Significance
Human blood provides a rich source of information about metabolites that reflects individual differences in health, disease, diet, and lifestyle. The coefficient of variation for human blood metabolites enriched in red blood cells or plasma was quantified after careful preparation. We identified 14 age-related metabolites. Metabolites that decline strikingly in the elderly include antioxidants and compounds involved in high physical activity, including carnosine, UDP-acetyl-glucosamine, ophthalmic acid,1,5-anhydroglucitol, NAD+, and leucine. Metabolites that increase significantly in the elderly include compounds related to declining renal and liver function. Statistical analysis suggests that certain age-related compounds that either increased or decreased in the elderly are correlated. Individual variability in blood metabolites may lead to identify candidates for markers of human aging or relevant diseases.
Keywords: red blood cells, antioxidants, urea cycle, aging markers, CV value
Abstract
Metabolites present in human blood document individual physiological states influenced by genetic, epigenetic, and lifestyle factors. Using high-resolution liquid chromatography-mass spectrometry (LC-MS), we performed nontargeted, quantitative metabolomics analysis in blood of 15 young (29 ± 4 y of age) and 15 elderly (81 ± 7 y of age) individuals. Coefficients of variation (CV = SD/mean) were obtained for 126 blood metabolites of all 30 donors. Fifty-five RBC-enriched metabolites, for which metabolomics studies have been scarce, are highlighted here. We found 14 blood compounds that show remarkable age-related increases or decreases; they include 1,5-anhydroglucitol, dimethyl-guanosine, acetyl-carnosine, carnosine, ophthalmic acid, UDP-acetyl-glucosamine, N-acetyl-arginine, N6-acetyl-lysine, pantothenate, citrulline, leucine, isoleucine, NAD+, and NADP+. Six of them are RBC-enriched, suggesting that RBC metabolomics is highly valuable for human aging research. Age differences are partly explained by a decrease in antioxidant production or increasing inefficiency of urea metabolism among the elderly. Pearson’s coefficients demonstrated that some age-related compounds are correlated, suggesting that aging affects them concomitantly. Although our CV values are mostly consistent with those CVs previously published, we here report previously unidentified CVs of 51 blood compounds. Compounds having moderate to high CV values (0.4–2.5) are often modified. Compounds having low CV values, such as ATP and glutathione, may be related to various diseases because their concentrations are strictly controlled, and changes in them would compromise health. Thus, human blood is a rich source of information about individual metabolic differences.
Human blood metabolites have been well-investigated to determine their abundance and biological significance, and for their potential use as diagnostic markers. For medical diagnosis, noncellular metabolites from plasma or serum are mostly commonly used due to the simplicity in collecting and examining them. Although mature human red blood cells (RBCs) lack nuclei and cellular organelles (1), RBCs use glycolysis for ATP production, maintain redox homeostasis, and osmoregulate (2). Their active metabolism supports cellular homeostasis and ensures lifespans of ∼4 mo (3). Their metabolites may reflect health status or environmental stresses differently than do metabolites of plasma. Because RBCs occupy about half the total blood volume (∼5 L), their metabolite profiles, which have scarcely been investigated, seemed worthy of investigation.
Metabolomics is a branch of chemical biology that profiles metabolites in cells and organisms, using techniques such as liquid chromatography (LC)-mass spectrometry (MS). It usually deals with molecules <1.5 kDa and is an important tool for studying metabolic regulation in combination with other comprehensive analyses, such as proteomics and transcriptomics. Recently, we reported that, among 133 compounds identified in human blood, 101 are also found in the fission yeast, Schizosaccharomyces pombe (4), implying that many metabolites might be evolutionarily conserved. Quantitative measurements of an array of compounds among individuals offer profound insights into health or disease conditions and the effects of nutrition, drugs, and stress. Moreover, comprehensive information about individual variation in metabolites could impact the future of medical science (5–11).
Although blood consists of noncellular (plasma or serum) and cellular components, most human blood metabolomics studies have focused on plasma or serum, for which large biobanks (curated collections of samples of plasma, urine, etc.) are now available (12–16). These studies are useful to understand disease mechanisms and to identify diagnostic markers for diseases, such as diabetes (17). Some genome-wide studies have also used metabolomics [reviewed in Kastenmüller et al. (18)]. In contrast, few comprehensive metabolomics reports exist regarding RBCs [e.g., Nishino et al. (19)] although RBCs constitute nearly half the blood volume. This situation is partly due to technical difficulties in stabilizing labile cellular metabolites (20).
Here, we report blood metabolites of 30 individuals in a study having three distinct facets. First, we collected samples from RBCs, plasma, and whole blood for metabolomics analysis. Combining the present quantitative data with previous analysis of RBCs and white blood cells (WBCs) carefully separated from RBCs (4), we now have ample knowledge of metabolites enriched in RBCs. RBC-enriched metabolites may reflect health status or environmental stresses differently than do metabolites of plasma.
Second, to quantify individual variation, we used a simple parameter, designated the coefficient of variation (CV), for each blood compound. The CV is the ratio of the SD of metabolite abundance (peak areas from LC-MS) divided by the mean. For stable and relatively invariant metabolites, SDs and CVs are low or negligible whereas CVs of variable metabolites may prove useful in the evaluation of metabolite variation among individuals. RBC and plasma metabolites from 30 volunteers were analyzed using LC-MS (4, 21). Hepes and piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) were spiked into all blood samples as internal standards. CVs of any compounds significantly larger than those CVs of Hepes and PIPES were candidates to be analyzed for individual metabolite variation.
Third, comparisons of blood metabolomes between young and elderly volunteers were performed with emphasis on RBC metabolites that have seldom been considered as targets of age analysis. We were able to identify a total of 14 metabolites statistically relevant to aging. Three of them were previously reported, but 11 others hitherto have not been. We discuss our findings in regard to human aging.
Results
Many Metabolite Levels Are Constant on a Daily Basis.
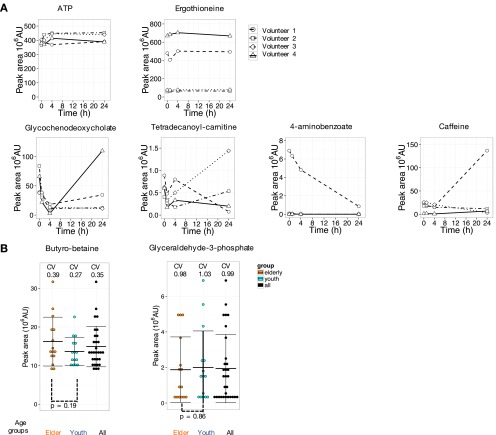
We first investigated diel variation of blood metabolites in four volunteers. Samples were taken after overnight fast without breakfast at 0900, 1000, 1300, and before lunch on the first day. Volunteers had lunches and dinners as usual on that day. On the second day after overnight fast, the blood was sampled again at 0900. During these short periods, the great majority of metabolites hardly fluctuated (117 from 126 metabolites varied less than 2.5-fold on average in four volunteers) (Fig. S1A). ATP and ergothioneine hardly varied although individual ergothioneine levels were distinct. In contrast, four variable compounds fluctuated considerably over 24 h. Metabolites, such as glycochenodeoxycholate, tetradecanoyl-carnitine, 4-aminobenzoate, and caffeine, vary widely, depending upon daily consumption of food, drink, supplements, and medications (22–24). Our results are consistent with those data previously reported. These daily variable compounds were found in both plasma and RBCs (4).
Fig. S1.
Many human blood metabolites exhibit diel constancy. For the great majority of 126 compounds, diel fluctuations in abundance among four volunteers were negligible. (A) Peak levels of ATP and ergothioneine, like most compounds, hardly changed. The abundance of ergothioneine varied from person to person, however. Furthermore, about 10 compounds, including glycochenodeoxycholate, tetradecanoyl-carnitine, 4-aminobenzoate, and caffeine, showed exceptional 24-h variation. (B) Raw abundance data for two example compounds [butyro-betaine and glyceraldehyde-3-phosphate (G-3-P)], both enriched in RBCs, determined from 30 individual metabolomes, are shown as dotplots (each dot represents a single individual). Coefficients of variation were 0.35 and 0.99, respectively, for butyro-betaine and G-3-P. Ratios of maximum to minimum abundance are 3.7 and 29, respectively. Peak areas of metabolites were divided into 10 bins in each group. Error bars represent means ± SD.
Determination of Individual CVs for Each Metabolite.
We performed metabolomic analyses of blood samples donated by 30 volunteers. Data on compound enrichment in RBCs were consistent with our previous report (4). The separation of RBCs from WBCs by Ficoll gradient centrifugation confirmed that metabolites and their levels were similar in RBCs and WBCs (4). Because WBCs make up only a small portion (<1%) of blood volume in healthy individuals, our current results should not be affected by WBC contamination.
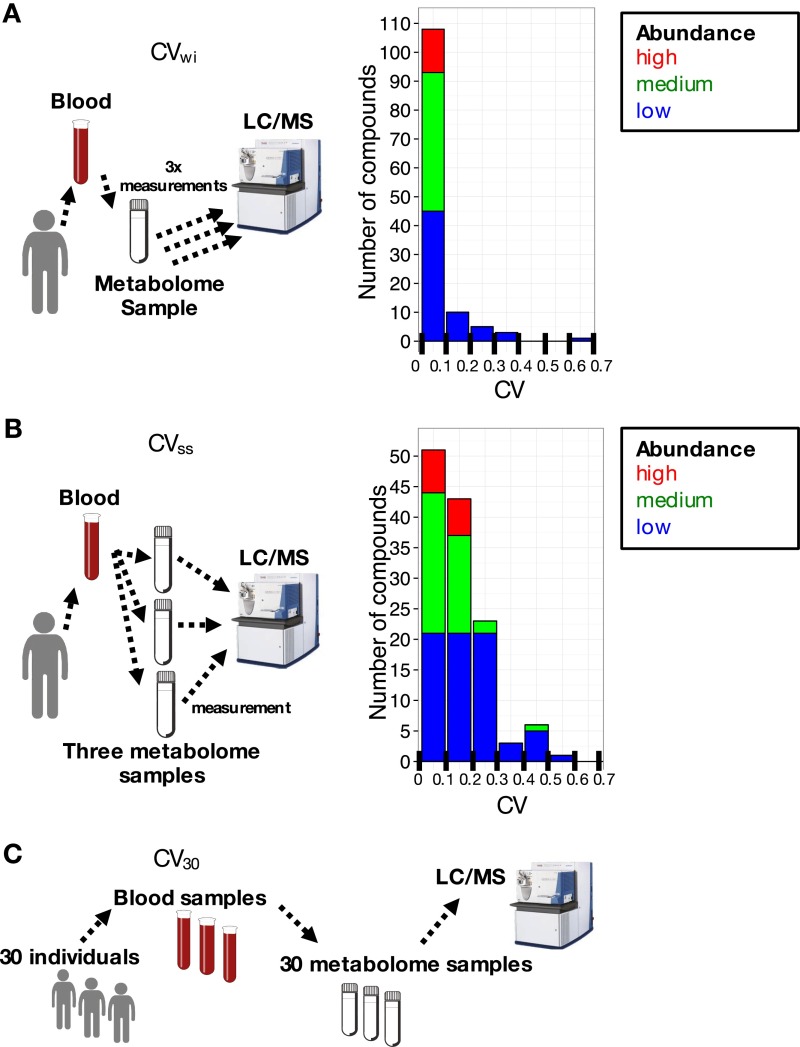
Procedures for LC-MS analysis and for obtaining and validating CVs are detailed in SI Materials and Methods. Methods used to determine CVs are briefly described below. First, we tested the effect of sample handling on within-sample variation. To accomplish this task, the same blood sample was injected three times into the LC-MS. The within-sample CVs (designated as CVwi) (Fig. S2A) were less than 0.1 in most cases. These exceptions seem to be labile during LC-MS.
Fig. S2.
Experimental variability for metabolite measurements is very small. CV distributions for 126 compounds are shown for (A) CVwi (three injections of the same blood sample preparation) and (B) CVss (three independently prepared samples from the same blood). (C) Coefficients of variation (CV30) for each compound from all 30 blood samples. Most compounds showed negligible CVwi whereas CVsss were more variable and considerably higher for certain metabolites.
Second, we examined sample-to-sample variation caused by sample preparation. For this purpose, three samples were independently prepared from the same blood sample, and CVs of compounds were determined (CVss) (Fig. S2B). CVsss of Hepes and PIPES (internal standards) in blood samples were very small (0.04∼0.08) because they are inert, nonreactive compounds. The great majority of blood compound CVsss were less than 0.3 (Table S1). CVsss of 10 compounds were exceptional, exceeding 0.3. They either may be affected by sample preparation techniques or may react with other blood metabolites during sample preparation.
Table S1.
List of 126 identified blood metabolites
| Category/compound | RBC-enriched | Reported concentration in plasma or blood, μM | Abundance | CVss | CV30 | CV reported | P value between age groups |
| Nucleotides (7/11) | |||||||
| ADP | X | 48 ± 13 | H-M | 0.05 | 0.27 | X | 0.64 |
| AMP | X | 6.2 ± 3.1 | H-M | 0.07 | 0.33 | X | 0.71 |
| ATP | X | 433 ± 73 | H-M | 0.07 | 0.17 | X | 0.31 |
| CTP | L | 0.1 | 0.33 | 0.078 | |||
| GDP | X | 15 ± 2 | L | 0.09 | 0.38 | 0.075 | |
| GTP | X | 25 ± 6 | H-M | 0.05 | 0.32 | X | 0.58 |
| GMP | X | 0 | L | 0.24 | 0.57 | 0.95 | |
| IMP | X | 14–100 | L | 0.06 | 0.38 | 0.55 | |
| UDP | X | 0 | L | 0.11 | 0.31 | 0.79 | |
| UMP | X | 0 | L | 0.58 | 0.88 | 0.86 | |
| UTP | L | 0.25 | 0.44 | 0.46 | |||
| Nucleosides, nucleobases and derivatives (1/12) | |||||||
| 1-Methyl-adenosine | 0.06 ± 0.006 | H-M | 0.45 | 0.29 | X | 0.63 | |
| 1-Methyl-guanosine** | 0.046 ± 0.019 | L | 0.23 | 0.25 | X | 0.25 | |
| Adenine | X | 0.3 ± 0.28 | L | 0.2 | 0.53 | X | 0.71 |
| Adenosine | 0.5 ± 0.1 | L | 0.38 | 0.49 | X | 0.052 | |
| Caffeine | 0–35 | H-M | 0.29 | 0.92 | X | 0.43 | |
| Cytidine | 0.25 ± 0.19 | L | 0.08 | 0.33 | X | 0.079 | |
| Dimethyl-guanosine | 0.029 ± 0.005 | L | 0.26 | 0.46 | X | 0.0081 | |
| Dimethyl-xanthine | 0–8 | H-M | 0.25 | 0.61 | 0.078 | ||
| Hypoxanthine** | 0.38 ± 0.18 | H-M | 0.32 | 0.35 | X | 0.98 | |
| Urate | X | 336 ± 94 | H-M | 0.07 | 0.28 | X | 0.15 |
| Uridine* | 3.12 ± 1.31 | H-M | 0.14 | 0.34 | X | 0.84 | |
| Xanthine* | 1.27 ± 0.78 | L | 0.03 | 0.56 | X | 0.87 | |
| Vitamins, coenzymes (2/5) | |||||||
| 4-Aminobenzoate | <1 ng/mL | L | 0.03 | 2.18 | 0.32 | ||
| NAD+ | X | 23.3 ± 6.9 | H-M | 0.08 | 0.3 | X | 0.046 |
| NADP+ | X | 19.6 ± 6.6 | H-M | 0.11 | 0.36 | X | 0.022 |
| Nicotinamide | X | 0.45 | H-M | 0.47 | 0.56 | 0.38 | |
| Pantothenate | X | 2.2 ± 1.02 | H-M | 0.2 | 0.82 | X | 0.022 |
| Nucleotide-sugar derivatives (4/4) | |||||||
| GDP-glucose | X | L | 0.09 | 0.53 | 0.26 | ||
| UDP-acetyl-glucosamine | X | L | 0.14 | 0.64 | 0.0073 | ||
| UDP-glucose | X | H-M | 0.18 | 0.24 | 0.088 | ||
| UDP-glucuronate | X | L | 0.24 | 0.63 | 0.12 | ||
| Sugar phosphates (8/9) | |||||||
| 6-Phosphogluconate | X | L | 0.25 | 0.3 | 0.49 | ||
| Diphospho-glycerate | X | 4,500 | H-M | 0.17 | 0.24 | 0.77 | |
| Fructose-6-phosphate | X | 16 | L | 0.21 | 0.24 | 0.61 | |
| Glucose-6-phosphate | X | 38 | H-M | 0.19 | 0.29 | 0.94 | |
| Glyceraldehyde-3-phosphate** | X | 6.7 ± 1.0 | L | 0.49 | 0.99 | 0.86 | |
| Glycerol-2-phosphate** | X | L | 0.16 | 0.31 | 0.32 | ||
| Pentose-phosphate | X | 13.2 ± 4.8 | L | 0.28 | 0.34 | 0.049 | |
| Phosphoglycerate | X | 58 ± 14 | H-M | 0.24 | 0.29 | 0.12 | |
| Sedoheptulose-7-phosphate | X | 0.89 ± 0.41 | L | 0.28 | 0.52 | X | 0.82 |
| Sugars and derivatives (3/6) | |||||||
| 1,5-Anhydroglucitol | 120 ± 39 | H-M | 0.08 | 0.46 | X | 0.0004 | |
| Gluconate | X | <5 | H-M | 0.16 | 0.33 | 0.32 | |
| Glucosamine | 0.23–0.68 | H-M | 0.19 | 0.47 | 0.21 | ||
| Glucose | 4700–6100 | H-M | 0.15 | 0.4 | X | 0.54 | |
| myo-Inositol* | 21–49 | L | 0.27 | 0.24 | X | 0.31 | |
| N-acetyl-d-glucosamine | X | H-M | 0.09 | 0.26 | 0.82 | ||
| Organic acids (6/10) | |||||||
| 2-Oxoglutarate | 8.6 ± 2.6 | L | 0.19 | 0.54 | X | 0.63 | |
| Chenodeoxycholate | 0.85 ± 0.88 | H-M | 0.05 | 1.33 | X | 0.77 | |
| Glycochenodeoxycholate | 0.06 ± 0.01 | H-M | 0.04 | 1.2 | X | 0.93 | |
| cis-Aconitate | L | 0.14 | 0.35 | 0.11 | |||
| Citramalate | X | L | 0.15 | 0.36 | 0.82 | ||
| Citrate | 79 ± 27 | H-M | 0.08 | 0.31 | X | 0.59 | |
| Glutarate | 0.0–1.8 | L | 0.16 | 0.29 | 0.65 | ||
| Glycerate | 0–24 | L | 0.02 | 0.43 | 0.39 | ||
| Malate | X | 0–21 | H-M | 0.17 | 0.2 | 0.76 | |
| Succinate | X | 8.8 ± 2.7 | L | 0.25 | 0.6 | 0.49 | |
| Standard amino acids (0/17) | |||||||
| Arginine | 80 ± 20 | H-M | 0.04 | 0.29 | X | 0.64 | |
| Asparagine | 41 ± 10 | L | 0.17 | 0.23 | X | 0.11 | |
| Aspartate | X | 3 ± 1, 400 ± 120 | H-M | 0.27 | 0.5 | X | 0.62 |
| Glutamate** | X | 25 ± 15, 1,110 ± 360 | H-M | 0.44 | 0.28 | X | 0.29 |
| Glutamine | 586 ± 84 | H-M | 0.16 | 0.2 | X | 0.14 | |
| Histidine | 82 ± 10 | H-M | 0.12 | 0.19 | X | 0.06 | |
| Isoleucine | 62 ± 14 | H-M | 0.18 | 0.32 | X | 0.012 | |
| Leucine | 123 ± 25 | H-M | 0.13 | 0.31 | X | 0.002 | |
| Lysine | 188 ± 32 | H-M | 0.24 | 0.39 | X | 0.87 | |
| Methionine | 25 ± 4 | H-M | 0.19 | 0.28 | X | 0.17 | |
| Phenylalanine | 57 ± 9 | H-M | 0.01 | 0.17 | X | 0.041 | |
| Proline | 168 ± 60 | L | 0.19 | 0.42 | X | 0.47 | |
| Serine | 114 ± 19 | L | 0.27 | 0.33 | X | 0.2 | |
| Threonine | 140 ± 33 | L | 0.17 | 0.42 | X | 0.63 | |
| Tryptophan | 44 ± 7 | H-M | 0.08 | 0.24 | X | 0.22 | |
| Tyrosine | 59 ± 12 | H-M | 0.06 | 0.27 | X | 0.051 | |
| Valine | 233 ± 43 | L | 0.2 | 0.48 | X | 0.95 | |
| Methylated amino acids (8/13) | |||||||
| Betaine | 47 ± 18 | H-M | 0.06 | 0.51 | X | 0.26 | |
| Butyro-betaine | X | 0.76 | H-M | 0.12 | 0.35 | 0.19 | |
| Dimethyl-arginine | 0.66 ± 0.19 | H-M | 0.05 | 0.31 | X | 0.079 | |
| Dimethyl-lysine | L | 0.04 | 0.44 | 0.078 | |||
| Dimethyl-proline (stachydrine) | X | 7 ± 10.8 | H-M | 0.03 | 0.79 | X | 0.68 |
| Methyl-histidine | 16.5 ± 10.1 | H-M | 0.04 | 0.3 | X | 0.45 | |
| Methyl-lysine | 7 ± 6 | H-M | 0.03 | 0.73 | 0.66 | ||
| S-methyl-ergothioneine | X | H-M | 0.05 | 0.63 | 0.36 | ||
| Trimethyl-histidine (hercynine) | X | L | 0.17 | 0.57 | 0.21 | ||
| Trimethyl-lysine | X | 0.56 ± 0.17 | H-M | 0.09 | 0.38 | X | 0.38 |
| Trimethyl-phenylalanine | X | L | 0.11 | 1.18 | 0.9 | ||
| Trimethyl-tryptophan (hypaphorine) | X | H-M | 0.03 | 1.67 | 0.96 | ||
| Trimethyl-tyrosine** | X | L | 0.25 | 2.5 | 0.21 | ||
| Acetylated amino acids (5/7) | |||||||
| N-acetyl-(iso)leucine | L | 0.36 | 0.63 | 0.056 | |||
| N-acetyl-arginine | 1.25 ± 0.28 | L | 0.13 | 0.62 | X | 0.0004 | |
| N-acetyl-aspartate | <0.35 | L | 0.11 | 0.58 | 0.18 | ||
| N-acetyl-glutamate | X | L | 0.07 | 0.57 | 0.16 | ||
| N-acetyl-ornithine* | 1.1 ± 0.4 | L | 0.05 | 1.17 | X | 0.98 | |
| N2-acetyl-lysine | L | 0.07 | 0.5 | 0.033 | |||
| N6-acetyl-lysine* | L | 0.13 | 0.43 | 0.012 | |||
| Other amino acid derivatives (4/16) | |||||||
| 4-Guanidinobutanoate* | <0.013–0.055 | L | 0.11 | 2.05 | 0.19 | ||
| Acetyl-carnosine | L | 0.09 | 1.07 | 0.001 | |||
| Arginino-succinate** | L | 0.27 | 0.4 | 0.61 | |||
| Carnosine* | X | 6.5 ± 2.8 | L | 0.19 | 0.79 | X | 0.003 |
| Citrulline | 40 ± 10 | H-M | 0.06 | 0.3 | X | 0.001 | |
| Creatine | X | 30.1 ± 12.3 | H-M | 0.12 | 0.29 | X | 0.58 |
| Creatinine | 82.6 ± 26.2 | H-M | 0.1 | 0.35 | X | 0.23 | |
| Hippurate | 4.28 ± 2.61 | H-M | 0.42 | 1.01 | X | 0.28 | |
| Indoxyl-sulfate | 2.5 ± 1.4 | H-M | 0.15 | 0.59 | X | 0.071 | |
| Kynurenine | 1.35 ± 0.26 | L | 0.06 | 0.48 | X | 0.91 | |
| Ornithine | 55 ± 16 | L | 0.26 | 0.48 | X | 0.32 | |
| Phosphocreatine | X | L | 0.07 | 0.48 | 0.65 | ||
| Quinolinic acid | 0.47 ± 0.22 | L | 0.3 | 1.18 | X | 0.23 | |
| S-adenosyl-homocysteine* | X | 0.46 ± 0.02 | L | 0.08 | 0.83 | X | 0.044 |
| S-adenosyl-methionine** | X | 0.68 ± 0.03 | L | 0.15 | 0.88 | X | 0.085 |
| Taurine | 55 ± 13 | H-M | 0.1 | 0.37 | X | 0.79 | |
| Carnitines (0/10) | |||||||
| Acetyl-carnitine | X | 10.2 ± 2.2 | H-M | 0.05 | 0.41 | X | 0.78 |
| Butyryl-carnitine | 0.267 ± 0.077 | H-M | 0.02 | 0.37 | X | 0.19 | |
| Carnitine | 30 ± 11 | H-M | 0.07 | 0.2 | X | 0.41 | |
| Decanoyl-carnitine | 0.141 ± 0.053 | H-M | 0.12 | 1.11 | X | 0.47 | |
| Dodecanoyl-carnitine | 0.052 ± 0.024 | H-M | 0.14 | 1.11 | X | 0.49 | |
| Hexanoyl-carnitine | 0.080 ± 0.035 | H-M | 0.04 | 0.76 | X | 0.8 | |
| Isovaleryl-carnitine | 0.138 ± 0.059 | H-M | 0.08 | 1.98 | X | 0.25 | |
| Octanoyl-carnitine | 0.121 ± 0.053 | H-M | 0.25 | 1.02 | X | 0.61 | |
| Propionyl-carnitine | X | 0.400 ± 0.124 | H-M | 0.02 | 0.41 | X | 0.68 |
| Tetradecanoyl-carnitine** | X | 0.024 ± 0.006 | L | 0.45 | 0.47 | X | 0.15 |
| Choline derivatives (2/3) | |||||||
| CDP-choline | <3 μM | H-M | 0.07 | 0.41 | 0.088 | ||
| CDP-ethanolamine* | <3 μM | L | 0.18 | 0.43 | 0.077 | ||
| Glycerophosphocholine | 32 ± 3 | H-M | 0.08 | 0.47 | X | 0.66 | |
| Antioxidant (1/3) | |||||||
| Ergothioneine | X | 56 ± 47 | H-M | 0.12 | 0.63 | X | 0.2 |
| Glutathione disulfide (GSSG) | X | 1.69 ± 0.38, 3,210 ± 1,500 | H-M | 0.19 | 0.18 | X | 0.21 |
| Ophthalmic acid* | X | 0.01–0.03, 11.8–16.4 | H-M | 0.12 | 0.43 | 0.0087 | |
| Standards | |||||||
| Hepes [M+H]+ | H-M | 0.06 | 0.29 | 0.761 | |||
| Hepes [M-H]− | H-M | 0.08 | 0.21 | 0.02 | |||
| PIPES [M+H]+ | H-M | 0.04 | 0.14 | 0.061 | |||
| PIPES [M-H]− | H-M | 0.08 | 0.23 | 0.113 |
The list of 126 compounds and two standards (Hepes, PIPES) detected by LC-MS are shown. Compounds with a single asterisk displayed within-sample variation (CVwi) equal to 0.1–0.2 whereas no asterisk indicates a CVwi less than 0.1. Those compounds with two asterisks had CVwi greater than 0.2. Compounds specifically enriched in RBCs, as opposed to plasma, are indicated (X). Compound concentrations previously reported in the literature (X) are shown (references in Dataset S1). Compound abundance (peak area) is indicated (H-M, high-medium; L, low). Coefficients of variation among three preparations from the same blood sample are denominated (CVss). Ten compounds had CVss values of >0.3, and 23 compounds had CVss values of >0.2 (highlighted in bold). Coefficients of variation for each compound among 30 subjects (CV30) are summarized in Fig. 1 and correspond roughly to other values in the literature. Compounds for which CVs have been reported previously are indicated (X). CV30 values of >0.4 are highlighted in bold. Levels of compounds between young and elderly subjects were considered to differ significantly at P < 0.05. Raw data are deposited in the MetaboLights database (see Data Availability).
CVs for the entire experimental population of 30 persons were determined for each blood compound (CV30) (Fig. S2C and Table S1). CV30s of blood metabolites from all 30 healthy volunteers (Table S2) were arranged into six different value ranges, with subcategories for compounds enriched in RBCs or present in whole blood (Fig. 1A). Many RBC-enriched compounds, such as ATP, glutathione, and sugar-phosphate, are virtually absent in plasma, but many plasma compounds are also present in RBCs (4).
Table S2.
Human subject characteristics
| Parameters | Youth (n = 15) | Elderly (n = 15) | All (n = 30) |
| Age (median, IQR) | 28.2 (26.1, 31.2) | 80.6 (76.0, 84.3) | 53.5 (28.2, 80.6) |
| Gender (male; female) | 10; 5 | 4; 11 | 14; 16 |
| Hb (median, IQR), g/dL | 15.3 (14.2, 16.0) | 13.5 (12.8, 14.1) | 14.1 (13.3, 15.6) |
| Hemoglobin A1c (median, IQR), % | 4.7 (4.5, 4.9) | 5.2 (4.9, 5.4) | 4.9 (4.7, 5.2) |
| WBC (median, IQR), × 109/L | 5.8 (5.1, 6.3) | 4.9 (4.2, 6.4) | 5.5 (4.5, 6.4) |
| RBC (median, IQR), × 1012/L | 5.1 (4.8, 5.2) | 4.2 (4.0, 4.6) | 4.7 (4.2, 5.2) |
| MCV (median, IQR), fL | 92.0 (89.5, 94.5) | 97.8 (92.1, 102.4) | 94.3 (90.5, 97.3) |
| Platelets (median, IQR), × 1011/L | 20.8 (20.0, 27.0) | 18.3 (15.1, 23.5) | 20.6 (16.4, 26.2) |
| HT hematocrite (median, IQR), % | 45.9 (44.0, 47.4) | 41.9 (41.1, 44.5) | 44.0 (41.7, 46.7) |
| Glucose (median, IQR), mg/dL | 88.5 (86.3, 92.8) | 96.0 (91.5, 100.3) | 92.5 (87.0, 95.5) |
| Creatinine (median, IQR), mg/dL | 0.8 (0.7, 0.8) | 0.7 (0.5, 0.8) | 0.7 (0.6, 0.8) |
HT, hematocrit; IQR, interquartile range; MCV, mean corpuscular volume.
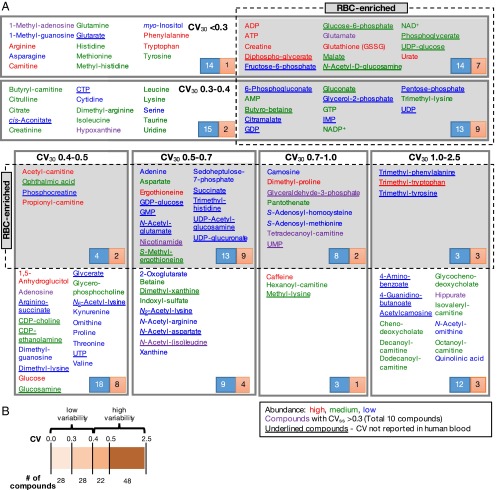
Fig. 1.
Summary of CV profiles for 126 human blood metabolites. (A) The 126 blood compounds with coefficients of variation (CV30s) in six different ranges. (Upper) Values of <0.3 and 0.3∼0.4. (Lower) Values of 0.4∼0.5, 0.5∼0.7, 0.7∼1.0, and 1.0∼2.5). The lowest CV30 (<0.3) group contains 28 compounds. RBC-enriched compounds are highlighted in gray. The abundance of compounds is indicated by their peak areas: red, compounds with high peak areas [>108 AU (arbitrary unit)]; green, medium peak areas (108 ∼107 AU); blue, with low peak areas (<107 AU). Compounds for which CVs have not previously been reported in the literature are underlined. The number in the blue box represents all compounds listed in one CV range whereas the number in the red box represents compounds for which CVs reported here are previously unidentified. (B) Overview of compound numbers in low and high variability groups.
Twenty-eight compounds having CV30s less than 0.30 constituted the least variable subset of 126 blood metabolites (Fig. 1B). An additional 28 compounds had CV30 values from 0.3 to 0.4 and belonged to the second least variable group. Butyrobetaine, a precursor of carnitine, was enriched in RBCs and belongs to this group (Fig. S1B). The remaining 70 compounds showed CV30 values from 0.4 to 2.5. We consider these compounds to be variable. Twenty-two compounds having CV30s from 0.4 to 0.5 were moderately variable. Glucose, 1,5-anhydroglucitol, CDP-choline, and glucosamine belong to this group. Creatinine, used for renal tests, belongs to the second group (CV30 = 0.3–0.4). The 48 compounds with CV30 > 0.5 are considered highly variable. They are often methylated or acetylated, or modified with bulky groups such as nucleotides or fatty acids.
Previously Unreported CVs for 51 Human Metabolites.
The CV30 values of compounds categorized above are, in many cases, well supported by evidence from the literature. CVs of 46 compounds, mostly standard amino acids and their derivatives, analyzed by LC-MS, GC-MS, and NMR have been previously reported (12, 15, 25) (Dataset S1). Of these 46 compounds, 36 had CVs within ±0.3 of our results (CV30). In the literature, we also found CVs for 71 of our 126 compounds (22, 26–69) (Dataset S1). In those reports, 72% of the CVs (51/71) were similar (±0.3) to ours. Overall, our CV30s for 75/126 compounds (60%) (Table S1) were reasonably consistent with the literature. CVs for the remaining 51 compounds were previously unidentified, so far as we know. Many of these 51 compounds (underlined in Fig. 1A and also listed in Table S3) are RBC-enriched.
Table S3.
List of 51 identified metabolite CVs not reported
| Category/compound | RBC enriched |
| Nucleotides (7) | |
| CTP | |
| GDP | X |
| GMP | X |
| IMP | X |
| UDP | X |
| UMP | X |
| UTP | |
| Nucleosides, nucleobases and derivatives (1) | |
| Dimethyl-xanthine | |
| Vitamins, Coenzymes (2) | |
| 4-Aminobenzoate | |
| Nicotinamide | X |
| Nucleotide-sugar derivatives (4) | |
| GDP-glucose | X |
| UDP-acetyl-glucosamine | X |
| UDP-glucose | X |
| UDP-glucuronate | X |
| Sugar phosphates (8) | |
| 6-Phosphogluconate | X |
| Diphospho-glycerate | X |
| Fructose-6-phosphate | X |
| Glucose-6-phosphate | X |
| Glyceraldehyde-3-phosphate | X |
| Glycerol-2-phosphate | X |
| Pentose-phosphate | X |
| Phosphoglycerate | X |
| Sugars and derivatives (3) | |
| Gluconate | X |
| Glucosamine | |
| N-acetyl-d-glucosamine | X |
| Organic acids (6) | |
| cis-Aconitate | |
| Citramalate | X |
| Glutarate | |
| Glycerate | |
| Malate | X |
| Succinate | X |
| Methylated amino acids (8) | |
| Butyro-betaine | X |
| Dimethyl-lysine | |
| Methyl-lysine | |
| S-methyl-ergothioneine | X |
| Trimethyl-histidine (hercynine) | X |
| Trimethyl-phenylalanine | X |
| Trimethyl-tryptophan (hypaphorine) | X |
| Trimethyl-tyrosine | X |
| Acetylated amino acids (5) | |
| N-acetyl-(iso)leucine | |
| N-acetyl-aspartate | |
| N-acetyl-glutamate | X |
| N2-acetyl-lysine | |
| N6-acetyl-lysine | |
| Other amino acid derivatives (4) | |
| 4-Guanidinobutanoate | |
| Acetyl-carnosine | |
| Arginino-succinate | |
| Phosphocreatine | X |
| Choline derivatives (2) | |
| CDP-choline | |
| CDP-ethanolamine | |
| Antioxidant (1) | |
| Ophthalmic acid | X |
Compounds enriched in RBCs, as opposed to plasma, are indicated (X).
We classified 126 compounds into 14 categories based on their molecular structures and functions (Table S1). CVs of 17 detectable standard amino acids and all 10 carnitines have been previously reported. Among 12 nucleosides, nucleobases, and their derivatives, only the CV for dimethyl-xanthine was previously unidentified. In contrast, all four nucleotide-sugar derivatives and most (8/9) sugar phosphate derivatives were likewise previously unidentified. Their novelty reflects the fact that these compounds are enriched in RBCs. For other categories, some CVs were new: methylated amino acids (8/13), nucleotides (7/11), vitamins and coenzymes (2/5), sugars and derivatives (3/6), organic acids (6/10), acetylated amino acids (5/7), other amino acid derivatives (4/16), choline derivatives (2/3), and antioxidants (1/3). Many of these metabolites are also RBC-enriched. It is curious that methylated amino acids are accumulated in RBCs.
Ergothioneine-Related, Glycolytic, and Methylated Compounds Are Correlated.
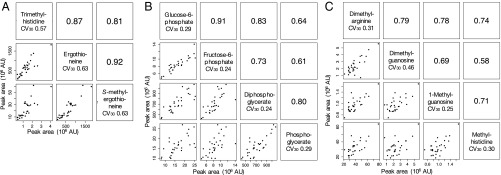
It is interesting that levels of some functionally related blood metabolites were correlated. We first examined whether correlations exist between trimethyl-histidine, ergothioneine, and S-methyl-ergothioneine because they are structurally related and the former two compounds are linked in a biochemical pathway. Abundance of these compounds was very strongly, positively correlated (r2 = 0.81∼0.92) (Fig. 2A).
Fig. 2.
Clusters of human blood metabolites, defined by structure or function, show similar CVs. Blood data from all 30 volunteers revealed several groups of compounds with Pearson correlation coefficients of >0.7. Among these clusters were compounds related to ergothioneine (A), glycolytic pathway metabolites (B), and methylated compounds (C). Pearson correlation coefficients between pairs of compounds are shown in the upper right corners of the panels. In the lower left corners, actual compound levels are plotted for each pair.
Second, potential correlations among RBC-enriched glucose-6-phosphate (G-6-P), fructose-6-phosphate (F-6-P), diphospho-glycerate (DG), and phosphoglycerate (PG) were examined (Fig. 2B). Very strong correlations were found between G-6-P and F-6-P, between DG and PG, between F-6-P and DG, and between G-6-P and DG. These RBC compounds are components of the glycolytic pathway.
Third, correlations among methylated compounds dimethyl-arginine (DA), dimethyl-guanosine (DGU), 1-methyl-guanosine (1MG), and methyl-histidine (MH) were also evaluated. DA abundance was strongly and positively correlated with that of DGU, 1MG, and MH (Fig. 2C). In addition, 1MG was also positively correlated to DGU and MH. These results suggest that the levels of some methylated compounds are linked to the same anabolic or catabolic pathways. Consistently, all these compounds are abundant in both RBCs and plasma. Metabolite variations among individuals were thus coordinated in terms of pathways such as for ergothioneine, glycolysis, and methylation.
Metabolites with Low CVs May Have Vital Functions.
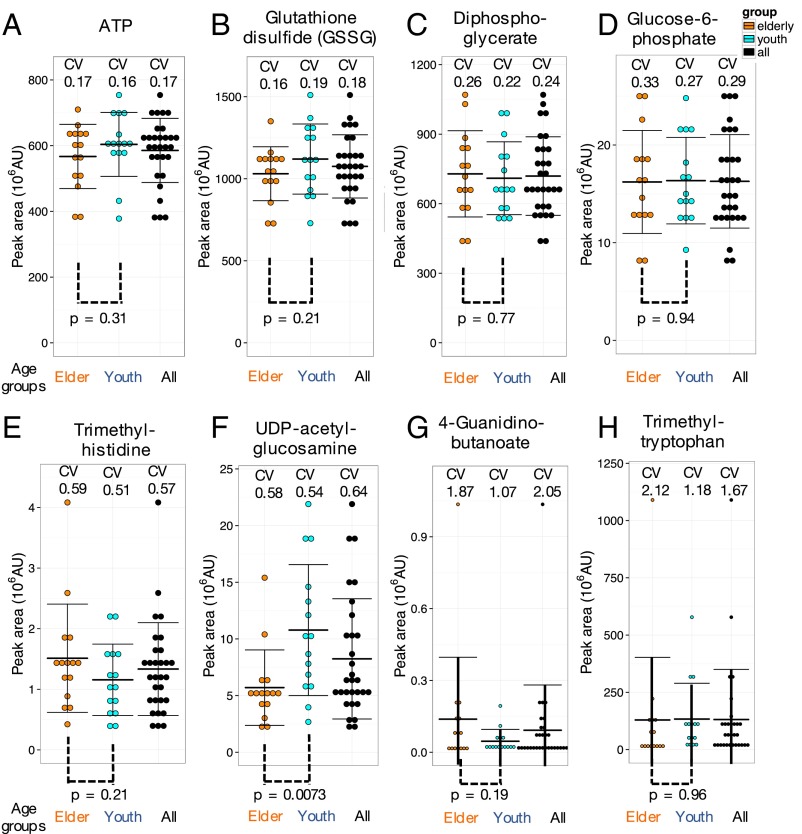
Among the 51 previously unidentified CV compounds, 19 showed low CV30 of <0.4; of these compounds, 16 were enriched in RBCs (Fig. 1A and Table S1). They include sugar phosphates, sugar-nucleotide derivatives, sugars and derivatives, and organic acids involved in ATP production. Compounds with low CVs likely support fundamental RBC functions. CVs of ATP (CV30 = 0.17) and glutathione disulfide (CV30 = 0.18) were low, and no significant difference was found between elderly and young subjects (Fig. 3 A and B). ATP and glutathione are vital as an energy source and an antioxidant, respectively, so their concentrations in RBCs may be tightly regulated, with little age-specific variation. A similar situation was seen for two sugar phosphates, diphosphoglycerate (CV30 = 0.24) and glucose-6-phosphate (CV30 = 0.29) (Fig. 3 C and D). It is likely that levels of these key metabolic compounds with small CVs, (ATP, NAD+, standard amino acids, and nucleotides) may be tightly regulated because they are essential to physiological homeostasis. In other words, small CV compounds might be good candidates for health check indices, provided that measurements are accurate.
Fig. 3.
Essential metabolites are almost invariant whereas modified metabolites (e.g., methylated amino acids) vary widely. Distributions of ATP (A), glutathione disulfide (GSSG) (B), diphosphoglycerate (C), glucose-6-phosphate (D), trimethyl-histidine (E), UDP-acetyl-glucosamine (F), 4-guanidinobutanoate (G), and trimethyl-tryptophan (H) in blood of 30 individuals. Black, orange, and azure dots represent all, elderly, and young subjects, respectively. Peak areas of metabolites were divided into 10 bins in each group. Error bars represent means ± SD.
Glyceraldehyde-3-phosphate (G-3-P), an essential glycolytic metabolite, may be an exception. It had a high CV30 (Fig. S1B). Levels of this compound varied considerably from individual to individual. It is an unstable compound (CVss, 0.49), however, so the high CV30 (0.99) has to be taken cautiously. The enzyme glyceraldehyde-3-phosphate dehydrogenase is known to be important in energy metabolism of cancer cells (70).
Unreported Compounds with High CVs Are Often Modified, Implicating Lifestyle Differences and Dietary Habits.
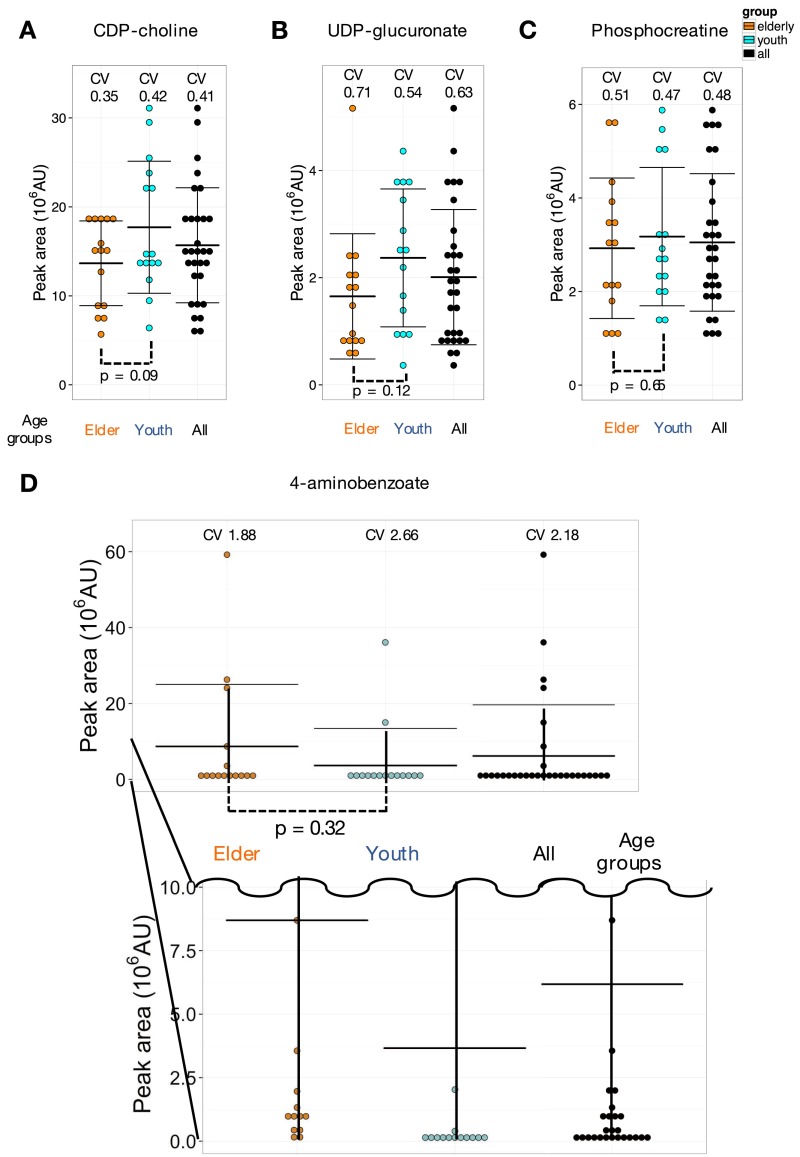
Ten blood metabolites, such as CDP-choline and phosphocreatine, which have not been reported previously, showed moderate 0.4∼0.5 CV30 variation (Fig. 1A and Fig. S3 A and C). Thirteen compounds showed still higher 0.5∼0.7 CV30 (trimethyl-histidine CV30 = 0.57) (Fig. 3E). Nine of them are RBC-enriched, containing nucleotide-sugar and trimethylated derivatives. Their CVs have not been reported previously in blood of healthy individuals. RBC-enriched UDP-glucuronate (CV30 = 0.64) (Fig. S3B) is an intermediate between glucuronides and UDP-glucose (71). UDP-acetyl-glucosamine (CV30 = 0.64) (Fig. 3F), a substrate for N-acetyl-glucosamine transferase, is a precursor for proteoglycan and glycolipid synthesis (72, 73). Abundances of UDP-acetyl-glucosamine and UDP-glucuronate showed some differences between young and elderly subjects (P value, 0.0073 and 0.12, respectively; see Age-Related Compounds Revealed by CV Measurements).
Fig. S3.
Variations of CDP-choline, UDP-glucuronate, phosphocreatinine, and 4-aminobenzoate. (A and B) Two moderately variable metabolites, CDP-choline and UDP-glucuronate, are candidate compounds possibly differing between the two age groups. These compounds are used in biosynthesis and may reflect higher activity levels in younger people. Metabolite peak areas were divided into 10 bins per group. Error bars represent means ± SD. (C) Phosphocreatinine shows moderate variation among individuals, but no significant difference between young and elderly. Peak areas were divided into 10 bins per group. Error bars represent means ± SD. (D) 4-Aminobenzoate is highly variable among individuals. Peak areas were divided into 10 bins per group. Error bars represent means ± SD.
Compounds showing higher CV30 (0.7–2.5) constituted the most variable group (e.g., 4-guanidino-butanoate CV30 2.05; trimethyl-tryptophan CV30 1.67) (Fig. 3 G and H). Nine of these compounds have not been reported previously. Four are methylated amino acids, three of which are trimethylated. Methylated amino acids were enriched in RBCs whereas acetylated amino acids were found in both plasma and RBCs. The reason for this distinction is unclear. Many of the most variable compounds found are modified amino acids, possibly appropriate as marker compounds related to lifestyle, especially dietary habits.
4-Aminobenzoate (also called PABA) data were curious. Its CV30 was very high (2.18). Five people had high levels of PABA whereas, in all others, the level was low or barely detectable (Fig. S3D). PABA is a precursor for vitamin B9 in animals, and in plants (74) and bacteria (75) for folate, but PABA is not essential for humans. This very large variable abundance may reflect dietary or other unknown individual differences.
Age-Related Compounds Revealed by CV Measurements.
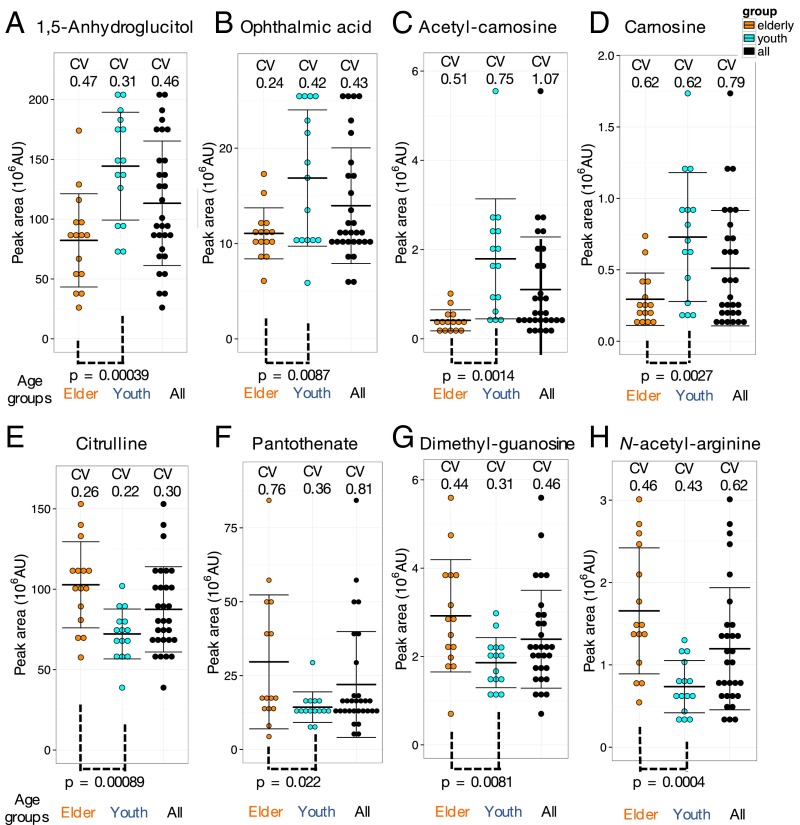
Among 126 compounds analyzed, the great majority showed similar CV levels in young and old people. We found 14 compounds that differed significantly between the two age groups. For example, 1,5-anhydroglucitol (Fig. 4A), known as a glycemic marker (76), showed strikingly lower levels in healthy elderly subjects compared with healthy youths (P = 0.00039). Note that none of 30 volunteers were diabetic patients (see the values of HbA1c and glucose in their blood test in Table S2). 1,5-Anhydroglucitol, a monosaccharide, is normally reabsorbed back into the blood via the kidneys, but this compound is competitive to glucose for reabsorption so that, in diabetic patients containing high glucose in blood, the abundance of 1,5-anhydroglucitol is low. A possible interpretation is that healthy elderly people may gradually lose the ability to reabsorb 1,5-anhydroglucitol, releasing it into urine, with a concomitant decrease in blood.
Fig. 4.
Identification of some blood metabolites that differ in abundance between young (29 ± 4 y of age) and elderly (81 ± 7 y of age) persons. 1,5-Anhydroglucitol (A), ophthalmic acid (B), acetyl-carnosine (C), and carnosine (D) are higher in young subjects whereas citrulline (E), pantothenate (F), dimethyl-guanosine (G), and N-acetyl-arginine (H) are higher in the elderly. Metabolite peak areas were divided into 10 bins in each group. Error bars represent means ± SD. P values between age groups are in the range of 0.022 and 0.00039.
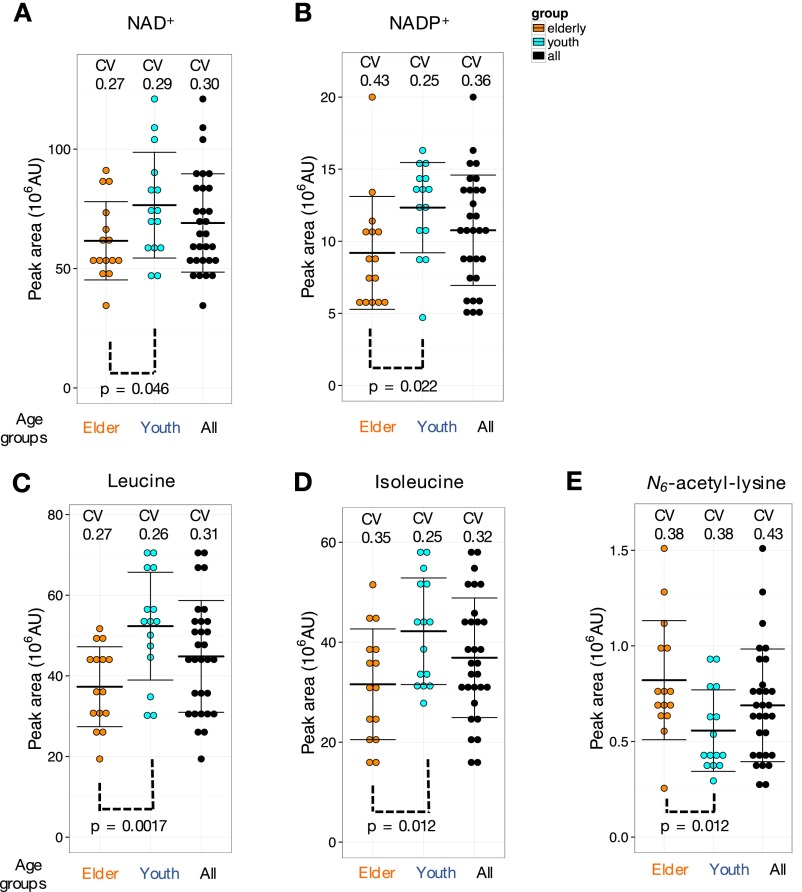
Ophthalmic acid, a tripeptide analog of glutathione, showed impressive difference between the young and the elderly (much less in elderly blood; P value of 0.0087) (Fig. 4B). Similarly, the levels of two oxidant scavengers, acetyl-carnosine (P = 0.0014) (Fig. 4C) and carnosine (P = 0.0027) (Fig. 4D), related dipeptides containing beta-alanine and histidine, were clearly less abundant in the elderly. The same holds true for two redox coenzymes enriched in RBCs, NAD+ (P = 0.046) and NADP+ (P = 0.022) (Fig. S4 A and B), suggesting that the redox metabolism in elderly RBCs might be somewhat declined. Leucine and isoleucine, however, may play a distinct role for supporting skeletal muscle activity in the elderly (77) so that their decrease in elderly blood metabolites (P = 0.0017 and 0.012, respectively) (Fig. S4 C and D) might suggest their decrease in blood due to aging. The level of UDP-acetyl-glucosamine that is probably also unrelated to antioxidants also decreased in elderly blood (P = 0.0073) (Fig. 3F). Because this compound is important for growth and proliferation, its decline might also accelerate aging. In short, elderly blood may have reduced antioxidants, redox, and nutrients required for vigorous body activities.
Fig. S4.
Additional metabolites showing different patterns of abundance between young and elderly groups. NAD+ (A), NADP+ (B), leucine (C), and isoleucine (D) showed higher levels in the youth. N6-acetyl-lysine is higher in elderly people (E). Peak areas of metabolites were divided into 10 bins in each group. Error bars represent means ± SD. The range of P values was 0.0017–0.046.
On the other hand, levels of citrulline (P = 0.00089) (Fig. 4E), pantothenate (P = 0.022) (Fig. 4F), dimethyl-guanosine (P = 0.0081) (Fig. 4G), N-acetyl-arginine (P = 0.0004) (Fig. 4H), and N6-acetyl-lysine (P = 0.012) (Fig. S4E) were clearly more abundant in the blood of elderly subjects. Pantothenate is a precursor of CoA, an important coenzyme involved in the TCA cycle and beta-oxidation. Citrulline is the initial metabolite of the urea cycle. Dimethyl-guanosine is a urinary nucleoside, presenting high levels in plasma of uremic patients (56). In patients deficient in arginase (the last enzyme of the urea cycle), N-acetyl-arginine concentrations are >4× higher than normal (78). Therefore, increased citrulline and N-acetyl-arginine suggest an impaired urea cycle. A possible interpretation of these results is that the excretion of urea cycle metabolites into urine may be somewhat compromised in the elderly. Decreased blood 1,5-anhydroglucitol may also be linked to weakened renal function. Abundant pantothenate in elderly subjects suggests that CoA biosynthesis may be slightly impaired.
Correlations Among Age-Related Compounds.
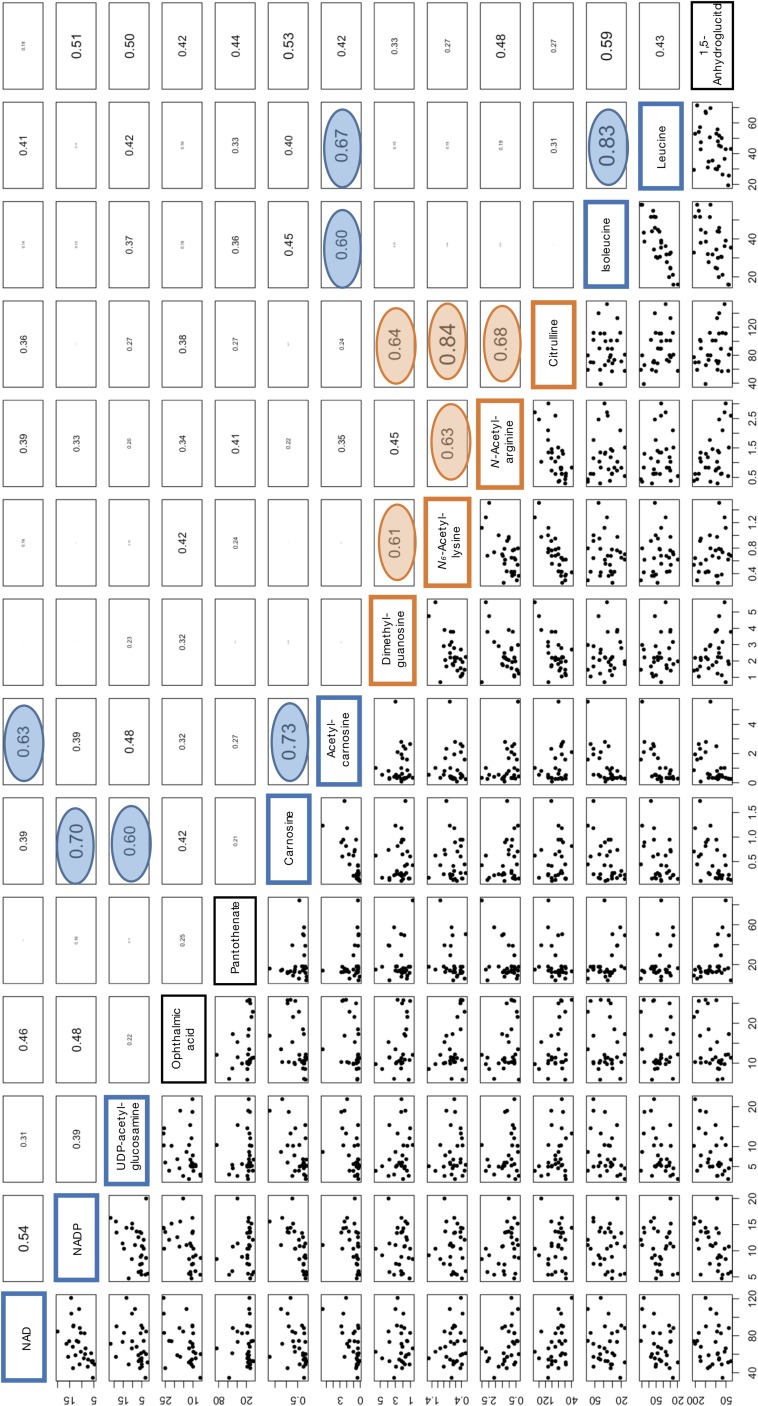
We found 12 pairs of 14 age-related compounds that showed relatively strong correlation coefficients (Pearson’s r) (0.60–0.84) (Table 1 and Fig. S5). Interestingly, such combinations occurred within groups of compounds that either increased or decreased among the elderly. Citrulline content was strongly correlated with N-acetyl-lysine (0.84), and less so with N-acetyl-arginine (0.68) and dimethyl guanosine (0.64) (Table 1). Correlations also existed between N-acetyl-arginine and N-acetyl-lysine (0.63) and between N-acetyl-arginine and dimethyl-guanosine (0.61). These four compounds showed increased blood levels in the elderly. We then found correlations (0.6–0.83) among seven compounds that decreased in the elderly. Correlations between leucine and isoleucine (0.83) and between carnosine and acetyl-carnosine (0.73) were strong, suggesting that these compounds are correlated because of their close functional relationships. Other closely correlated combinations included carnosine and NADP+, and leucine and acetyl carnosine (Table 1 and Fig. S5). These results are consistent with a notion that abundances of two distinct groups of age-related compounds (decrease or increase in the elderly) are internally correlated, but no correlation exists between the groups. For example, elderly volunteers who have abundant leucine would have also high isoleucine in a high probability whereas those elderly who have abundant citrulline would have high N6-acetyl-lysine also in a high probability. However, there is no correlation for leucine and citrulline abundances among individuals.
Table 1.
The pairs of age-related compounds that show relatively high correlation values
| Age-related | Age-related | Correlation |
| Citrulline | N-acetyl-arginine | 0.68 |
| Citrulline | N-acetyl-lysine | 0.84 |
| Citrulline | Dimethyl-guanosine | 0.64 |
| N-acetyl-arginine | N-acetyl-lysine | 0.63 |
| N-acetyl-lysine | Dimethyl-guanosine | 0.61 |
| Leucine | Isoleucine | 0.83 |
| Leucine | Acetyl-carnosine | 0.67 |
| Isoleucine | Acetyl-carnosine | 0.60 |
| Carnosine | Acetyl-carnosine | 0.73 |
| Acetyl-carnosine | NAD+ | 0.63 |
| Carnosine | UDP-acetylglucosamine | 0.70 |
| Carnosine | NADP+ | 0.70 |
The first five pairs of compounds show higher levels in the elderly whereas the other seven pairs of compounds are more abundant in young persons (Fig. S5).
Fig. S5.
Correlation values for all 14 age-related human blood compounds. See Correlations Among Age-Related Compounds.
SI Materials and Methods
Human Subject Characteristics.
Thirty healthy male and female volunteers participated in this study (Table S2). Blood samples for metabolomics analysis and clinical blood parameters were taken in the morning, and subjects were asked not to eat breakfast to ensure at least 12 h of fasting.
Blood Sample Preparation for Metabolomics Analysis.
Metabolomic samples were prepared as reported previously (4). All blood samples were drawn in a hospital laboratory to ensure rapid sample preparation. Briefly, venous blood samples for metabolomics analysis were taken into 50-mL heparinized tubes (Terumo). Immediately, 0.2 mL of blood (8–12 × 108 RBCs) were quenched in 1.8 mL of 55% (vol/vol) methanol at −40 °C. This quick-quenching step immediately after blood sampling ensured accurate measurement of many labile metabolites. The use of whole blood samples also allowed us to observe cellular metabolite levels that might otherwise have been affected by lengthy cell separation procedures. During Ficoll separation or leukodepletion by filtration, blood cells are exposed to nonphysiological conditions for prolonged periods (4).
The remaining blood sample from each donor was centrifuged at 120 × g for 15 min at room temperature to separate plasma and RBCs. After centrifugation, 0.2 mL each of separated plasma and RBCs (14–20 × 108 RBCs) were quenched in 1.8 mL of 55% methanol at −40 °C. Two internal standards (10 nmol Hepes and PIPES) were added to each sample. After brief vortexing, samples were transferred to Amicon Ultra 10-kDa cutoff filters (Millipore) to remove proteins and cellular debris. Thus, from each blood sample, three different subsamples—whole blood, RBCs, and plasma—were prepared. The white blood cell (WBC) content was less than 1% of the cellular volume in our preparations (4). Full metabolomics analysis of WBCs using a Ficoll gradient confirmed that WBCs should not affect our present metabolomics results regarding RBCs. After sample concentration by vacuum evaporation, each sample was resuspended in 40 μL of 50% acetonitrile, and 1 μL was used for each injection into the LC-MS system.
LC-MS Analysis.
LC-MS data were obtained using a Paradigm MS4 HPLC system (Michrom Bioresources) coupled to an LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific), as previously described (21). Briefly, LC separation was performed on a ZIC-pHILIC column (150 mm × 2.1 mm, 5-μm particle size; Merck SeQuant). The HILIC column is quite useful for separating many hydrophilic blood metabolites, which were previously not assayed by others (4). Acetonitrile (A) and 10 mM ammonium carbonate buffer, pH 9.3 (B) were used as the mobile phase, with a gradient elution from 80% to 20% A in 30 min, at a flow rate of 100 μL⋅mL−1. Peak areas of metabolites of interest were measured using MZmine 2 software (87). Detailed data analytical procedures and parameters have been described previously (21). Metabolomic datasets are deposited in the MetaboLights database (Data Availability).
CVs of Blood Metabolites Analyzed by LC-MS.
We analyzed 126 blood compounds confirmed by standards or MS/MS analysis (4). For each metabolite, we chose a singly charged, [M+H]+ or [M-H]−, peak (Table S1; more information is in Dataset S1). Metabolites were classified into three groups (H, M, and L), according to their peak areas. H denotes compounds with high peak areas (>108 AU), M with medium peak areas (108 ∼107 AU), and L with low peak areas (<107 AU).
As previously reported, some standards at identical molar concentrations, such as AMP and ATP, ionize with different efficiencies, thereby affecting quantification of their peak areas (21). Thus, in some cases, peak areas could not be reliably converted into actual molar amounts due to different ionization efficiencies of certain compounds between pure sample and metabolite sample mixtures. In this study, however, individually different relative ratios of peak areas were relevant to obtain CVs so that actual molar concentrations of compounds were not needed.
Determination of CVs for Each Metabolite.
Validation of experimental procedures was performed as follows. First, we evaluated the contribution of sample handling to within-sample variation. The same blood sample preparation was injected 3× into the LC-MS at 80-min intervals (Fig. S2A). We thus obtained within-sample CVs (designated as CVwi), which were less than 0.1 in 107 of 126 compounds (85%). Only 10 compounds showed CVwi of 0.1–0.2 whereas 9 had CVwi of >0.2 (Table S1). Most of the variable compounds belonged to the low-peak-area (L) group, suggesting that some low-abundance compounds may be labile during LC-MS.
Second, we also examined sample-to-sample variation caused by sample preparation. Three samples were independently prepared from the same blood sample (one person), and CVs thus determined were designated as CVss (Fig. S2B). CVss values of Hepes and PIPES in the blood samples were very small (0.06∼0.08 for Hepes and 0.04∼0.08 for PIPES). The great majority (116/126 = 92%) of CVsss were <0.3 (Table S1). Among 10 compounds with CVsss of >0.3, 9 compounds belonged to the low peak area (L) group. Nicotinamide, a vitamin with M peak area, had CVss = 0.47. Three injections of the nicotinamide standard had a CV = 0.05, however. Similar situations were observed for 9 other compounds, such as UMP and 1-methyl-guanosine, with high CVss values. CVsss of these 10 compounds in blood may be affected by sample preparation or they may react with other metabolites during sample preparation. Third, we obtained CVs for each blood compound from all 30 volunteers (CV30) (Table S1 and Fig. S2C). CV30s were grouped into six different value ranges (Fig. 1A).
Data Availability.
Raw LC-MS data in mzML format are accessible via the MetaboLights repository (www.ebi.ac.uk/metabolights). Data from three injections of the same sample and three samples prepared from the same donated blood are available under accession number MTBLS263. Blood samples drawn from four volunteers four times within 24 h are available under accession number MTBLS264. Whole blood metabolomic data from all 30 subjects are available under accession number MTBLS265. Plasma and RBC data from all 30 subjects can be found under MTBLS266 and MTBLS267, respectively.
Discussion
Metabolomics of RBCs.
In this study, untargeted metabolomics of human blood by LC-MS (4) was performed to evaluate individual variation among healthy subjects, using the coefficient of variation (CV). Our technique, including rapid quenching of samples, whole blood analysis without centrifugation, and use of a hydrophilic interaction liquid chromatography (HILIC) column, partly explains why we succeeded in identifying hitherto unreported CVs for many metabolites. We emphasized the importance of RBC metabolomics. This significance is not simply due to the scarcity of such studies, but because RBCs serve such a crucial function. For example, abundant antioxidants in blood, such as glutathione, are exclusively enriched in RBCs over 1,000× . In addition, we show that ophthalmic acid and carnosine, both related to antioxidants, are RBC-enriched and that their abundance seems age-dependent. RBCs thus seem to play the central role in antioxidation in blood. Many cellular compounds, such as sugar phosphates, nucleotides, and nucleotide-sugar derivatives for energy production, are enriched in RBCs. Because half the blood volume is occupied by RBCs, RBC metabolomics may be as important as that of plasma to understand the diverse functions of human blood.
Blood Metabolites with High CVs as Personal Markers.
We identified 48 metabolites showing moderate to very high CV30s (0.5∼2.3). To our knowledge, CVs of 22 of these compounds have not been previously reported. For the most part, these compounds do not fluctuate on a diel basis; thus, we suppose that individual variability may reflect (epi)genetic differences or chronic states. To fully explore their potential as personal markers, further investigation of the physiological roles of these compounds is required. Compounds with low CVs may support physiological homeostasis in vivo. Indeed, anomalous glutathione levels are reported in many diseases, such as Parkinson’s disease, HIV, liver disease, and cystic fibrosis, as well as aging. A number of diseases are reportedly relevant to degradation pathways for leucine, valine, and isoleucine. Thus, low CV compounds might be good candidates as health markers.
Increases of Certain Age-Related Compounds in Blood of the Elderly.
Our metabolomic comparisons of human blood, including RBCs, between young and elderly subjects revealed 14 age-related compounds. Six of them are RBC-enriched. Our results regarding CV30 for three of the 14 compounds (1,5-anhydroglucitol, pantothenate, and citrulline) are confirmatory to the previous study: 1,5-Anhydroglucitol is higher in young people (57) whereas pantothenate and citrulline are more abundant in healthy elderly persons (14, 79). The design of our approach might help us to identify these novel aspects with statistical significance, even though the population for the study was not large (n = 30). Exclusion of middle-aged people (40∼70 y old) from the study gave us clearer age-difference between two groups. Samples were also collectively analyzed at one time for accurate measurement.
Eight of the remaining previously unidentified age-related 11 compounds are lower in elderly subjects. Our results suggest that the blood of elderly subjects shows reduced levels of some compounds related to antioxidants (ophthalmic acid, carnosine, etc.) and redox metabolites (NAD+, NADP+), as well as compounds that support muscle maintenance and reinforcement (leucine, isoleucine).
In contrast, three plasma-enriched compounds (N-acetyl-arginine, dimethyl-guanosine, and N6-acetyl-lysine) increase in the elderly. N-acetyl-arginine and citrulline, the by-products of the urea cycle, might increase due to impaired efficiency of this cycle. Indeed, deficiencies of urea cycle enzymes are known to cause the accumulations of these compounds (78, 80). Dimethyl-guanosine is known to increase in the plasma of uremic patients (56). These results suggest that gradual, progressive decay of liver or renal function may be typical among elderly people generally, resulting in a gradual rise in these blood metabolites.
Certain Compounds Supporting Vigorous Activity Decline in the Elderly.
It is also noteworthy that several age-related compounds, including carnosine, are identified in RBC analysis. Carnosine (beta-alanyl-l-histidine), a possible scavenger of oxidants, is highly concentrated in muscle and brain (81). Our data demonstrate that carnosine is a highly variable metabolite enriched in RBCs. These findings allow us to reconsider the physiological role of RBCs in blood. RBCs may also serve to transport carnosine and other metabolites to distant tissues. Consistently acetyl-carnosine, which is resistant to degradation (82), is plasma-enriched. The RBC/plasma ratios among 30 subjects are 10.8 (carnosine) and 0.13 (acetyl-carnosine). Carnosine is clearly RBC-enriched whereas acetyl-carnosine is clearly a plasma compound. Our study demonstrated that both compounds decline in the elderly. Further study to elucidate the role of carnosine in RBCs is of considerable interest.
Antioxidants, and Compounds Related to Energy and Cellular Maintenance in Blood.
Compounds required for vigorous activity during youth may decline in the elderly. Ophthalmic acid is related to glutathione, and both are generated by the same biosynthetic enzymes. Therefore, ophthalmic acid is thought to be related to antioxidants; it also decreases in the elderly. The level of UDP-acetyl-glucosamine was twofold higher in young subjects than in elderly subjects. This compound is required for cell signaling during proteoglycan and glycolipid synthesis and for the formation of nuclear pores (83). These functions are compatible with the hypothesis that synthesis of antioxidants and cellular maintenance compounds declines with age. Consistently, leucine, isoleucine, NAD+, and NADP+ are more abundant in youth. These data may suggest that these compounds are more vigorously consumed in the body, particularly in muscle, when physical activity is higher (84, 85). It is unclear whether lower levels of these compounds result in diminished muscle and possibly brain activity, or whether they reflect reduced activity. Scavengers of oxidants may be required to restore energy-related biochemical reactions in RBCs (86).
Future Prospects of Human Metabolomics.
It is noteworthy that 11 of these 14 age-related compounds (except for 1,5-anhydroglucitol, carnosine, and acetyl-carnosine) are also present in fission yeast. In the near future, genetics of these compounds in fission yeast and other organisms may be helpful to dissect their physiological and cytological significance. If so, the present analysis of RBCs, plasma, and whole blood will support the development of human metabolomics
Materials and Methods
Ethics Statement.
Written, informed consent was obtained from all donors, in accordance with the Declaration of Helsinki. All experiments were performed in compliance with relevant Japanese laws and institutional guidelines. All protocols were approved by the Ethical Committee on Human Research of Kyoto University Hospital and by the Human Subjects Research Review Committee of the Okinawa Institute of Science and Technology Graduate University (OIST).
Human Subject Characteristics and Blood Metabolomics Analysis.
Thirty healthy male and female volunteers participated in this study (Table S2). Metabolomic samples were prepared as reported previously (4). Detailed procedures of LC-MS measurements and determination of CVs for each metabolite can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Eri Shibata and Akemi Yoshioka for providing excellent technical assistance and Dr. Tomáš Pluskal and Professor Shigeaki Saitoh for valuable comments. We gratefully acknowledge the editorial help of Dr. Steven D. Aird. This work was supported by grants from the Okinawa Intellectual Cluster Program (to M.Y. and H.K.), from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and from the Japan Science and Technology Agency (to H.K.). We acknowledge generous support from the Okinawa Institute of Science and Technology Graduate University. R.C. was supported by a Japanese government predoctoral scholarship (Monbukagakusho). I.M. was supported by a postdoctoral fellowship from the Okinawa Intellectual Cluster Program.
Footnotes
The authors declare no conflict of interest.
Data deposition: Raw LC-MS data in the mzML format have been deposited in the MetaboLights repository, www.ebi.ac.uk/metabolights [accession nos. MTBLS263 (data from three injections of the same sample and three samples prepared from the same donated blood), MTBLS264 (blood samples drawn from four volunteers four times within 24 h), MTBLS265 (whole blood metabolomic data from all 30 subjects), MTBLS266 (plasma from all 30 subjects), and MTBLS267 (RBC data from all 30 subjects)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603023113/-/DCSupplemental.
References
- 1.Rapoport SM, Schewe T, Thiele B-J. Maturational breakdown of mitochondria and other organelles in reticulocytes. In: Harris JR, editor. Erythroid Cells. Springer; New York: 1990. pp. 151–194. [Google Scholar]
- 2.van Wijk R, van Solinge WW. The energy-less red blood cell is lost: Erythrocyte enzyme abnormalities of glycolysis. Blood. 2005;106(13):4034–4042. doi: 10.1182/blood-2005-04-1622. [DOI] [PubMed] [Google Scholar]
- 3.Bax BE, Bain MD, Talbot PJ, Parker-Williams EJ, Chalmers RA. Survival of human carrier erythrocytes in vivo. Clin Sci (Lond) 1999;96(2):171–178. [PubMed] [Google Scholar]
- 4.Chaleckis R, et al. Unexpected similarities between the Schizosaccharomyces and human blood metabolomes, and novel human metabolites. Mol Biosyst. 2014;10(10):2538–2551. doi: 10.1039/c4mb00346b. [DOI] [PubMed] [Google Scholar]
- 5.Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Metabolite profiling: From diagnostics to systems biology. Nat Rev Mol Cell Biol. 2004;5(9):763–769. doi: 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- 6.Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends Biotechnol. 2004;22(5):245–252. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Hirai MY, et al. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2004;101(27):10205–10210. doi: 10.1073/pnas.0403218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kell DB. Metabolomics and systems biology: Making sense of the soup. Curr Opin Microbiol. 2004;7(3):296–307. doi: 10.1016/j.mib.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Nicholson JK, Lindon JC. Systems biology: Metabonomics. Nature. 2008;455(7216):1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 10.Patti GJ, Yanes O, Siuzdak G. Metabolomics: The apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramautar R, Berger R, van der Greef J, Hankemeier T. Human metabolomics: Strategies to understand biology. Curr Opin Chem Biol. 2013;17(5):841–846. doi: 10.1016/j.cbpa.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 12.Dunn WB, et al. Molecular phenotyping of a UK population: Defining the human serum metabolome. Metabolomics. 2015;11:9–26. doi: 10.1007/s11306-014-0707-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guertin KA, et al. Metabolomics in nutritional epidemiology: Identifying metabolites associated with diet and quantifying their potential to uncover diet-disease relations in populations. Am J Clin Nutr. 2014;100(1):208–217. doi: 10.3945/ajcn.113.078758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawton KA, et al. Analysis of the adult human plasma metabolome. Pharmacogenomics. 2008;9(4):383–397. doi: 10.2217/14622416.9.4.383. [DOI] [PubMed] [Google Scholar]
- 15.Psychogios N, et al. The human serum metabolome. PLoS One. 2011;6(2):e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Z, et al. Human serum metabolic profiles are age dependent. Aging Cell. 2012;11(6):960–967. doi: 10.1111/j.1474-9726.2012.00865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suhre K. Metabolic profiling in diabetes. J Endocrinol. 2014;221(3):R75–R85. doi: 10.1530/JOE-14-0024. [DOI] [PubMed] [Google Scholar]
- 18.Kastenmüller G, Raffler J, Gieger C, Suhre K. Genetics of human metabolism: An update. Hum Mol Genet. 2015;24(R1):R93–R101. doi: 10.1093/hmg/ddv263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishino T, et al. In silico modeling and metabolome analysis of long-stored erythrocytes to improve blood storage methods. J Biotechnol. 2009;144(3):212–223. doi: 10.1016/j.jbiotec.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 20.Gil A, et al. Stability of energy metabolites—an often overlooked issue in metabolomics studies: A review. Electrophoresis. 2015;36(18):2156–2169. doi: 10.1002/elps.201500031. [DOI] [PubMed] [Google Scholar]
- 21.Pluskal T, Nakamura T, Villar-Briones A, Yanagida M. Metabolic profiling of the fission yeast S. pombe: Quantification of compounds under different temperatures and genetic perturbation. Mol Biosyst. 2010;6(1):182–198. doi: 10.1039/b908784b. [DOI] [PubMed] [Google Scholar]
- 22.Kaya M, et al. Plasma concentrations and urinary excretion of purine bases (uric acid, hypoxanthine, and xanthine) and oxypurinol after rigorous exercise. Metabolism. 2006;55(1):103–107. doi: 10.1016/j.metabol.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Benowitz NL. Clinical pharmacology of caffeine. Annu Rev Med. 1990;41:277–288. doi: 10.1146/annurev.me.41.020190.001425. [DOI] [PubMed] [Google Scholar]
- 24.Campbell CB, McGuffie C, Powell LW, Roberts RK, Stewart AW. Postprandial changes in serum concentrations of individual bile salts in normal subjects and patients with acute viral hepatitis. Am J Dig Dis. 1978;23(7):599–608. doi: 10.1007/BF01072594. [DOI] [PubMed] [Google Scholar]
- 25.Breier M, et al. Targeted metabolomics identifies reliable and stable metabolites in human serum and plasma samples. PLoS One. 2014;9(2):e89728. doi: 10.1371/journal.pone.0089728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arends J, Chiu F, Bier DM. Analysis of plasma hippurate in humans using gas chromatography-mass spectrometry: Concentration and incorporation of infused [15N]glycine. Anal Biochem. 1990;191(2):401–410. doi: 10.1016/0003-2697(90)90239-6. [DOI] [PubMed] [Google Scholar]
- 27.Armstrong MD. N-delta-acetylornithine and S-methylcysteine in blood plasma. Biochim Biophys Acta. 1979;587(4):638–642. doi: 10.1016/0304-4165(79)90015-1. [DOI] [PubMed] [Google Scholar]
- 28.Bene J, et al. Plasma carnitine ester profile in adult celiac disease patients maintained on long-term gluten free diet. World J Gastroenterol. 2005;11(42):6671–6675. doi: 10.3748/wjg.v11.i42.6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bishop C, Rankine DM, Talbott JH. The nucleotides in normal human blood. J Biol Chem. 1959;234(5):1233–1237. [PubMed] [Google Scholar]
- 30.Capogrossi MC, Holdiness MR, Israili ZH. Determination of adenosine in normal human plasma and serum by high-performance liquid chromatography. J Chromatogr A. 1982;227(1):168–173. doi: 10.1016/s0378-4347(00)80367-9. [DOI] [PubMed] [Google Scholar]
- 31.Cheng H, et al. Levels of L-methionine S-adenosyltransferase activity in erythrocytes and concentrations of S-adenosylmethionine and S-adenosylhomocysteine in whole blood of patients with Parkinson’s disease. Exp Neurol. 1997;145(2 Pt 1):580–585. doi: 10.1006/exnr.1997.6466. [DOI] [PubMed] [Google Scholar]
- 32.Conway KJ, Orr R, Stannard SR. Effect of a divided caffeine dose on endurance cycling performance, postexercise urinary caffeine concentration, and plasma paraxanthine. J Appl Physiol (1985) 2003;94(4):1557–1562. doi: 10.1152/japplphysiol.00911.2002. [DOI] [PubMed] [Google Scholar]
- 33.Costa A, Igualá I, Bedini J, Quintó L, Conget I. Uric acid concentration in subjects at risk of type 2 diabetes mellitus: Relationship to components of the metabolic syndrome. Metabolism. 2002;51(3):372–375. doi: 10.1053/meta.2002.30523. [DOI] [PubMed] [Google Scholar]
- 34.Creeke PI, et al. Whole blood NAD and NADP concentrations are not depressed in subjects with clinical pellagra. J Nutr. 2007;137(9):2013–2017. doi: 10.1093/jn/137.9.2013. [DOI] [PubMed] [Google Scholar]
- 35.Crenn P, Coudray-Lucas C, Thuillier F, Cynober L, Messing B. Postabsorptive plasma citrulline concentration is a marker of absorptive enterocyte mass and intestinal failure in humans. Gastroenterology. 2000;119(6):1496–1505. doi: 10.1053/gast.2000.20227. [DOI] [PubMed] [Google Scholar]
- 36.Curless R, French JM, James OF, Wynne HA. Is caffeine a factor in subjective insomnia of elderly people? Age Ageing. 1993;22(1):41–45. doi: 10.1093/ageing/22.1.41. [DOI] [PubMed] [Google Scholar]
- 37.Cynober LA. Plasma amino acid levels with a note on membrane transport: Characteristics, regulation, and metabolic significance. Nutrition. 2002;18(9):761–766. doi: 10.1016/s0899-9007(02)00780-3. [DOI] [PubMed] [Google Scholar]
- 38.Dello SA, et al. Ophthalmate detection in human plasma with LC-MS-MS. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;903:1–6. doi: 10.1016/j.jchromb.2012.06.023. [DOI] [PubMed] [Google Scholar]
- 39.Eells JT, Spector R. Purine and pyrimidine base and nucleoside concentrations in human cerebrospinal fluid and plasma. Neurochem Res. 1983;8(11):1451–1457. doi: 10.1007/BF00965000. [DOI] [PubMed] [Google Scholar]
- 40.Fleck C, Schweitzer F, Karge E, Busch M, Stein G. Serum concentrations of asymmetric (ADMA) and symmetric (SDMA) dimethylarginine in patients with chronic kidney diseases. Clin Chim Acta. 2003;336(1-2):1–12. doi: 10.1016/s0009-8981(03)00338-3. [DOI] [PubMed] [Google Scholar]
- 41.Fonteh AN, Harrington RJ, Harrington MG. Quantification of free amino acids and dipeptides using isotope dilution liquid chromatography and electrospray ionization tandem mass spectrometry. Amino Acids. 2007;32(2):203–212. doi: 10.1007/s00726-006-0370-6. [DOI] [PubMed] [Google Scholar]
- 42.Fonteh AN, Harrington RJ, Tsai A, Liao P, Harrington MG. Free amino acid and dipeptide changes in the body fluids from Alzheimer’s disease subjects. Amino Acids. 2007;32(2):213–224. doi: 10.1007/s00726-006-0409-8. [DOI] [PubMed] [Google Scholar]
- 43.Hervé C, Beyne P, Jamault H, Delacoux E. Determination of tryptophan and its kynurenine pathway metabolites in human serum by high-performance liquid chromatography with simultaneous ultraviolet and fluorimetric detection. J Chromatogr B Biomed Appl. 1996;675(1):157–161. doi: 10.1016/0378-4347(95)00341-x. [DOI] [PubMed] [Google Scholar]
- 44.Hoffmann GF, et al. Physiology and pathophysiology of organic acids in cerebrospinal fluid. J Inherit Metab Dis. 1993;16(4):648–669. doi: 10.1007/BF00711898. [DOI] [PubMed] [Google Scholar]
- 45.Hoppe B, Kemper MJ, Hvizd MG, Sailer DE, Langman CB. Simultaneous determination of oxalate, citrate and sulfate in children’s plasma with ion chromatography. Kidney Int. 1998;53(5):1348–1352. doi: 10.1046/j.1523-1755.1998.00891.x. [DOI] [PubMed] [Google Scholar]
- 46.Huck JH, Struys EA, Verhoeven NM, Jakobs C, van der Knaap MS. Profiling of pentose phosphate pathway intermediates in blood spots by tandem mass spectrometry: Application to transaldolase deficiency. Clin Chem. 2003;49(8):1375–1380. doi: 10.1373/49.8.1375. [DOI] [PubMed] [Google Scholar]
- 47.Huck JH, et al. Ribose-5-phosphate isomerase deficiency: New inborn error in the pentose phosphate pathway associated with a slowly progressive leukoencephalopathy. Am J Hum Genet. 2004;74(4):745–751. doi: 10.1086/383204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ilcol YO, Ozbek R, Hamurtekin E, Ulus IH. Choline status in newborns, infants, children, breast-feeding women, breast-fed infants and human breast milk. J Nutr Biochem. 2005;16(8):489–499. doi: 10.1016/j.jnutbio.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 49.Kikuchi T, et al. Liquid-chromatographic determination of guanidino compounds in plasma and erythrocyte of normal persons and uremic patients. Clin Chem. 1981;27(11):1899–1902. [PubMed] [Google Scholar]
- 50.Laurence AD, Layton M, Duley JA, Simmonds HA. Elevated erythrocyte CDP-choline levels associated with beta-thalassaemia in patients with transfusion independent anaemia. Nucleosides Nucleotides Nucleic Acids. 2004;23(8-9):1265–1267. doi: 10.1081/NCN-200027535. [DOI] [PubMed] [Google Scholar]
- 51.Lawson N, Berg JD, Chesner I. Liquid-chromatographic determination of p-aminobenzoic acid in plasma to evaluate exocrine pancreatic function. Clin Chem. 1985;31(6):1073–1075. [PubMed] [Google Scholar]
- 52.Lehman LJ, Olson AL, Rebouche CJ. Measurement of epsilon-N-trimethyllysine in human blood plasma and urine. Anal Biochem. 1987;162(1):137–142. doi: 10.1016/0003-2697(87)90018-2. [DOI] [PubMed] [Google Scholar]
- 53.Lever M, Sizeland PC, Bason LM, Hayman CM, Chambers ST. Glycine betaine and proline betaine in human blood and urine. Biochim Biophys Acta. 1994;1200(3):259–264. doi: 10.1016/0304-4165(94)90165-1. [DOI] [PubMed] [Google Scholar]
- 54.Marescau B, et al. Guanidino compounds in serum and urine of cirrhotic patients. Metabolism. 1995;44(5):584–588. doi: 10.1016/0026-0495(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 55.Nakayama Y, Kinoshita A, Tomita M. Dynamic simulation of red blood cell metabolism and its application to the analysis of a pathological condition. Theor Biol Med Model. 2005;2:18. doi: 10.1186/1742-4682-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niwa T, Takeda N, Yoshizumi H. RNA metabolism in uremic patients: Accumulation of modified ribonucleosides in uremic serum. Technical note. Kidney Int. 1998;53(6):1801–1806. doi: 10.1046/j.1523-1755.1998.00944.x. [DOI] [PubMed] [Google Scholar]
- 57.Ouchi M, et al. Effects of sex and age on serum 1,5-anhydroglucitol in nondiabetic subjects. Exp Clin Endocrinol Diabetes. 2012;120(5):288–295. doi: 10.1055/s-0032-1304604. [DOI] [PubMed] [Google Scholar]
- 58.Persiani S, et al. Synovial and plasma glucosamine concentrations in osteoarthritic patients following oral crystalline glucosamine sulphate at therapeutic dose. Osteoarthritis Cartilage. 2007;15(7):764–772. doi: 10.1016/j.joca.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 59.Rocchiccioli F, Leroux JP, Cartier PH. Microdetermination of 2-ketoglutaric acid in plasma and cerebrospinal fluid by capillary gas chromatography mass spectrometry: Application to pediatrics. Biomed Mass Spectrom. 1984;11(1):24–28. doi: 10.1002/bms.1200110105. [DOI] [PubMed] [Google Scholar]
- 60.Sandberg DH, Sjoevall J, Sjoevall K, Turner DA. Measurement of human serum bile acids by gas-liquid chromatography. J Lipid Res. 1965;6:182–192. [PubMed] [Google Scholar]
- 61.Smythe GA, et al. ECNI GC-MS analysis of picolinic and quinolinic acids and their amides in human plasma, CSF, and brain tissue. Adv Exp Med Biol. 2003;527:705–712. doi: 10.1007/978-1-4615-0135-0_83. [DOI] [PubMed] [Google Scholar]
- 62.Sotgia S, et al. Quantification of L-ergothioneine in whole blood by hydrophilic interaction ultra-performance liquid chromatography and UV-detection. J Sep Sci. 2013;36(6):1002–1006. doi: 10.1002/jssc.201201016. [DOI] [PubMed] [Google Scholar]
- 63.Stratford MR, Dennis MF. High-performance liquid chromatographic determination of nicotinamide and its metabolites in human and murine plasma and urine. J Chromatogr A. 1992;582(1-2):145–151. doi: 10.1016/0378-4347(92)80313-f. [DOI] [PubMed] [Google Scholar]
- 64.Tatidis L, Vitols S, Gruber A, Paul C, Axelson M. Cholesterol catabolism in patients with acute myelogenous leukemia and hypocholesterolemia: Suppressed levels of a circulating marker for bile acid synthesis. Cancer Lett. 2001;170(2):169–175. doi: 10.1016/s0304-3835(01)00592-4. [DOI] [PubMed] [Google Scholar]
- 65.Tavazzi B, et al. Simultaneous high performance liquid chromatographic separation of purines, pyrimidines, N-acetylated amino acids, and dicarboxylic acids for the chemical diagnosis of inborn errors of metabolism. Clin Biochem. 2005;38(11):997–1008. doi: 10.1016/j.clinbiochem.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 66.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140(1):1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 67.Ujhelyi L, et al. Hemodialysis reduces inhibitory effect of plasma ultrafiltrate on LDL oxidation and subsequent endothelial reactions. Kidney Int. 2006;69(1):144–151. doi: 10.1038/sj.ki.5000007. [DOI] [PubMed] [Google Scholar]
- 68.Vernez L, Wenk M, Krähenbühl S. Determination of carnitine and acylcarnitines in plasma by high-performance liquid chromatography/electrospray ionization ion trap tandem mass spectrometry. Rapid Commun Mass Spectrom. 2004;18(11):1233–1238. doi: 10.1002/rcm.1470. [DOI] [PubMed] [Google Scholar]
- 69.Wittwer CT, et al. Enzymes for liberation of pantothenic acid in blood: Use of plasma pantetheinase. Am J Clin Nutr. 1989;50(5):1072–1078. doi: 10.1093/ajcn/50.5.1072. [DOI] [PubMed] [Google Scholar]
- 70.Tokunaga K, et al. Enhanced expression of a glyceraldehyde-3-phosphate dehydrogenase gene in human lung cancers. Cancer Res. 1987;47(21):5616–5619. [PubMed] [Google Scholar]
- 71.Linster CL, Van Schaftingen E. Vitamin C: Biosynthesis, recycling and degradation in mammals. FEBS J. 2007;274(1):1–22. doi: 10.1111/j.1742-4658.2006.05607.x. [DOI] [PubMed] [Google Scholar]
- 72.Hirschberg CB, Snider MD. Topography of glycosylation in the rough endoplasmic reticulum and Golgi apparatus. Annu Rev Biochem. 1987;56(1):63–87. doi: 10.1146/annurev.bi.56.070187.000431. [DOI] [PubMed] [Google Scholar]
- 73.Wells L, Vosseller K, Hart GW. Glycosylation of nucleocytoplasmic proteins: Signal transduction and O-GlcNAc. Science. 2001;291(5512):2376–2378. doi: 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- 74.DellaPenna D. Biofortification of plant-based food: Enhancing folate levels by metabolic engineering. Proc Natl Acad Sci USA. 2007;104(10):3675–3676. doi: 10.1073/pnas.0700640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Camilo E, et al. Folate synthesized by bacteria in the human upper small intestine is assimilated by the host. Gastroenterology. 1996;110(4):991–998. doi: 10.1053/gast.1996.v110.pm8613033. [DOI] [PubMed] [Google Scholar]
- 76.Dungan KM. 1,5-anhydroglucitol (GlycoMark) as a marker of short-term glycemic control and glycemic excursions. Expert Rev Mol Diagn. 2008;8(1):9–19. doi: 10.1586/14737159.8.1.9. [DOI] [PubMed] [Google Scholar]
- 77.Katsanos CS, Kobayashi H, Sheffield-Moore M, Aarsland A, Wolfe RR. A high proportion of leucine is required for optimal stimulation of the rate of muscle protein synthesis by essential amino acids in the elderly. Am J Physiol Endocrinol Metab. 2006;291(2):E381–E387. doi: 10.1152/ajpendo.00488.2005. [DOI] [PubMed] [Google Scholar]
- 78.Mizutani N, et al. Guanidino compounds in hyperargininemia. Tohoku J Exp Med. 1987;153(3):197–205. doi: 10.1620/tjem.153.197. [DOI] [PubMed] [Google Scholar]
- 79.Pitkänen HT, Oja SS, Kemppainen K, Seppä JM, Mero AA. Serum amino acid concentrations in aging men and women. Amino Acids. 2003;24(4):413–421. doi: 10.1007/s00726-002-0338-0. [DOI] [PubMed] [Google Scholar]
- 80.Leonard JV, Morris AA. Urea cycle disorders. Semin Neonatol. 2002;7(1):27–35. doi: 10.1053/siny.2001.0085. [DOI] [PubMed] [Google Scholar]
- 81.Park YJ, Volpe SL, Decker EA. Quantitation of carnosine in humans plasma after dietary consumption of beef. J Agric Food Chem. 2005;53(12):4736–4739. doi: 10.1021/jf047934h. [DOI] [PubMed] [Google Scholar]
- 82.Hipkiss AR, Cartwright SP, Bromley C, Gross SR, Bill RM. Carnosine: Can understanding its actions on energy metabolism and protein homeostasis inform its therapeutic potential? Chem Cent J. 2013;7(1):38. doi: 10.1186/1752-153X-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li B, Kohler JJ. Glycosylation of the nuclear pore. Traffic. 2014;15(4):347–361. doi: 10.1111/tra.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Buse MG, Reid SS. Leucine: A possible regulator of protein turnover in muscle. J Clin Invest. 1975;56(5):1250–1261. doi: 10.1172/JCI108201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285(5432):1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 86.Hunt T, Herbert P, Campbell EA, Delidakis C, Jackson RJ. The use of affinity chromatography on 2‘5’ ADP-sepharose reveals a requirement for NADPH, thioredoxin and thioredoxin reductase for the maintenance of high protein synthesis activity in rabbit reticulocyte lysates. Eur J Biochem. 1983;131(2):303–311. doi: 10.1111/j.1432-1033.1983.tb07263.x. [DOI] [PubMed] [Google Scholar]
- 87.Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw LC-MS data in mzML format are accessible via the MetaboLights repository (www.ebi.ac.uk/metabolights). Data from three injections of the same sample and three samples prepared from the same donated blood are available under accession number MTBLS263. Blood samples drawn from four volunteers four times within 24 h are available under accession number MTBLS264. Whole blood metabolomic data from all 30 subjects are available under accession number MTBLS265. Plasma and RBC data from all 30 subjects can be found under MTBLS266 and MTBLS267, respectively.