Significance
Transcription termination of rRNA genes by RNA polymerase I (pol I) in fission yeast requires the binding of the Reb1 protein to a terminator site (Ter). Termination is physiologically necessary because its elimination can cause replication–transcription collision and induction of genome instability. Furthermore, without termination, pol I can become unproductively sequestered on the DNA templates. We have determined the crystal structure of fission yeast terminator protein Reb1-Ter complex revealing its functional architecture. Structure-guided functional analysis revealed that it is not just tight binding of the protein to Ter but protein–protein interactions with the Rpa12 subunit of RNA polymerase I that causes transcriptional arrest.
Keywords: crystal structure, RNA polymerase I, transcription termination, protein–DNA interaction, replication termination
Abstract
Reb1 of Schizosaccharomyces pombe represents a family of multifunctional proteins that bind to specific terminator sites (Ter) and cause polar termination of transcription catalyzed by RNA polymerase I (pol I) and arrest of replication forks approaching the Ter sites from the opposite direction. However, it remains to be investigated whether the same mechanism causes arrest of both DNA transactions. Here, we present the structure of Reb1 as a complex with a Ter site at a resolution of 2.7 Å. Structure-guided molecular genetic analyses revealed that it has distinct and well-defined DNA binding and transcription termination (TTD) domains. The region of the protein involved in replication termination is distinct from the TTD. Mechanistically, the data support the conclusion that transcription termination is not caused by just high affinity Reb1-Ter protein–DNA interactions. Rather, protein–protein interactions between the TTD with the Rpa12 subunit of RNA pol I seem to be an integral part of the mechanism. This conclusion is further supported by the observation that double mutations in TTD that abolished its interaction with Rpa12 also greatly reduced transcription termination thereby revealing a conduit for functional communications between RNA pol I and the terminator protein.
Eukaryotic rDNAs are found as multiple tandem copies encoding pre-rRNA and upstream and downstream regulatory elements (1–3) including DNA sequences (Ter) that promote site-specific termination of transcription catalyzed by RNA polymerase I (pol I) from yeast to humans (4–14). Specialized transcription terminator proteins bind to Ter sites and not only arrest transcription by RNA polymerase I (pol I) in a polar mode but also replication forks approaching from the opposite direction. Several studies have suggested that pol I transcription termination is a multistep process that requires (i) pausing of chain elongation by the terminator protein and (ii) dissociation and release of pol I and the primary transcript from the template. In mice, dissociation of pol I and release of the transcript require the release factor PTRF, a 44-kDa protein that interacts with the largest subunit of pol I (15). In yeast, additional factors for the processing of the end include the endonuclease Rnt1, the 5′-3′ Rat1 exonuclease, Sen1 helicase, and the kinase Grc3. They are part of an alternate pathway for termination by transcriptional coprocessing (4, 16–19). In addition, in vivo analyses of Saccharomyces cerevisiae have shown a requirement for Rpa12, a component of pol I necessary for transcription termination (20). Whether the polar arrest of transcription is caused by an interaction between Rpa12 with the terminator protein is unknown.
The terminator proteins that mediate transcription termination have been identified in multiple organisms and include Reb1 and Nsi1 [also called yeast transcription terminator (Ytt1)] of S. cerevisiae (21–24), Reb1 of Schizosaccharomyces pombe (8), Rib2 of Xenopus (25), and mammalian TTF I (26, 27). In S. pombe, studies using an in vitro system consisting of Reb1 protein and partially fractionated cell extracts confirmed orientation-dependent transcription termination (8). In S. cerevisiae, an in vitro system consisting of purified pol I, Reb1 (ScReb1), the Ter site, and an upstream AT-rich release element were necessary and sufficient for transcription termination (7). However, recent work has shown that Nsi1 (Ytt1) is also responsible for transcription termination in S. cerevisiae (28).
In addition to their aforementioned role, many of these proteins are also involved in replication termination and transcription activation. For instance, in S. pombe, the Reb1 (Sp.Reb1) protein binds to tandem pairs of terminator sites (Ter2 and Ter3) located on the spacer regions of each rDNA repeat in chromosome III and to Ter sites located in the other two chromosomes. This interaction mediates at least four different functions namely termination of DNA replication (29–31), activation of transcription (8, 32), termination of transcription, and a long-range chromosome-to-chromosome interaction called “chromosome kissing” (31). The mammalian ortholog TTF-I also performs similar functions (26, 27, 33, 34).
Replication termination is of critical importance, especially during the S phase of the cell cycle, when chromosomal DNA is often transcribed and replicated at the same time. Ter sites are needed to prevent collisions between the replication and transcription machineries approaching each other from opposite directions, thereby preventing R-loop formation and potential genome instability (35, 36). Sequence and molecular analyses revealed that the terminator proteins have multiple domains with a variable N-terminal region that may be involved in oligomerization (27, 37–40) and two or more Myb-like sequence-specific DNA binding domains at the C terminus. However, additional regions located immediately N-terminal to the predicted Myb domains are also needed for full binding affinity (37, 39).
To assign the various known functions of Sp.Reb1 to its particular structural domains and to perform structure–function analyses, we determined the crystal structure of Reb1 bound to its cognate Ter3 site at 2.7 Å. Guided by the structure and using yeast molecular genetics, we have resolved the protein into a composite DNA binding domain (DBD) that includes the replication termination domain (RTD) and a separate C-terminal transcription termination domain (TTD). Guided by the structural information, this work has uncovered that the TTD interacts with the Rpa12 subunit of RNA pol I. Our data show that either deletion of the TTD or mutational disruption of its interaction with Rpa12 was accompanied by significant reduction of transcription termination. Therefore, contrary to the previously suggested model that just tight binding of Reb1 to Ter sites possibly acted as a passive road block to stall pol I (7), our data suggest that the mechanism is an active one involving the aforementioned protein–protein interaction to establish functional communication between pol I and Reb1. The ubiquitous presence of the Reb1 class of proteins in many eukaryotic systems and a general conservation of RNA polymerase structure strongly suggest that interaction between the terminator protein and component(s) of pol I is likely to be a conserved feature of this DNA transaction.
Results
Overview of the Reb1-Ter3 Complex.
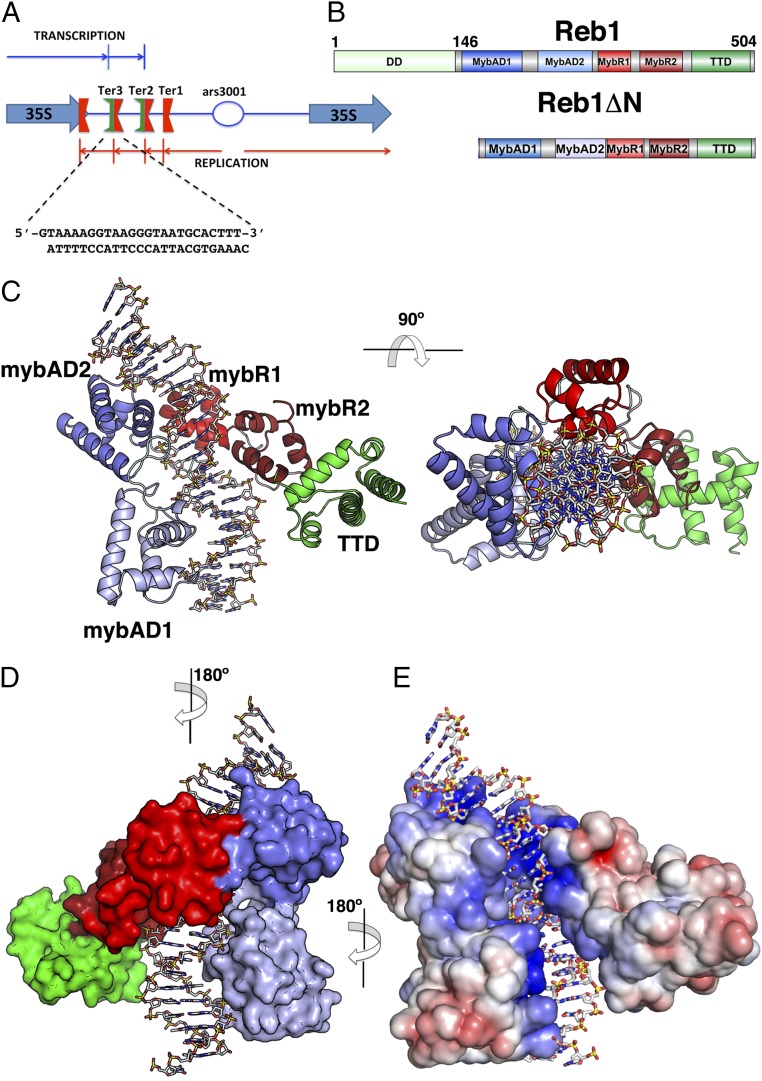
We and others have previously reported that an N-terminally truncated version of the Reb1 protein from residues 146–504 is necessary and sufficient for both replication and transcription termination on binding to the Ter2 and/ or Ter3 sites (8, 29, 30, 39). This truncated form of Reb1 (Reb1ΔN) was cocrystallized with a 26-mer double-stranded DNA containing the complete Ter3 sequence (Fig. 1 A and B). Experimental phases were determined by single wavelength anomalous dispersion (SAD) using selenomethionine-substituted protein, and the complex was refined to a resolution of 2.7 Å (Table S1). There are two molecules in the asymmetric unit with almost identical conformations, with main chain atoms of protein and DNA superimposing with a RMS deviation (RMSD) of 1.09 and 0.95 Å, respectively (Fig. S1 and SI Materials and Methods). The structure showed that Reb1ΔN is almost entirely α-helical with five distinct domains that straddle the DNA forming a “saddle”-shaped structure (Fig. 1C). Two Myb-associated domains (MybADs) bind to contiguous major grooves separated by a full turn on one face of the DNA. Each MybAD consists of four α-helices, αA–αD for mybAD1, and αA′–αD′ for MybAD2. A short linker leads to two Myb repeats (MybRs), with MybR1 sharing the same major groove with MybAD2 resulting in a change of direction of the polypeptide chain, which results in the second Myb repeat (MybR2) to be positioned almost opposite MybAD1 (Fig. 1C). The final C-terminal domain labeled TTD from here on steers away from DNA and consists of five α-helices (α1–α5). This unique domain arrangement covers ∼23 bp of DNA in agreement with previous chemical footprinting studies (39). A distance-matrix alignment (DALI) search did not find similar structures containing the complete quadripartite DNA binding domain (41). The structure shows that, although the MybADs make the majority of the phosphate contacts with the Ter3 site (∼90%), the Myb repeats make most of the base-specific contacts with the central bases. Overall, Reb1 binding to the Ter3 site occupied ∼4,700 Å2 of surface area inducing significant bending of the DNA of ∼56° toward the two MybADs (Fig. 1 D and E).
Fig. 1.
Structure of the Reb1-DNA complex. (A) Diagram showing the S. pombe rRNA nontranscribed spacer region containing three Ter sites and the fork-pausing site RFP4. Reb1 binds to Ter2 and Ter3 sites arresting replication approaching from one direction (red arrows). Transcription is terminated in the opposite direction (blue arrows). Also shown is the sequence of the Ter3 DNA used for crystallization of the complex. (B) Modular domain organization of Reb1FL and Reb1ΔN proteins. DD, dimerization domain (light green); MybAD1, Myb-associated domain 1 (blue); MybAD2, Myb-associated domain2 (light blue); MybR1, Myb repeat 1 (red); MybR2, Myb repeat 2 (dark red); TTD, transcription termination domain (green). Limits of domains are based on the crystal structure. (C) Crystal structure of Reb1ΔN-Ter3 DNA complex in two orientations (rotation of 90° along the indicated axis). Domains are colored according to diagram shown in B. Orientation of DNA in the complex shows that transcription will be approaching from left and replication from right. (D) Surface representation of Reb1ΔN-Ter3 complex rotated by 180° with relative to the view shown in C. (E) Electrostatic potential surface of Reb1ΔN-Ter3 complex. The electropositive DNA binding surface is blue, neutral is white and negatively charged shown in red. DNA is shown as a stick representation.
Table S1.
Data collection and refinement statistics
| Data collection | Native | Peak (Se-SAD) |
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 68.9, 162.9, 71.1 | 68.5, 162.8, 70.8 |
| α, β, γ (°) | 90, 94.7, 90 | 90, 95.2, 90 |
| Wavelength (Å) | 0.9762 | 0.979 |
| Resolution (Å) | 50–2.70 (2.75–2.70) | 50–3.0 (3.05–3.0) |
| No. of measured | 640,762 | 237,990 |
| No. of unique | 42,853 | 31,158 |
| Data coverage (%)* | 99.2 (100) | 99.8 (100.0) |
| Rmerge (%)*,† | 0.059 (0.686) | 0.105 (0.66) |
| I/σ* | 20.8 (2.2) | 18.2 (3.6) |
| Refinement statistics | ||
| Resolution range | 32.5–2.7 | |
| Reflections | 42,149 | |
| Rcryst (%)‡ | 21.0 | |
| Rfree (%)§ | 24.3 | |
| Nonhydrogen atoms | 7,626 | |
| Protein | 5,377 | |
| DNA | 2,120 | |
| Water | 129 | |
| Average B-factors (Å2) | 76.8 | |
| Protein | 83.7 | |
| DNA | 82.1 | |
| Water | 64.5 | |
| RMSD | ||
| Bonds (Å) | 0.003 | |
| Angles (Å) | 0.572 | |
| Ramachandran plot quality | ||
| Most favored (%) | 93.3 | |
| Additional allowed (%) | 5.8 | |
| Generously allowed (%) | 0 | |
| Disallowed (%) | 0 |
Values for the outmost shells are given in parentheses.
Rmerge = Σ|I−<I>|/Σ I, where I is the integrated intensity of a given reflection.
Rcryst = Σ [||Fo|−|Fc|| Σ |Fo|.
For Rfree calculations, 5% of data were excluded from refinement.
Fig. S1.
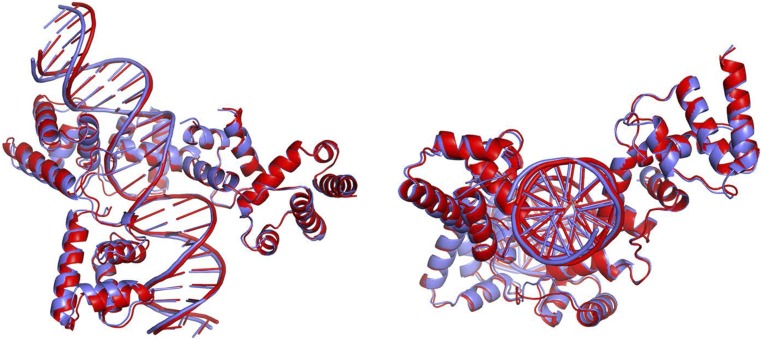
Superposition of the two complexes in the asymmetric unit. Superposition of the two complexes was carried out using the program LSQMAN. Reb1 molecules superimposed with an RMSD of 1.09 Å, whereas the two Ter3 DNA molecules superimpose with an RMSD of 0.95 Å. Complexes A and B are colored blue and red, respectively.
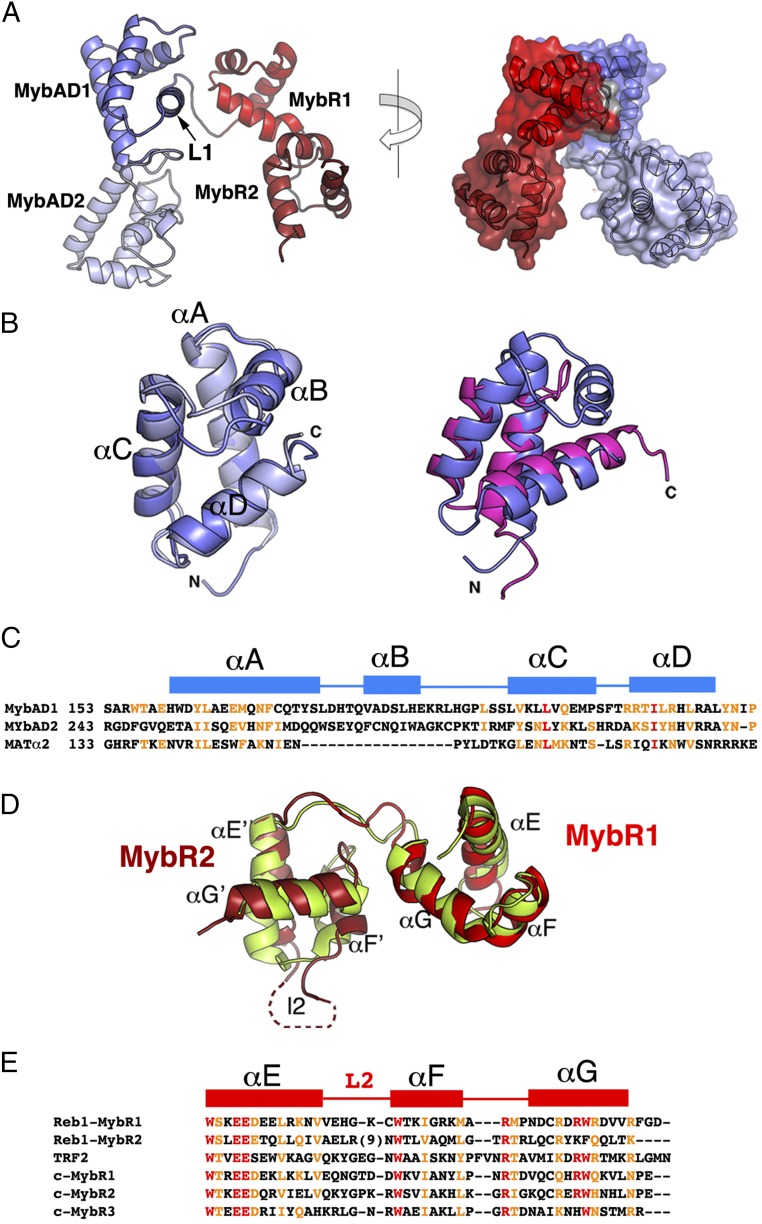
Reb1 MybADs.
The two MybADs are related by a twofold axis of symmetry perpendicular to the DNA axis that reflects their docking positions into the contiguous major grooves (Fig. 2A). They are connected by a 20 residue linker (L1) with a small helical region at its N-terminal end that interacts with the DNA backbone. The second half of the linker forms a hairpin that docks into MybAD2 and interacts with the DNA backbone of the minor groove (Fig. 1C). Surprisingly, although the two MybADs shared an overall sequence identity of only 15%, they superimposed well with an RMSD of 1.36 Å (Fig. 2B). Structural alignment shows that MybAD helices A, C, and D superimpose with the three helices of a typical HTH protein such as MAT-α2, with helix D corresponding to the recognition helix (Fig. 2B, Right). The topology and arrangement of the helices suggested that the MybAD represented a new variation of the HTH proteins containing an insertion of an α-helix between helix1 and helix2 (Fig. 2C).
Fig. 2.
Structure of the quadripartite Reb1 DNA-binding domain. (A) Domain structure of quadripartite Reb1 DBD consisting in MybAD1 (teal), MybAD2 (blue), MybR1 (red), and MyR2 (dark red). The four domains form a U-shaped structure. (Left) Ribbon representation. (Right) Surface rendering. (B) (Left) Superposition of MybAD domains. Although sequence homology is only 15%, the two domains superimposed with an RMSD of 1.36 Å. (Right) Superposition of Reb1 MybAD2 (blue) and MATα2 (magenta) reveals that MybAD αB helix is an addition to the classical HTH motif. (C) Sequence alignment of Reb1 MybADs with MATα2. Sequence coloring highlights regions that are highly conserved (red) and moderately conserved (orange); positions of the α-helices are shown above the sequences as blue bars. (D) Structural alignment of Reb1 Myb repeats (red) and c-Myb repeats (green) represented as ribbon diagrams. MybAD2 loop 2 (l2) is shown as a dotted connection. (E) Sequence alignment of Reb1 Myb repeats with c-Myb repeats and TRF2. Residues in red indicate highly conserved; orange, moderate conserved.
Reb1 Myb Repeats.
Individual Reb1 Myb repeats have a higher sequence homology with the first repeat of c-Myb and superimposed it with an RMSD of 1.8 Å (Fig. 2D). The major difference with classical Myb repeats is found in the second Myb repeat that has an insertion of ∼11 residues in loop l2 connecting helix E′ and helix F′ (Fig. 2 D and E). In addition, it lacks the signature tryptophan residue in the third helix, being replaced instead by phenylalanine. The size of loop l2 is one of the most divergent features among this family of proteins. In S. cerevisiae Reb1, the loop contains 128 residues, whereas in murine TTF-I, it is 25 residues long (Fig. S1).
Recognition and Specificity for Ter3 DNA Site.
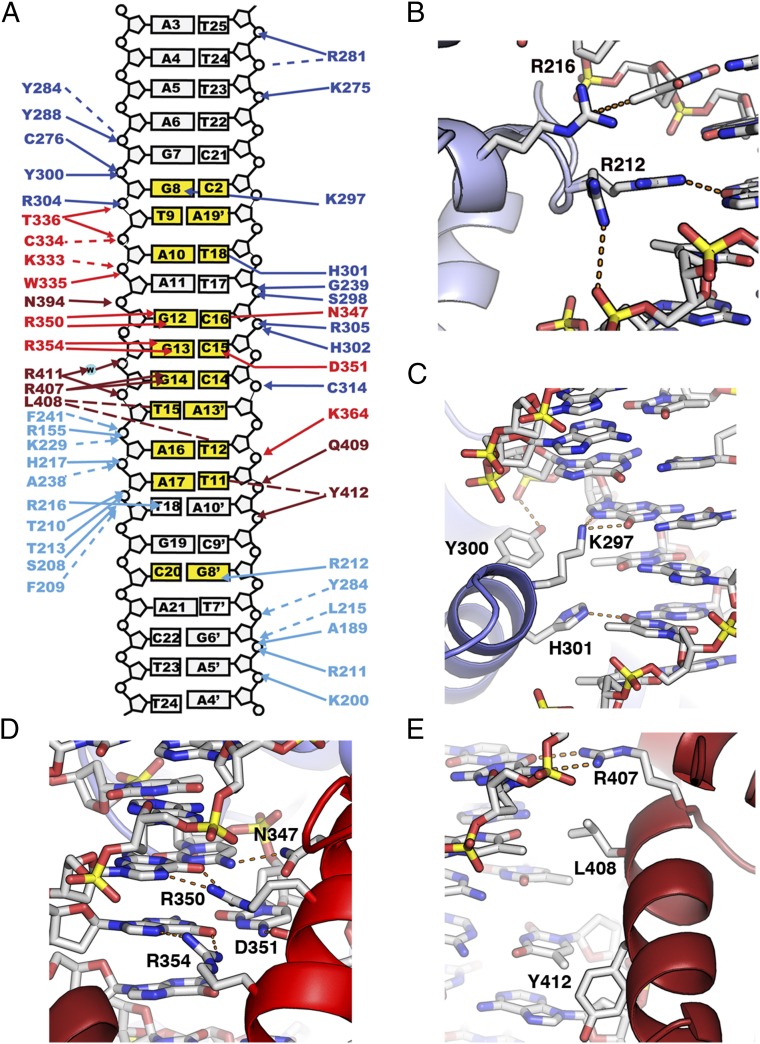
The quadripartite Reb1 DBD shows a unique mode of interaction with DNA in which four domains containing two versions of the helix-turn-helix motif cooperate in binding to a 23-bp stretch of DNA that defines the transcription and replication termination site Ter3 (Fig. 1B). This large DNA footprint appears to be a signature of the Reb1 family of proteins. For instance, the Sal boxes (mammalian equivalent of Ter) are recognized by TTF-I protein and are 18 bp long (26). The specific contacts that SpReb1 makes with the Ter3 site define the recognition sequence as 5′-GTANGGGTAANNC-3′, where the specificity of the central core 5′-GGGTAA-3′ is determined by the Myb repeats, whereas the upstream or downstream flanking sequences are recognized by MybAD2 and MybAD1, respectively.
MybAD-Ter3 Interactions.
The MybAD domains contact one face of the DNA making a large number of interactions with the sugar-phosphate backbone atoms and delineate two contiguous major grooves and three minor grooves over a region of 21 bp (Figs. 1C and 3A). Helices αD and αD′ dock into the major grooves, whereas the linker that connects the MybADs interacts with the minor groove. In addition, the loops between the second and third helices of each MybAD contact the minor grooves at both ends on the Ter3 site with 12 residues interacting with opposite strands of the phosphate backbone atoms, thus stabilizing the docking of MybAD1 into the major groove. The number of protein–phosphate backbone contacts is similar in MybAD2, with 13 residues contacting a DNA region spanning 11 bp. The MybAD domains make few base-specific DNA contacts. In the case of MybAD1, it makes only two specific interactions (underline) downstream of the core recognition sequence 5′-GGGTAANNC-3′, where R216 contacts the methyl group of the paired thymine via van der Waals interactions and R212 hydrogen bonds with N7 of Guanine 8′ (Fig. 3B). The small number of specific contacts is a reflection of the docking mode of the recognition helix that only inserts its C-terminal end into the major groove. MybAD2 specifies the first three bases of the core sequence (underline) GTANGGGTAA with residue K297, making a bidentate interaction with Guanine N7 and the adenine in the opposite strand (Fig. 3C). At the same time, H301 donates a hydrogen bond to the O4 of adenine of the A-T pair. In addition, several residues from the linker connecting the MybADs interact with the minor groove backbone.
Fig. 3.
Reb1-Ter3 interactions. (A) Contacts are colored according to the different domains: MybAD1, teal; MybAD2, blue; MybR1, red; MybR2, orange. Bases directly contacted are colored yellow. Dotted lines depict hydrophobic interactions. (B) Close-up view of MybAD1-Ter3 interaction. R216 is shown making a hydrophobic interaction with the methyl group of thymine18. R212 is shown in two conformations, one is making interaction a phosphate group and the second makes hydrogen bonds with Guanine8’. (C) Details of MybAD2-Ter3 interface showing the residues involved in DNA recognition. K297 makes a bidentate interaction with Guanine8, whereas H301 interacts with thymine18’. Also shown is Tyr-300 that interacts with the phosphate backbone. (D) MybR1 sequence-specific interaction of Ter3 DNA. R350 makes bidentate hydrogen bonds with Guanine12, whereas R354 interacts in a similar way with Guanine13. Paired cytosines at positions 16’ and 15’ from the opposite strand make interactions with N347 and D351, respectively. (E) A close-up view of MybR2 recognition helix interacting with the Ter3 site. R407 makes bidentate hydrogen bonds with Guanine14. L408 interacts via hydrophobic interactions with the methyl group of both Thymine15 and Thymine12’.
Myb-Ter3 Interactions.
The Myb repeats specify the central core GGGTAA (underline denotes direct base contacts by Myb repeats) of the Ter3 sequence as shown in Fig. 3D. The first Myb repeat binds two G-C pairs with R350 and R354, making bipartite hydrogen bonds with Guanine12 and Guanine13, respectively, whereas D351 makes a hydrogen bond with Cytosine40 and N347 with Cytosine41 in the complementary strand. MybR1 makes only four contacts with the phosphate backbone. The second Myb repeat (MybR2) specifies the final 4 bp (underline) of the recognition sequence GTAGGGTAA. Residue R407 makes a bidentate contact with Guanine14, whereas L408 makes van der Waals contacts with the methyl group of both Thymine15 and Thymine37 in the opposite strand (Fig. 3E). The final specific contact is made by Y412, which makes van der Waal contacts with the methyl group of Thymine36. The protein–DNA interactions are consistent with base–protein contacts as previously reported by site-directed mutagenesis of Ter3, deletions in Reb1, and chemical footprinting data (39).
Role of Individual Domains in DNA Binding.
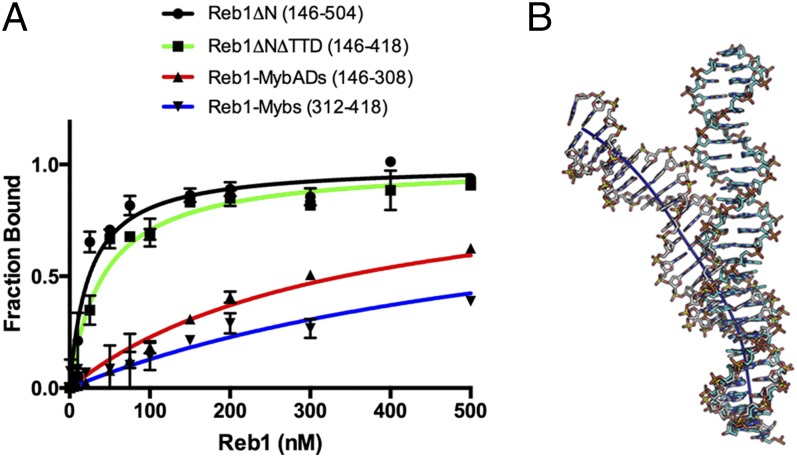
The four HTH domains synergistically bind to the Ter3 site over a region of 23 bp. There is, however, a specialization of function that divides affinity/specificity among the domains. The MybADs make up about 85% of all of the backbone contacts. In contrast, the Myb repeats supply about 80% of all of the base-specific contacts. To determine the contribution of each pair of domains to the DNA binding affinity of the Reb1 protein, we carried out DNA binding assays of Reb1ΔN and three other Reb1 constructs namely Reb1ΔN-ΔTTD (146–418), Reb1-MybADs (146–308), and the Reb1-Myb repeats (314–418). The measurements were carried out using fluorescent polarization and are shown in Fig. 4A. Whereas the Reb1ΔN construct binds with an affinity of 30 nM, deletion of the TTD has only a small effect on the binding affinity (42 nM). In contrast, Reb1-MybADs bind with almost fivefold lower affinity (∼200 nM) and the Myb repeats with an affinity close to 1 μM. Our results are in agreement with our previous data showing that any Reb1 construct that is truncated beyond residue 156 is critically impaired in its ability to bind DNA and to promote fork arrest. In addition, the Reb1 DNA-specific contacts are consistent with binding studies on mutated Ter3 sites and DNA interference studies (30).
Fig. 4.
Role of Reb1 domains in DNA binding. (A) Binding isotherms of Reb1ΔN (black circles), Reb1ΔNΔTTD (green squares), Reb1MybAds (red triangles), and Myb repeats (blue triangles). Fluorescein-labeled Ter3 DNA site was titrated with different protein concentrations. Fluorescence anisotropy was read, and the DNA fraction bound was plotted as a function of Reb1 concentration. For the experiments of MybADs and Myb repeats, titration was done up to a concentration of 3 μM to achieve saturation. (B) Superposition of Ter3 DNA from the crystal structure (white sugar backbone) onto an ideal B-DNA Ter3 (blue sugar backbone). Dark blue line represents the calculated DNA axis.
The binding of Reb1 to the Ter3 site induces an overall DNA curvature of 56° as calculated by the program Curves+ (Fig. 4B) (42). The DNA bending angle is directed toward the MybAD domains presumably due to neutralization of negative charges on one face of the complex but not of the opposite one (Fig. 1 A and B). The conformations of the DNA molecules in the two complexes present in the asymmetric unit are essentially the same and superimpose with an RMSD of <1 Å for all atoms, suggesting that the DNA curvature is not produced by crystal contacts. This result is in agreement with previous analysis using the circular permutation method that determined Reb1 induced bending of the Ter3 site (39).
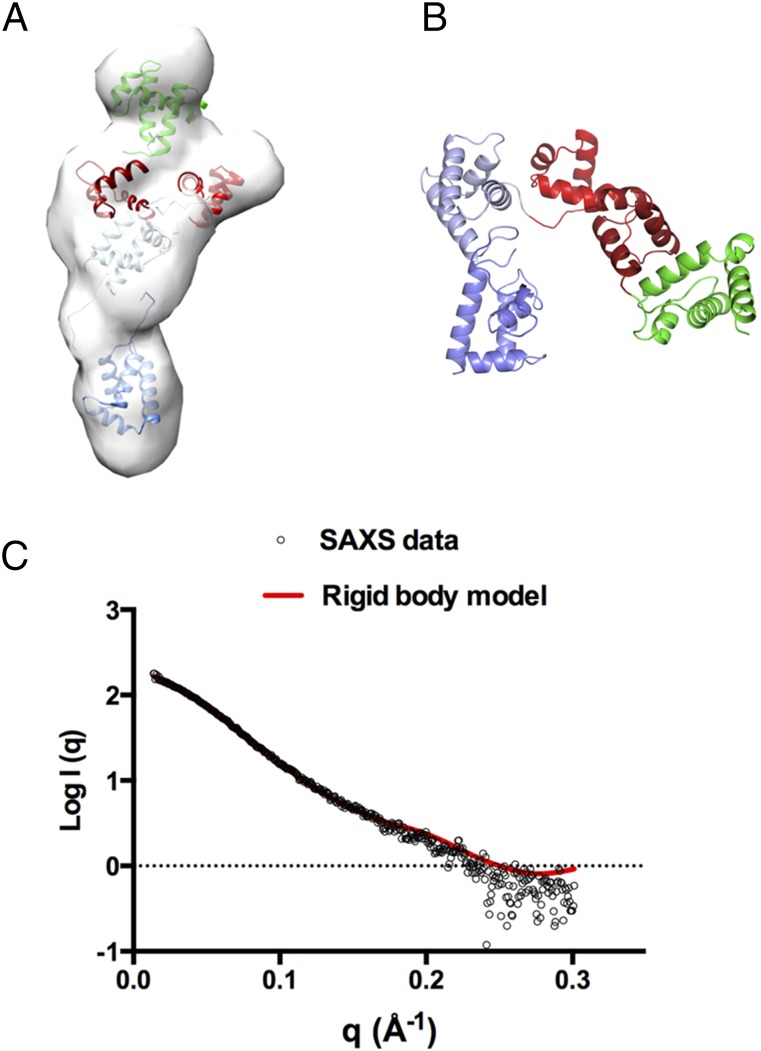
To determine whether the conformation of Reb1 in the complex is also found in the apoprotein, which could not be crystallized without the DNA ligand, we performed small-angle X-ray scattering (SAXS) experiments of Reb1ΔN at different protein concentrations. The experimental parameters including the radius of gyration (Rg) and the maximum particle dimension (Vmax) calculated from experimental scattering profiles suggests that apo-Reb1ΔN is more elongated than the Ter3 bound protein (Table S2). To validate these results, we calculated molecular envelopes using the program DAMMIN (43). The ab initio calculated envelope of Reb1ΔN is very elongated compared with the Reb1 conformation in the complex (Fig. 5 A and B), suggesting that on DNA binding, Reb1 undergoes a large conformational change. We divided the structure into five rigid bodies (Myb-ADs, MybR1, MybR2, and TTD) for modeling with CORAL (44). The best model fits the experimental data with a χ of 0.3 (Fig. 5C) and shows an almost linear structure (Fig. 5A).
Table S2.
Summary of hydrodynamic data
Experimentally determined from Guinier analysis.
Experimentally determined from p(r) analysis by PRIMUS.
Fig. 5.
Reb1 has an extended conformation in absence of DNA. (A) apo-Reb1ΔN ab initio molecular envelope calculated with Dammin with the docked model calculated with the program CORAL. (B) Structure of Reb1ΔN in the bound conformation. (C) Fit of the theoretical scattering profile of the CORAL rigid model shown in A (red), with the experimental scattering profile (circles).
The Reb1 C-Terminal Region Contains a TTD.
The C-terminal domain (TTD; residues 419–504) consists of five α-helices (α1–α5) that form a new fold (Fig. 6A). A DALI search failed to find any structural homologs (41). The domain has three of the helices (α1, α2, and α5) forming a plane, with helix 3 sitting perpendicularly across them (Fig. 6A). The majority of the hydrophobic residues in the TTD are conserved among members of this family, with the highest region of conservation located in α-helix 1 (Fig. S2). The TTD helix 3 wedges loosely against MybR2 helices E′ and G′, burying ∼400 Å2 of accessible surface area. Superposition of the two Reb1 molecules in the asymmetric unit shows a small movement of TTD helix 3 with respect to the core of the protein of ∼2.5°, suggesting a small degree of flexibility.
Fig. 6.
TTD is a transcription termination domain. (A) Ribbon diagrams showing two views of the TTD domain (residues 425–504). (B) Schematic representation of the template and the probes used to measure the extent of transcriptional read through past the Ter site. (C) Transcription run on assay showing that in the absence of the C-terminal region of Reb1 caused significant loss of termination at the Ter site. (D) Bar chart of results shown in C.
Fig. S2.
Structure-based sequence alignment of the eukaryotic transcription and/or replication terminator family. Secondary structure elements are shown above the sequence and color-coded according to Fig. 1 with MybAD1(αA–αD), MybAD2(αA′–αD′), MybR1(α1–α3), MybR2(α4–α6), and TTD(αE–αI). Sequence coloring highlights regions that are highly conserved (red) and moderately conserved (orange). Reb1_Sp, S. pombe Reb1; RTF1_Sp, S. pombe replication termination factor 1; Nsi1_Sc, S. cerevisiae NTS1 silencing protein 1 also known as Ydr026c; Reb1p_Sc, S. cerevisiae Reb1 protein; TTFI_Hs, Homo sapiens transcription termination factor I; TTFI_Mm, Mus musculus transcription termination factor I; Rib2_Xt, X. tropicalis.
To investigate the function of the TTD, we first proceeded to measure transcription termination in the homologous host promoted by the WT Reb1 and its C-terminally truncated form by transcription run-on (TRO) experiments (Fig. 6 B–D). Incomplete rRNA transcripts generated in S. pombe in vivo were further extended in vitro in permeabilized cells in the presence of α[32P]-CTP and α[32P]-UTP. The labeled RNA was hybridized to a set of unlabeled oligonucleotide probes that were slot blotted on to a nitrocellulose membrane, and the intensity of the image of each slot was quantified (Fig. 6 C and D). The data unequivocally showed that transcription was terminated at or very close to the Ter site when full-length Reb1 was present (Fig. 6 A–C). However, a significant proportion of the transcripts passed through the Ter site in cells expressing Reb1 lacking its C terminus (ΔTTD). It should be noted that even in the absence of TTD, there was significant termination of transcription at Ter. We attribute this to the previously reported parallel mechanism of termination caused by transcriptional coprocessing (16, 45, 46). Even in a reb1Δ strain, a similar result was obtained, suggesting that the other domains of Reb1 had minimal contributions to the process excepting perhaps in anchoring the protein to the Ter sequence.
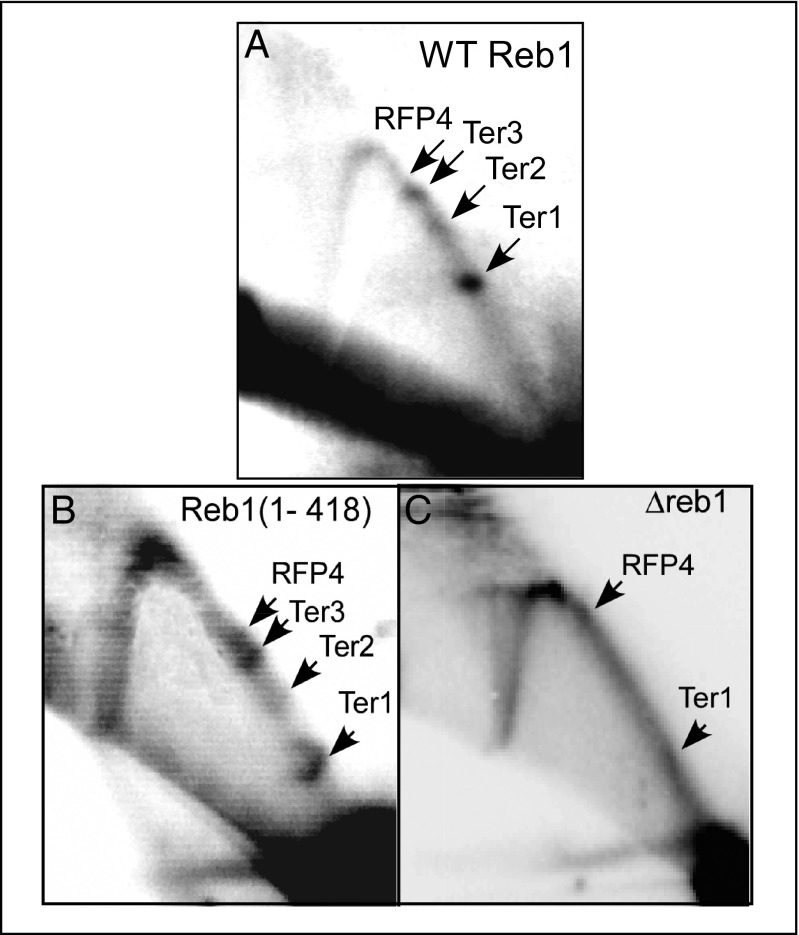
Two pieces of evidence argue against the possibility that deletion of the TTD caused misfolding of the remainder of Reb1. First, the DNA binding affinities of Reb1 with or without the TTD were very similar (Fig. 4A), and second, the protein without the TTD was still capable of arresting replication forks in vivo as revealed by Brewer–Fangman 2D gels (Fig. 7). Thus, the data not only showed that the protein with or without the TTD was biologically functional but also was separable from its replication termination and DNA-binding domain(s) (DBD/RTD). Therefore, the data supported the conclusion that the C-terminal residues 419–504 of Reb1 contained a TTD that is loosely packed against the core of the protein.
Fig. 7.
Brewer–Fangman 2D gels. 2D gels of the (A) WT, (B) truncated Reb1 (1–418) lacking the TTD, and (C) the reb1Δ control showing that deletion of the C-terminal region (419–504) that contains the TTD did not detectably alter the fork arrests at Ter2 and Ter3 that are caused by Reb1 binding.
Reb1 Interacts with RNA pol I Rpa12 Subunit Through the TTD.
How does the Reb1 TTD terminate transcription? Our data suggest an active mechanism promoted by physical interactions between Reb1 TTD and RNA pol I rather than a passive roadblock imposed by the tight binding of Reb1 to Ter DNA sites (7, 47, 48). To determine which subunit(s) of RNA pol I interacted with Reb1, we took advantage of the S. pombe genetic database (www.pombase.org) that reports genetic interactions between Reb1 and other component proteins of RNA pol I. This search revealed Rpa12 as a possible candidate for protein–protein interaction with Reb1. Furthermore, the structurally and functionally related Rpa12 of S. cerevisiae is known to be involved in termination of transcription catalyzed by pol I (20). Although, there are currently no reports of any other genetic or physical interaction between TTD and component proteins of S. pombe RNA pol I, such interactions cannot be precluded at this time.
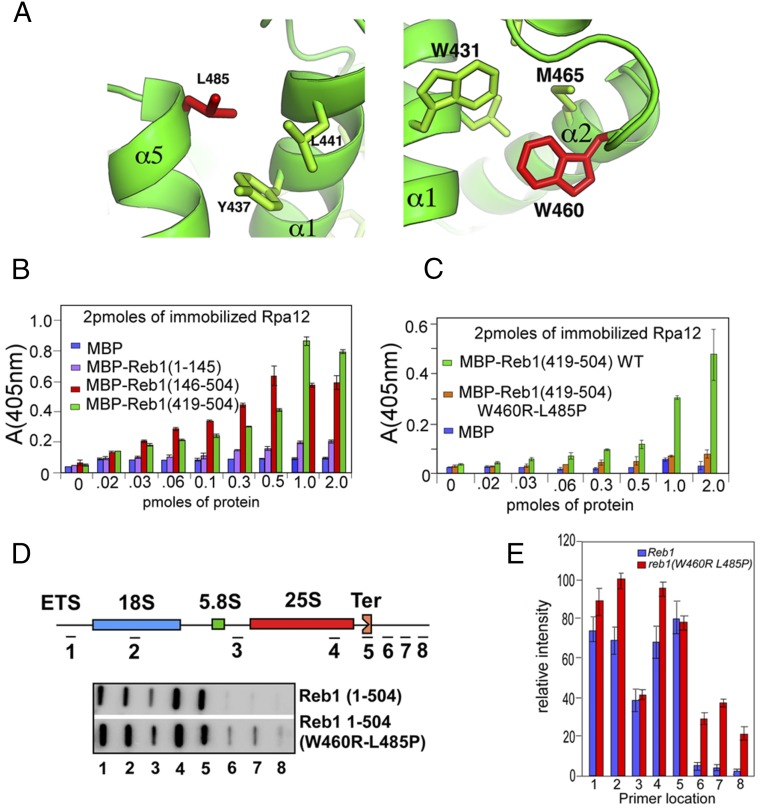
We analyzed the possibility of a physical interaction between Rpa12/Reb1-TTD by using ELISA. We functionally confirmed it further by isolation of mutations that disrupted the protein–protein interaction, using a yeast reverse two-hybrid technique (YR2H; SI Materials and Methods). For technical reasons, we found that evidence of interactions between purified proteins by the ELISA, in this case, was more reliable than the yeast forward two-hybrid procedure. The mutants causing noninteraction between the two proteins were investigated functionally for their ability to terminate transcription by the TRO analyses. Rpa12 immobilized on plastic surfaces was challenged with potential interacting proteins (peptides) and control protein [maltose binding protein (MBP)]. The data showed that both Reb1ΔN and Reb1-TTD interacted with Rpa12 (Fig. 8B). The interaction with the Reb1 N-terminal domain (1–145) was low, and the control MBP of Escherichia coli showed only background levels of interaction (Fig. 8B).
Fig. 8.
Disruption of protein–protein interaction between the TTD and Rpa12 by double point mutations in TTD significantly reduced termination of transcription at Ter. (A) Ribbon diagram of the TTD showing with the location of the L485 (Left) and Trp465 (Right) point mutants that disrupted its interaction with Rpa12. (B) ELISA data showing that TTD and to a lesser extent the N-terminal domain (1–145) of Reb1 interacted with Rpa12. (C) ELISA data showing that Reb1 double mutant failed to interact with Rpa12. (D and E) Representative TRO analysis showing that in the WT Reb1, transcription termination occurred at the Ter site, whereas in the double mutant, a significant amount of transcripts passed through the Ter site.
The authenticity of the interaction was further verified by the reverse yeast two-hybrid (YR2H) method that selected for noninteracting mutants and functional analysis of the mutants as described below. Using the Y2RH selection, we isolated a double mutant W460R and L485P that was efficient in knocking down the protein–protein interaction between Rpa12 and TTD of Sp.Reb1 (Fig. 8C). It should be noted that both W460 and L485 residues are solvent exposed and therefore should be available for protein–protein interactions in vivo (Fig. 8A).
The mutant form of TTD showed significant reduction in interaction with Rpa12 (Fig. 8C and Fig. S3). Subsequent analyses were performed using only the double mutant that was introduced by site-directed mutagenesis into the WT ORF of full-length Reb1. We performed TRO analyses of the WT and the mutant form of Reb1 by comparing and contrasting transcription termination by the TRO assay. The results repeatedly and unequivocally showed that the noninteracting mutant form of Reb1 caused transcriptional read through at the Ter site (Fig. 8 D and E), although a complete deletion of the TTD elicited a stronger read through in comparison with that of the double mutant (Fig. 6 C and D). The data suggested that protein–protein interactions between the TTD with Rpa12 played a critical role in transcription termination by establishing communication between the TTD and the RNA pol I through this portal for protein–protein interaction.
Fig. S3.
Rpa12 interaction with WT and mutant form of 419–504 Reb1. Y2H data (β-galactosidase activities) showing that single mutants, W460R and L485P of 419–504 reb1, mostly retain their ability to interact with Rpa12 (>80%); in contrast, double mutant W460R-L485P has greatly reduced (just above background level) interaction with Rpa12.
SI Materials and Methods
Y2H and YR2H Analysis.
Y2H interactions were carried out using the yeast strain PJ69-4A as described previously (65). WT genes were cloned in pGAD424 and pGBT9 vectors. The plasmids were transformed in pairs into PJ69-4A and colonies containing the plasmid pair were patched on SD/Leu−Trp− and SD/Leu−Trp−Ade− plates. Colonies that grew on SD/Leu−Trp−Ade− were considered putative positive interactors and their counterparts from SD/Leu−Trp− plates were analyzed further by β-galactosidase assays as described in the Clontech manual. YR2H selections were carried out by mutagenizing one of the ORFs (e.g., 419–504Reb1) of an interacting pair by low fidelity PCR. Each mutagenesis reaction was carried out by vent exo− DNA polymerase in four reaction tubes containing three deoxynucleotide triphosphates (dNTPs) at 0.25 mM each and one dNTP at 0.0625 mM. After PCR amplification four PCR products were mixed to generate a pool of mutants. Yeast competent cells containing pGBT9Rpa12 were transformed with mutagenized 419–504Reb1 PCR products along with linearized pGAD424 vector for in vivo recombination and circularization. The transformed cells were diluted and plated on SD/Leu−Trp−. Colonies that grew on the SD/Leu−Trp− but not on the SD/Leu−Trp−Ade− were expected to contain either a putative noninteracting mutant of 419–504Reb1 or a blank vector that circularized without an insert. Extraction of plasmid followed by PCR amplification and sequencing was done to confirm whether the pGAD424 plasmid contained the correct 419-504Reb1 ORF with potential mutant.
Expression and Purification of Reb1.
Reb1ΔN(146–504) cDNA fragment was generated using the PCR as described previously (55). In short, protein was expressed at 18 °C and purified using Sp-Sepharose and Ni-NTA affinity column. The N-terminal His6 tag was proteolytically cleaved using human α-thrombin. The concentrated protein was purified on a HiLoad 16/60 Superdex200 prep grade (GE Healthcare) column pre-equilibrated with size exclusion buffer [20 mM Tris⋅HCl, pH 7.5, 200 mM NaCl, 5 mM tris(2-carboxyethyl)phosphine (TCEP), 1 mM EDTA, 10% (vol/vol) glycerol]. The fractions with target protein were pooled, and protein was spin concentrated to ∼5 mg/mL and stored at −80 °C.
Fluorescence Anisotropy DNA Binding Assay.
Binding assays were performed using 5 nM fluorescein-labeled 26-mer Ter3 DNA. Reb1 constructs at different concentrations were mixed with DNA at a final volume of 300 µL using the following buffer: 25 mM Hepes, pH 7.0, 100 mM NaCl, and 1 mM TCEP. Fluorescence readings were taken on a PC1 fluorimeter (ISS) with excitation and emission filters at 490 and 520 nm. Tubes were equilibrated at 20 °C for 20 min before measurement. Each anisotropy point is the average of 10 measurements. Anisotropy was calculated as the ratio of the difference between vertical and horizontal emission intensities over the total normalized intensity. The fraction of DNA bound (B) was calculated using
| [S1] |
where [A]x represents the anisotropy measured at protein concentration X, [A]DNA is the anisotropy of free fluorescence DNA, and [A]FINAL is the anisotropy at saturation. Data were fit to a single binding site model using the program Origin (Origin Labs). Each experiment was done in triplicate. Titrations for MybADs and Myb repeats proteins were carried out to a concentration of 3 µM to reach saturation.
2D Gel Analysis of Replication Intermediates.
Preparation and separation of replication intermediates by 2D gel electrophoresis were performed following modifications of published procedures (64). Exponentially growing yeast cell (3.5 × 107 cells/mL) were lysed in 12 mL sorbitol (0.9 M) and EDTA (0.1 M) with 200 µL lyticase, stock solution (5,000 U/mL), and 14 mM β mercaptoethanol, and incubated at 30 °C for 45 min. Lysed cell pellets were suspended in 15 mL buffer I (800 mM guanidine HCl, 50 mM Tris·HCl, pH 8.0, 30 mM EDTA, pH 8.0, 4% Tween-20, and 1% Triton X-100) with RNase A (400 µg/mL), and mix by inverting the tube several times. The lysate was treated with proteinase K, incubated at 50 °C for 45 min, and centrifuged at 5,000 × g for 10 min at 4 °C. The supernatant was equilibrated with buffer II (750 mM NaCl, 50 mM Mops, pH 7.0, 15% ethanol, and 0.15% Triton X-100, pH 7.0), loaded on pre-equilibrated QIAGEN column (500/G) and allowed to pass through by gravity flow. The column was washed with 2 × 15 mL of buffer III (1.0 M NaCl, 50 mM Mops, pH 7.0, 15% ethanol) and eluted with prewarmed (50 °C) 15 mL buffer IV (1.25 M NaCl, 50 mM Tris·HCl, pH 8.5, 15% ethanol). The eluent was then precipitated with equal volume of iso- propanol, and the DNA pellet was washed with 70% ethanol. Genomic DNA intermediates were digested with BamHI and resolved on 2D agarose gel electrophoresis.
TRO Analysis.
Transcription run on assay was carried out with a modification of published procedure (19). Cells (2–4 × 107 cells/mL) were harvested by centrifugation at 2,000 × g for 10 min at 4 °C. They were washed once with water and then with 15 mL ice-cold TMN buffer (10 mM Tris⋅HCl, pH 7.5, 5 mM MgCl2, and 100 mM NaCl) and centrifuged at 3,000 × g for 5 min at 4 °C. Cell pellets were resuspeded in 950 µL ice-cold double distilled H2O, and 50 µL 10% N-lauroylsarcosine was added. Tubes were placed on a roller wheel at 4 °C and gently mixed for 30 min. After 30 min of permeabilization, cells were centrifuged for 1 min at 12,000 × g, and supernatant was removed. Cell pellets were resuspended in 60 µL ice-cold 2.5× transcription buffer (50 mM Tris⋅HCl, pH 8.0, 500 mM KCl, 80 mM MgCl2, 2 mM DDT), 1.5 µL ice-cold 200 mM DTT, and 32.5 µL ice-cold 4.5× 32P-NTP premix (2.3 mM ATP, GTP, 1 µM α-32P-UTP, 1 µM α-32P-CTP), and incubated to a 30 °C water bath for 10 min. Reaction was stopped by adding 950 µL TMN buffer and spinning at 3,000 × g for 1 min to remove all residual supernatant. RNA extraction was done using hot phenol (equilibrated at pH 5.2), chloroform and ethanol precipitation, and was dissolved in 80 µL DEPC-treated water. DNA fragments of 10 pmol each (PCR fragment of primer 1002/1004, 1006/1008, 1014/1016, 1018/1020, 1022/1024, 1030/1032, 1034/1036, 1038/1040) used for detection of TRO transcripts were denatured by adding 1 µL 1 M NaoH and incubated for 5 min at 37 °C. Probes were immobilized on a NytranTM SPC membrane (GE Healthcare) using a slot blot apparatus (Hoofer) and UV cross-linked. The membrane was transferred in a hybridization bottle with the addition of 10 mL hybridization buffer [50% formamide (vol/vol), 5× SSC, 5× Denhardt’s solution, 0.2% SDS (wt/vol), 5.0 mM EDTA, and SS DNA (0.5 mg/mL)]. Radiolabeled RNA was partially hydrolyzed by adding 10 µL 1 M NaOH and incubated on ice for 5 min.
Immediately, 6 µL neutralization solution (1.36 M sodium acetate, pH 5.2, 0.77 M glacial acetic acid) was added and mixed. RNA was added to the hybridization solution, and membranes were incubated overnight at 42 °C. After washing two times with washing solution I (2× SSC, 0.5% SDS) and one time with washing solution II (0.5× SSC, 0.05% SDS), autoradiographs were visualized using phosphoimager and quantified using ImageJ software (National Institutes of Health).
Purification of a Different Fragment of Reb1 and pol I Subunit Rpa12.
The 1–145AA, 146–504AA, 419–504AA, and 419–504AA (W460R-L485P) pieces of Reb1 were cloned in pMAL C kinase vector as an MBP fusion protein and Rpa12 in pGEX2TK vector as a GST fusion protein. Briefly, 10 mL overnight culture was inoculated in 1 L Luria–Bertani (LB) medium + amp and grown at OD600 of 0.5. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added at a final concentration of 0.5 mM and incubated for another 3 h. Cells were harvested at 4,000 × g for 10 min and resuspended in 25 mL osmotic buffer (50 mM Tris, pH 8.0, 20% sucrose, 1 mM EDTA, 200 mM Nacl, 10 mM DTT, 1 mM PMSF, and complete PI). After doing freeze-thaw cycles thrice, samples were sonicated in short pulses of 15 s or less (total 2 min) and centrifuged at 30,000 × g for 30 min. Supernatants were incubated with amylose resin (For MBP fusion protein) or glutathione Sepharose 4B (For GST fusion protein), washed in five column volume (CV) of column buffer (50 mM Tris, pH 8.0, 1 mM EDTA, 200 mM Nacl, 10 mM DTT, 1 mM PMSF, and complete PI), washed in another five CV of column buffer with 500 mM NaCl, and eluted with column buffer + 10 mM maltose (For MBP fusion protein) and column buffer + glutathione reduced (184 mg glutathione reduced in 12 mL column buffer; for GST fusion protein).
Protein–Protein Interaction by ELISA.
S. pombe RNA pol I subunit Rpa12 (2 pmol) was adsorbed onto a plastic microtiter plate, washed, blocked, and overlaid separately with a range (0–2 pmol) of MBP, 1–145Reb1-MBP, 146–504Reb1-MBP, 419–504Reb1-MBP, and 419–504(W460R-L485P)Reb1-MBP and allowed to interact with the immobilized Rpa12. The binding was quantified by adding mouse anti-MBP IgG, anti-mouse IgG alkaline phosphatase antibody, and the absorbance was measured at 405 nm with a plate reader.
Discussion
In this work, we presented the first structure, to our knowledge, of a eukaryotic replication and transcription terminator protein and determined its functional architecture and the protein–protein interaction involved in its communication with RNA pol I, which mediates transcription termination. We will discuss first the structural aspects of the terminator protein followed by a delineation of the protein–protein interaction with the Rpa12 subunit of RNA pol I and its biological significance.
Structure of Reb1 Defines the Prototypic Architecture of a Eukaryotic Transcription/Replication Termination Family.
The occurrence of a quadripartite, composite DBD (two MybADs, two MybRs) and a TTD appears to be a signature of multifunctional eukaryotic proteins involved in termination of transcription of RNA pol I and of replication termination in rRNA genes. Structural alignment of Reb1 with other terminator proteins such as S. pombe Rtf1, S. cerevisiae Reb1p, S. cerevisiae Nsi1 (Ytt1), Xenopus tropicalis Rib2, and mammalian TTF-I is shown in Fig. S2. Secondary structure prediction of the protein sequences was also calculated, confirming that most of the α-helices found in Reb1 appear to be conserved in all other family members. Sequence conservation is high in the MybR and lower in the MybAD domains, with MybAD1 having the lowest sequence conservation. However, even between the two Reb1 MybADs, the sequence identity is only 15%.
Reb1 shows an unexpected arrangement of four HTH motifs that form a composite DNA binding domain. This composite DBD arrangement appears to be conserved in all of the different members of the terminator protein family. Secondary structure prediction and domain mapping studies of TTF-I and Rtf1 showed that protein constructs with impaired DNA binding properties have deletions in regions that we now predict to have the first MybAD (27, 38). The alignment shows structural differences among family members in the composite DBD. One occurs in the linker region between MybAD1 and MybAD2, with almost 30 additional residues in the case of Sc.Reb1. The second is found as a large loop between the first and second helices of MybR2. The size of the loop varies from 12 residues in Sp.Reb1 to 128 in Sc.Reb1. The presence of this loop in the second Myb repeat is one of the signatures of this family of terminator proteins. Mammalian TTF-I also has an insertion in the TTD domain between α2 and α3. These differences represent regions that could be involved in protein–protein interactions or protein–DNA interactions that are species specific.
The DNA Terminator Sequence Motif Is Modular and Is Conserved Across Species.
The quadripartite DBD interaction with DNA delineates a modular, composite termination site that can be divided into three sections (Fig. S4). The central section (box 2) with sequence 5′-GGGTNN-3′ is recognized primarily by the two Myb repeats with minor contributions from MybAD1 (Fig. 3A and Fig. S4). The sequence of this element is highly conserved across species from yeast to human and is characterized by the GGG triplet. The sequences found in the upstream box 1 and downstream box 3 are highly diverse among species and are recognized by MybAD2 and MybAD1, respectively. The lack of sequence conservation in these regions reflects the fact that the MybADs make few base-specific contacts with DNA. The quadripartite nature of the DBD and different binding modes used by the MybAD and Myb domains make the composite DBD flexible enough to accommodate large variations in the sequence of their terminator binding sites. For instance, TTF-I binds to a series of 10 termination sites found at the downstream end of the rRNA transcriptional unit that is needed for transcription termination (26). These termination sites (T1–T10) have the conserved Sal box (5′-AGGTCGACCAG-3′; underlined nucleotides are contacted by myb repeats) with adenine instead of guanine in the first position. This core sequence is followed by a 3-bp spacer and the 5′-TCCG-3′ sequence. The distance between the TCCG sequence and the Sal box suggests that the first MybAD domain in TTF-I may make additional interactions with the TCCG sequences.
Fig. S4.
DNA terminator sequence motif across species. Alignment of the different terminator DNA sequences recognize by Terminator proteins. Shown is from top to bottom: Ter3 from S. pombe recognize by Reb1; Ter from S. cerevisiae recognized by Reb1p; REP4 from S. pombe recognize by RTF1; T0 and T1 from M. musculus recognized by murine TTF-I and T1 from H. sapiens recognized by TTF-I. Binding sites can be divided in three modules that interact with MybAD2 (box 1), MybRepeats (box 2), and MybAD1 (box 3).
Our results showed that binding of Reb1 to the Ter3 site induces a large bend in the DNA, which confirms our previous results that were derived from the measurements of the mobility of circularly permuted linear DNA containing a Ter3 bound to purified Reb1 protein (39). The bend appears to be conserved in this family of terminator proteins on binding to their cognate sites (Fig. 4B). Indeed, a previous report showed that binding of TTF-I induces bending of the Sal box motif of about 40°, although no structural data corroborating this observation are available for any other member of this family (49). Moreover, it was reported that S. cerevisiae Reb1p also induces a strong bend in the DNA (50). The functional significance of DNA bending is not clear, but the latter report suggests that it may be involved in nucleosome positioning. In S. pombe, two Ter sites are separated by ∼157 bp of DNA, and it is not clear whether the local DNA bending would be enough to bring the two sites together. We previously reported that all point mutations and deletions of a Ter site that reduce Reb1-induced DNA bending are also defective in fork arrest, an observation that suggests its possible functional significance, namely promotion of optimal DNA–protein interaction (39).
Role of Reb1 in Transcription Termination.
What are the structural determinants that cause the polarity of transcriptional arrest by Reb1? Our results suggest that the relative stereo-chemical orientation of the TTD with respect to the DBD could generate polarity by making the domain accessible to interaction with the transcription apparatus in one orientation of the Ter-Reb1 complex but not in the opposite one. Another possibility is that a particular orientation is required during the higher-order interaction between the promoter and the termination sites. Experiments are in progress to test these hypotheses.
This work showed that pol I transcription termination is not caused by a passive Reb1-Ter barrier but instead suggests that interaction between Reb1-TTD and Rpa12 is important for this function. This conclusion is also consistent with recent data showing that binding of S. cerevisiae terminator Nsi1 (also known as Ytt1) to its DNA binding site is sufficient to terminate RNA pol I transcription (28). However, at this time, we do not know the details of exactly how the TTD-Rpa12 interaction causes transcription termination. However, on the basis of the known biochemical functions of Rpa12 of S. cerevisiae and of other proteins, we suggest the following potential mechanisms. It is known that Rpa12 and its functional homologs Rpb9, TFIIS (both specific for pol II), and Rpc11 of pol III promote cleavage of the nascent transcript and that the C termini of Rpa12 and TFIIS are part of the catalytic center of RNA pol I and pol II (51). In the case of pol II, TFIIS stimulates endonuclease activity when the polymerase encounters an obstacle that causes the enzyme to backtrack (52–54). We speculate that the interactions of Reb1 with Rpa12 induces an allosteric conformational change that leads to the suppression of the endonuclease and/or the polymerase activity. Prevention of backtracking and trapping the enzyme at the U-rich release element could destabilize the RNA–DNA hybrid, causing enzyme and transcript release.
In summary, our present work provides the structure of a prototypical eukaryotic transcription and replication terminator protein at high resolution and identifies its functional domains. It also clearly identifies protein–protein interaction between Rpa12 and Reb1 to be an integral part of the mechanism of termination of transcription catalyzed by RNA pol I. Furthermore, it provides us with hypotheses that can be tested in the immediate future to further unravel the mechanistic details of the functions of Reb1.
Materials and Methods
Strains and Plasmids.
A complete list of strains, plasmids, and oligos is shown in Tables S3 and S4.
Table S3.
Yeast strains and plasmids used in this study
| Yeast strains and plasmids | Genotype | Source |
| S. pombe strains | ||
| SP976 | h+ ade6-M210 Ura4-D18 leu1-32 | (38) |
| SPGK100 | h+ ade6-M210 Ura4-D18 leu1-32 reb1Δ:: kanMX | Lab stock |
| S. cerevisiae | ||
| PJ69-4A | MATa trp1-Δ901 leu2-3,112 901 ura3-52 his3-Δ200 gal4Δ gal8Δ | (65) |
| GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | ||
| Plasmids | ||
| pREP41-FReb1 pREP41-146- | Full-length Reb1 cloned in pREP41 vector | This study |
| 504Reb1 pREP41- | 146–504 Reb1 cloned in pREP41 vector | This study |
| Reb1Δ418 pREP41- | 1–418 Reb1 cloned in pREP41 vector | This study |
| FReb1 | Full length Reb1(W 460R-L485P) cloned in pREP41 vector | This study |
| pGBT9-FReb1 | Full length Reb1 cloned in pGBT9 vector | This study |
| pGBT9-146–504Reb1 | 146–504 Reb1 cloned in pGBT9 vector | This study |
| pGBT9-1–145Reb1 | 1–145 Reb1 cloned in pGBT9 vector | This study |
| pGAD-146–504Reb1 | 146–504 Reb1 cloned in pGAD424 vector | This study |
| pGAD-419–504Reb1 | 419–504 Reb1 cloned in pGAD424 vector | This study |
| pGAD-W460R-L485PReb1 | 419–504 Reb1(W460R- L485P) cloned in pGAD424 vector | This study |
| pGBKT7Rpa12 | Rpa12 cloned in pGBKT7 vector | This study |
| pMAL C Kin 1–145Reb1 | 1–145 Reb1 cloned in pMAL C kinase vector | This study |
| pMAL C Kin 146–504Reb1 | 146–504 Reb1 cloned in pMAL C kinase vector | This study |
| pMAL C Kin 419–504Reb1 | 419–504 Reb1 cloned in pMAL C kinase vector | This study |
| Reb1 (W460R- L485P) | 419–504 Reb1(W460R-L485P) cloned in pMAL | This study |
| pGEX2TK Rpa12 | Rpa12 cloned in pGEX2TK vector | This study |
Table S4.
Oligonucleotides used in this study
| Oligos | Sequence (5′–3′) |
| 1002 | GTTGGCATCGCGGTGCATTGG3 |
| 1004 | CCCCTTCTTCTCGTTCAACC |
| 1006 | CAACTACTTGGATAACCGTGG |
| 1008 | CGAAAAGTTATTATGACTCACC |
| 1014 | CAGCAACGGATCTCTTGGCTC |
| 1016 | CTCAAACAGGCATGCCTTTGG |
| 1018 | GAAGACTGAA GTGGGGAAAGG |
| 1020 | GTTGATTCCACCTTCTGGTTCC |
| 1022 | GGTAGTAGTAGTAGTAGAAG |
| 1024 | GACAATTCCAACTCACCAAC |
| 1030 | GAATGAATAACAGTTTTACAAAC |
| 1032 | 5′GTTTATCTTGTTTCTGCTTTATTC |
| 1034 | CATACATATAGTAGTGTAG |
| 1036 | CCATTTCCATCC GTTGTTTGC |
| 1038 | GGTCAGGTATCGTGGCTGG |
| 1040 | CCTATCGAATTTCGTAAAG |
| 146Reb1F | GGCGGGGTCGACGGATGGATCCATTTTTGAAAGGTAGTGC |
| Reb1R | GGCGGGCTGCAGTCAATTGGAGAATCCAGAAAGTA CCAC |
| 419REB1F | GGCGGGGGATCCGGATGAAGGCTGCTTCAAAATTTGAACTTCAAG |
| S.PRPA12F | GGCGGGGGATCCGGATGTC GGCAATAGGGTCTTTGATATTTTGC |
| S.PRPA12R | GGCGGGGTCGACCTAATTGTTTGTACTAAAT TTATAGCAC |
| pMALRPA12F | GGCGGGGGATC CATGTCGGCAATAGGGTCTTTGATATTTTGC |
| pMALRPA12R | GGCGGGGAATTCCTAATTGTTTGTACTAAATTTATAAGCACATC |
| pMALReb1F | GGCGGGGAATTCATGGATACCTCCGTATTAAATCCGG |
| pMAL146Reb1F | GGCGGGGAATTCATGGATCCATTTTTGAAAGGTAGTGC |
| pMAL419Reb1F | GGCGGGGAATTCATGAAGGCTGCTTCAAAATTTGAACTTC |
| pMAL418Reb1R | GGCGGGGTCGACCGTCAATTGCTGAAATTTATAACGAC |
| pMALReb1R | GGCGGGGTCGACATTGGAGAATCCAGAAAGTACCAC |
| pMAL145Reb1R | GGCGGGGTCGACATTGGTAATTTCCGAAGGGC |
Expression, Purification, and Crystallization of Reb1ΔN.
The N-terminally truncated Reb1 146–504 cDNA fragment was generated using PCR and subcloned in pET-15b vector (Novagen). Details of the methods for protein purification and crystallization can be found in SI Materials and Methods and previously published information (55).
Data Collection and Structure Determination.
Seleno-methionine SAD data were collected at beam line X6a (Brookhaven National Laboratories). Data were indexed, integrated, and scaled with the program HKL2000 (56). Phases were determined using the program SHELX (57). Refinement, density modification, and model building were done using the program package PHENIX (58). Manual model rebuilding was performed with the program COOT (59). The quality of the model was checked with the program PROCHECK (60). Figures representing atomic models were generated using PYMOL (61). Further details are presented in SI Materials and Methods. Analysis of the buried surface area (BSA) was calculated using the following equation:
where ASA is the solvent accessible area.
SAXS Measurements.
SAXS data were collected at the SIBYLS beam line in the Lawrence Berkeley National Laboratory and at X9 beam line at Brookhaven National Laboratory. Scattering patterns were collected in triplicate at different concentrations (1–10 mg/mL). The data were analyzed with the program SCATTER, and merged datasets were analyzed using the PRIMUS program (62). Ab initio molecular shapes were calculated using the programs DAMMIN (43) and GASBOR (63). SAXS-based rigid body modeling was carried out using the program CORAL (64).
Brewer–Fangman 2D Agarose Gel Electrophoresis.
Preparation and separation of replication intermediates by 2D gel electrophoresis were performed according to modifications of published procedures (64) (SI Materials and Methods).
Purification of S. pombe Rpa12, Putative Domains of Sp.Reb1, and Protein–Protein Interaction.
Details for Rpa12 and Sp.Reb1 purification and protein–protein interactions are described in SI Materials and Methods.
Y2H and YR2H Analysis.
Y2H interactions were carried out using the yeast strain PJ69-4A as described previously (65). YR2H selection was carried out by mutagenizing the DNA encoding the peptide (419–504 Reb1), testing its interaction with Rpa12, and isolating noninteracting mutants. See SI Materials and Methods for further details.
DNA-Binding Assays.
Fluorescence anisotropy measurements were performed to detect the Reb1–DNA interaction. A detailed description can be found in SI Materials and Methods.
TRO Assay.
This process is described in detail in SI Materials and Methods.
Acknowledgments
We thank Mikhail Kashlev, Joachim Griesenbeck, and Jon Warner for useful strains and valuable discussions. This work was supported by National Institute of General Medical Sciences Grant 5R01GM098013-04 (to D.B.), South Carolina Clinical & Translational Research Institute (SCTR) Grant 1211 (to D.B.), NIH Grants 1R01GM092854 and R21CA179008 (to C.R.E.), and American Cancer Society Institutional Research Grant 11997-IRG-73-001-34-IRG (to C.R.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 5EYB).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1525465113/-/DCSupplemental.
References
- 1.Sollner-Webb B, Tower J. Transcription of cloned eukaryotic ribosomal RNA genes. Annu Rev Biochem. 1986;55:801–830. doi: 10.1146/annurev.bi.55.070186.004101. [DOI] [PubMed] [Google Scholar]
- 2.Clos J, Normann A, Ohrlein A, Grummt I. The core promoter of mouse rDNA consists of two functionally distinct domains. Nucleic Acids Res. 1986;14(19):7581–7595. doi: 10.1093/nar/14.19.7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reeder RH. Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog Nucleic Acid Res Mol Biol. 1999;62:293–327. doi: 10.1016/s0079-6603(08)60511-5. [DOI] [PubMed] [Google Scholar]
- 4.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23(11):1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Németh A, et al. RNA polymerase I termination: Where is the end? Biochim Biophys Acta. 2013;1829(3-4):306–317. doi: 10.1016/j.bbagrm.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Kang JJ, Yokoi TJ, Holland MJ. Binding sites for abundant nuclear factors modulate RNA polymerase I-dependent enhancer function in Saccharomyces cerevisiae. J Biol Chem. 1995;270(48):28723–28732. doi: 10.1074/jbc.270.48.28723. [DOI] [PubMed] [Google Scholar]
- 7.Lang WH, Morrow BE, Ju Q, Warner JR, Reeder RH. A model for transcription termination by RNA polymerase I. Cell. 1994;79(3):527–534. doi: 10.1016/0092-8674(94)90261-5. [DOI] [PubMed] [Google Scholar]
- 8.Zhao A, Guo A, Liu Z, Pape L. Molecular cloning and analysis of Schizosaccharomyces pombe Reb1p: Sequence-specific recognition of two sites in the far upstream rDNA intergenic spacer. Nucleic Acids Res. 1997;25(4):904–910. doi: 10.1093/nar/25.4.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McStay B, Reeder RH. An RNA polymerase I termination site can stimulate the adjacent ribosomal gene promoter by two distinct mechanisms in Xenopus laevis. Genes Dev. 1990;4(7):1240–1251. doi: 10.1101/gad.4.7.1240. [DOI] [PubMed] [Google Scholar]
- 10.Lang WH, Reeder RH. The REB1 site is an essential component of a terminator for RNA polymerase I in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13(1):649–658. doi: 10.1128/mcb.13.1.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Labhart P, Reeder RH. A 12-base-pair sequence is an essential element of the ribosomal gene terminator in Xenopus laevis. Mol Cell Biol. 1987;7(5):1900–1905. doi: 10.1128/mcb.7.5.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kempers-Veenstra AE, et al. -End formation of transcripts from the yeast rRNA operon. EMBO J. 1986;5(10):2703–2710. doi: 10.1002/j.1460-2075.1986.tb04554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson SP, Warner JR. Termination of transcription of ribosomal RNA in Saccharomyces cerevisiae. Mol Cell Biochem. 1991;104(1-2):163–168. doi: 10.1007/BF00229816. [DOI] [PubMed] [Google Scholar]
- 14.La Volpe A, et al. Molecular analysis of the heterogeneity region of the human ribosomal spacer. J Mol Biol. 1985;183(2):213–223. doi: 10.1016/0022-2836(85)90214-1. [DOI] [PubMed] [Google Scholar]
- 15.Jansa P, Grummt I. Mechanism of transcription termination: PTRF interacts with the largest subunit of RNA polymerase I and dissociates paused transcription complexes from yeast and mouse. Mol Gen Genet. 1999;262(3):508–514. doi: 10.1007/s004380051112. [DOI] [PubMed] [Google Scholar]
- 16.Braglia P, Kawauchi J, Proudfoot NJ. Co-transcriptional RNA cleavage provides a failsafe termination mechanism for yeast RNA polymerase I. Nucleic Acids Res. 2011;39(4):1439–1448. doi: 10.1093/nar/gkq894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Németh A, Guibert S, Tiwari VK, Ohlsson R, Längst G. Epigenetic regulation of TTF-I-mediated promoter-terminator interactions of rRNA genes. EMBO J. 2008;27(8):1255–1265. doi: 10.1038/emboj.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rondon AG, Mischo HE, Proudfoot NJ. Terminating transcription in yeast: Whether to be a ‘nerd’ or a ‘rat’. Nat Struct Mol Biol. 2008;15(8):775–776. doi: 10.1038/nsmb0808-775. [DOI] [PubMed] [Google Scholar]
- 19.Kawauchi J, Mischo H, Braglia P, Rondon A, Proudfoot NJ. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev. 2008;22(8):1082–1092. doi: 10.1101/gad.463408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prescott EM, et al. Transcriptional termination by RNA polymerase I requires the small subunit Rpa12p. Proc Natl Acad Sci USA. 2004;101(16):6068–6073. doi: 10.1073/pnas.0401393101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ju QD, Morrow BE, Warner JR. REB1, a yeast DNA-binding protein with many targets, is essential for growth and bears some resemblance to the oncogene myb. Mol Cell Biol. 1990;10(10):5226–5234. doi: 10.1128/mcb.10.10.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrow BE, Ju Q, Warner JR. Purification and characterization of the yeast rDNA binding protein REB1. J Biol Chem. 1990;265(34):20778–20783. [PubMed] [Google Scholar]
- 23.Reiter A, et al. The Reb1-homologue Ydr026c/Nsi1 is required for efficient RNA polymerase I termination in yeast. EMBO J. 2012;31(16):3480–3493. doi: 10.1038/emboj.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ha CW, Sung MK, Huh WK. Nsi1 plays a significant role in the silencing of ribosomal DNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40(11):4892–4903. doi: 10.1093/nar/gks188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McStay B, Reeder RH. A DNA-binding protein is required for termination of transcription by RNA polymerase I in Xenopus laevis. Mol Cell Biol. 1990;10(6):2793–2800. doi: 10.1128/mcb.10.6.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grummt I, Rosenbauer H, Niedermeyer I, Maier U, Ohrlein A. A repeated 18 bp sequence motif in the mouse rDNA spacer mediates binding of a nuclear factor and transcription termination. Cell. 1986;45(6):837–846. doi: 10.1016/0092-8674(86)90558-1. [DOI] [PubMed] [Google Scholar]
- 27.Evers R, Grummt I. Molecular coevolution of mammalian ribosomal gene terminator sequences and the transcription termination factor TTF-I. Proc Natl Acad Sci USA. 1995;92(13):5827–5831. doi: 10.1073/pnas.92.13.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merkl P, et al. Binding of the termination factor Nsi1 to its cognate DNA site is sufficient to terminate RNA polymerase I transcription in vitro and to induce termination in vivo. Mol Cell Biol. 2014;34(20):3817–3827. doi: 10.1128/MCB.00395-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sánchez-Gorostiaga A, López-Estraño C, Krimer DB, Schvartzman JB, Hernández P. Transcription termination factor reb1p causes two replication fork barriers at its cognate sites in fission yeast ribosomal DNA in vivo. Mol Cell Biol. 2004;24(1):398–406. doi: 10.1128/MCB.24.1.398-406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krings G, Bastia D. swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 2004;101(39):14085–14090. doi: 10.1073/pnas.0406037101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh SK, Sabatinos S, Forsburg S, Bastia D. Regulation of replication termination by Reb1 protein-mediated action at a distance. Cell. 2010;142(6):868–878. doi: 10.1016/j.cell.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dominguez M, Ferres-Marco D, Gutierrez-Aviño FJ, Speicher SA, Beneyto M. Growth and specification of the eye are controlled independently by Eyegone and Eyeless in Drosophila melanogaster. Nat Genet. 2004;36(1):31–39. doi: 10.1038/ng1281. [DOI] [PubMed] [Google Scholar]
- 33.Gerber JK, et al. Termination of mammalian rDNA replication: Polar arrest of replication fork movement by transcription termination factor TTF-I. Cell. 1997;90(3):559–567. doi: 10.1016/s0092-8674(00)80515-2. [DOI] [PubMed] [Google Scholar]
- 34.Längst G, Schätz T, Langowski J, Grummt I. Structural analysis of mouse rDNA: Coincidence between nuclease hypersensitive sites, DNA curvature and regulatory elements in the intergenic spacer. Nucleic Acids Res. 1997;25(3):511–517. doi: 10.1093/nar/25.3.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeuchi Y, Horiuchi T, Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 2003;17(12):1497–1506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brewer BJ. When polymerases collide: Replication and the transcriptional organization of the E. coli chromosome. Cell. 1988;53(5):679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- 37.Evers R, Smid A, Rudloff U, Lottspeich F, Grummt I. Different domains of the murine RNA polymerase I-specific termination factor mTTF-I serve distinct functions in transcription termination. EMBO J. 1995;14(6):1248–1256. doi: 10.1002/j.1460-2075.1995.tb07108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eydmann T, et al. Rtf1-mediated eukaryotic site-specific replication termination. Genetics. 2008;180(1):27–39. doi: 10.1534/genetics.108.089243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biswas S, Bastia D. Mechanistic insights into replication termination as revealed by investigations of the Reb1-Ter3 complex of Schizosaccharomyces pombe. Mol Cell Biol. 2008;28(22):6844–6857. doi: 10.1128/MCB.01235-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sander EE, Mason SW, Munz C, Grummt I. The amino-terminal domain of the transcription termination factor TTF-I causes protein oligomerization and inhibition of DNA binding. Nucleic Acids Res. 1996;24(19):3677–3684. doi: 10.1093/nar/24.19.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37(17):5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76(6):2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89(2):1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Braglia P, Heindl K, Schleiffer A, Martinez J, Proudfoot NJ. Role of the RNA/DNA kinase Grc3 in transcription termination by RNA polymerase I. EMBO Rep. 2010;11(10):758–764. doi: 10.1038/embor.2010.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El Hage A, Koper M, Kufel J, Tollervey D. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev. 2008;22(8):1069–1081. doi: 10.1101/gad.463708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reeder RH, Lang W. The mechanism of transcription termination by RNA polymerase I. Mol Microbiol. 1994;12(1):11–15. doi: 10.1111/j.1365-2958.1994.tb00989.x. [DOI] [PubMed] [Google Scholar]
- 48.Lang WH, Reeder RH. Transcription termination of RNA polymerase I due to a T-rich element interacting with Reb1p. Proc Natl Acad Sci USA. 1995;92(21):9781–9785. doi: 10.1073/pnas.92.21.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smid A, Finsterer M, Grummt I. Limited proteolysis unmasks specific DNA-binding of the murine RNA polymerase I-specific transcription termination factor TTFI. J Mol Biol. 1992;227(3):635–647. doi: 10.1016/0022-2836(92)90213-4. [DOI] [PubMed] [Google Scholar]
- 50.Angermayr M, Oechsner U, Bandlow W. Reb1p-dependent DNA bending effects nucleosome positioning and constitutive transcription at the yeast profilin promoter. J Biol Chem. 2003;278(20):17918–17926. doi: 10.1074/jbc.M301806200. [DOI] [PubMed] [Google Scholar]
- 51.Ruan W, Lehmann E, Thomm M, Kostrewa D, Cramer P. Evolution of two modes of intrinsic RNA polymerase transcript cleavage. J Biol Chem. 2011;286(21):18701–18707. doi: 10.1074/jbc.M111.222273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states By translocating back and forth along the DNA and the RNA. J Biol Chem. 1997;272(24):15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 53.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149(7):1438–1445. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471(7337):249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 55.Jaiswal R, Singh SK, Bastia D, Escalante CR. Crystallization and preliminary X-ray characterization of the eukaryotic replication terminator Reb1-Ter DNA complex. Acta Crystallogr F Struct Biol Commun. 2015;71(Pt 4):414–418. doi: 10.1107/S2053230X15004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 57.Sheldrick GM. Experimental phasing with SHELXC/D/E: Combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 59.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 60.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26(2):283–291. [Google Scholar]
- 61.Schrodinger LLC. 2010. The PyMOL Molecular Graphics System, Version 1.3r1.
- 62.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J Appl Cryst. 2003;36(5):1277–1282. [Google Scholar]
- 63.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80(6):2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51(3):463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 65.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144(4):1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]