Significance
Wild bumble bees are experiencing population declines globally. Causes of declines in North American populations are unclear, although declining species are more frequently infected by the pathogen Nosema bombi. A widely accepted hypothesis suggests that contact with European species during domestication led to the introduction of exotic N. bombi. By screening museum specimens, we show that N. bombi prevalence increased significantly in declining species in the early to mid-1990s, coincident with N. bombi outbreaks in North American commercial stocks. There is no evidence that exotic Nosema strains were introduced from Europe. Regardless of geographic origins, the temporal connection between N. bombi epizootics in commercial Bombus stocks and increases in wild populations suggests a substantial risk of pathogen transmission with domestication.
Keywords: Bombus, Microsporidia, Nosema bombi, pollinator, conservation
Abstract
Emergent fungal diseases are critical factors in global biodiversity declines. The fungal pathogen Nosema bombi was recently found to be widespread in declining species of North American bumble bees (Bombus), with circumstantial evidence suggesting an exotic introduction from Europe. This interpretation has been hampered by a lack of knowledge of global genetic variation, geographic origin, and changing prevalence patterns of N. bombi in declining North American populations. Thus, the temporal and spatial emergence of N. bombi and its potential role in bumble bee decline remain speculative. We analyze Nosema prevalence and genetic variation in the United States and Europe from 1980, before an alleged introduction in the early 1990s, to 2011, extracting Nosema DNA from Bombus natural history collection specimens from across this time period. Nosema bombi prevalence increased significantly from low detectable frequency in the 1980s to significantly higher frequency in the mid- to late-1990s, corresponding to a period of reported massive infectious outbreak of N. bombi in commercial bumble bee rearing stocks in North America. Despite the increased frequency, we find no conclusive evidence of an exotic N. bombi origin based on genetic analysis of global Nosema populations; the widespread Nosema strain found currently in declining United States bumble bees was present in the United States before commercial colony trade. Notably, the US N. bombi is not detectably different from that found predominantly throughout Western Europe, with both regions characterized by low genetic diversity compared with high levels of diversity found in Asia, where commercial bee breeding activities are low or nonexistent.
There is growing concern that emerging infectious diseases in wild animals pose increasing risks to biodiversity and ecosystem services (1). Although quantitative data are accumulating on the deteriorating status of bumble bee (Bombus) pollinator populations throughout North America (2–7), the factors causing species decline remain uncertain and controversial. In the United States, shrinking or disappearing populations have been ascribed principally to an invasive virulent strain of fungal pathogen, Nosema bombi (Microsporidia), hypothesized to have been introduced from Europe in the early 1990s via commercial development of bumble bee colonies for pollination [“European Nosema invasion hypothesis” (ENIH)] (2, 4, 8, 9). However, this important hypothesis has remained largely untested, despite a highly publicized report documenting significant positive correlations between decline status of bumble bees in the United States and N. bombi prevalence (4). Because Nosema diminishes bumble bee colony fitness by reducing reproductive performance of sexuals (males and gynes) and lowering the survival rate of workers (e.g., ref. 10), there are compelling reasons to investigate exotic pathogen release as a causal factor in North American bumble bee decline.
The ENIH was based on circumstantial evidence from multiple associated observations. First, large-scale commercial bumble bee pollination of crops began in Europe in the late 1980s, using Europe’s native Bombus terrestris. From 1992 through 1994, with the goal of expanding US markets, queens of Bombus occidentalis and Bombus impatiens were exported to Europe for colony rearing, grown in facilities used for rearing B. terrestris. Subsequently, these colonies were imported back into the United States for use in open-field and greenhouse pollination (11). Around this time, commercial colony production by other companies began in eastern Canada (1990), using wild queens of B. impatiens, and in California (1992) using B. occidentalis. However, B. occidentalis was abandoned by both major producers in North America shortly after 1997 because of infestation of the rearing stock with N. bombi (11), and soon afterward, wild B. occidentalis populations began to decline precipitously (2) along with this species’ close western US relative Bombus franklini (2). Both species belong to the subgenus Bombus sensu stricto, which includes B. terrestris. Parallel declines were soon detected in the two eastern species of Bombus s. s., Bombus affinis and Bombus terricola (3, 12, 13). These declines were confirmed by more recent surveys, which also showed steep population declines among eastern Bombus s. s. species and members of a second subgenus (Thoracobombus), including Bombus pensylvanicus and Bombus fervidus (4–6, 13, 14). Finally, these declines were occurring in the absence of a general decline among all bumble bee species, suggesting a targeted cause rather than more general environmental causes (8). Indeed, of the species examined to date, the declining North American bumble bees exhibit significantly elevated N. bombi prevalence compared with more stable codistributed species (4). Collectively, these observations are consistent with the hypothesis that N. bombi jumped from the European commercial B. terrestris—during a 3-y period of export of North American queens to Europe and import of colonies reared from those queens—and was subsequently transmitted into wild bumble bee populations in North America. However, the observations to date do not provide quantitative data to test the Nosema invasion hypothesis.
An analysis of the ENIH would require genetic comparisons of Nosema strains from bumble bees collected before and subsequent to commercial introduction in 1992. Data on genetic variation in Nosema from both the native range and the invasive range can be used to test whether an invasion event actually occurred. Furthermore, evaluating Nosema frequencies in host populations over time, before and subsequent to the population declines, could reveal whether N. bombi prevalence has increased in declining populations in association with commercial production for pollination. To reveal the presence/absence of Nosema before and after the hypothesized early-1990s invasion, we use a technique to extract DNA nondestructively from museum specimens of Bombus species that currently exhibit moderate to high N. bombi prevalence in wild populations (SI Methods). We show that N. bombi was present in the United States historically but increased in incidence concomitantly with commercial Bombus trade. We also show that US and European Nosema are highly genetically similar relative to the genetically diverse Nosema found in Asian Bombus.
SI Methods
Screening for Nosema bombi in Bumble Bee Museum Specimens.
We screened 2,048 bumble bee specimens (collected from 1979 to 2011 from 20 institutions and museums) of five North American species known to be declining (B. affinis, B. franklini, B. occidentalis, B. pensylvanicus, and B. terricola) and one European species (B. terrestris) (Table S1). Bombus terrestris, whose history of general infection with N. bombi in Europe is well documented and not disputed, was screened primarily to determine that Nosema could be detected effectively in decades-old pinned specimens (47, 48). We focus on these declining species because surveys of contemporary populations show high to moderate infection prevalence (e.g., ∼15–37%), whereas populations of nondeclining North American species exhibit negligible infection rates (e.g., ∼0.3–1%) (4) and would thus fail to produce meaningful detection rates with realistic museum specimen sample sizes. Our study, therefore, was not designed to compare Nosema prevalence in stable vs. declining species but rather to make a temporal analysis of prevalence in species known to harbor Nosema.
DNA from pinned specimens was extracted using the following protocol, preapproved by individual institutions. Importantly, all specimens remained on their pins to avoid damaging the specimen. First, one to two holes were made in the underside of the abdomen using a sterilized no. 2 insect pin (0.45 mm diameter). Each specimen was then immersed intact in a 50-mL centrifuge tube in a mixture containing 4 mL of 5% (wt/vol) Chelex 100 Resin solution (Bio-Rad) and 600 mg of Proteinase K (Life Technologies) and maintained at 55 °C inside an incubator overnight. Samples were gently hand-shaken periodically. Following gentle mixing and resuspension, 1 mL of the mixture was placed in a shaking incubator at 99 °C and 800 rpm for 30 min. The resulting extractions were stored at −20 °C until further use. Following immersion and incubation of pinned specimens, each specimen was cleaned using a dilute detergent solution, rinsed in water and then 95% (vol/vol) ethanol, dried, and reassociated with the specimen’s original museum label. This approach proved excellent for maintaining specimen integrity. Specimen label data, including species identity, collection locality, and date, were recorded (Table S1).
To detect N. bombi, we conducted a minimum of three PCR replicates for each extracted specimen (average, 3.3; maximum, 12). We used the published primers ITS-f2 and ITS-r2, which are sensitive for Nosema taxa (25) and yield short products (118–120 bp) likely to be amplified from degraded DNA of museum specimens. We periodically confirmed positive detections with a second set of N. bombi-specific primers that target a 159-bp region of the SSU rRNA (25). Note that these sequences may weakly amplify other Nosema (e.g., N. apis and N. ceranae) that can infect or be carried by bumble bees (41), but more specific primers targeting a slightly larger fragment (Nbombi-SSU-Jf1/r1) (25) failed to detect Nosema in all “known” infections. In this study, we were more concerned with false negatives than false positives from the old museum specimens. We thus used the more sensitive ITS primers in favor of greater specificity of the SSU primers. Importantly, we have never previously observed N. apis or N. ceranae in North American bumble bees with microscopy or DNA sequencing (4, 24), and sequencing of bumble bees with Sanger or pyrosequencing here never produced N. apis or N. ceranae sequences in North American specimens, with only three sequenced bees exhibiting unusual non-N. bombi SSU sequences (Results). PCR reagents for 10 μL reactions were as follows: 5 μL of template, 2 μL of 5× Promega Green GoTaq Buffer (Promega), 0.75 U of Promega GoTaq, 0.2 mM each dNTP, 1.5 mM MgCl2, and 0.4 μM each forward and reverse primers. Thermocycling conditions for ITS were slightly modified from Klee et al. (25): denaturing at 95 °C for 3 min, 40 cycles of 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s, followed by a final extension step of 72 °C for 5 min. Each batch of PCR experiments included three reactions for each of the following: 28 historical bee samples, one positive control, and two negative controls (total number of reactions per PCR batch, 93). One of the two negative controls contained molecular grade water instead of the DNA template; the other contained a negative DNA extraction from previously screened samples. To avoid cross-contamination, DNA extractions were conducted in a laminar flow hood, PCRs were prepared in a clean room (no PCR products introduced), dedicated reagents and filtered tips were used, and we regularly applied surface disinfectants (e.g., DNA away and bleach). We visualized PCR products via gel electrophoresis, with 1 μL of each reaction on a 1.5% (wt/vol) agarose gel stained with GelRed (Biotium), running at 90 V for 1.5 h. We considered a bee infected with Nosema when at least two of three of the PCRs showed a band of the expected size (conservative threshold) (Table S1); we also examined results based on a lower threshold, when at least one reaction exhibited a positive band (Table S2 and Fig. S3).
Fig. S3.
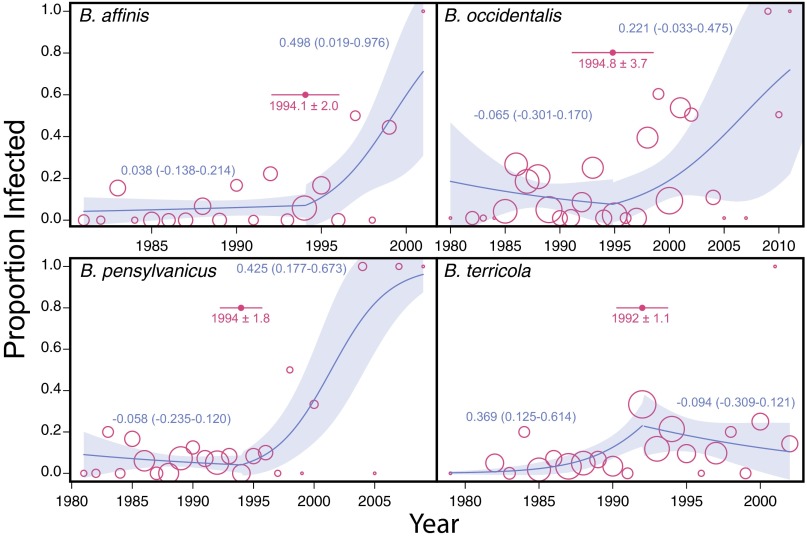
Proportion of four declining bumble bee species infected by Nosema (open circles) based on a less-stringent PCR detection criterion (at least one of three PCRs showed product of correct size on a gel). The size of each circle is proportional on a logarithmic scale to the number of bees screened in a given year. Yearly infection rate data are fitted with a piecewise quasibinomial regression curve and its 95% CI (shaded area). For each species, the change point in time (i.e., year) with SE is indicated by a filled circle, with error bar on either side. Numbers beside curves are estimated slope parameters for each segment of the piecewise regression (95% CI in parentheses). For B. affinis and B. pensylvanicus, the 95% CI of the slope parameter of post-1990 segment does not include 0.
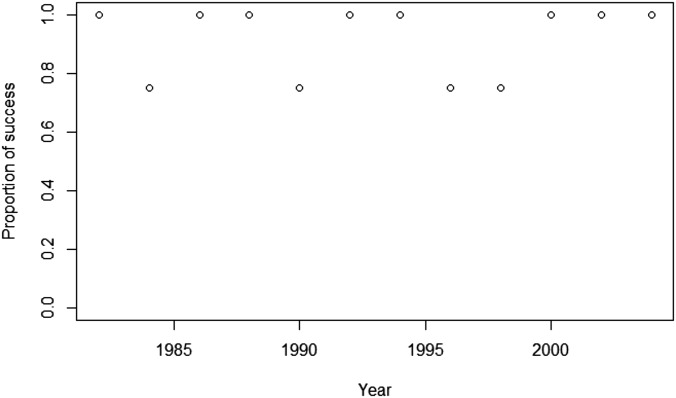
To ascertain whether specimen age had significant adverse effects on PCR success, we conducted a control experiment with a 117-bp portion of the long-wavelength opsin (LW Rh) gene for 47 North American bumble bees collected from 1982 to 2004 (Fig. S2). Primer sequences were as follows: bb.opsin1f, 5′-TCCAGGCGGTAGCTGCCCACGAG; and bb.opsin1r, 5′-CTTTTGCCAATTTACACTCGGCAC. We conducted one PCR for each specimen and defined a successful LW Rh detection for each bee as having observed PCR product of the correct size on an agarose gel. PCR conditions were the same as above.
Fig. S2.
Success rate of amplifying the LW Rh gene in bumble bee specimens of different ages. General linear modeling (binomial errors: dependent variable, success rate; independent variable, year of specimen collection) showed that year as an independent variable contributed little to model fit (regression coefficient = 0.0182, P = 0.814). Except for 2004, when only three bees were screened, four bees were screened in each study year.
Statistical Analysis of Nosema Prevalence.
All statistical analyses, unless stated otherwise, were conducted in R-2.15.3 (49). For each of the North American bee species with sufficient sample size (B. affinis, B. occidentalis, B. pensylvanicus, and B. terricola), we determined whether the prevalence of Nosema changed significantly over time (and across the putative invasion event of 1992) by conducting χ2 tests of independence of proportions, using 5,000 Monte Carlo simulations to compute P values. We divided bee collection years into four time periods (1979 to 1987, 1988 to 1991, 1992 to 1994 and 1995 to 2011) selected to yield bins with similar numbers of bees in each period surrounding the hypothetical introduction date (1992). We also conducted a multiple comparison test for each species using arcsine transformation of prevalence data and Tukey’s honest significant difference test (conducted in Minitab 15 using the macro multiprop.mac).
In addition to tests of independence of proportions for binned data, we modeled the prevalence of Nosema through time in each species by conducting piecewise GLM (family, quasibinomial) (40) using the R-package “segmented,” which is useful if there are “breakpoints” in the data indicating that the relationship between two variables sharply changes, such as we hypothesized for Nosema prevalence over time. Using year as the independent variable and Nosema prevalence as the dependent variable, we used “segmented” to determine the most likely location of a single time point where regression slope parameters changed (maximum number of bootstrap iterations, 500).
We applied GLM (binomial errors) to the LW Rh (control) gene data to test the significance of the effect of museum specimen age on the proportion of successful PCRs observed.
SSU rRNA: Taxon Sampling, Laboratory Protocols, and Library Preparation.
For phylogenetic analysis of global strain variation, we sequenced the SSU rRNA of Nosema from infected North American and European Bombus species.
European samples.
To determine infection status of European bumble bees, we screened a total of 528 individuals from 18 species collected in France, Denmark, and Sweden in 2010 or 2011 (Table S5). Bees were identified to species in the field by experts (P. Rasmont, Université de Mons, Mons, Belgium; B. Cederberg, Swedish University of Agricultural Sciences, Uppsala; and C. Rasmussen, Aarhus University, Aarhus, Denmark). Bombus terrestris accounted for 83.3% (440) of the bees collected. Field-collected bees were caught with hand nets, placed into 90% ethanol, shipped to the S.A.C. laboratory and stored at −20 °C. We used PCR to detect the presence of Nosema in each bee. We extracted the entire gut and abdominal tissue using sterilized forceps, added the tissue to 200 μL of molecular-grade water, and homogenized using a pipette tip. Approximately 75 μL of this mixture was added to 150 μL of 5% Chelex 100 Resin and 5 μL of Proteinase K (30 mg/μL). We incubated the mixture at 55 °C for 1 h, 99 °C for 15 min, 37 °C for 1 min, and 99 °C for 15 min. Nosema detection from this extraction was determined via PCR, using the same ITS-f2 and ITS-r2 primers and PCR conditions used for the museum samples. We periodically confirmed our results using the general Nosema primers SSUrRNA-f1/r1c or through phase-contrast microscopy on a portion of the remaining abdominal content (25). Overall, 18.0% of the screened European bees were infected (95 of 528), with males infected more frequently (40.2%) than females (12.0%) (Fisher’s exact test, P < 0.0001).
North American samples.
Infection status of recently collected North American taxa for SSU analysis was determined by Cordes et al. (24) for the Continental United States and by Koch and Strange (50) for Alaska. We sequenced a total of 35 infected North American specimens representing 16 species (Table S3). In addition to the contemporary European and North American samples, we also included seven of the 135 infected historical samples from North American museums (Table S3) for SSU sequencing. The seven samples were collected from 1986 to 1988, before any putative Nosema invasion event. DNA was extracted from museum specimens using the protocol described above for screening museum specimens. We were limited to obtaining SSU sequence for only seven of the infected historical samples because of the relatively few individuals from 1986 to 1988 that were infected and the difficulty of obtaining sufficient good Nosema DNA from specimens of that age.
To sequence a portion of the SSU rRNA that amplified across divergent clades and in potentially degraded museum DNA, we designed primers (SSU.PriFi-1f and SSU.PriFi-298r) that target a ∼298-bp section of the SSU based on priming sites conserved across nine divergent Nosema species and a related taxon (Oligosporidium occidentalis). We designed primers using the online utility PriFi (cgi-www.daimi.au.dk/cgi-chili/PriFi/main) and the following 10 GenBank sequences: accession nos. HM370543, U27359, AF495379, JF443592, GU131083, JN872238, JN872253, JN872292, JN872254, JN872219 (51).
We used two methods to sequence the region of interest. The first was PCR, using the primers Nb.SSU-PriFi-1f (5′-CACCAGGTTGATTCTGCCTGACG-3′) and Nb.SSU-PriFi-298r (5′-CGCCCCTGCTSCAATCCTTAGAC-3′), followed by TA cloning with the pGEM-T Vector Systems kit (Promega) and NEB Turbo Competent Cells (New England Biolabs) (see Table S3 for species list). PCR reagents for 12 μL reactions were as follows: 4.7 μL of H2O, 2 μL of 5× NEB OneTaq Buffer, 0.4 μL of 25 mM MgCl2 (3–4 μL for museum specimens), 0.2 μL of 40 mM dNTP, 0.1 μL of DNA polymerase, 0.8 μL of 100× BSA, 0.4 μL of each primer, 1 μL of template DNA (5 μL for museum specimens). Thermocycling conditions were as follows: denaturation at 95 °C for 3 min; denaturation, annealing, and elongation for 35 cycles (40 cycles for museum samples) of 95 °C for 40 s plus 54 °C for 40 s plus 72 °C for 30 s; and final elongation at 72 °C for 5 min and then 95 °C for 3 min. For each sample, we sequenced 1–16 clones using the Big Dye v3.1 Cycle Sequencing Kit and an AB 3730xl capillary sequencer operated by the University of Illinois W. M. Keck Center for Comparative and Functional Genomics.
The second SSU sequencing method involved Roche 454 pyrosequencing to deep sequence a subset of samples (Table S3). The deep sequencing coverage afforded by pyrosequencing enables determination of coinfection by multiple Nosema strains or species, as commonly seen in the honey bee Apis cerana (41). A previous study (52) also showed that multiple rRNA sequence variants coexist in a single Nosema spore, indicating the lack of perfect concerted evolution. To generate sequencing libraries, we modified the basic cloning primers (Nb.SSU-PriFi-1f and Nb.SSU-PriFi-298r) with a 5′ 20-bp universal tail. Reagents for this first round of 12-μL PCR reactions were: 4.7 μL of H2O, 2 μL of 5× HF (Phusion High-Fidelity) Buffer, 0.4 μL of 25 mM MgCl2 (3–4 μL for museum specimens), 0.2 μL of 40 mM dNTP, 0.1 μL DNA polymerase, 0.8 μL of 100× BSA, 0.4 μL of each primer, 1 μL of template DNA (5 μL for museum specimens). Thermocycling conditions were as follows: denaturation at 98 °C for 3 min; denaturation, annealing, and elongation for 12 cycles of 98 °C for 1 min plus 70 °C for 1 min plus 72 °C for 40 s; second denaturation and elongation for 28 cycles of 98 °C for 30 s plus 72 °C for 40 s; and final elongation at 72 °C for 5 min. A second round of PCR incorporated HPLC-cleaned, barcoded Roche 454 Lib-L sequencing primers (A and B) that targeted these universal tails. The first and second rounds of PCR used the same reagent volumes and concentrations. Thermocycling conditions for the second round of PCR were as follows: denaturation at 95 °C for 3 min; denaturation, annealing, and elongation for 35 cycles (40 cycles for museum samples) of 95 °C for 40 s plus 54 °C for 40 s plus 72 °C for 30 s; and final elongation at 72 °C for 5 min and then 95 °C for 3 min. In all PCRs, high-fidelity Phusion Taq polymerase (New England Biolabs) was used. The final products were cleaned with AMPure XP beads (Beckman Coulter), checked for quality and quantity using a Bioanalyzer 2100 (Agilent Technologies) and Qubit fluorometer (Life Technologies), and sent for sequencing on a Roche 454 GS FLX+ [3 × 1/8 Pico Titer Plate (PTP) single-end amplicon sequencing runs] at the University of Illinois W. M. Keck Center.
Nosema SSU rRNA Sequence Editing and Phylogenetic Analysis.
Sanger sequences were edited in Geneious Pro-5.3.4. Pyrosequencing reads (comprising .fasta and .qual files) were handled primarily using commands in the Mothur (version 1.28) package (42). For the pyrosequences, we first carried out demultiplexing (assigning reads to individuals) and aggressive quality control using the trim.seqs tool [settings: remove reads with average quality score <25; maximum number of primer mismatches, 3; maximum number of barcode mismatches, 1; maximum number of ambiguous bases (i.e., N), 2; maximum number of bases in a homopolymer run, 8; and minimum length, 200 bp]. To further remove sequencing errors, we used Mothur’s split.abund function to remove sequences that are found 10 times or less across all samples in each sequencing run. [Note that because the raw data were generated using the latest Roche software, which uses an acyclic flow pattern during sequencing (version 2.8 flow pattern B), applying common data filtering/cleaning methods, such as flowgram-based sequence denoising (53), was unnecessary.]
Overall, we generated 41 unique sequences through molecular cloning, 136 unique sequences through Roche 454 sequencing, and 4 four unique sequences using both methods (Table S3). Unique sequences were generated by collapsing identical reads using unique.seqs in Mothur. The samples used consisted of 10 European bumble bees from three species, 35 recently collected North American bumble bees from 16 species, and 7 museum samples of North American bumble bees from three species (Table S3). To provide additional phylogenetic context, we supplemented our sequence data with 113 nonredundant GenBank sequences derived from a variety of Nosema spp. and related species, giving us a combined sequence dataset representing 35 bumble bee species, several other insects (Lepidoptera, Hymenoptera, Coleoptera) and one mite (Mesostigmata) (Table S3).
To align the 294 Nosema sequences, we first selected 32 major sequence variants from the Sanger dataset of 41 sequences and simultaneously folded and aligned those rRNA sequences using a realistic energy model, implemented in the application LocARNA (54). This alignment was used as the template for subsequent alignment of the remaining 262 sequences using the online version of MAFFT version 7 (alignment strategy: FFT-NS-i) (55), leaving us with a total of 294 aligned sequences for analysis.
From these aligned Nosema rRNA sequences, we generated a phylogenetic network from distances using a pipeline implemented in SplitsTree version 4.12.6 (43) (Fig. 3, Fig. S4, and Table S3). The first step was to calculate pairwise uncorrected p-distances between sequences. A nucleotide site with an ambiguous state (e.g., W and M) was treated as an average of all possible resolutions of the ambiguity. Following distance calculation, the NeighborNet algorithm (default settings; variance, ordinary least square) was used to generate a splits network representing all splits in the dataset, in which a split is a partition of the taxa into two nonempty sets implied by haplotype relationships (44). Because Nosema from a single bee may have multiple haplotypes, we were able to show the phylogenetic relationships and positions of all haplotypes in the various clades and nodes. A splits network is a superior representation of evolution relative to a less realistic bifurcating phylogenetic tree, especially when sequences have evolved recently. Furthermore, whereas splits networks realistically represent incompatibilities in the underlying data as parallel branches, phylogenetic trees (because of the need to constrain the tree to a bifurcating or multifurcating graph) may represent misleading resolved topologies (56). To improve clarity (i.e., reduce the number of parallel branches) and further reduce noise from sequencing errors, we filtered the splits by weights (which are equivalent to branch lengths in a phylogenetic tree) and retained the most important splits (67.9% out of the total splits weight of 1.022 was retained).
Fig. 3.
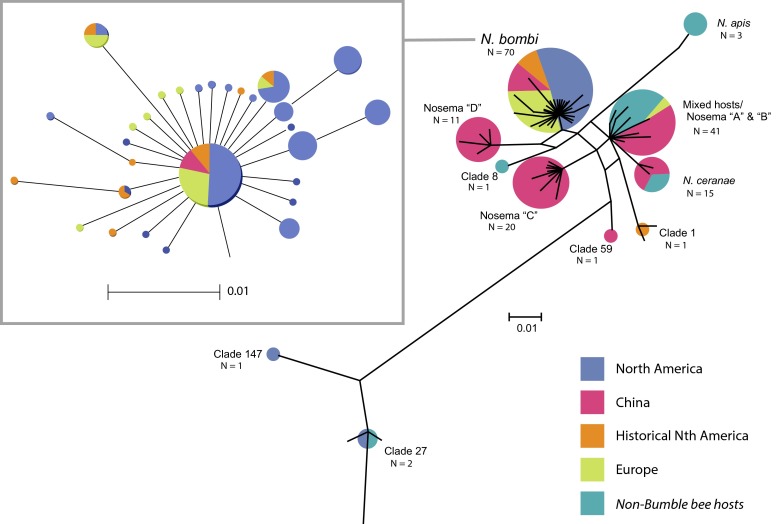
Filtered splits phylogenetic network showing relationships among Nosema clades. Clades may contain members that are attributable to known species (e.g., N. apis) that have been previously named in the literature (e.g., Nosema “D”) or newly delineated (e.g., clade 1). For each clade, the number of host individuals (represented both numerically and by size of circle at the tips) and their geographic origins are also shown. Clade memberships can be found in Table S3.
Fig. S4.
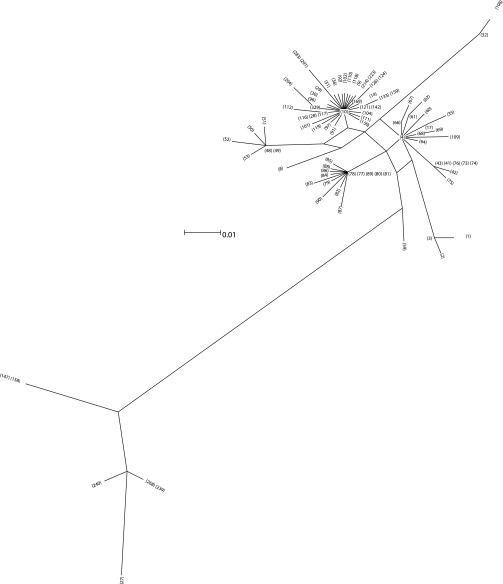
Filtered splits phylogenetic network showing node designations. Refer to Table S3 for samples represented by each node.
Nosema Marker Discovery Using Reduced Genome Sequencing.
PCR primers designed to target additional loci from the available genome of Nosema ceranae routinely failed to amplify or lacked sequence variation in N. bombi samples during initial screens. Moreover, the SSU sequencing revealed almost no variation across the European and North American taxa, relative to the diversity found among the Asian taxa. To look for additional Nosema genomic variation, we thus elected to develop new N. bombi-specific polymorphic markers by performing reduced representation genome sequencing from pools of N. bombi isolated from nine wild-caught bumble bees (Table S4A).
Ludox density gradient spore purification.
The gut content of N. bombi infected bees were individually homogenized in 0.5 mL of sterile distilled water using a micropestle and inside a 1.5-mL tube. The mixture was then filtered through a cotton mesh to remove large tissue pieces. Following this procedure, we flushed the mesh with 10 mL of distilled water. All filtrate from each bee was collected inside a 15-mL conical tube, which was subsequently subjected to centrifugation at 3,000 × g for 5 min to pellet the tissues and spores. Next, the tissue/spore pellet and a small amount of water were aliquoted and placed in density gradient comprising 10 mL of 50% (wt/vol) Ludox HS 40 solution (Sigma-Aldrich). The density gradient was made by first mixing 5 mL of water with 5 mL of Ludox HS 40 solution. The tube containing the mixture was laid on its side for 1 h and then gently turned upright. After the pellet was gently placed at the top of the density gradient, the mixture was spun at 1,500 × g for 10 min. We then pipetted the resultant band of organic materials into a 1.5-mL tube. After two rounds of centrifugation (10,000 × g, 5–10 min) and pipetting, the materials that gathered at the bottom of the tube were aliquoted and stored in 100 μL of water at −20 °C.
DNA extraction.
To isolate DNA, we added 500 μL of cetyltrimethyl ammonium bromide (CTAB) buffer and ∼50 glass beads (425–600 μm; Sigma-Aldrich) to the purified spores. The mixture was vortexed at maximum speed for 2–3 min and incubated overnight at 55 °C with 30 μL of proteinase K (20 mg/μL) and 5 U of chitinase added (New England Biolabs). The resulting viscous supernatant was transferred into a fresh tube for standard phenol-chloroform DNA purification. The DNA pellet obtained was resuspended in tris-EDTA (TE) buffer and stored at −20 °C.
Genome reduction.
We followed the genome reduction protocol described by McCormack et al. (57). Briefly, 50–100 ng of DNA was subjected to simultaneous restriction digest (EcoRI and MseI) and sticky-end adaptor ligation. PCR preamplification with EcoRI and MseI primers that targeted the adaptors was performed with Phusion Taq. A second selective amplification was conducted using a fusion reverse primer (consisting of Roche 454 Lib-L primer B and MseI primer) and forward fusion primer made up of Roche 454 Lib-L primer A, sample-specific barcode (10 bp) and Eco-RI primer. Following electrophoresis (1.5% agarose gel with 100-bp size ladder), PCR fragments in the range of 400–550 bp were excised and column purified with QIAquick gel extraction kit (Qiagen). The eluted products were cleaned with AMPure XP beads to remove all fragments below the size of 300 bp (Beckman-Coulter). The final products were quantified and assessed for quality before being submitted for 1 × 1/8 PTP sequencing with a Roche 454 GS FLX+ machine at the University of Illinois W. M. Keck Center.
Bioinformatics.
The total number of raw reads that passed initial quality control was 45,250 (average length, 272.6 bp). Most bioinformatics processing was carried out in an eight-core Mac Pro using the Mothur suite of programs. First, we demultiplexed the samples by individual barcodes using the command trim.flows (settings: bdiffs, 2; pdiffs, 6; tdiffs, 8; minflows, 100; maxflows, 720; maxhomop, 9). We then used the command shhh.flows to denoise the reads. Next, barcodes were removed, and reads shorter than 80 bp were discarded, resulting in 28,650 reads (average length, 248.2 bp; range, 80–503 bp). We used 454 CD-hit (weizhong-lab.ucsd.edu/cd-hit) to cluster reads at 90% similarity, which resulted in 4,143 clusters (average of 3.01 reads per cluster) and 11,882 singletons. Using default settings, we conducted blastx searches (conducted in August 2012) against the GenBank nr database using consensus sequences or reads from singleton clusters (maximum of one hit per query). We retained sequences or clusters with blastx hits matching the Microsporidia species Encephalitozoon cuniculi, Encephalitozoon intestinalis, Enterocytozoon bieneusi, N. apis, Nosema bombycis, or N. ceranae at an e value of no more than 0.001, resulting in 309 relevant hits representing 209 unique National Center for Biotechnology Information accessions. Using Batch Entrez, we extracted feature information for each of these hits and excluded those with feature terms suggesting that the hits were transposable or repeat elements (e.g., reverse transcriptases from retrotransposons and retroviruses, transposase). This procedure produced a final set of 287 hits and 200 unique blast accessions. The .fasta files of the 287 consensus sequences and blastx results (database searched: all nonredundant GenBank coding sequence translations plus Protein Data Bank plus UniProt plus Protein Information Resource plus Protein Research Foundation, excluding environmental samples from Whole-Genome Shotgun project results) are available in the Dryad Digital Repository (dx.doi.org/10.5061/dryad.83fb8). Using the relevant aligned sequences, we designed and tested primers that were likely to yield polymorphic markers.
Multilocus amplicon sequencing and analysis.
Based on our reduced genome sequencing work and preliminary screening, we designed and tested primers that amplify six loci (Loc127, Loc228, Loc326, Loc381, Loc416, and Loc430) in N. bombi. By sequencing these loci from 31 N. bombi isolates from 16 host bumble bee species (Table S4B), we aimed to decipher additional population genetic structure within N. bombi. To sequence each locus with a Roche 454 GS FLX+, we conducted two rounds of PCR, the first to amplify each locus using locus-specific primers and the second to attach Roche 454 sequencing primers and barcodes. The amplicons were sequenced in 4 × 1/8 PTP, producing a total of 244,635 raw reads. We used the function trim.seq in the Mothur package to conduct initial quality control, (settings: qaverage, 20; qwindowsize, 10; pdiffs, 3; bdiffs, 1; maxambig, 2; maxhomop, 12; minlength, 120–200 bp, depending on the locus), which removed 44.3% of the raw reads. We then took the remaining reads and assembled them against reference reads in Geneious (assembly option: medium sensitivity, no fine-tuning) and obtained 1,547–41,702 mapped reads per locus (average, 16,817.5 reads). To further reduce errors, we removed sequences that occurred at a frequency that was less than 0.1% of the total number of reads for the locus. The sequences for each locus were then realigned using Muscle [default settings, except gap opening penalty (0.5)] (58), followed by Geneious’s own alignment algorithm (settings: cost matrix, 70% similarity; gap open, 18; gap extend, 10). Using Mothur’s filter.seqs function and manual editing, we then removed indel errors commonly found in Roche 454 data, producing a final alignment for each locus. Following this, for each locus, we used the functions count.seqs and dist.seqs to calculate the number of haplotypes in each population and the number of nucleotide substitution steps among haplotypes.
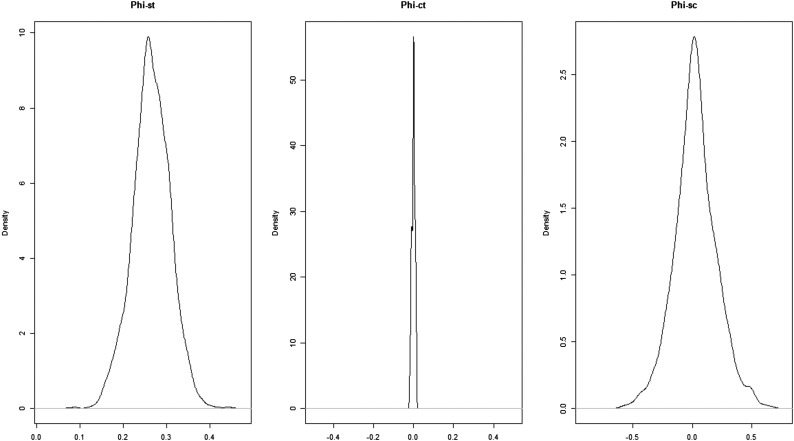
This information was used in a hierarchical Bayesian analysis of molecular variance implemented in the software bamova (17), used to estimate the amount of genetic variation accounted for when N. bombi populations were grouped as European or North American (indicated by parameter ΦCT). We used the next-generation sequencing (NGS)-population model (appropriate when identities of individuals in a population are unknown) and ran the analysis for 500,000 steps (thinning parameter, 10), discarding the first 10% as burn-in. Because the number of individual N. bombi spores/meronts in the gut of each bee was unknown, we ran the analysis specifying a range of population sizes (N = 1000, 10,000, 1,000,000) and obtained similar results. Here, we report the results when N = 10,000 was used. Kernel density plots for this analysis are presented in Fig. S5. To further explore interisolate relationships, we used POPTREEW to construct a bootstrapped neighbor-joining tree based on isolate allele frequency data and pairwise DA distance matrix (45). The P value of the Skillings–Mack test of difference in heterozygosity between North American and European N. bombi isolates was obtained using 1,000 Monte Carlo simulations.
Fig. S5.
Kernel density plots of ΦST (among-isolate genetic variation), ΦCT (between-group genetic variation), and ΦSC (within-group, among-isolate genetic variation) calculated by hierarchical analysis of molecular variance. The analysis was based on data from six variable loci from 31 N. bombi isolates.
Results
Temporal Patterns of Nosema Prevalence.
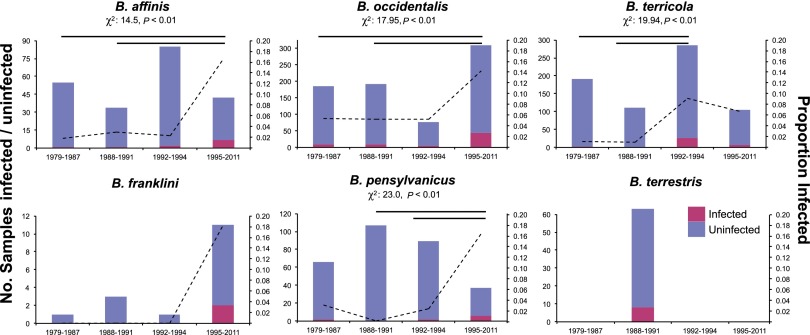
To examine changing patterns of Nosema prevalence over time in five declining North American bumble bee species (B. affinis, B. franklini, B. occidentalis, B. terricola, and B. pensylvanicus) (4), we used PCR to detect Nosema in pinned museum specimens collected from 1979 to 2011 (details are provided in SI Methods). We focus on these declining species because surveys of contemporary populations show moderate to high infection prevalence (e.g., ∼15–37%), whereas populations of nondeclining North American species exhibit negligible infection rates [e.g., ∼0.3–1% (4)] and are not of concern in our investigation. Nosema detection techniques were worked out initially by successful PCR screens of pinned historical European B. terrestris collected from populations with high known N. bombi prevalence (collected 1988 to 1991; n = 63; Table S1). Following successful retrieval and PCR detection of N. bombi DNA from dry European specimens, we screened historical collections of the five declining North American species (total samples: 2,048; Table S1), examining at least 216 individuals per species (mean: 492.3), except B. franklini (N = 16), which was difficult to obtain in large numbers because of its recent precipitous collapse (2), historically small geographic range (2), and rarity in collections and is thus excluded from most statistical analyses (Fig. 1).
Fig. 1.
Number of specimens screened and prevalence level per time period of Nosema for five target North American bumble bee species and the European B. terrestris (pink, infected bees; blue, uninfected bees; left y axis and bars, number of bees; right y axis and dashed lines, proportion out of the total screened). For species with sufficient sample sizes, results of χ2 tests of independence of proportions are shown above each bar graph. Horizontal lines indicate pairs of time periods in which Nosema prevalence is significantly different at P = 0.05 based on arcsine-transformed prevalence data.
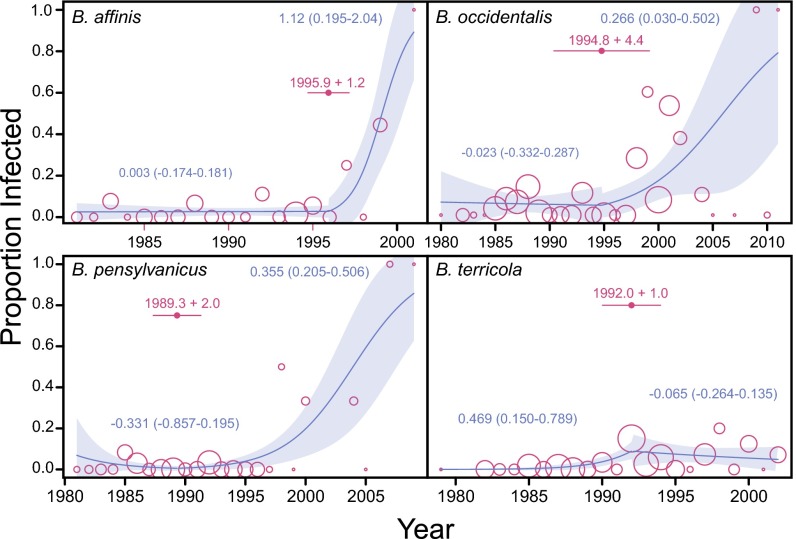
Comparisons of Nosema prevalence in bumble bees sampled over >30 y revealed nonrandom changes over time. In all examined North American Bombus species with substantial sample sizes, Nosema was detected before 1992 (Figs. 1 and 2 and Fig. S1), but significant increases in prevalence occurred after 1992 (Figs. 1 and 2, SI Methods, and Fig. S1). Change-point analysis indicates shifts in year–prevalence regression slopes between 1990 and 1995 (Fig. 2). All species show a positive trend in prevalence over time after 1992, except B. terricola, which shows a prevalence peak about 1992, followed by stability thereafter. In B. franklini, which was listed as severely threatened in 2003 (15), Nosema was detected only during the period of 1995 to 2011. The high Nosema levels amplified from the more recently collected Bombus samples are unlikely the result of greater sensitivity of PCR detection in younger samples based in part on the fact that all control B. terrestris samples exhibited constant prevalence over time—the 20- to 30-y-old historical samples exhibited similar detection frequencies (12.7%, n = 63) to those of freshly collected samples (this study, 18.4%; n = 440) (binomial test, P = 0.15). Furthermore, no temporally correlated amplification success was found for the nuclear long-wavelength opsin (LW Rh) gene in Bombus samples collected between 1982 and 2004 (n = 47 bees; generalized linear model, binomial errors, null deviance vs. residual deviance: χ21 = 0.0558, P = 0.8133; Fig. S2). A less-stringent N. bombi PCR detection threshold (at least one-third of samples had positive PCR) produced qualitatively comparable results (Fig. S3 and Table S2).
Fig. 2.
Yearly proportion of four declining bumble bee species infected with Nosema (open circles). The size of each circle is proportional on a logarithmic scale to the number of bees screened in a given year. Yearly infection rate data are fitted with a piecewise quasibinomial regression curve and its 95% CI (shaded area). For each species, the change point in time (i.e., year) with SE is indicated by a filled circle, with an error bar on either side. Numbers beside curves are estimated slope parameters for each segment of the piecewise regression (95% slope CI in parentheses). For B. affinis, B. pensylvanicus, and B. occidentalis, Nosema prevalence did not vary with time before the estimated change points (i.e., 95% CI of slope parameters include 0), but the prevalence–time relationship became significantly positive (with 95% CI of slope parameters excluding 0) after the change points.
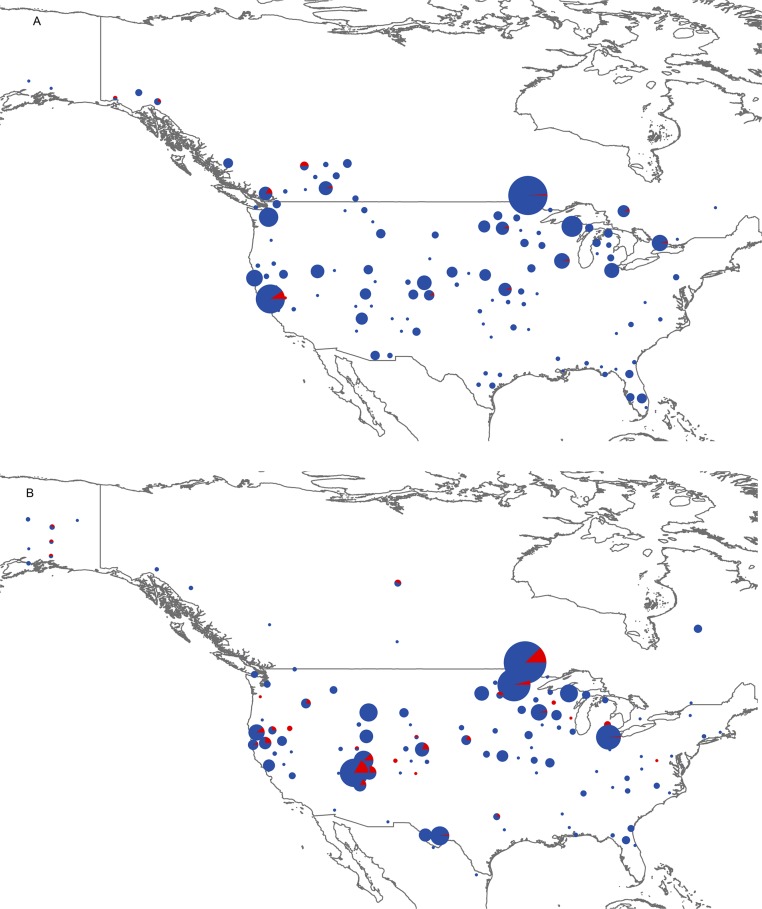
Fig. S1.
Pie charts showing spatial distributions of infected (red) or uninfected (blue) bumble bees for all five North American study species combined. Bees were collected between 1979 and 1991 (A) and between 1992 and 2011 (B). The largest circle in A represents 157 bees, and the largest circle in B represents 187 bees.
Global Pattern of Nosema Diversity.
To obtain a global perspective of Nosema diversity in bumble bees, we used pyrosequencing and Sanger sequencing to obtain a 298-bp portion of the small subunit (SSU) rRNA of Nosema from our North American samples (n = 35 modern and 7 historical samples; 16 species) and three European bumble bee species (N = 10 bees) (SI Methods and Table S3). We combined these data with those from 113 corresponding SSU rRNA GenBank sequences of Asian taxa (27 species sampled in China) and nonredundant European and North American taxa (Table S3). Filtered splits phylogenetic network analysis produced 11 Nosema SSU clades (Fig. 3 and Fig. S4), but the majority (97.1%, n = 34) of both the historical and modern North American Nosema isolates fell into a single clade (N. bombi s. s.), along with 91% of European isolates (n = 22) (Fig. 3). Only two North American samples, both from nondeclining Bombus (Pyrobombus) species (4), produced distinct haplotypes (Table S3 and Fig. S4; B. impatiens, clade 147; Bombus mixtus, clade 27), and only 2 of 22 European samples (Bombus lapidarius and Bombus lucorum) yielded Nosema sequences that fell outside the N. bombi clade (Fig. 3, Mixed hosts/Nosema “A” & B”). This “mixed-hosts” clade comprises Nosema from highly diverse hosts [western honey bee Apis mellifera, bumble bees, and several moth species (Table S3)] and appears closely related to Nosema ceranae, commonly found in honey bees. All Nosema from historical North American specimens fell into the N. bombi clade, except for sequences from a single B. terricola, which produced a highly distinct sequence (clade 1) more closely related to isolates from the mixed-hosts and N. ceranae clades (Fig. 3 and Fig. S4). In contrast to the extremely low Nosema diversity found in North American and European samples, Nosema from Chinese Bombus is highly diverse, falling into six clades distributed across the network (16).
Genetic Differentiation Between Nosema bombi from Europe and North America.
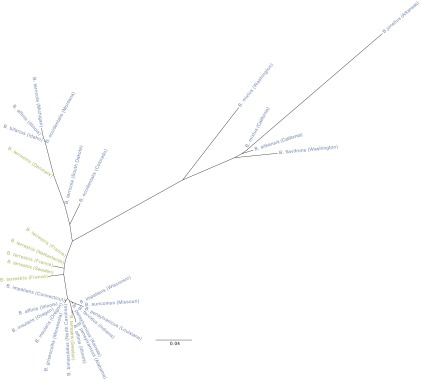
To probe more deeply into possible genetic differentiation between European and North American N. bombi, we conducted deep amplicon pyrosequencing of 31 N. bombi isolates (SI Methods and Table S4) using six previously unidentified genomic markers [246.3 ± 96.4 bp per locus (±SD); >1,000 reads per locus (SI Methods)]. The small number of discovered putative polymorphisms out of a preliminary screen of 287 alignments from reduced representation sequencing of nine N. bombi isolates (SI Methods and Table S4) suggests low variation across the genome. Despite high sequence coverage of polymorphic loci (>1,000 reads per locus), aimed at saturating detection of variation within individual bees, the overall level of N. bombi genetic variation was low for both North American and European isolates (average per locus heterozygosity, 0–0.456) and similar between North American and European samples (locus-specific allelic heterozygosity: Skillings–Mack statistic, 127.67; P = 0.109; 1,000 simulations). Hierarchical Bayesian analysis of molecular variance (17) revealed no significant differentiation between N. bombi from Europe and North American [ΦCT = 0.00; 95% confidence interval (CI), −0.01 to 0.01] but showed that most existing variation occurs among isolates within each region (ΦST = 0.26; 95% CI, 0.19–0.46) (SI Methods and Fig. S5). The lack of differentiation among isolates between continents is supported by the clustering of European N. bombi with the majority of North American N. bombi on a neighbor-joining tree constructed from the genomic allele frequency data (Fig. 4, DA distances).
Fig. 4.
Neighbor-joining tree showing interisolate relationships based on allele frequency data from six genomic loci. Blue text represents North American Bombus species from across the United States; green text represents European B. terrestris taxa from different countries of western Europe.
Discussion
Significantly higher N. bombi prevalence in US Bombus species undergoing population declines relative to healthy species has been documented nationwide (4) and at local and regional levels (2, 3, 5–7, 12, 13). However, resolving the essential question of cause and effect has proven elusive: Did N. bombi infection actually cause the precipitous declines in Bombus populations, or is N. bombi naturally more prevalent in species experiencing decline from other causes? Our examination of N. bombi prevalence over a 30-y time span suggests that prevalence has not been continuously high in declining US species, although infections have been historically present and widely dispersed. Notably, infection rates were low historically (before reported population declines) in all sampled North American host species and began to increase markedly in the mid-1990s, just before the first documented observations of US Bombus decline.
These results align with the hypothesis that N. bombi is a factor in US Bombus decline (18). Our findings link the onset of Bombus population declines with increasing N. bombi infections in wild populations and indicate a temporal connection between historical infections in wild populations and the late 1990s N. bombi-induced collapse of commercial B. occidentalis production in North America—the hypothesized “smoking gun” of Bombus decline and a central tenet of the ENIH (2). These temporal associations add support to the hypothesis that N. bombi escaped into wild populations from heavily infected commercial colonies (2, 3, 18, 19). Although the transmission mechanisms are unknown, it is known that worker bees from commercially reared colonies foraged at high frequencies outside of greenhouses (20, 21), presumably preferring pollen and nectar from wild flowers over the pollen provided by cultivated tomato flowers (18, 22). Commercial pollination of greenhouse tomatoes occurred mostly in the Pacific West region of North America and in Eastern Canada, which comprise the ranges of the declining Bombus s. s. species. Colonies derived from those produced in Europe during the early 1990s were also used in open-field pollination and for field research in those regions (11). Moreover, B. impatiens, a stable species (4) currently reared for commercial pollination in eastern Canada and north central United States, is also known to have escaped commonly from greenhouses into the wild (18, 20, 23). Although overall infection rates are low in B. impatiens (4, 24), infections do occur (6); thus, the species could still be a carrier of N. bombi. Because disease transmission risks were likely unknown in the earliest days of commercial colony production, it is possible that by the time producers became aware of their Nosema problems (11), heavily infected workers servicing greenhouse and open-field pollination throughout North America had already transmitted the pathogen to wild bees. One caveat is that our detection primers were selected for their high sensitivity (25) and can weakly amplify honey bee-associated Nosema (Nosema apis and N. ceranae) that can also be carried occasionally by Bombus in Europe (26). It is thus possible that positive PCR results could reflect infections with these Microsporidia. However, we are unaware of any evidence for N. ceranae or N. apis in North American bumble bees, and all sequencing studies conducted to date by ourselves and others have identified only N. bombi, apart from the small number of outlier sequences from this study. Thus, even if other Nosema are present occasionally, N. bombi is the dominant species in both historical and contemporary samples, and such detections have minimal impact on conclusions, especially given the greater risk of false-negative detections in museum specimens. It would be nearly impossible to rule out with 100% certainty that the temporal increase in Nosema could reflect age-dependent DNA degradation; however, our positive B. terrestris and LW Rh controls suggest the patterns are robust.
The last decade has seen rising acceptance of the view that bumble bee decline in North America is the result of exposure to an exotic European N. bombi strain introduced into native North American populations from European-bred colonies in the 1990s (2, 9). Despite the temporal increase we found in Nosema prevalence in declining species, there is no evidence that a distinct strain of N. bombi was introduced from Europe in the 1990s. The same SSU genotype currently found in host populations throughout Europe and North America was in the United States long before any known commercial trade began between Europe and North America. Moreover, the low, near identical Nosema genetic diversity found throughout North America and Europe precludes the introduction of a distinct N. bombi strain. Both historical and current North American isolates are essentially undifferentiated from those in Europe. The low SSU diversity is in stark contrast to the extensive Nosema diversity found in Chinese bumble bees (16). Although not all of the Chinese Nosema sequences necessarily represent actual infections, the greater diversity of Nosema SSU sequences detected in Asia nonetheless lies in stark contrast to Europe and North America. Our random genomic markers further support the low genetic differentiation of Nosema across Europe and North America. Such low diversity outside of Asia could have arisen through multiple mechanisms, including the natural spread of a Holarctic Nosema lineage before Bombus domestication, or more recently as a result of intensive Bombus domestication in Europe and North America, which has not occurred in China (27). Given the high parasite levels found today in European commercial bumble bees (28), together with the knowledge that commercial foraging bees come and go from “leaky” greenhouses (18, 21) and have been used in open-field pollination, a sweep of a common N. bombi strain is possible. In this scenario, the N. bombi strain resulting in increasing North American prevalence could conceivably reflect a secondary introduction from Europe. If North American and European N. bombi are similar because of recent shared evolutionary history, it will be challenging to identify the signatures of a recent secondary introduction even if one were to have occurred. Regardless of N. bombi's geographic origin, the strong temporal connection between N. bombi epizootics in commercial Bombus stock in the mid-1990s and escalating prevalence of a genetically similar Nosema in declining wild host populations suggests a substantial risk of pathogen transmission from commercial stocks.
In contrast to an invasion hypothesis, increased Nosema prevalence in North America could simply reflect a natural increase in native N bombi. A natural increase in the wild of native N. bombi during the period of increased prevalence could have contributed to the collapse of commercial B. occidentalis breeding following stock replenishment with infected wild B. occidentalis queens (27). Alternatively, if native queens had even low N. bombi prevalence, infections could spread rapidly among high-density colonies in breeding facilities and get transmitted back into wild populations as an epizootic because bumble bees are excellent dispersers (4). Moreover, the artificial conditions of intense breeding of a single narrowly collected species grown in dense facilities are ideal for selection for increased virulence of microbial parasites (29). Given the lack of genetic diversity in Microsporidia genomes in general (30), higher-resolution genomic data will be needed to fully tease apart all signatures of genetic variation in N. bombi. Indeed, genotype-by-genotype host–parasite interactions could be important in bumble bee Nosema infection (31). Identifying such highly localized functional variation in North American and European N. bombi isolates will require fine resolution whole-genome data.
The observations that declining bumble bee species exhibit higher N. bombi prevalence, and that increases in prevalence appear temporally correlated with the onset of Bombus declines in North America, are reminiscent of reports of other Nosema pathogens known to cause widespread threats to honey bees, including N. ceranae, which has been implicated as one factor in Colony Collapse Disorder worldwide (32). Other fungal pathogens are decimating populations of frogs (Batrachochytrium dendrobatidis) (33) and bats (Geomyces destructans) (34) throughout their native ranges. However, confirming a direct causal link between N. bombi and North American bumble bee decline will require further research. Increased prevalence does not necessarily lead to increased pathogen impact on the host (35), and our detection methods do not distinguish individuals carrying infectious and noninfectious Nosema. Although stable and declining species clearly exhibit significantly different prevalences (4), patterns among declining species may differ. Temporal increases were observed across declining species but were more dramatic in B. occidentalis, B. pensylvanicus, and B. affinis than in B. terricola (4). Comparative studies of susceptibility in declining and stable species are needed to reveal whether the increased prevalence in declining species is the result of higher susceptibility to the pathogen or greater N. bombi virulence in some species.
Understanding the transmission mode of N. bombi is also essential for understanding whether Nosema could have caused the observed population declines. Finally, additional factors may also have been involved. Recent reviews of pollinator declines are leaning toward the position that multiple stressors acting in concert are likely causing pollinator decline worldwide (36). These stressors include other pathogens reported from commercially produced bumble bee colonies (28, 37), loss of floral and nesting resources, agrochemicals, and changing climate (36, 38). Increasing physiological stress attributable to environmental degradation is likely to enhance the effects of pathogens. Enhanced procedures put into place by the commercial pollination industry to reduce pathogen infection and transmission, and to curtail intercontinental and interregional trade in commercial colonies, will lower the elevated health risks to wild pollinators witnessed globally over the last two decades.
Methods
Screening for Nosema in Bumble Bee Museum Specimens.
We screened 2,048 bumble bee specimens from five declining North American species (B. affinis, B. franklini, B. occidentalis, B. pensylvanicus, and B. terricola) collected from 1979 to 2011. Historical (1988 to 1991) European B. terrestris and freshly collected samples from regions known to harbor N. bombi were screened as positive controls (Tables S1 and S5). DNA was extracted nondestructively from pinned specimens (SI Methods). An ∼120-bp fragment of the Nosema internal transcribed spacer (ITS) region of the rRNA gene was PCR-amplified with dedicated primers (ITS-f2 and ITS-r2) (25) to detect the presence of Nosema. Both extraction and PCR preparation were conducted in a clean environment, free from PCR products and other contaminants. All amplifications were replicated at least three times. A sample was considered Nosema-positive when at least 50% of PCR replicates successfully amplified the expected product (see Table S2 and Fig. S3 for results with a lower-stringency threshold). To examine whether specimen age adversely affected PCR success, besides using historical B. terrestris with high Nosema prevalence as a control, we amplified a 117-bp fragment of the Bombus opsin (LW Rh) gene from 47 bumble bee specimens of varying ages (1982 to 2004) (Table S1 and Fig. S2).
To detect temporal changes in Nosema prevalence in the four North American bumble bee species with large sample sizes, we conducted χ2 tests of independence of the proportion of infected bees across four time periods (SI Methods), as well as multiple comparison tests. We also conducted piecewise general linear modeling (GLM) (family, quasibinomial), a regression model with change points, to determine whether the year–Nosema prevalence relationship changed significantly for each species around 1992 (39). In this analysis, we used the R-package “segmented” to determine the time point at which the regression slope parameters changed (40). We also conducted analysis of deviance to determine whether the piecewise models provided significantly better fit to the data compared with models without change points (SI Methods).
Small Subunit rRNA Sequencing.
We sequenced a 298-bp fragment of the Nosema SSU rRNA gene from infected North American (n = 35) and European (n = 10) bumble bees collected recently (2009 to 2011), using custom primers targeting conserved sites (SI Methods). In addition, we included seven infected North American museum specimens (Table S3). PCR products were cloned and Sanger-sequenced and/or pyrosequenced. Cloning and sequencing allowed detection of potential coinfections, as commonly seen in the honey bees (26, 41). Demultiplexing and quality control of pyrosequencing reads were performed with Mothur version 1.28 (42). To provide additional geographic context of global Nosema diversity, we supplemented the dataset with 113 nonredundant GenBank sequences derived from a variety of Nosema and related Nosema taxa [species]. SplitsTree version 4.12.6 (43) was used to construct a network using the neighbor-net method and uncorrected p-distances (44) (SI Methods).
Multilocus Amplicon Sequencing.
We used reduced representation genome pyrosequencing of nine N. bombi isolates [each from a single infected Bombus individual (Table S4]) to identify potentially informative genetic loci beyond the rRNA gene (SI Methods). The amount of genetic variation uncovered was low, although we identified six loci for application of deep amplicon pyrosequencing on N. bombi isolates from seven European B. terrestris specimens and 24 North American specimens (15 species) (Table S4). Using POPTREEW, we constructed a neighbor-joining tree of among-isolate relationships based on combined allele frequency data and pairwise DA distance matrix (45). We also tested whether heterozygosity of North American N. bombi isolates was significantly lower than that of isolates from European species using the Skillings–Mack test (46). To estimate differentiation among European or North American N. bombi isolates at multiple levels, we applied a hierarchical Bayesian analysis of molecular variance at three levels: within isolates (ΦST), among isolates-within regions (ΦSC), and between regions (ΦCT) (SI Methods).
Supplementary Material
Acknowledgments
We especially thank L. McIntyre, as well as R. McClure, A. Hudson, and M. Honkanen for DNA extractions of museum samples. We are indebted to P. and A. Rasmont for hosting field collections in France. We also thank J. Strange for Alaska specimens, the many curators from the institutions listed in Table S1, Alvaro Hernandez and Chris Wright for technical support at the University of Illinois W. M. Keck Center, and J. Whitfield for critical reading of the manuscript and helpful discussions. This research was supported by the National Institute of Food and Agriculture, US Department of Agriculture, Agriculture and Food Research Initiative Grant 2010-65104-05992 (to S.A.C., J.D.L., and R.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Nine Roche 454 libraries sequenced for locus discovery have been deposited in the Sequence Read Archive (SRA) database (BioProject, www.ncbi.nlm.nih.gov/bioproject, accession no. PRJNA289884). Sequences and alignments for rRNA and genomic loci and additional data information tables have been deposited in the Dryad Digital Repository (dx.doi.org/10.5061/dryad.83fb8).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1525266113/-/DCSupplemental.
References
- 1.Jones KE, et al. Global trends in emerging infectious diseases. Nature. 2008;451(7181):990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thorp R, Shepherd MD. Subgenus Bombus Latreille, 1802. In: Shepherd MD, Vaughan DM, Black SH, editors. Red List of Pollinator Insects of North America. Xerces Society for Invertebrate Conservation; Portland, OR: 2005. p. 5. [Google Scholar]
- 3.Colla S, Packer L. Evidence for decline in eastern North American bumblebees (Hymenoptera: Apidae), with special focus on Bombus affinis Cresson. Biodivers Conserv. 2008;17(6):1379–1391. [Google Scholar]
- 4.Cameron SA, et al. Patterns of widespread decline in North American bumble bees. Proc Natl Acad Sci USA. 2011;108(2):662–667. doi: 10.1073/pnas.1014743108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillespie S. Factors affecting parasite prevalence among wild bumblebees. Ecol Entomol. 2010;35:737–747. [Google Scholar]
- 6.Malfi RL, Roulston T. Patterns of parasite infection in bumble bees (Bombus spp.) of northern Virginia. Ecol Entomol. 2014;39:17–29. [Google Scholar]
- 7.Tripodi AD, Cibils-Stewart X, McCornack BP, Szalanski AL. Nosema bombi (Microsporidia: Nosematidae) and trypanosomatid prevalence in spring bumble bee queens (Hymenoptera: Apidae: Bombus) in Kansas. J Kans Entomol Soc. 2014;87:225–233. [Google Scholar]
- 8.Goulson D, Lye GC, Darvill B. Decline and conservation of bumble bees. Annu Rev Entomol. 2008;53:191–208. doi: 10.1146/annurev.ento.53.103106.093454. [DOI] [PubMed] [Google Scholar]
- 9.Williams PH, Osborne JL. Bumblebee vulnerability and conservation world-wide. Apidologie (Celle) 2009;40:367–387. [Google Scholar]
- 10.Otti O, Schmid-Hempel P. Nosema bombi: A pollinator parasite with detrimental fitness effects. J Invertebr Pathol. 2007;96(2):118–124. doi: 10.1016/j.jip.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 11.Flanders RV, Wheling WF, Craghead AL. 2003. Laws and regulations on the import, movement and release of bees in the United States. For Nonnative Crops, Whence Pollinators of the Future? eds Strickler K, Cane JH (Thomas Say Publications in Entomology, Lanham, MD), pp 99–111.
- 12.Giles V, Ascher JS. A survey of the bees of the Black Rock Forest Preserve, New York (Hymenoptera: Apoidea) J Hym Res. 2006;15:208–231. [Google Scholar]
- 13.Grixti JC, Wong LT, Cameron SA, Favret C. Decline of bumble bees (Bombus) in the North American Midwest. Biol Conserv. 2009;142:75–84. [Google Scholar]
- 14.Colla SF, Gadallah F, Richardson L, Wagner D, Gall L. Assessing declines of North American bumble bees (Bombus spp.) using museum specimens. Biodivers Conserv. 2012;21:3585–3595. [Google Scholar]
- 15.Kevan PG. 2008 Bombus franklini. The IUCN Red List of Threatened Species 2008: e.T135295A4070259. Available at dx.doi.org/10.2305/IUCN.UK.2008.RLTS.T135295A4070259.en. Accessed March 14, 2016.
- 16.Li J, et al. Diversity of Nosema associated with bumblebees (Bombus spp.) from China. Int J Parasitol. 2012;42(1):49–61. doi: 10.1016/j.ijpara.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Gompert Z, Buerkle CA. A hierarchical Bayesian model for next-generation population genomics. Genetics. 2011;187(3):903–917. doi: 10.1534/genetics.110.124693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colla SR, Otterstatter MC, Gegear RJ, Thomson JD. Plight of the bumble bee: Pathogen spillover from commercial to wild populations. Biol Conserv. 2006;129:461–467. [Google Scholar]
- 19.Thorp RW. 2003. Bumble bees (Hymenoptera: Apidae): Commercial use and environmental concerns. For Nonnative Crops, Whence Pollinators of the Future? eds Strickler K, Cane JH (Thomas Say Publications in Entomology, Lanham, MD), pp 21–40.
- 20.Whittington R, Winston ML. Comparison and examination of Bombus occidentalis and Bombus impatiens (Hymenoptera: Apidae) in tomato greenhouses. J Econ Entomol. 2004;97(4):1384–1389. doi: 10.1093/jee/97.4.1384. [DOI] [PubMed] [Google Scholar]
- 21.Kraus FB, et al. Greenhouse bumblebees (Bombus terrestris) spread their genes into the wild. Conserv Genet. 2011;12:187–192. [Google Scholar]
- 22.Whittington R, Winston ML. Effects of Nosema bombi and its treatment fumagillin on bumble bee (Bombus occidentalis) colonies. J Invertebr Pathol. 2003;84(1):54–58. doi: 10.1016/s0022-2011(03)00123-x. [DOI] [PubMed] [Google Scholar]
- 23.Murray T, Coffey MF, Kehoe E, Horgan FG. Pathogen prevalence in commercially reared bumble bees and evidence of spillover in conspecific populations. Biol Conserv. 2013;159:269–276. doi: 10.1016/j.biocon.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cordes N, et al. Interspecific geographic distribution and variation of the pathogens Nosema bombi and Crithidia species in United States bumble bee populations. J Invertebr Pathol. 2012;109(2):209–216. doi: 10.1016/j.jip.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Klee J, Tek Tay W, Paxton RJ. Specific and sensitive detection of Nosema bombi (Microsporidia: Nosematidae) in bumble bees (Bombus spp.; Hymenoptera: Apidae) by PCR of partial rRNA gene sequences. J Invertebr Pathol. 2006;91(2):98–104. doi: 10.1016/j.jip.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 26.Fürst MA, McMahon DP, Osborne JL, Paxton RJ, Brown MJF. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature. 2014;506(7488):364–366. doi: 10.1038/nature12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Velthuis HHW, van Doorn A. A century of advances in bumblebee domestication and the economic and environmental aspects of its commercialization for pollination. Apidologie (Celle) 2006;37:421–451. [Google Scholar]
- 28.Graystock P, et al. The trojan hives: Pollinator pathogens, imported and distributed in bumblebee colonies. J Appl Ecol. 2013;50:1207–1215. [Google Scholar]
- 29.Schmid-Hempel P. Evolutionary Parasitology: The Integrated Study of Infections, Immunology, Ecology, and Genetics. Oxford University Press; New York: 2011. [Google Scholar]
- 30.Cornman RS, et al. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS Pathog. 2009;5(6):e1000466. doi: 10.1371/journal.ppat.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmid-Hempel P, Loosli R. A contribution to the knowledge of Nosema infections in bumble bees, Bombus spp. Apidologie (Celle) 1998;29:525–535. [Google Scholar]
- 32.Higes M, et al. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ Microbiol. 2008;10(10):2659–2669. doi: 10.1111/j.1462-2920.2008.01687.x. [DOI] [PubMed] [Google Scholar]
- 33.Berger L, et al. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc Natl Acad Sci USA. 1998;95(15):9031–9036. doi: 10.1073/pnas.95.15.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blehert DS, et al. Bat white-nose syndrome: An emerging fungal pathogen? Science. 2009;323(5911):227. doi: 10.1126/science.1163874. [DOI] [PubMed] [Google Scholar]
- 35.Anderson RM, May RM. Infectious Diseases of Humans: Dynamics and Control. Oxford Univ Press; Oxford, New York: 1991. [Google Scholar]
- 36.Goulson D, Nicholls E, Botías C, Rotheray EL. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science. 2015;347(6229):1255957. doi: 10.1126/science.1255957. [DOI] [PubMed] [Google Scholar]
- 37.Sachman-Ruiz B, Narváez-Padilla V, Reynaud E. Commercial Bombus impatiens as reservoirs of emerging infectious diseases in central Mexico. Biol Invasions. 2015;17:2043–2053. [Google Scholar]
- 38.Kerr JT, et al. CLIMATE CHANGE. Climate change impacts on bumblebees converge across continents. Science. 2015;349(6244):177–180. doi: 10.1126/science.aaa7031. [DOI] [PubMed] [Google Scholar]
- 39.Ulm K. A statistical method for assessing a threshold in epidemiological studies. Stat Med. 1991;10(3):341–349. doi: 10.1002/sim.4780100306. [DOI] [PubMed] [Google Scholar]
- 40.Muggeo V. Segmented: An R package to fit regression models with broken-line relationships. R News. 2008;8:20–25. [Google Scholar]
- 41.Chen Y, et al. Asymmetrical coexistence of Nosema ceranae and Nosema apis in honey bees. J Invertebr Pathol. 2009;101(3):204–209. doi: 10.1016/j.jip.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 42.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23(2):254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 44.Bryant D, Moulton V. Neighbor-net: An agglomerative method for the construction of phylogenetic networks. Mol Biol Evol. 2004;21(2):255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- 45.Takezaki N, Nei M, Tamura K. POPTREEW: Web version of POPTREE for constructing population trees from allele frequency data and computing some other quantities. Mol Biol Evol. 2014;31(6):1622–1624. doi: 10.1093/molbev/msu093. [DOI] [PubMed] [Google Scholar]
- 46.Chatfield M, Mander A. The Skillings-Mack test (Friedman test when there are missing data) Stata J. 2009;9(2):299–305. [PMC free article] [PubMed] [Google Scholar]
- 47.Shykoff JA, Schmid-Hempel P. Incidence and effects of four parasites in natural-populations of bumble bees in Switzerland. Apidologie (Celle) 1991;22:117–125. [Google Scholar]
- 48.Durrer S, Schmid-Hempel P. Parasites and the regional distribution of bumblebee species. Ecography. 1995;18:114–122. [Google Scholar]
- 49.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2013. [Google Scholar]
- 50.Koch JB, Strange JP. The status of Bombus occidentalis and B. moderatus in Alaska with special focus on Nosema bombi incidence. Northwest Sci. 2012;86:212–220. [Google Scholar]
- 51.Fredslund J, Schauser L, Madsen LH, Sandal N, Stougaard J. PriFi: Using a multiple alignment of related sequences to find primers for amplification of homologs. Nucleic Acids Res. 2005;33(Web Server issue):W516–W520. doi: 10.1093/nar/gki425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Mahony EM, Tay WT, Paxton RJ. Multiple rRNA variants in a single spore of the microsporidian Nosema bombi. J Eukaryot Microbiol. 2007;54(1):103–109. doi: 10.1111/j.1550-7408.2006.00232.x. [DOI] [PubMed] [Google Scholar]
- 53.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA Tools: A web server integrating INTARNA, EXPARNA and LOCARNA. Nucleic Acids Res. 2010;38(Web Server issue):W373–W377. doi: 10.1093/nar/gkq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morrison DA. Introduction to Phylogenetic Networks. RJR Productions; Uppsala: 2011. [Google Scholar]
- 57.McCormack JE, et al. Next-generation sequencing reveals phylogeographic structure and a species tree for recent bird divergences. Mol Phylogenet Evol. 2012;62(1):397–406. doi: 10.1016/j.ympev.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 58.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.