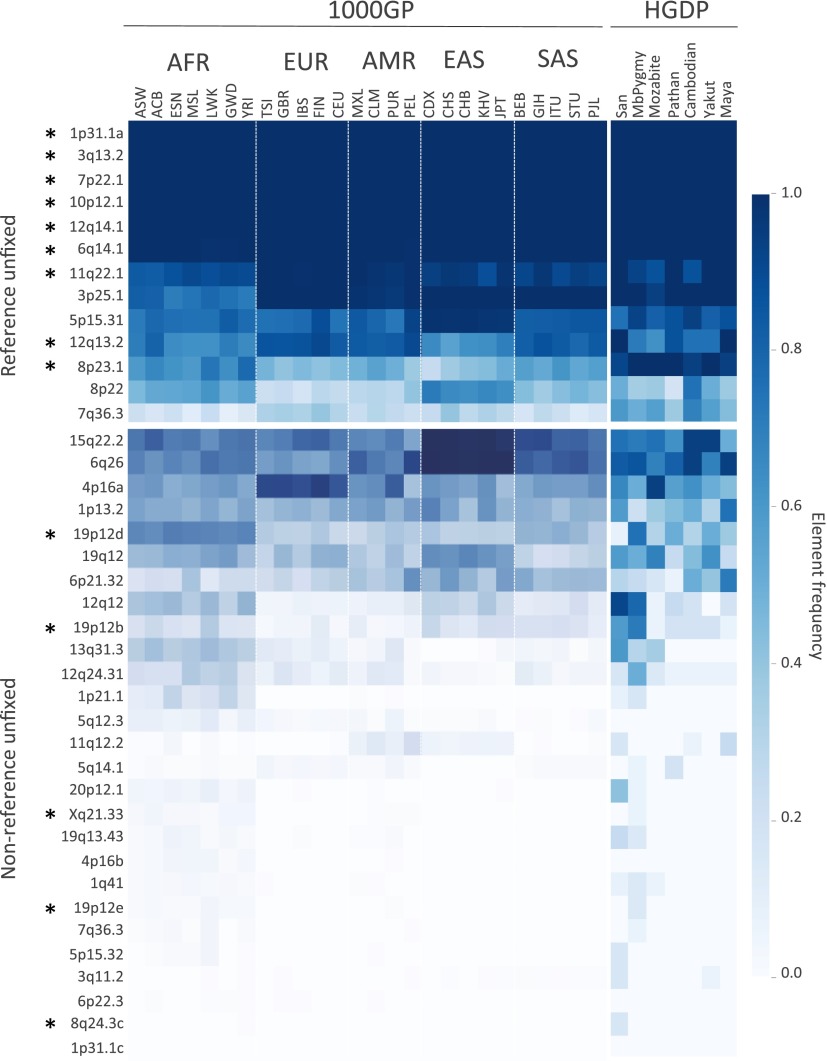
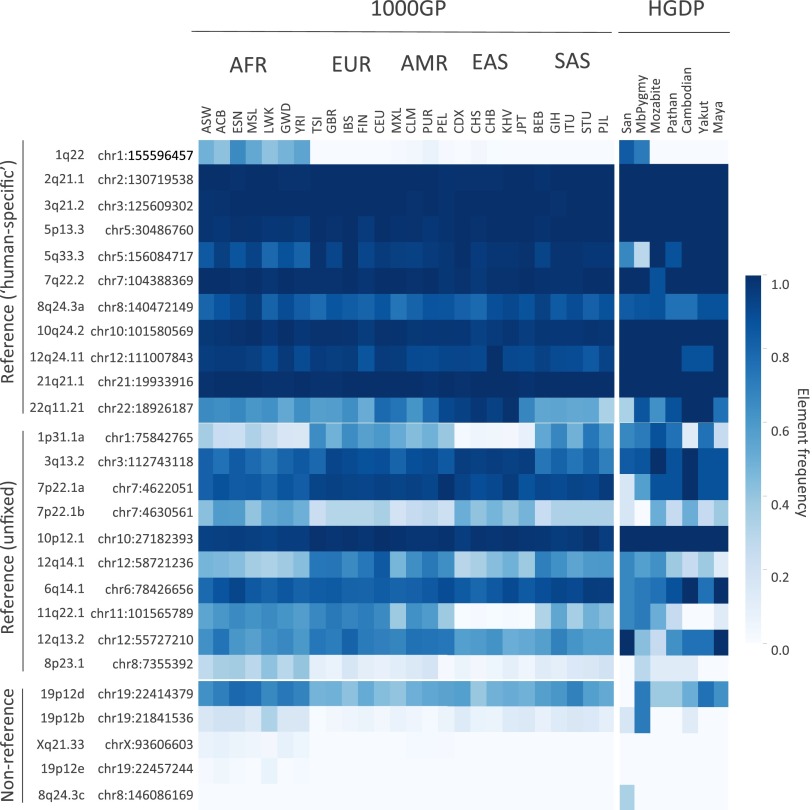
Estimated frequencies of the variable HML-2 insertions present in the reference genome ranged from ∼0.25 to >0.99 in genotyped samples (Fig. 2, Upper). Sites with the highest estimated frequencies corresponded to those loci previously reported with a solo-LTR or provirus present, but not a preinsertion site, based on limited PCR screens of those sites (6) (at 1p31.1, 3q13.2, 7p22.1, 12q14.1, and 6q14.1 in Fig. 2). This pattern is consistent with variability at these sites based predominantly on the 2-LTR and solo-LTR states. Genotyping of the insertions at 11q22.1 and 8p23.1a (K115) implied the presence of both insertion and preinsertion alleles, also consistent with PCR screens in other reports (6, 9, 33, 45), noting the higher frequency of K115 within our samples (∼53%) than in those reports (up to ∼34% depending on ancestry). Four unfixed reference solo-LTRs ranged in frequencies from ∼0.25 to as high as ∼0.93, also consistent with previous analysis of these sites (5). Extending the analysis to the 85 remaining human-specific HML-2 insertions that are suitable for genotyping in the human reference (81 solo-LTRs and four full-length proviruses) (5, 8) was consistent with sample-wide fixation among the vast majority of these loci; just eight loci had evidence of the nonreference allele among genotyped samples (Fig. S4 and Dataset S3).
Fig. 2.
Estimated insertion allele frequencies of unfixed HML-2 insertions in humans. A total of 40 HML-2 loci were subjected to in silico genotyping: 13 sites represented the unfixed HML-2 loci from the hg19 reference, and 27 sites corresponded to nonreference polymorphic HML-2 reported here. Genotypes were inferred for each unfixed HML-2 locus across samples based on remapping of Illumina reads to reconstructed insertion or empty alleles corresponding to each site. Samples lacking remapped reads at a particular site were excluded from genotyping at that site. Allele frequencies were then calculated for each population as the total number of insertion alleles divided by total alleles. Allele frequencies are depicted as a heat map according to the color legend to the right. The 1KGP (1000GP) and HGDP populations are labeled above (also refer to Dataset S1 for population descriptors and other information). The locus of each of the unfixed HML-2 loci is labeled to the left according to its cytoband position. An asterisk is used to indicate insertions that have confirmed full-length copies. (Upper) Estimated distribution of reference unfixed HML-2 [from loci reported by Subramanian et al. (11) and Belshaw et al. (5)]. (Lower) Estimated distribution of nonreference HML-2 insertions. AFR, African; AMR, Admixed American; EAS, East Asian; EUR, European; SAS, South Asian.
Fig. S4.
Inferred frequencies of additional human-specific HML-2 insertions in humans. A total of 85 additional HML-2 loci were subjected to in silico genotyping based on previous reports of human-specific HML-2 elements from Belshaw et al. (5) and Subramanian et al. (11). Of these loci, 81 insertions are present as solo-LTRs and four (indicated to the left by an asterisk) are present as proviruses in the hg19 reference genome, excluding insertions with unmatched TSDs and requiring the presence of cognate LTRs for full-length proviruses. Three solo-LTRs corresponded to duplicated loci and were excluded (chr8_91696140, chrY_25039185, and chrY_26672934). Genotypes were inferred by remapping Illumina reads to reconstructed reference (i.e., insertion) or alternate (i.e., empty) alleles for each site. Population allele frequencies were calculated as the total number of insertion alleles divided by total alleles and shown by heat map. The 1KGP and HGDP populations are labeled above (also refer to Dataset S1). The locus of each of the unfixed HML-2 loci is labeled to the left according to its chromosomal position.
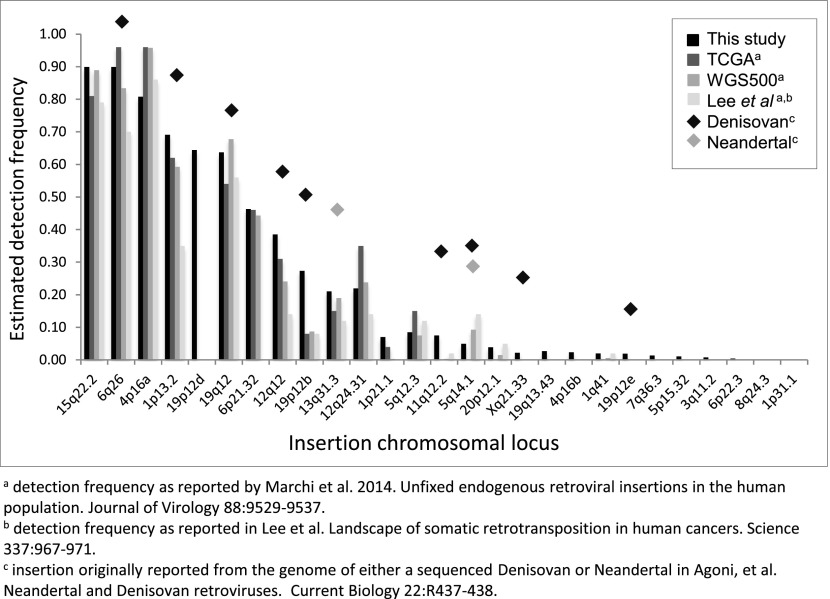
Estimated frequencies of the nonreference HML-2 insertions were inferred to be from <0.0005 (the insertion having clear support in one or few individuals) to >0.75 of genotyped samples (Fig. 2, Lower). More than half of the nonreference HML-2 insertions were rare, with 15 insertions detected at frequencies of <5% and six insertions in <1% of all samples; just four of these loci have been previously reported (12). Sites with the lowest allele frequencies were predominantly in individuals of African ancestry, with nine of 13 loci inferred in <5% of all samples but mostly limited to African populations, although insertions were also detected in non-African samples at ∼0.005 to ∼0.016 in those populations (e.g., at 5q14.1 and Xq21.33 in Fig. 3). The solo-LTR insertion at 1p31.1c was only identified in a single sample and was not detected in any other sample by genotyping; however, this observation does not exclude the possibility of its presence in some individuals, given the variability in read coverage between samples (Discussion). Nine of the 10 common insertions (detected in >10% of all samples), including the K113 provirus, have been previously reported in searches of WGS data (12, 41). A comparison of the overall presence of each HML-2 insertion, calculated as the proportion of individuals with evidence of the insertion, was generally in agreement with those reports (Fig. S5). The presence of K113 was estimated at a higher prevalence across samples here than in previous reports, in ∼27% of all samples and as high as ∼52% in African populations, consistent with the prevalence of this insertion varying with ancestry (45).
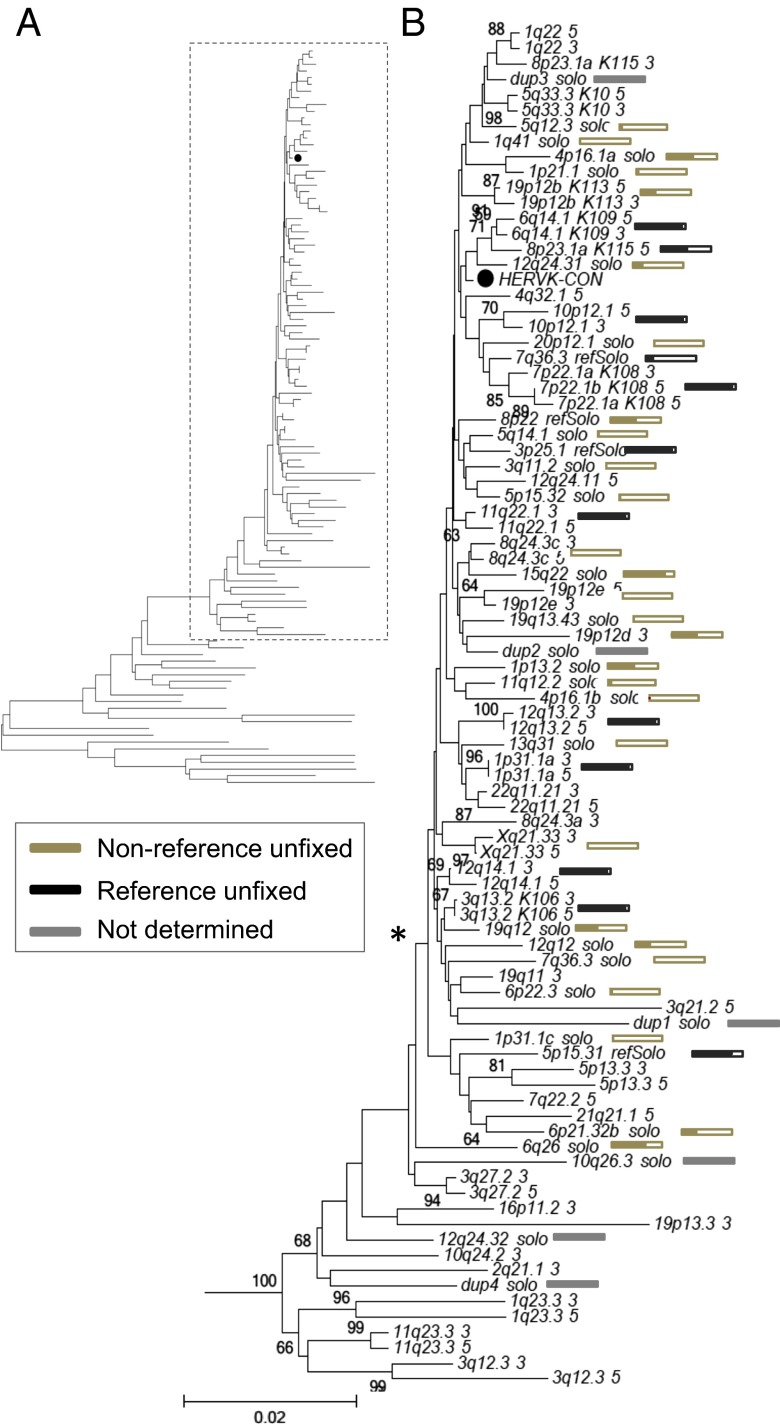
Fig. 3.
Phylogenetic construction of HML-2 LTRs within humans. (A) Neighbor-joining tree was constructed based on the aligned nucleotide sequences corresponding to HML-2 LTRs from the LTR5Hs group, specifically including those nucleotide sequences considered to be human-specific and/or polymorphic. The LTRs were extracted from (i) all reference HML-2 proviruses previously inferred as belonging to the LTR5Hs HML-2 subgroup [as reported by Subramanian et al. (11)], (ii) unfixed reference solo-LTRs [as reported by Belshaw et al. (5), and (iii) unfixed nonreference insertions as reported here. Both 5′ and 3′ LTRs were used for full-length insertions, when present. The closed circle (●) indicates the taxon corresponding to the HERV-KCON LTR within the tree. Classic nomenclature has been included in taxon names for the better studied insertions: K113 (19p12b), K108 (7p22.1), K115 (8p23.1a), K106 (3q13.2), and K109 (6q14.1). (B) Detailed view of branches representing unfixed HML-2 insertions. Individual HML-2 loci are indicated for each branch as follows: the cytoband followed by a 5′ or 3′ for the 5′ or 3′ LTRs from full-length insertions, solo for nonreference unfixed solo-LTR insertions, or refSolo for reference unfixed loci. An asterisk is used to indicate the position of the clade containing the majority of unfixed insertions. Boxes are used to indicate estimated allele frequencies for each unfixed insertion at the end of each respective branch. The filled area within each box is shown as proportional to the estimated frequency of the insertion in all samples; the derived values are provided in Dataset S3. Gold and black boxes are used to represent nonreference and reference unfixed insertions, and gray bars indicate the elements for which the frequency could not be determined.
Fig. S5.
Comparison of the inferred presence of nonreference HML-2 insertions with other reports. Insertion frequencies were calculated as the proportion of individuals with evidence for the insertion, as estimated by remapping of local reads to reconstructed alleles representing the insertion and empty states across samples. Samples without read support at a given site were not included in calculations for that site. The obtained values are plotted for 27 nonreference HML-2 insertions identified in our analysis. Insertion detection frequencies from this analysis are plotted in black; frequencies inferred from similar analysis of sequenced genomes from 26 TCGA and 332 WGS500 samples [Marchi et al. (12)], and 44 sequenced clinical samples [Lee et al. (41)] are shown in gray as indicated in the key. Shaded diamonds are used to indicate elements also detected in the sequenced Neandertal and Denisovan genomes [Agoni et al. (42) and Lee et al. (51)].
Because the above analysis cannot distinguish between solo-LTR and 2-LTR alleles, we used a k-mer counting approach to infer the presence of 2-LTR HML-2 sequences in each sample. WGS data from each sample were queried using a catalog of 30-mers unique to each proviral sequence. Elements with a normalized depth ≥0.25 were inferred to be present in a sample. We find large variation in the proportion of individuals estimated to carry 2-LTR alleles for different elements, with 2-LTR alleles for elements present in the reference genome detected in a high proportion of individuals, whereas the nonreference 2-LTR insertions discovered in this study are extremely rare (Fig. S6 and Dataset S3). For example, we identified 44 samples with k-mer counts consistent with the presence of a 2-LTR allele for the Xq21.33 insertion.
Fig. S6.
Presence of full-length unfixed HML-2 proviruses in the human genome. The presence of provirus (2-LTR) insertions was inferred based on counting of unique k-mers identified from the full nucleotide sequences corresponding to unfixed full-length insertions. Cytoband information and chromosomal positions corresponding to each locus are shown to the left. Sample carrier frequencies are represented as a heat map as shown by the color legend to the right. Samples are labeled by population according to the 1KGP (1000GP) or HGDP dataset (also refer to Dataset S1). The estimated distribution of reference unfixed HML-2 with confirmed 2-LTR proviruses [from loci reported by Subramanian et al. (11)] and the estimated distribution of nonreference HML-2 2-LTR insertions reported here are shown. AFR, African; AMR, Admixed American; EAS, East Asian; EUR, European; SAS, South Asian.
LTR-Based Analysis of Unfixed HML-2 Proviruses.
Using sequence information obtained for each insertion, we performed an LTR-based phylogenetic analysis (Fig. 3). Because proviral LTRs are identical at integration, the two LTRs on the same provirus will always pair in a phylogenetic tree, barring recombination between elements (46). Their unique source is further supported by the presence of TSDs, which are preserved during solo-LTR formation (6, 46, 47). To create the most informative tree, we added the LTRs from 21 human-specific proviruses, including 11 polymorphic 2-LTR insertions as reported by Subramanian et al. (8) and four unfixed solo-LTRs as reported by Belshaw et al. (5), to our validated set of LTRs from three 2-LTR proviruses (insertions at 8q24.3c, 19p12e, and Xq21.33), the 3′ LTR of a truncated provirus (19p12d), and 30 solo-LTRs.
The analysis revealed a major lineage leading to a well-supported clade that contained all human-specific and polymorphic HML-2 sequences (Fig. 3A, boxed); a minor lineage included HML-2 elements that are fixed in humans (8) and did not contain any newly identified insertions. This phylogeny is consistent with previous analyses (8, 13, 46); however, the addition of our 34 unfixed loci permitted a more detailed examination of variable insertions (Fig. 3B). The majority of unfixed insertions clustered with the HML-2 consensus LTR (● in Fig. 3B) in a clade (* in Fig. 3B) whose members tended to have the shortest branches, consistent with their relatively recent integration and insertionally variable presence within humans (also refer to filled boxes in Fig. 3B). The human-specific reference elements were also distributed within this clade, consistent with previous observations (8). The remaining insertions were on branches with longer lengths, reflecting changes that accrued before insertion as well as during their longer existence as endogenous elements. We searched for, but did not observe, mispaired LTRs from the 2-LTR proviruses reported here. Subsequent examination of the TSDs from these proviruses confirmed all were intact, indicating these elements have not seeded past rearrangements (46). Extending this comparison with the TSDs of the identified solo-LTRs also verified their intact state.
Properties of Nonreference 2-LTR HML-2 Integrations.
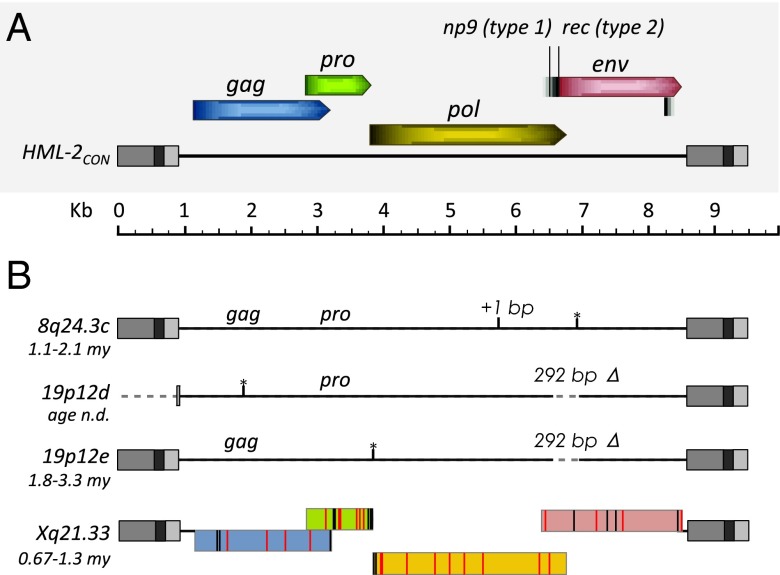
Assuming a constant mutation rate, the nucleotide divergence between cognate 5′-3′ LTR pairs may be used to estimate the time since integration (47). Using this method, we previously estimated the average age of the human-specific 2-LTR insertions to within ∼2.7 (±1.1) My (8). Applying this method to the 2-LTR proviruses identified here suggests these insertions were formed within ∼0.67–1.8 Mya (Table 1). Further refinement for the youngest elements is limited, because the variance for age estimates increases significantly for insertions with very little or no LTR divergence. The 19p12d provirus was excluded from this analysis due to deletion of the majority of its 5′ LTR (Fig. 4). This truncation has also been observed in a few reference LTR5Hs 2-LTR insertions (8q24.3a, 10q24.2a, and 19q11) (8), which all possess unique, intact TSDs and do not share any flanking sequence, supporting their classification as independent integrations. It is likely that this common rearrangement occurred as a result of aberrant strand transfer during RT, as has been discussed (48).
Fig. 4.
Features of newly identified HML-2 proviruses in humans. (A) Schematic representation of the consensus HML-2 provirus, including the viral gene positions and frames to scale. Splice sites for np9 (type 1 insertion, 292 bp Δ) and rec (type 2) are indicated. Regions within the LTRs are colored in gray: U3, medium; R, dark; U5, light. (B) Features of nonreference identified proviruses are shown to scale. The region of 292 bp is labeled for type 1 insertions. Age estimations are shown for each site. n.d., not determined. The black vertical line indicates a frameshift mutation (as indicated “+1 bp”); black lines with asterisks are used to indicate positions of stop codons where present. Reading frames are shown for the Xq21.33 2-LTR provirus as colored as in A. Black vertical lines within the frames indicate the positions of base changes that are observed in other full-length HML-2 proviruses. Red vertical lines are used to indicate base changes that are unique to the sequenced Xq21.33 provirus.
HML-2 proviruses are classified by the presence or absence of a 292-bp deletion at the pol-env boundary, designated “type 1” or “type 2,” respectively. Deletion of a splice site in type 1 elements obliterates env and rec expression and results in mRNA encoding an ∼9-kD protein, Np9, of possible cellular function (17, 19, 49). Sequence comparison of the 2-LTR proviruses identified here classifies the 19p12d and 19p12e elements as type 1 and 8q24.3c and Xq21.33 as type 2. The 19p12d and 19p12e insertions were found to have intact pro and gag ORFs, respectively, and the 8q24.3c insertion had both gag and pro ORFs. The Xq21.33 2-LTR element was found to be intact with ORFs for all HML-2 encoded genes (i.e., gag, pro, pol, env, rec) (Fig. 4). Indeed, it differs by only 39 of a total of 2,820 amino acids (98.6% amino acid identity) in all genes from the infectious consensus provirus HERV-KCON (21). We searched for, but did not observe, any substitutions that would alter conserved sequence motifs (including the YIDD motif in reverse transcriptase), making this element a candidate for activity. This provirus is only the second naturally occurring intact HERV to be described, with the other being the noninfectious K113 (19p12b) that shares 98.9% amino acid identity to HERV-KCON (9). Its potential for generation of infectious virus is currently under investigation.
Discussion
We report 36 nonreference HML-2 insertions, including 19 previously identified loci, from analysis of WGS read data from more than 2,500 globally sampled individuals. Seventeen of the 36 sites were recently reported in humans (12, 41), although with limited validation or element characterization. Here, we take full advantage of the 1KGP and HGDP WGS read data to identify nonreference viral-genome junctions from assembled anchored read pairs and individual unmapped reads, and use these data to estimate the presence of each of these elements within our sampled populations. We validated the presence of 34 of the 36 loci, including five loci with 2-LTR proviruses (including K113) and 29 solo-LTRs, and report the complete sequences for 30 of these insertions, including a 2-LTR provirus at Xq21.33 that appears to be intact. We provide a thorough analysis of unfixed HML-2 insertions that complements and builds on previous studies, and should enable future examination of the HML-2 group.
We used the available reads from each sample for in silico genotyping of a subset of sites to infer the population-wide frequencies of unfixed HML-2 elements, which is impractical on this scale in standard PCR-based screens. The inferred allele frequencies of the nonreference insertions ranged from 0.05 to >75% of genotyped samples and varied between populations, generally with the highest presence in African populations. With the exception of two previously sequenced sites in our set (dup1 and 12q24.11), all nonreference insertions were validated in samples of African ancestry, as has been observed for all HERV-K loci characterized to date, implying their insertion before the human migration out of Africa ∼45,000–60,000 y ago (50). These two insertions could not be confidently mapped to the hg19 reference, and were therefore excluded from genotyping. All but one nonreference insertion was identified in more than one individual, with the exception being the 1p31.1c solo-LTR validated in NA18867. Genotyping of that site failed to reveal (but does rule out) its presence in other individuals. Analysis of the surrounding region revealed the presence of several SNPs that were unique to NA18867 within the 1KGP panel, suggesting that 1p31.1c may be associated with a very rare haplotype, rather than a de novo event, in the absence of comprehensive screening. These observations support the utility of short read data for element discoveries and sequence-based analysis, but also underscore the necessity of additional experimental validation steps and characterization of candidate proviruses.
Eight of our validated loci have been recently reported in the genomes of two sequenced archaic samples (42, 51) in addition to modern humans (12). We confirmed an additional three reported “archaic” sites in our data [19p12e and 10q24.2b, respectively: “De11” and “De12” in the study by Agoni et al. (42); 19q13.43: “Ne5” in the study by Lee et al. (51)], but found no evidence of the remaining eight reported archaic events. Properties of these 11 HML-2 loci are more consistent with insertion before the most recent ancestor with modern humans ∼0.6–0.8 Mya (52) than with introgression. For example, the 2-LTR insertions at 19p12e and Xq21.33 are most prevalent in samples of African ancestry, and LTR divergences indicate their respective insertions to have been ∼1.8–3.3 Mya and ∼0.67–1.3 Mya, consistent with this time frame. Both sites are rare, with sample-wide allele frequencies estimated at 0.0103 and 0.0157 (∼0.026–0.069 in the African sample) in our data (Dataset S3). Of the remaining genotyped loci also in archaic genomes, each was also most represented in African ancestry, with exception of the insertions at 11q12.2 and 5q14.1 (sample-wide allele frequencies estimated at ∼0.046 and 0.026) that appeared most frequently in populations from the Americas or of East Asian ancestries but are also present in African populations, again implying ancient events (50). Given their overall distribution, it is likely these insertions are also older, although our ability to estimate insertion times is limited, given their presence as solo-LTRs.
We confirmed the presence of full-length proviruses at four loci, including the Xq21.33 provirus, which appears to be intact and without obvious defects, which implies the potential for replication competence and is now under further investigation. Given a genomic mutation rate of ∼2.2 × 10−9 changes per site per year (53), an ERV could maintain infectivity over very long periods, and a number of infectious ERVs are known in other species, including mice, cats, and some birds (3). Although such elements are likely to regulated by silencing and downstream host mechanisms, disease states causing prolonged reactivation of HERVs could drive expression of such proviruses or the generation of recombinant infectious chimeras, as has been shown to occur in Ab-deficient mice (54) and as claimed for HML-2–derived transcripts in the blood of HIV-infected individuals (44). Indeed, a “recombinant” HML-2 provirus engineered from just three well-studied defective reference loci is infectious (derived from portions of K109, K115, and K108, respectively, at 6q14.1, 8p23.1a, and 7p22.1) (20), as are the HML-2 consensus genomes (20, 21). We anticipate that sequence-based comparisons and future experimental interrogation of intact proviruses, such as Xq21.33 and others that have yet to be discovered, will give insight into the functionality of similar HML-2 members.
Expression of HML-2 elements has been studied foremost in the context of disease, particularly from tumor-derived tissues (reviewed in 3, 16, 17), but also in otherwise normal human tissues (19). This expression has been shown to exhibit tissue specificity in the form of proviral RNAs from both type 1 and 2 proviruses that have no apparent match to annotated loci (16, 17), implying the presence of transcriptionally active but not yet characterized loci. Although the consequence of such expression is not fully understood, the correlation of HML-2 expression with certain disease states suggests that such variable expression may provide a useful biomarker. Because RNAs corresponding to all previously known polymorphic HML-2 proviruses (and the majority of human-specific elements) have been identified in such assays, additional polymorphic HML-2 copies are likely to be transcribed under certain conditions, justifying the continued characterization of “new” loci and the regulation of their expression. Our analysis of four additional nonreference HML-2 proviruses indicates the presence of discriminatory nucleotide positions within each of these elements (Fig. S6); such sites should aid their assignment in future experimental assays.
Several HML-2 insertions were nearby or within genic regions (Table 1). For example, the 8q24.3c provirus is situated within COMMD5, a gene involved in hypertension-related renal repair whose expression is elevated in the kidneys of hypertension-resistant rat models (55). The 4p16d LTR, in ∼1.5% of all samples (with highest prevalence in African ancestry), was within the SLCA29 (GLUT9) gene; the GLUT9 uric transporter is a direct target of p53 and is implicated in antioxidant functions (56). Also, the 5q14.1 LTR lies within the RASGFR gene associated with regulation of dopamine neuron activity and reward sensitivity in alcohol use (57). Previous mining of cohort-based WGS data has inferred the 5q14.1 LTR in ∼14 to ∼30% of those samples (12, 41), although it was detected in ∼2.7% of all samples here, possibly explained by the global survey in the data used. Genotyping of this site did permit population distribution estimates, in which we found the highest prevalence in European and American ancestry (up to ∼7% of samples), with apparent absence in East Asian individuals (Fig. 2 and Dataset S3). The relationship between a particular LTR and a biological effect requires further investigations, but these observations serve as a reminder that such insertions may be associated with phenotypic effects in some individuals.
Low levels of replication have been suggested based on the presence of unfixed HML-2 loci (13). Coalescent analysis of globally sequenced LTRs from the K106 provirus (the reference insertion at 3q13.2) has produced an age estimation of 0.15 My based on sequence conservation of that site across sampled individuals (11). Other studies suggest replication until at least 0.25 Mya (12) based on modeling estimations of an expected number of loci, given the number of observed unfixed sites and the proportion of individuals predicted to carry those insertions (12). Although we cannot rule out the possibility of ongoing replication, our comparison of 2-LTR sites suggests a most recent time of insertion at least ∼0.67 Mya for those proviruses and we find no evidence of insertions with evidence of more recent formation, noting limitations in properly estimating integration times for recombinant solo-LTRs. The number of rare insertions in our data (15 insertions in <5% and six in <1% of all samples), including the 1p31.1c LTR detected in a single individual and the 2-LTR provirus at 8q24.3c in just a few samples (from the HGDP San and 1KGP Yoruba populations), suggests that additional remaining HML-2 loci are likely to be very rare, specific to groups not yet surveyed, or within low coverage regions of the genome. The rarity of such proviruses, however, is likely to reflect more recently integrated proviruses as well as less time for selective removal of deleterious, pathogenic ones. Continued efforts to analyze additional genome sequences, particularly from previously unstudied populations (particularly of African ancestry), will contribute to the identification of intact and potentially replication-competent proviruses, as is supported by this study.
Our approach shares limitations common to all read-based discovery methods. Given the variability in per-sample coverage, we have likely missed other sites that may be present in one or a few samples or insertions located in otherwise inaccessible regions of the genome. Likewise, other read-based analyses, such as genotyping and derived frequency estimates of each site, must be interpreted with caution, given requirements for read support over each site. Continued improvements in sequencing technologies (longer read lengths) and costs will ameliorate such issues in the future. Such changes will also increase the feasibility of assembly-based approaches, permitting the direct reconstruction of full insertion, ultimately contributing to a more complete picture of all types of genomic variation.
Supplementary Material
Acknowledgments
We thank Ryan Mills for assistance in accessing 1KGP WGS data and Amanda Pendleton, Neeru Bhardwaj, and Farrah Roy for helpful discussions and editorial comments. This work was supported by NIH Research Grant 1DP5OD009154 (to J.M.K.) and Research Grant R37CA089441 from the National Cancer Institute (to J.M.C.). J.M.C. was an American Cancer Society Research Professor with support from the F. M. Kirby Foundation. J.H.W. was a recipient of National Research Service Award F32GM112339 from the NIH.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KU054242–KU054309).
See Commentary on page 4240.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602336113/-/DCSupplemental.
References
- 1.Boeke JD, Stoye JP. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Hughes S, Varmus H, editors. Retroviruses. Cold Spring Harbor Laboratory Press; Plainview, NY: 1997. pp. 343–435. [PubMed] [Google Scholar]
- 2.McPherson JD, et al. International Human Genome Mapping Consortium A physical map of the human genome. Nature. 2001;409(6822):934–941. doi: 10.1038/35057157. [DOI] [PubMed] [Google Scholar]
- 3.Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 4.Barbulescu M, et al. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr Biol. 1999;9(16):861–868. doi: 10.1016/s0960-9822(99)80390-x. [DOI] [PubMed] [Google Scholar]
- 5.Belshaw R, et al. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): Implications for present-day activity. J Virol. 2005;79(19):12507–12514. doi: 10.1128/JVI.79.19.12507-12514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hughes JF, Coffin JM. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: Implications for human and viral evolution. Proc Natl Acad Sci USA. 2004;101(6):1668–1672. doi: 10.1073/pnas.0307885100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medstrand P, Mager DL. Human-specific integrations of the HERV-K endogenous retrovirus family. J Virol. 1998;72(12):9782–9787. doi: 10.1128/jvi.72.12.9782-9787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subramanian RP, Wildschutte JH, Russo C, Coffin JM. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology. 2011;8:90. doi: 10.1186/1742-4690-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner G, et al. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol. 2001;11(19):1531–1535. doi: 10.1016/s0960-9822(01)00455-9. [DOI] [PubMed] [Google Scholar]
- 10.Ross SR. MMTV infectious cycle and the contribution of virus-encoded proteins to transformation of mammary tissue. J Mammary Gland Biol Neoplasia. 2008;13(3):299–307. doi: 10.1007/s10911-008-9090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jha AR, et al. Human endogenous retrovirus K106 (HERV-K106) was infectious after the emergence of anatomically modern humans. PLoS One. 2011;6(5):e20234. doi: 10.1371/journal.pone.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchi E, Kanapin A, Magiorkinis G, Belshaw R. Unfixed endogenous retroviral insertions in the human population. J Virol. 2014;88(17):9529–9537. doi: 10.1128/JVI.00919-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belshaw R, et al. Long-term reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci USA. 2004;101(14):4894–4899. doi: 10.1073/pnas.0307800101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costas J. Evolutionary dynamics of the human endogenous retrovirus family HERV-K inferred from full-length proviral genomes. J Mol Evol. 2001;53(3):237–243. doi: 10.1007/s002390010213. [DOI] [PubMed] [Google Scholar]
- 15.Reus K, et al. Genomic organization of the human endogenous retrovirus HERV-K(HML-2.HOM) (ERVK6) on chromosome 7. Genomics. 2001;72(3):314–320. doi: 10.1006/geno.2000.6488. [DOI] [PubMed] [Google Scholar]
- 16.Hohn O, Hanke K, Bannert N. HERV-K(HML-2), the Best Preserved Family of HERVs: Endogenization, Expression, and Implications in Health and Disease. Front Oncol. 2013;3:246. doi: 10.3389/fonc.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magiorkinis G, Belshaw R, Katzourakis A. ‘There and back again’: Revisiting the pathophysiological roles of human endogenous retroviruses in the post-genomic era. Philos Trans R Soc Lond B Biol Sci. 2013;368(1626):20120504. doi: 10.1098/rstb.2012.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flockerzi A, et al. Expression patterns of transcribed human endogenous retrovirus HERV-K(HML-2) loci in human tissues and the need for a HERV Transcriptome Project. BMC Genomics. 2008;9:354. doi: 10.1186/1471-2164-9-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmitt K, Heyne K, Roemer K, Meese E, Mayer J. HERV-K(HML-2) rec and np9 transcripts not restricted to disease but present in many normal human tissues. Mob DNA. 2015;6:4. doi: 10.1186/s13100-015-0035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dewannieux M, et al. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 2006;16(12):1548–1556. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007;3(1):e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci USA. 2004;101(Suppl 2):14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee WP, Wu J, Marth GT. Toolbox for mobile-element insertion detection on cancer genomes. Cancer Inform. 2014;13(Suppl 4):45–52. doi: 10.4137/CIN.S13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sudmant PH, et al. 1000 Genomes Project Consortium An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526(7571):75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cann HM, et al. A human genome diversity cell line panel. Science. 2002;296(5566):261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 26.Martin AR, et al. Transcriptome sequencing from diverse human populations reveals differentiated regulatory architecture. PLoS Genet. 2014;10(8):e1004549. doi: 10.1371/journal.pgen.1004549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keane TM, Wong K, Adams DJ. RetroSeq: Transposable element discovery from next-generation sequencing data. Bioinformatics. 2013;29(3):389–390. doi: 10.1093/bioinformatics/bts697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110(1-4):462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 30.Smit AFA, Hubley R, Green P. 2013 RepeatMasker Open-4.0. Available at www.repeatmasker.org. Accessed August 5, 2013.
- 31.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9(9):868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wildschutte JH, Baron A, Diroff NM, Kidd JM. Discovery and characterization of Alu repeat sequences via precise local read assembly. Nucleic Acids Res. 2015;43(21):10292–10307. doi: 10.1093/nar/gkv1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wildschutte JH, Ram D, Subramanian R, Stevens VL, Coffin JM. The distribution of insertionally polymorphic endogenous retroviruses in breast cancer patients and cancer-free controls. Retrovirology. 2014;11(1):62. doi: 10.1186/s12977-014-0062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 35.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scally A, Durbin R. Revising the human mutation rate: Implications for understanding human evolution. Nat Rev Genet. 2012;13(10):745–753. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- 37.Stewart C, et al. 1000 Genomes Project A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet. 2011;7(8):e1002236. doi: 10.1371/journal.pgen.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27(6):764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hach F, et al. mrsFAST: A cache-oblivious algorithm for short-read mapping. Nat Methods. 2010;7(8):576–577. doi: 10.1038/nmeth0810-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee E, et al. Cancer Genome Atlas Research Network Landscape of somatic retrotransposition in human cancers. Science. 2012;337(6097):967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agoni L, Golden A, Guha C, Lenz J. Neandertal and Denisovan retroviruses. Curr Biol. 2012;22(11):R437–R438. doi: 10.1016/j.cub.2012.04.049. [DOI] [PubMed] [Google Scholar]
- 43.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453(7191):56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Contreras-Galindo R, et al. Characterization of human endogenous retroviral elements in the blood of HIV-1-infected individuals. J Virol. 2012;86(1):262–276. doi: 10.1128/JVI.00602-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moyes DL, et al. The distribution of the endogenous retroviruses HERV-K113 and HERV-K115 in health and disease. Genomics. 2005;86(3):337–341. doi: 10.1016/j.ygeno.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 46.Hughes JF, Coffin JM. Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nat Genet. 2001;29(4):487–489. doi: 10.1038/ng775. [DOI] [PubMed] [Google Scholar]
- 47.Johnson WE, Coffin JM. Constructing primate phylogenies from ancient retrovirus sequences. Proc Natl Acad Sci USA. 1999;96(18):10254–10260. doi: 10.1073/pnas.96.18.10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hughes JF, Coffin JM. A novel endogenous retrovirus-related element in the human genome resembles a DNA transposon: Evidence for an evolutionary link? Genomics. 2002;80(5):453–455. [PubMed] [Google Scholar]
- 49.Downey RF, et al. Human endogenous retrovirus K and cancer: Innocent bystander or tumorigenic accomplice? Int J Cancer. 2015;137(6):1249–1257. doi: 10.1002/ijc.29003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henn BM, Cavalli-Sforza LL, Feldman MW. The great human expansion. Proc Natl Acad Sci USA. 2012;109(44):17758–17764. doi: 10.1073/pnas.1212380109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee A, et al. Novel Denisovan and Neanderthal retroviruses. J Virol. 2014;88(21):12907–12909. doi: 10.1128/JVI.01825-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reich D, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468(7327):1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar S, Subramanian S. Mutation rates in mammalian genomes. Proc Natl Acad Sci USA. 2002;99(2):803–808. doi: 10.1073/pnas.022629899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young GR, et al. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature. 2012;491(7426):774–778. doi: 10.1038/nature11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsuda H, Hamet P, Tremblay J. Hypertension-related, calcium-regulated gene (HCaRG/COMMD5) and kidney diseases: HCaRG accelerates tubular repair. J Nephrol. 2014;27(4):351–360. doi: 10.1007/s40620-014-0054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Itahana Y, et al. The uric acid transporter SLC2A9 is a direct target gene of the tumor suppressor p53 contributing to antioxidant defense. Oncogene. 2015;34(14):1799–1810. doi: 10.1038/onc.2014.119. [DOI] [PubMed] [Google Scholar]
- 57.Stacey D, et al. IMAGEN Consortium RASGRF2 regulates alcohol-induced reinforcement by influencing mesolimbic dopamine neuron activity and dopamine release. Proc Natl Acad Sci USA. 2012;109(51):21128–21133. doi: 10.1073/pnas.1211844110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parson J. Miropeats: Graphical DNA sequence comparisons. Comput Appl Biosci. 1995;11(6):615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.