Significance
This study demonstrates that both primary human basal and luminal epithelial cells are cells of origin for prostate cancer through the use of a prostate organoid culture system. This technology enables the monitoring of early stages of prostate tumorigenesis in vitro and the interrogation of human prostate epithelial populations with synonymous oncogenic stimuli. The combination of c-Myc overexpression and activation of the PI3K/AKT pathway drives high-grade prostate adenocarcinoma in basal cell-derived tumors; however, the same oncogenic stress causes low-grade prostate adenocarcinoma in luminal cell-derived tumors. These findings indicate inherent context-specific and lineage-dependent differences in the response of human prostate epithelial cells to oncogenic stimuli.
Keywords: human prostate cancer, cells of origin, luminal cell, cancer differentiation, oragnoid culture
Abstract
The cell of origin for prostate cancer remains a subject of debate. Genetically engineered mouse models have demonstrated that both basal and luminal cells can serve as cells of origin for prostate cancer. Using a human prostate regeneration and transformation assay, our group previously demonstrated that basal cells can serve as efficient targets for transformation. Recently, a subpopulation of multipotent human luminal cells defined by CD26 expression that retains progenitor activity in a defined organoid culture was identified. We transduced primary human prostate basal and luminal cells with lentiviruses expressing c-Myc and activated AKT1 (myristoylated AKT1 or myrAKT1) to mimic the MYC amplification and PTEN loss commonly detected in human prostate cancer. These cells were propagated in organoid culture before being transplanted into immunodeficient mice. We found that c-Myc/myrAKT1–transduced luminal xenografts exhibited histological features of well-differentiated acinar adenocarcinoma, with strong androgen receptor (AR) and prostate-specific antigen (PSA) expression. In contrast, c-Myc/myrAKT1–transduced basal xenografts were histologically more aggressive, with a loss of acinar structures and low/absent AR and PSA expression. Our findings imply that distinct subtypes of prostate cancer may arise from luminal and basal epithelial cell types subjected to the same oncogenic insults. This study provides a platform for the functional evaluation of oncogenes in basal and luminal epithelial populations of the human prostate. Tumors derived in this fashion with defined genetics can be used in the preclinical development of targeted therapeutics.
The human prostate has two main epithelial cell types, basal and luminal, as well as a minor population of neuroendocrine cells. Primary prostate cancer nearly always has a luminal phenotype characterized by atypical glands, strong androgen receptor (AR) signaling, and an absence of basal cells (1). This histological description suggests that prostate cancer has a luminal cell of origin. Animal studies in genetically engineered mouse models have shown that basal and luminal populations can both serve as cells of origin for prostate cancer (2, 3). Isolation and in vivo transplantation of oncogene-transduced epithelial populations has produced similar results (4, 5). In the human prostate, however, only basal cells have been shown to be efficient targets for transformation (6, 7). In this study, we sought to establish whether human prostate luminal cells could also serve as cells of origin for prostate cancer in an organoid culture assay with enforced oncogene expression.
The development of organotypic culture conditions has greatly aided the study of normal tissue development in diverse epithelial tissues. The use of 3D ex vivo culture systems of purified epithelial cells have made it possible to define stem-like characteristics of cellular subpopulations. Organoid culture has allowed the identification of minimal sets of signaling molecules required for normal growth, self-renewal, and differentiation (8–11).
Along with providing insight into developmental processes, organoid systems also have facilitated studies of carcinogenesis. One distinct advantage of these assays is that they begin with primary benign cells, removing much of the genetic complexity in traditional cell line xenograft assays. Organoid culture has allowed the functional validation of carcinogenic loci identified in genomic studies of pancreatic, gastric, and colon cancers. In one study, APC, TP53, KRAS (G12D), and SMAD4 mutations were shown to be required for progressive transformation to adenocarcinoma-like phenotypes in organoid culture and for tumorigenicity in vivo (9).
Recent work has established organoid culture conditions for mouse and human prostate epithelial cells (12). These conditions allow the continuous propagation of basal cells (CD49fHi) and luminal cells (CD26+). Purified populations of each cell type were cultured separately, but after expansion in vitro, both populations generated mixtures of CK5+ basal cells and CK8+ luminal cells. However, only purified luminal cells were able to generate organoids with glandular architecture. Consistent with previous mouse studies (2, 13), Karthaus et al. (12) postulated the existence of luminal stem/progenitor cells capable of regenerating the normal glandular architecture of the human prostate.
In the present study, we demonstrate that luminal cells can be propagated after oncogene transduction in organoid culture. These transduced cells produce atypical glandular structures when xenografted in immunodeficient mice [NSG; NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (14)]. The structures display strong AR and prostate-specific antigen (PSA) expression, but lack tumor protein p63 (p63) expression. Xenografts derived from c-Myc/AKT1–transduced basal cells showed histological features of poorly differentiated adenocarcinoma, whereas xenografts from luminal cell organoids transduced with the same two oncogenes exhibited features of low-grade prostate adenocarcinoma. Our findings suggest that prostate epithelial lineages respond differentially to the same oncogenic insults to generate distinct types of human prostate cancer.
Results
Establishment of Oncogene-Transduced Human Primary Prostate Basal and Luminal Organoids.
We previously demonstrated that human prostate basal cells are efficient targets for transformation upon the ectopic expression of selected oncogenes (6, 7). In those studies, we transduced primary human basal cells with oncogenes and immediately implanted them s.c. into NSG mice. Overexpression of AR, ERG, and myrAKT1 in the basal cells produced a transformed phenotype of low-grade adenocarcinoma; however, we failed to transform primary human luminal cells (6). We surmised that the different transformation potentials of the cell types in our assay could be either cell-intrinsic or modified by cell-extrinsic environmental cues that preferentially facilitate transformation of basal cells. To address these issues, we adapted a recently described prostate organoid culture system (12) to provide a permissive environment for recovery and growth after the introduction of oncogenes in epithelial populations freshly isolated from primary human prostate tissues.
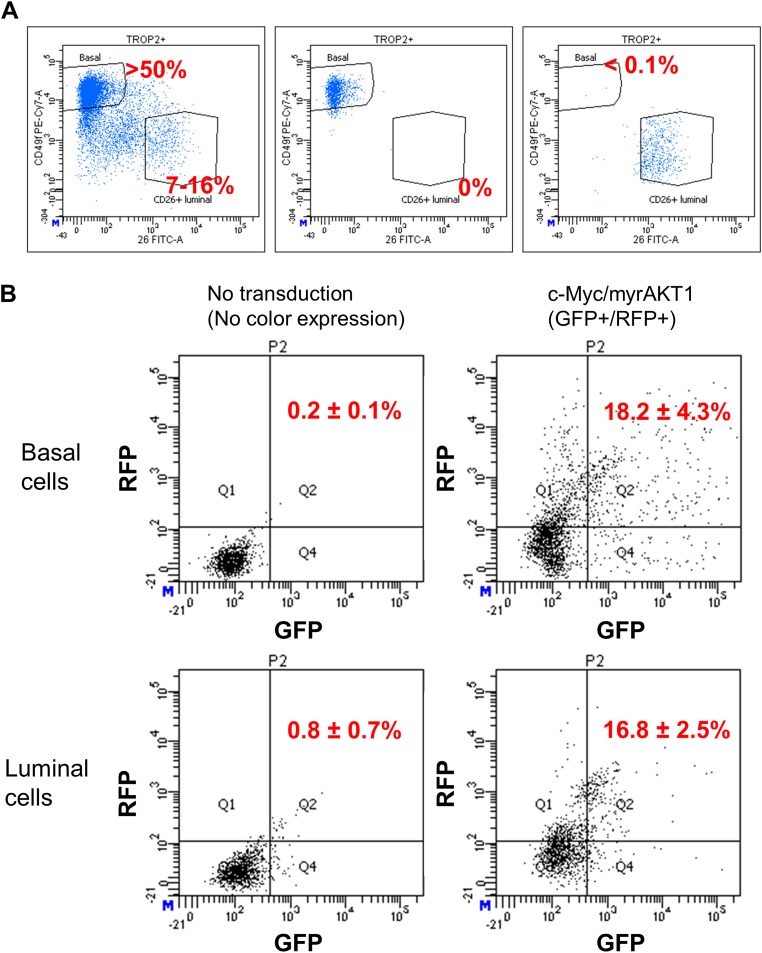
To isolate basal and luminal cells from benign human prostate tissues, we used antibodies targeting the cell surface markers CD49f and CD26, to differentiate the populations by fluorescence-activated cell sorting (FACS). More than 50% of the dissociated epithelial cells from human prostates were CD49fHi basal cells, and only 7–16% were CD26+ luminal cells (Fig. S1A). A postsort analysis of the basal and luminal cell populations was performed to measure cross-contamination from each population. The cross-contamination rate of basal cells in the sorted luminal population was <0.1%. No cross-contamination of luminal cells was detected in the purified basal cell population (0 out of 2,000 total events) (Fig. S1A).
Fig. S1.
Isolation of primary human prostate basal and luminal epithelial cells and assessment of lentiviral transduction efficiency. (A) Flow cytometry analysis for CD49f and CD26 dissociated human prostate tissue as gated during the sort (Left) and postsort of basal (Middle) and luminal (Right) cells before organoid culture. (B) Representative plots of dissociated organoids at 2 d after lentivial transduction cells with no lentiviral transduction were used as a negative control to make a positive gate for GFP/RFP doubly transduced cells.
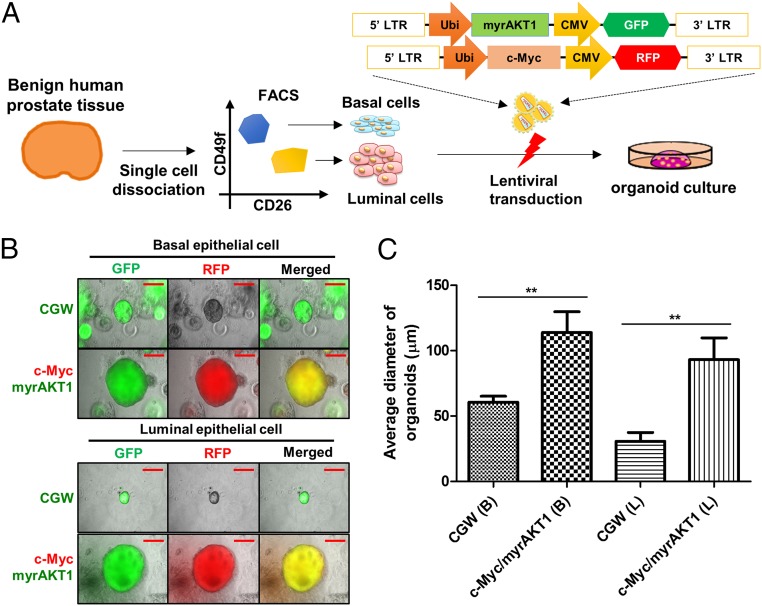
We transduced basal and luminal cells with an empty vector or vectors bearing c-Myc and myrAKT1. This oncogene pair mimics MYC amplification and PTEN loss, two alterations commonly seen in prostate cancer (15–17). PTEN loss in basal and luminal cells drives tumor development in a genetically engineered mouse model (18). We previously showed that c-Myc/myrAKT1 can transform human prostate basal cells to poorly differentiated adenocarcinoma and squamous cell carcinoma in vivo in immune-defective mice (7). After transduction of isolated basal and luminal cells, the populations were propagated separately in organoid culture for 2 wk (Fig. 1A and Fig. S2A).
Fig. 1.
Expression of c-Myc and myrAKT1 induced growth of basal and luminal cells in organoid culture. (A) Schematic of an organoid culture of human primary basal and luminal cells with lentiviral transduction. Ubi, human ubiquitin C promoter; CMV, cytomegalovirus promoter; LTR, long terminal repeat. (B) Representative images of c-Myc and myrAKT1–transduced basal or luminal organoids. CGW, empty vector. (Scale bar, 50 μm.) (C) Average diameter of c-Myc/myrAKT1–transduced basal and luminal organoids. B, basal cell; L, luminal cell. **P < 0.05.
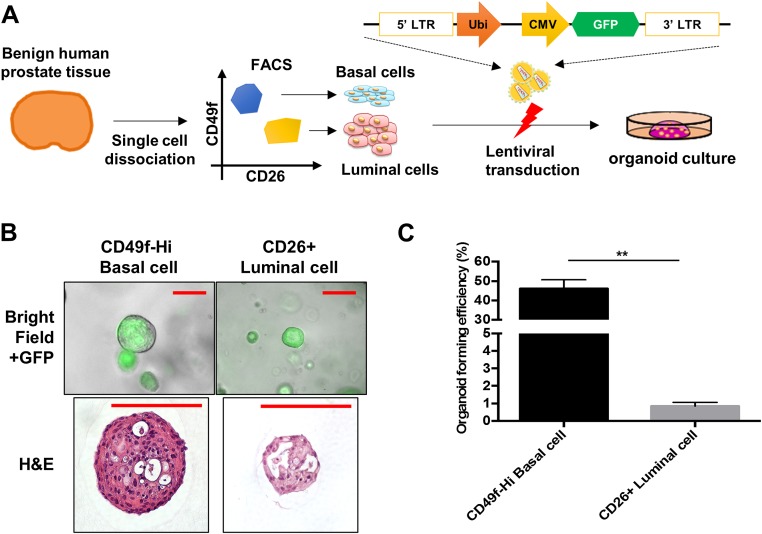
Fig. S2.
Basal cells are more efficient than luminal cells at forming organoids. (A) Schematic of organoid culture of human primary basal and luminal cells lentivirally transduced with empty vector. (B) Representative merged images (bright and GFP fluorescence fields) of organoids of basal and luminal cells. (Scale bar, 50 µm.) (C) Organoid-forming efficiency of basal and luminal cells transduced with empty vector. **P < 0.05.
In the empty vector condition, basal cells grew as solid spherical structures, whereas luminal cells developed gland-like structures with a central lumen (Fig. S2B). Despite starting with the same number of cells on culture initiation, basal cells were approximately 50-fold enriched in organoid-forming capacity compared with luminal cells (461 ± 90 out of 1,000 basal cells and 9 ± 2 out of 1,000 luminal cells; Fig. S2C). This finding is consistent with a previous report (12).
To assess transduction efficiency, we measured green fluorescent protein (GFP) or red fluorescent protein (RFP) as a surrogate marker for oncogene expression, because these are coexpressed from the proviral sequence (Fig. 1A). On average, 18% of the basal organoids and 16% of the luminal organoids were GFP- and RFP-positive at 3 d after transduction (Fig. S1B). After 2 wk in culture, the diameter of the double-transduced basal or luminal organoids was significantly larger than that of the empty vector organoids, suggesting a growth advantage after the introduction of c-Myc and myrAKT1 (Fig. 1C).
We measured the organoid-forming efficiency of c-Myc/myrAKT1–transduced basal or luminal cells from four independent patient specimens. Initial experiments starting with 1,000 luminal cells did not yield any GFP/RFP-positive organoids. After increasing the number of starting luminal cells to 10,000 per assay, we detected 5–22 GFP/RFP doubly transduced luminal organoids (0.05–0.22% in Table 1). In the basal population, 1,000 initiating cells yielded 11–30 GFP/RFP doubly transduced basal organoids (1.1–3.0% in Table 1).
Table 1.
Efficiency of GFP/RFP double-positive luminal and basal cell organoids
| Type | No. of cells seeded | Average no. of GFP/RFP double-positive organoids (range) |
| CD26+ luminal cell | 10,000 | 12 (5–22) |
| CD49fHi basal cell | 10,000 | >200 |
| CD26+ luminal cell | 1,000 | 0 |
| CD49fHi basal cell | 1,000 | 21 (11–30) |
c-Myc/myrAKT1–Transduced Basal and Luminal Organoids Display Histological and Molecular Features of Human Prostate Cancer.
c-Myc/myrAKT1–transduced basal organoids grew as solid spherical structures (Figs. 1B and 2A). We observed that oncogene-transduced luminal cells also grew as solid spherical structures with loss of the central lumen seen in the empty vector condition (Fig. S2B). Both oncogene-transduced basal and luminal cell organoids displayed vague/absent glandular structures with poorly formed lumens. Some c-Myc/myrAKT1–transduced luminal organoids showed adenosquamous differentiation (Fig. 2A).
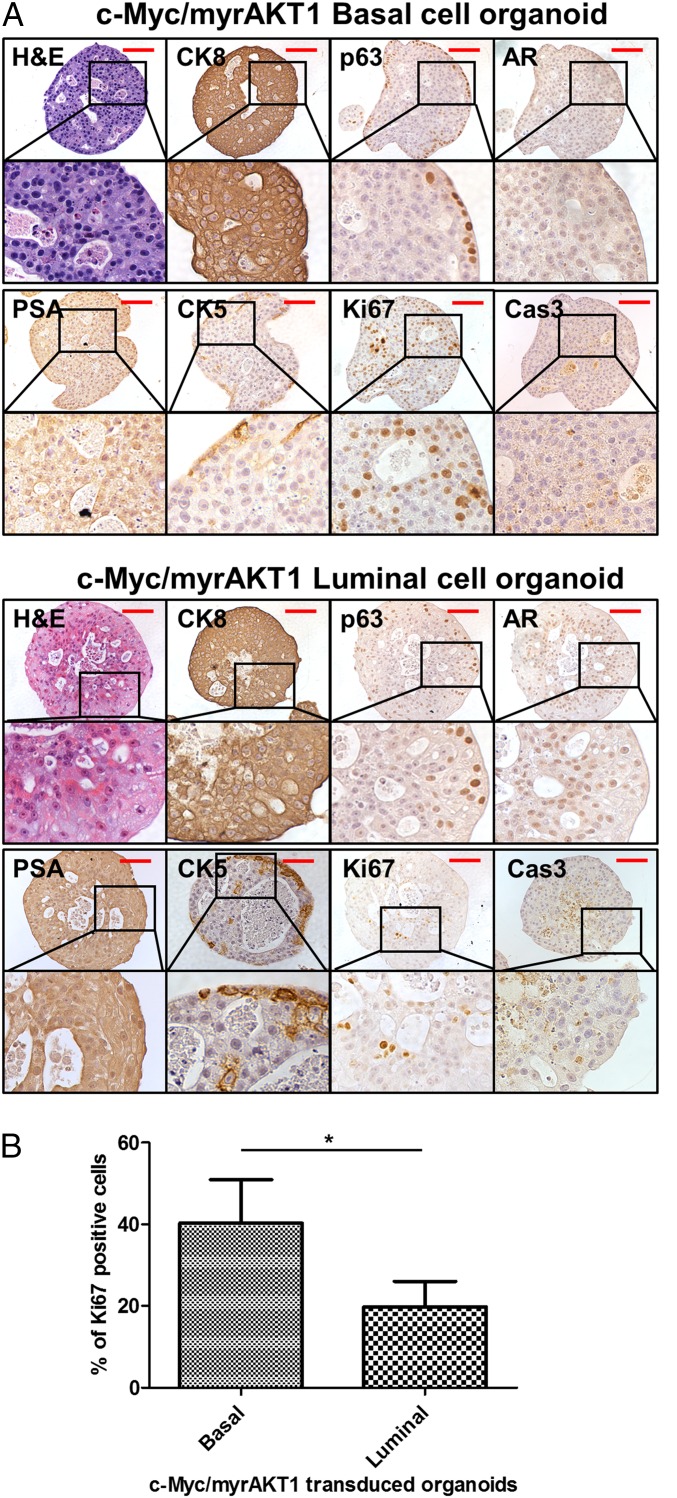
Fig. 2.
Molecular characterization of c-Myc/myrAKT1–transduced basal and luminal organoids. (A) Representative images of c-Myc/myrAKT1–transduced basal and luminal organoids with H&E and IHC staining for CK8, p63, AR, PSA, CK5, Ki67, and Cas3. (B) Percentage of Ki67+ cells in c-Myc/myrAKT1 basal, and luminal cell organoids. *P < 0.05. (Scale bar, 100 μm.)
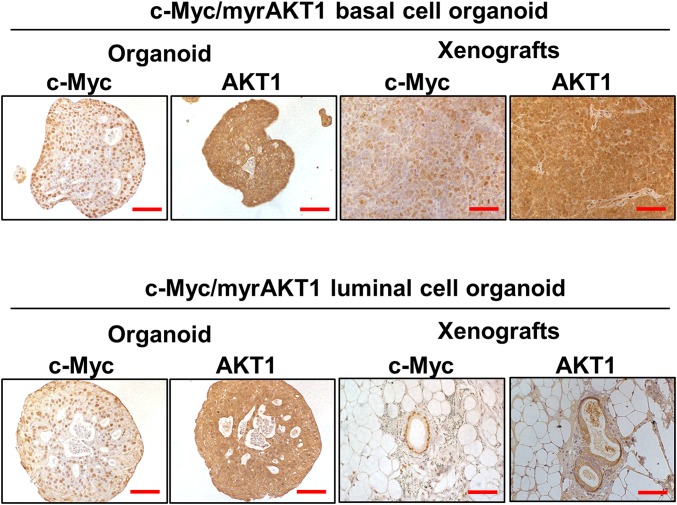
We confirmed the expression of c-Myc and myrAKT1 oncogenes in the basal and luminal organoids by immunohistochemistry (IHC) (Fig. S3). c-Myc/myrAKT1–transduced basal and luminal organoids displayed molecular phenotypes of human prostate adenocarcinoma, including strong expression of cytokeratin 8 (CK8) and low/absent expression of p63 and cytokeratin 5 (CK5). Focal expression of p63 and CK5 was detected along the rim of the oncogene-transduced basal and luminal organoids (Fig. 2A). This could represent basal cell differentiation that may be activated by direct contact with the Matrigel basal membrane matrix in the organoid culture system. AR and PSA expression was low in the oncogene-transduced basal and luminal organoids (Fig. 2A). The observation of basal-to-luminal differentiation during tumorigenesis is consistent with previous studies (4, 6, 7). Measurement of the Ki67 cell proliferation marker showed that c-Myc/myrAKT1 basal organoids harbored more Ki67-positive cells compared with c-Myc/myrAKT1 luminal organoids (40% vs. 20%; Fig. 2B). We found no significant difference in expression of cleaved-caspase 3 (Cas3), a marker of apoptosis, between c-Myc/myrAKT1–transduced basal and luminal organoids (Fig. 2A). We were able to maintain the oncogene-transduced basal and luminal organoids for at least three passages, with 2 wk of culture between each passage.
Fig. S3.
Expression c-Myc and myrAKT1 oncogenes in xenografts of oncogene-transduced basal and luminal organoids. Representative IHC images for anti–c-Myc and AKT1 are shown. (Scale bar, 100 µm.)
Human Luminal Cells Develop a Less Aggressive Phenotype Than Basal Cells with the Same Oncogenic Stimuli.
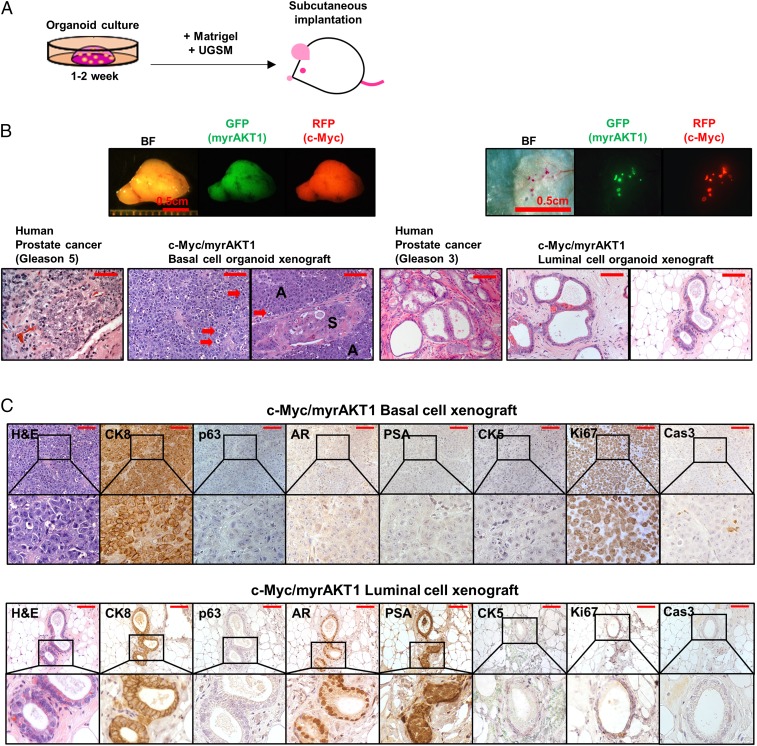
We collected intact c-Myc/myrAKT1–transduced basal and luminal cell organoids by enzymatic digestion of the Matrigel. We combined these organoids with murine urogenital sinus mesenchyme (UGSM) cells and implanted these cell grafts s.c. into NSG mice (19) (Fig. 3A).
Fig. 3.
Comparison of tumors derived from c-Myc/myrAKT1 basal and luminal cell organoids. (A) Schematic of the process of establishing xenografts by s.c. injection. (B) Representative human prostate cancers and c-Myc/myrAKT1 xenografts. (Scale bar, 100 μm.) Shown are photomicrographs of tumor sections of c-Myc/myrAKT1 basal and luminal xenografts. Red arrows indicate vague/absent glandular structure with poorly formed lumens. A, adenocarcinoma; S, squamous cell carcinoma. (C) IHC staining for CK8, p63, AR, PSA, CK5, Ki67, and Cas3. (Scale bar, 100 μm.)
Xenografts of the c-Myc/myrAKT1–transduced basal organoids formed large outgrowths (>1 cm in diameter) within 8 wk of transplantation. In contrast, the oncogene-transduced luminal organoids developed small tumor grafts (<0.5 mm) with a longer latency of 5–8 mo. The xenografts derived from c-Myc/AKT1–transduced basal and luminal cell organoids showed expression of GFP and RFP (Fig. 3B). Whereas basal cell-derived xenograft tumors were uniformly fluorescent, only two to six fluorescent foci were found in the luminal cell-derived xenografts from four independent human specimens. Necropsy revealed that all tumors were limited to the s.c. space, with no evidence of gross metastasis.
c-Myc/myrAKT1 xenografts derived from basal cells showed histological features of a poorly differentiated Gleason score 9 or 10 (4 + 5 or 5 + 5) adenocarcinoma. They exhibited a vague/absent glandular structure with poorly formed lumens in a diffuse pattern with solid cell nests. Adenosquamous differentiation was observed as well (Fig. 3B). This mixed tumor phenotype was also seen in our previous study (7). The basal organoid xenografts displayed histological features of human prostate cancer, with strong CK8 expression, absent p63 and CK5 expression, and low/absent AR and PSA expression (Fig. 3C). Expression of the oncogenes c-Myc and myrAKT1 in the xenografts was confirmed by IHC (Fig. S3).
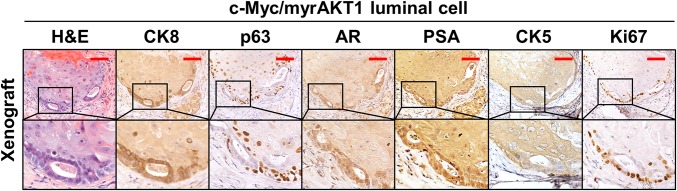
In contrast to the basal xenografts, the c-Myc/myrAKT1 luminal xenografts exhibited clear glandular structure with well-formed lumens. We found a single cell layer and absence of basal cells on the microscopic appearance of the luminal xenografts stained using H&E. Loss of the basal cell layer is an essential diagnostic feature of prostate carcinoma (20–23). The tumors were well differentiated and exhibited histological features of a well-differentiated Gleason score 6 (3 + 3) adenocarcinoma (Fig. 3B).
IHC analysis of the luminal cell-derived tumors showed uniform expression of the luminal cell marker CK8, as well as the absence of basal cell markers CK5 and p63 (Fig. 3C). Unlike the basal cell-derived tumors, the luminal cell-derived tumors showed high levels of nuclear AR staining and evidence of AR pathway activation, with strong staining for PSA (Fig. 3C). Xenografts produced from c-Myc/myrAKT1–transduced luminal organoids using the tissue of a second patient produced a mixed adenosquamous carcinoma in the luminal xenografts (Fig. S4). These results suggest that luminal cells respond to the oncogenes c-Myc and myrAKT1 similarly to basal cells in terms of their ability to differentiate to adenocarcinoma and squamous cell carcinoma during tumorigenesis.
Fig. S4.
Mixed adenosquamous cell carcinoma in c-Myc/myrAKT luminal xenografts and IHC for luminal cell markers (CK8, AR, and PSA), basal cell markers (p63 and CK5), and an index of cell proliferation (Ki67). (Scale bar, 100 µm.)
We found a significantly higher frequency of Ki67-positive cells in basal cell organoid xenografts than in luminal cell organoid xenografts (>80% vs. 5–10%; Fig. 3C). This finding implies that the differences in tumor latency and tumor size between c-Myc/myrAKT–transduced basal- and luminal-derived xenografts may be explained in part by differences in cellular proliferation rate. We detected Cas3-positive cells in basal cell organoid xenografts, but not in luminal cell organoid xenografts (Fig. 3C).
Discussion
Our previous studies have shown that primary basal epithelial cells from human prostate tissues can be readily transformed by select oncogenes in a transplantation assay. Until now, we have been unable to show that human prostate luminal cells also can be a direct target of transformation (6, 7). We have adapted an organoid culture system that enables the propagation of basal cells and rare luminal progenitor cells to transform both populations of cells. Our studies provide evidence that human prostate luminal cells can serve as cells of origin of prostate cancer, as has been suggested by previous work with mouse models (24, 25).
Intriguingly, human basal- and luminal-derived tumors demonstrate histologically distinct phenotypes when challenged with the same oncogenes, MYC and AKT1. Tumors derived from basal cells are of a higher grade than tumors derived from luminal cells. This finding is consistent with previous observations of slower disease progression and decreased cellular proliferation and tumor invasion in luminal-derived prostate tumors compared with basal-derived tumors in the context of PTEN loss (18). It also coincides with our recent report that aggressive human prostate cancers are enriched for a basal stem cell expression signature (26). In contrast, however, another study found that tumors of luminal origin were more aggressive than tumors of basal origin based on cross-species bioinformatics analyses (3). A similar cell-of-origin effect has been demonstrated in breast cancer. One study identified human CD10+ basal cells of the breast as precursors to rare metaplastic tumors, and EpCAM+/CD49f− luminal cells as leading to common forms of estrogen receptor-positive and -negative human breast cancer (27). Other recent studies have demonstrated that basal and luminal cells of the mouse mammary gland can be transformed by mutant PIK3CAH1047R overexpression (28, 29). Mutant PIK3CA in basal cells evoked benign tumors, such as adenomyoepithelioma and fibroadenoma, in contrast to the mostly aggressive mammary tumors, including adenosquamous carcinoma and carcinosarcoma, when it was expressed in luminal cells. Mutant PIK3CA caused a different spectrum of tumor types when expressed in basal or luminal cells. These findings suggest that the cell of origin could dictate the aggressiveness and heterogeneity of various tumors driven by the same oncogenic event (28, 29). Although it is widely accepted that combinations of oncogenic events have a major role in determining tumor phenotype, our findings and those of other studies suggest that the differentiation state and cell context in which oncogenic events are expressed play significant roles in defining the molecular subtype of the resulting tumor in human prostate cancer.
Previous efforts to transform prostate epithelial cells have been limited to basal and transit-amplifying or intermediate basal cells, owing to their enhanced ability to endure and proliferate in a variety of in vitro and in vivo conditions (6, 7). Although current growth factor-enriched prostate organoid culture conditions have begun to overcome this limitation, the low frequency of luminal progenitor cell propagation suggests the need for further optimization. In future work, single-cell RNA sequencing technology will allow us to better identify distinct epithelial cell subpopulations within the basal and luminal cells. We suspect that multiple cells of origin within the classic basal and luminal epithelial dichotomy may play a significant role in explaining the heterogeneity of human prostate cancer. As we identify new subpopulations within the epithelial hierarchy of the human prostate, it also will be necessary to understand and mimic in organoid culture the critical stromal interactions and signaling pathways that promote the survival and growth of these cells.
Next-generation sequencing technology has revealed significant genetic heterogeneity in human prostate cancer (30–32). Importantly, Baca et al. (33) defined sequential somatic DNA alterations in the natural history of human prostate cancer development and progression, and identified mutations in FOXA1 and SPOP, loss of NKX3.1, and rearrangement of the ERG gene as among the earliest events in prostate cancer development. Mutations of TP53, CDKN1A, CDKN1B, CHD1, and PTEN follow these early events (33). The genomic amplification of MYCN has been associated with neuroendocrine prostate cancer (34). Using a tissue recombination model, N-Myc and myrAKT1 overexpression in primary human basal cells was able to initiate a mixed phenotype of neuroendocrine carcinoma and adenocarcinoma (35). Defining the functional consequences of sequential oncogenic events in human prostate cancer development will provide insight into human prostate cancer progression and aggressiveness at the molecular level.
In summary, we have provided evidence of the direct transformation of human prostate luminal cells using an organoid culture. This culture system enables the real-time visualization of early events during tumorigenesis and a direct comparison of human prostate basal and luminal epithelial cell transformation, which was not possible with previous technologies. Our finding that basal- and luminal-derived tumors demonstrate different phenotypes when challenged with the same oncogenic stimuli suggests that identifying alternative cells of origin for prostate cancer may provide a way to subclassify prostate cancers and facilitate investigation of human prostate cancer heterogeneity.
Methods
Lentiviral Vectors.
The myristoylated AKT vector has been described previously (4), as has the c-Myc vector (7).
Organoid Culture of Primary Human Prostate Cells.
Patient tissue was provided in a deidentified manner and thus was exempt from Institutional Review Board approval. Acquisition and processing of human tissue, dissociation and isolation of distinct epithelial subsets, and lentiviral transduction have been described in detail previously (19). Between 1,000 and 10,000 FACS-sorted cells were plated in 20–30 μL of growth factor-reduced Matrigel (Corning) after lentiviral transduction. Organoid culture was performed as described previously (36).
In Vivo Implantation of Organoids.
Organoids were harvested by dissociation of Matrigel with 1 mg/mL Dispase. The organoids were washed three times with PBS and then mixed with 100,000 UGSM cells in 20–30 μL of Matrigel. The preparation of UGSM cells has been described previously (37). The organoid-Matrigel mixture was implanted s.c. in immunodeficient mice using a 28-gauge syringe.
IHC.
Organoids and xenografts were fixed in 10% buffered formalin for 6–24 h and then embedded in HistoGel (Thermo Fisher Scientific) and paraffin, sectioned (4 μm thickness), and mounted on glass slides (Thermo Fisher Scientific). IHC was performed as described previously (6).
Antibodies.
Antibodies used for flow cytometry included CD49f-PE and HLA-A/B/C-biotin (eBiosciences), CD49f-Alexa Fluor 647 and CD26-FITC (BioLegend), and Trop2-APC (R&D Systems). Antibodies used for IHC included CK5 (PRB-160P; Covance), CK8 (MMS-162P; Covance), p63 and AR (SC-8431 and SC-816; Santa Cruz Biotechnology), CD26/DPP4 (LS-C122983; LifeSpan Biosciences), c-Myc (ab32072, Abcam), pAKT (9271; Cell Signaling), PSA (A056201-2; Dako), Ki67 (ab16667; Abcam), and Cas3 (9664; Cell Signaling).
Animal Work.
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were originally purchased from the Jackson Laboratories and were housed and bred under the care of the Division of Laboratory Animal Medicine at the University of California, Los Angeles. Subcutaneous injections were performed according to protocols approved by the university’s Animal Research Committee.
Acknowledgments
J. W. Park is supported by the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at University of California, Los Angeles (UCLA) Training Program. J.K.L. is supported by the Tower Cancer Research Foundation Career Development Award, a Prostate Cancer Foundation Young Investigator Award, and the UCLA Subspecialty Training and Advanced Research Program. J. W. Phillips is supported by UCLA Tumor Cell Biology Training Grant T32CA09056. J.H. is supported by the National Institutes of Health (Grants 5R01 CA172603-02, 2P30 CA016042-39, 1R01 CA181242-01A1, and 1R01 CA195505), the Department of Defense Prostate Cancer Research Program (Grant W81XWH-12-1-0206), UCLA Specialized Program of Research Excellence (SPORE) in Prostate Cancer, the Prostate Cancer Foundation’s Honorable A. David Mazzone Special Challenge Award, and a UCLA Jonsson Comprehensive Cancer Center Impact Grant. O.N.W. is an Investigator of the Howard Hughes Medical Institute supported by National Institutes of Health Grant U01 CA164188-01A and a Prostate Cancer Foundation Challenge Award. J.H. and O.N.W. are supported by Stand Up To Cancer/Prostate Cancer Foundation/Prostate Dream Team Translational Cancer Research Grant SU2C-AACR-DT0812. This research grant is made possible by the generous support of the Movember Foundation. Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603645113/-/DCSupplemental.
References
- 1.Humphrey PA. Histological variants of prostatic carcinoma and their significance. Histopathology. 2012;60(1):59–74. doi: 10.1111/j.1365-2559.2011.04039.x. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461(7263):495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang ZA, et al. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat Cell Biol. 2013;15(3):274–283. doi: 10.1038/ncb2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xin L, Lawson DA, Witte ON. The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis. Proc Natl Acad Sci USA. 2005;102(19):6942–6947. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci USA. 2007;104(1):181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. Identification of a cell of origin for human prostate cancer. Science. 2010;329(5991):568–571. doi: 10.1126/science.1189992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoyanova T, et al. Prostate cancer originating in basal cells progresses to adenocarcinoma propagated by luminal-like cells. Proc Natl Acad Sci USA. 2013;110(50):20111–20116. doi: 10.1073/pnas.1320565110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shin K, et al. Hedgehog/Wnt feedback supports regenerative proliferation of epithelial stem cells in bladder. Nature. 2011;472(7341):110–114. doi: 10.1038/nature09851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat Med. 2014;20(7):769–777. doi: 10.1038/nm.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCracken KW, et al. Modelling human development and disease in pluripotent stem cell-derived gastric organoids. Nature. 2014;516(7531):400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, Wrana JL. Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature. 2015;526(7575):715–718. doi: 10.1038/nature15382. [DOI] [PubMed] [Google Scholar]
- 12.Karthaus WR, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159(1):163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korsten H, Ziel-van der Made A, Ma X, van der Kwast T, Trapman J. Accumulating progenitor cells in the luminal epithelial cell layer are candidate tumor-initiating cells in a Pten knockout mouse prostate cancer model. PLoS One. 2009;4(5):e5662. doi: 10.1371/journal.pone.0005662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shultz LD, Ishikawa F, Greiner DL. Humanized mice in translational biomedical research. Nat Rev Immunol. 2007;7(2):118–130. doi: 10.1038/nri2017. [DOI] [PubMed] [Google Scholar]
- 15.Gurel B, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21(9):1156–1167. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King JC, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41(5):524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carver BS, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41(5):619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu TL, et al. Conditionally ablated Pten in prostate basal cells promotes basal-to-luminal differentiation and causes invasive prostate cancer in mice. Am J Pathol. 2013;182(3):975–991. doi: 10.1016/j.ajpath.2012.11.025. [DOI] [PubMed] [Google Scholar]
- 19.Goldstein AS, et al. Purification and direct transformation of epithelial progenitor cells from primary human prostate. Nat Protoc. 2011;6(5):656–667. doi: 10.1038/nprot.2011.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humphrey PA. Diagnosis of adenocarcinoma in prostate needle biopsy tissue. J Clin Pathol. 2007;60(1):35–42. doi: 10.1136/jcp.2005.036442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Algaba F, et al. Assessment of prostate carcinoma in core needle biopsy: Definition of minimal criteria for the diagnosis of cancer in biopsy material. Cancer. 1996;78(2):376–381. doi: 10.1002/(SICI)1097-0142(19960715)78:2<376::AID-CNCR32>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 22.Epstein JI. Diagnostic criteria of limited adenocarcinoma of the prostate on needle biopsy. Hum Pathol. 1995;26(2):223–229. doi: 10.1016/0046-8177(95)90041-1. [DOI] [PubMed] [Google Scholar]
- 23.Epstein JI. Diagnosis and reporting of limited adenocarcinoma of the prostate on needle biopsy. Mod Pathol. 2004;17(3):307–315. doi: 10.1038/modpathol.3800050. [DOI] [PubMed] [Google Scholar]
- 24.Wang ZA, Toivanen R, Bergren SK, Chambon P, Shen MM. Luminal cells are favored as the cell of origin for prostate cancer. Cell Reports. 2014;8(5):1339–1346. doi: 10.1016/j.celrep.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chua CW, et al. Single luminal epithelial progenitors can generate prostate organoids in culture. Nat Cell Biol. 2014;16(10):951–961. doi: 10.1038/ncb3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith BA, et al. A basal stem cell signature identifies aggressive prostate cancer phenotypes. Proc Natl Acad Sci USA. 2015;112(47):E6544–E6552. doi: 10.1073/pnas.1518007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller PJ, et al. Defining the cellular precursors to human breast cancer. Proc Natl Acad Sci USA. 2012;109(8):2772–2777. doi: 10.1073/pnas.1017626108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Keymeulen A, et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature. 2015;525(7567):119–123. doi: 10.1038/nature14665. [DOI] [PubMed] [Google Scholar]
- 29.Koren S, et al. PIK3CA(H1047R) induces multipotency and multi-lineage mammary tumours. Nature. 2015;525(7567):114–118. doi: 10.1038/nature14669. [DOI] [PubMed] [Google Scholar]
- 30.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baca SC, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153(3):666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beltran H, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1(6):487–495. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JK, et al. N-Myc drives neuroendocrine prostate cancer initiated from human prostate epithelial cells. Cancer Cell. 2016 doi: 10.1016/j.ccell.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drost J, et al. Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc. 2016;11(2):347–358. doi: 10.1038/nprot.2016.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukacs RU, Goldstein AS, Lawson DA, Cheng D, Witte ON. Isolation, cultivation and characterization of adult murine prostate stem cells. Nat Protoc. 2010;5(4):702–713. doi: 10.1038/nprot.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]