Significance
Homologous pairing occurs in budding yeast during vegetative growth, but its function in gene regulation is unknown. In this work, we revealed that a GAL1 reporter gene can interact with its homologous allele and influence its activity. These results are significant because (i) it shows that the 3D organization of yeast chromosomes can contribute to gene regulation, (ii) it goes against the dogma that gene regulation in budding yeast only occurs over short distance via cis regulatory elements, and (iii) it contributes to the mechanistic understanding of long-distance gene regulation in yeast, as well as related phenomena in higher eukaryotes.
Keywords: homologous pairing, interallelic interaction, interallelic regulation, transvection, single-cell gene expression
Abstract
In Drosophila, homologous chromosome pairing leads to “transvection,” in which the enhancer of a gene can regulate the allelic transcription in trans. Interallelic interactions were also observed in vegetative diploid budding yeast, but their functional significance is unknown. Here, we show that a GAL1 reporter can interact with its homologous allele and affect its expression. By ectopically inserting two allelic reporters, one driven by wild-type GAL1 promoter (WT GAL1pr) and the other by a mutant promoter with delayed response to galactose induction, we found that the two reporters physically associate, and the WT GAL1pr triggers synchronized firing of the defective promoter and accelerates its activation without affecting its steady-state expression level. This interaction and the transregulatory effect disappear when the same reporters are located at nonallelic sites. We further demonstrated that the activator Gal4 is essential for the interallelic interaction, and the transregulation requires fully activated WT GAL1pr transcription. The mechanism of this phenomenon was further discussed. Taken together, our data revealed the existence of interallelic gene regulation in yeast, which serves as a starting point for understanding long-distance gene regulation in this genetically tractable system.
Cell proliferation and differentiation depend on rigorously controlled gene activities. The mechanism of gene regulation is best understood at the level of linear organization of the genome, including the primary DNA sequences and arrays of closely associated regulatory proteins. Three-dimensional (3D) organization of chromosomes also plays an important role in gene regulation. For example, distant cis-regulatory elements can be looped to the target promoters and regulate the corresponding genes, affecting both the average expression level and the cell-to-cell variability (1–4). In yeast, promoters and terminators can form loops to allow rapid reactivation of the gene following a period of repression (5, 6). Highly transcribed genes tend to associate with nuclear pore complex, and this association helps to maintain the transcriptional memory (7–9). Elucidating the regulatory roles of high-order chromosome organization is thus a key step toward the understanding of eukaryotic gene regulation.
One type of high-order interaction involves pairing between homologous chromosomes in somatic diploid or polyploid cells. Homologous pairing is most prominent in the salivary gland cells in Drosophila, where many copies of homologous chromosomes bundle along their entire length to form a polytene chromosome. Such pairing underlies a phenomenon called “transvection,” in which the regulatory region of a gene influences the transcription of the paired gene in “trans” (10, 11). For a pair of essential genes, one with a defective open reading frame (ORF) and the other with a defective enhancer, the functional enhancer can activate the WT ORF in trans and lead to phenotypic rescue (10, 11). These observations demonstrate the physiological importance of transvection.
Homologous pairing was also observed in diploid budding yeast during vegetative growth, although the extent of the pairing is controversial among literature (12–15). Unlike the Drosophila homologous chromosomes that stably associate with each other, the yeast chromosomes tend to interact more dynamic and sporadic at interstitial locations (12, 14). Similarly, in interphase mammalian cells, homologous chromosomes occupy different territories (16), but discrete regions on these chromosomes may loop out of the territories and interact with each other dynamically (17–20). Whether such dynamic interactions play a role in gene regulation is not clear.
In this study, we picked the promoter of a galactose metabolism gene GAL1 (GAL1pr) as a model to investigate the potential interallelic trans-regulatory effect in budding yeast. According to a statistical analysis of the Hi-C data, the Gal4 binding sites (GAL1 activator) cluster in the nucleus (21). β-estradiol–induced GAL1pr on two different chromosomes could form trans interactions under the activating condition in haploid yeast (22). The upstream activation sequence (UAS) of GAL1pr was able to activate transcription in trans when introduced into the Drosophila genome (23). Based on this evidence, we reasoned that GAL1pr is a good candidate for long-distance gene regulation. Indeed, our findings below support the notion that GAL1 promoter can interact with its homologous allele and influence its activity during galactose induction.
Results
The Activation of a Defective GAL1 Promoter Is Accelerated by a Wild-Type Allele.
To mimic the classic transvection experiment, we generated two reporter genes, one with a truncated GAL1pr (Δ1pr) driving WT GFP, and the other with WT GAL1pr driving inactive gfp (GFP with frame-shift mutation) (Fig. 1A and Methods). The WT GAL1pr contains the entire GAL1-10 intergenic region and is flanked by a 170-bp segment of the GAL10 ORF from the 5′ end (but not the rest of the GAL10). The Δ1pr deletes one of the four Gal4 binding sites and a few putative Rsc3 binding sites and shows delayed activation during galactose induction (24). We inserted these reporters either as a single copy (WT-GFP/- and Δ1pr-GFP/-) or as alleles (WT-gfp/Δ1pr-GFP) into a ChrII location in diploid yeast (Methods). This location was chosen to be far away from the endogenous GAL1-10 locus and other galactose-related genes so that the insertion would not perturb the galactose metabolism. Indeed, these strains have the same growth rate as wild type in both glucose and galactose media.
Fig. 1.
Interallelic gene regulation measured by FACS. (A) Strain constructs. We constructed diploid strains containing WT GAL1pr driving gfp (GFP with frame-shift mutation) and mutant promoter (Δ1 or Δ2) driving WT GFP. The two reporters were integrated at allelic loci on ChrII. Yellow bar, Gal4 binding site; parentheses, deleted sequence; arrow, TSS; red bar, integration site; purple circle, centromere. Same notations apply to the figures below. (B) Histograms of Δ1pr-GFP FACS data at three time points of induction. Δ1pr-GFP/-, diploid cells with one ChrII containing Δ1pr-GFP and the other intact; Δ1pr-GFP/wt-gfp, diploid cells with one ChrII containing Δ1pr-GFP and the other containing wt-gfp at the allelic locus. To enhance the difference between the WT and mutant promoter, the stains were induced with 3% galactose + 2% raffinose + 0.1% glucose (for the 4 h and 6 h data). For the steady-state measurement, the strains were induced with 3% galactose + 2% raffinose (no glucose) for 15 h. (C) Fraction of activated cells containing Δ1pr-GFP or Δ2pr-GFP in the presence or absence of the allelic WT GAL1pr-gfp. (D) The steady-state level of GFP expression in these strains (normalized by that of WT GAL1pr-GFP) (D). Note that the induction rate was increased, but not the steady-state level. (E) The same as in C except that the Δ1pr-GFP and WT GAL1pr-gfp are at nonallelic location.
We induced the GAL1pr by switching the media from glucose to galactose [3% (wt/wt)] + raffinose [2% (wt/wt)] + glucose (0.1%) and measured the GFP intensity using flow cytometry (FACS) at different induction times. The small amount of glucose was used to slow down the induction and enhance the difference in the activation rates between the WT and mutated GAL1pr. Consistent with previous reports that GAL1pr turns on stochastically in individual cells (25, 26), GFP intensity showed an “on or off” bimodal distribution before reaching the steady state (Fig. 1B, 4 h and 6 h). At each time point, we quantified the activated fraction by fitting the distribution with two skewed Gaussians.
As a single copy (Δ1pr-GFP/-), Δ1pr showed both slower activation and lower steady-state expression comparing with the WT (Fig. 1 B–D) (24). However, in the presence of allelic WT GAL1pr-GFP (Δ1pr-GFP/WT-gfp), the activation of Δ1pr-GFP was accelerated to a near-wild type level (Fig. 1 B and C). In contrast, there was no significant change in the steady-state level with or without the WT allele (Fig. 1D). Similar results were obtained with another mutant GAL1 promoter where two of the four Gal4 binding sites were deleted (Δ2pr) (Fig. 1 A, C, and D). Importantly, when we inserted the two reporters into nonallelic locations by keeping Δ1pr-GFP on ChrII and moving WT-gfp to ChrV, this effect disappeared (Fig. 1E and Methods). Therefore, this trans-regulatory effect is sensitive to the relative location of the two reporters.
The Activation of Δ1pr Is Highly Synchronized with the WT Allele in Each Single Cell.
A “snap-shot” method like FACS cannot follow gene activation dynamics in single cells. To visualize the acceleration effect in real time, we performed time-lapse live cell imaging in strains containing Δ1pr-Venus (a version of YFP) and/or WT GAL1pr-GFP at allelic or nonallelic locations (Fig. 2A). GFP and Venus have comparable brightness and maturation rate in yeast, allowing us to directly compare their intensity (27, 28). Using a flow cell device that allows rapid media exchange, we pregrew the cells in glucose, switched to galactose + raffinose + 0.05% glucose, and monitored the GFP and YFP intensity during the induction process (Fig. 2B and Methods). We then mathematically removed the cross-talk between the GFP and YFP fluorescence and analyzed their activation dynamics separately (Methods).
Fig. 2.
Interallelic gene regulation measured by time-lapse fluorescence microscopy. (A) Strain constructs. The diploid strains contain Δ1pr-YFP and WT GAL1pr-GFP at allelic (II/II′) or nonallelic (II/XVI) locations. (B) Time-lapse live-cell imaging of the allelic strain during induction with 3% galactose + 2% raffinose + 0.05% glucose. (C) GFP and YFP intensity (after cross-talk elimination) as a function of time in three single cells, each with a different color. The solid/stippled arrows point to the activation time in the GFP/YFP traces, respectively. (D) Histogram of the difference in GFP and YFP activation time (ΔT) in the allelic vs. the nonallelic strain. The activation of GFP and YFP are more synchronized in the allelic case. Total trace number: allelic, 82; nonallelic, 128. (E) Activated fraction as a function of time. The plot shows the cumulative distribution of the activation time derived from the single-cell traces in the allelic (Left) and the nonallelic (Right) strain. The GFP activation follows similar rate in these two strains, but YFP activation is faster in the allelic case. (F) Steady-state YFP intensity in Δ1pr-YFP(II)/-, Δ1pr-YFP(II)/wt-GFP(II), and Δ1pr-YFP(II)/wt-GFP(XVI) strains. The box plot shows the distribution of the steady-state level in single cells, which was quantified as the average YFP intensity >4 h after activation.
Consistent with previous reports (25, 26), we observed a large cell-to-cell variability in GFP and YFP activation: Some cells were turned on within 3 h after the media exchange, and others remained “off” for 8 h. However, when the two reporters are at allelic sites, the activation of GFP and YFP in the same cell are almost perfectly synchronized (Fig. 2 C–E). For nonallelic reporters, their activation timings are also correlated, but the Δ1pr-YFP activation tends to have a 20- to 60-min delay relative to WT GAL1pr-GFP, so the activation curve of YFP lags behind that of GFP (Fig. 2 C–E). We also analyzed the steady-state Δ1pr-YFP activity (YFP intensity > 4 h after activation), and they are essentially the same with or without the allelic WT copy (Fig. 2F). Overall, the data in Figs. 1 and 2 indicate that (i) the main rate-limiting step of the GAL1pr activation is the switch of the GAL network that affects Gal4-regulated genes globally, (ii) Δ1pr has an intrinsic slower response to the global GAL activation signal, and (iii) WT GAL1pr allele causes a faster response of Δ1pr by triggering synchronized transcription without affecting the steady-state Δ1pr activity.
Two Allelic Reporters Interact in trans Under the Inducing Condition.
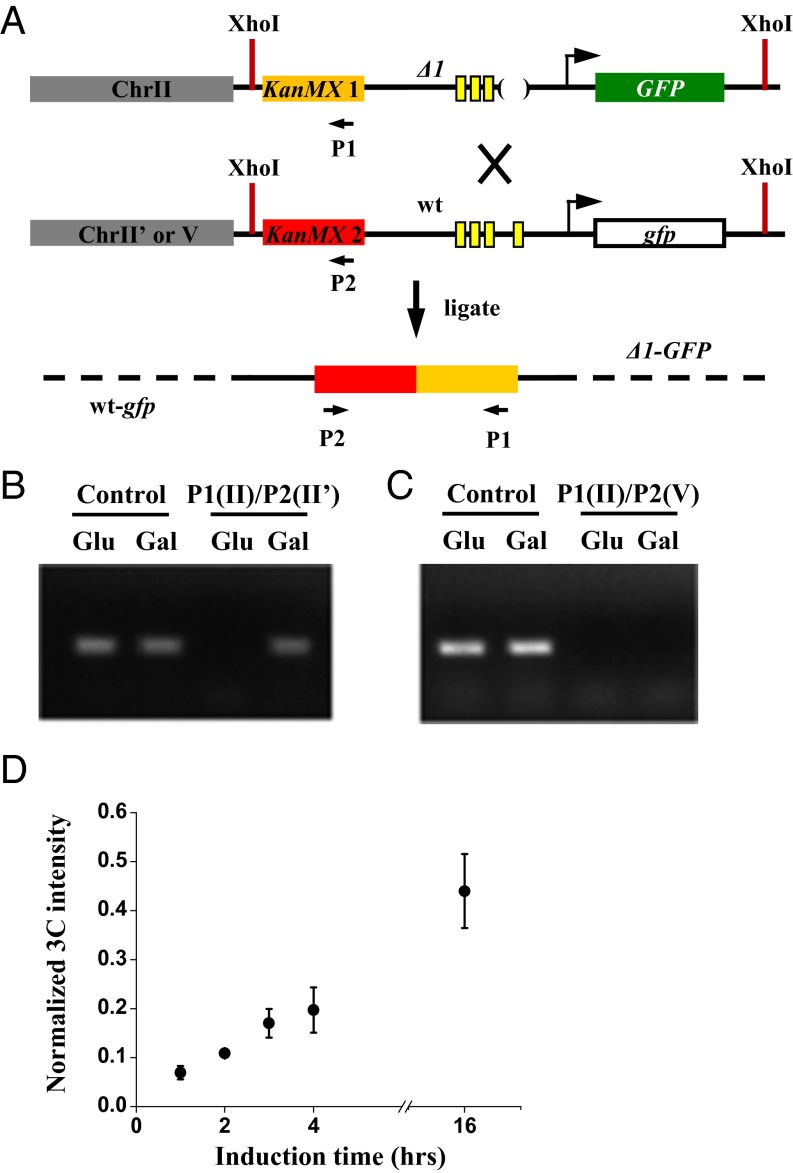
For WT GAL1pr to influence the activation of Δ1pr, they presumably need to form physical interactions. To directly probe such trans interaction, we performed a chromosome configuration capture (3C) measurement between either allelic or nonallelic Δ1pr and WT GAL1pr in both glucose and galactose media (Fig. 3A and Methods). We also carried out the same measurement by using two cis segments 13 kb apart on ChrXV as a positive control (22).
Fig. 3.
Inducible trans interaction between Δ1 and the WT GAL1 promoter. (A) Design of the 3C assay. Two flanking KanMx segments and XhoI cutting sites were inserted for PCR purpose. Cross-linked chromosomes were digested with XhoI, ligated, and subjected to PCR between primers P1 and P2. The two alleles were either inserted at allelic (II/II′) or nonallelic loci (II/V). (B and C) Representative gel showing the result of the 3C assay. The P1/P2 PCR product was only visible between alleles after galactose induction. The interaction between two cis regions on ChrXV was detected as a positive control. (D) The intensity of the P1/P2 3C signal as a function of induction time. The intensity was quantified by qPCR and normalized by the 3C signal of the positive control.
As shown in Fig. 3 B and C, the 3C signal was only detected between the allelic Δ1pr and WT GAL1pr under the inducing condition, whereas the control signal remained constant. When we repeated the 3C measurements between the allelic Δ1pr-GFP and WT-gfp at different induction times and quantified the 3C signal with quantitative PCR (qPCR), we found that the interaction gradually increased over time and continued to be present after GAL1pr reached the steady state (Fig. 3D and Methods). These results show that, at least among the integration sites we selected, the chromosomal interaction can only be established between two allelic reporters. The interaction strength is well-correlated with transcription, indicating that the association may be mediated by proteins involved in GAL1pr activation.
The Gal4 Binding Is Required for the Interaction Between the Two Alleles, but Not Sufficient for the Transactivation Effect.
GAL1pr activation starts with the binding of transcription activator Gal4. To determine whether Gal4 binding is required for the interallelic interaction and trans regulation, we constructed a GAL1pr with all four Gal4 binding sites deleted (Δ4pr) and integrated the Δ4pr-gfp and Δ1pr-GFP into allelic locations on ChrII (same location as in Fig. 1A). The Δ4pr does not bind to Gal4 and has no galactose-induced activity (29). The 3C signal was completely abolished between these two alleles under the activating condition (Fig. 4A), demonstrating that Gal4 is indispensable for the interallelic interaction. No acceleration effect was observed for Δ1pr-GFP in the presence of allelic Δ4pr-gfp (Fig. 4A).
Fig. 4.
Interallelic interaction and regulation between varied alleles. (A) Interallelic interaction and regulation between Δ1pr-GFP and Δ4pr-gfp. In Δ4pr, the entire UAS from the WT GAL1pr was deleted. (Lower Left) Gel image of the 3C measurement between the two alleles. (Lower Right) Activated fraction of Δ1pr-GFP as a function of time with or without allelic Δ4pr-gfp. (B) The same as A except that the Δ4pr was replaced by ΔTATApr, where the TATA element of the WT GAL1pr was mutated (Methods).
The absence of the interallelic interaction and regulation between Δ1 and Δ4 promoters may be due to the loss of Gal4 binding per se or the loss of transcription. To differentiate between these two possibilities, we mutated the TATA box of the WT GAL1pr (ΔTATApr) to selectively abolish the transcription without affecting the Gal4 binding. This particular TATA mutation was shown to severely decrease the GAL1pr activity (29), and our FACS measurement of the ΔTATApr-GFP/- strain confirmed that the steady-state GFP level driven by the ΔTATApr is less than 10% of that by the WT GAL1pr. Interestingly, allelic ΔTATApr-gfp and Δ1pr-GFP can still form physical contacts, but Δ1pr-GFP activation is no longer accelerated (Fig. 4B). These findings suggest that the WT level of GAL1pr transcription is not required for the allelic interaction, but is essential for the trans-regulatory effect on gene expression.
Discussion
Genes in budding yeast are traditionally thought to be regulated over short distances through cis regulatory elements immediately upstream the core promoters. However, numerous long-distance intrachromosomal and interchromosomal interactions have been observed in the yeast genome (30), raising the question as to whether these interactions play a role in gene regulation. In this study, we focused on the interaction and trans regulation between allelic/nonallelic reporter genes in diploid budding yeast. We found that a defective GAL1 promoter can make contacts with an allelic WT GAL1pr during galactose induction and acquires a faster activation rate.
Our findings have implications for the 3D organization of the genome. First, the interaction between the GAL1 reporters was only detected under the activating condition. This observation is consistent with previous statistical analysis, indicating that coregulated genes tend to cluster, and genes that are physically proximal tend to coexpress (21, 31, 32). Coactivated genes can also form “transcription factories” or “multigene complexes” in higher eukaryotic species (33, 34), so our findings here are likely to be one example of a more general phenomenon. This result can also explain why there have been conflicting observations on homologous pairing in yeast (12–15): The experimental outcome likely depends on the inspected regions and their transcription status. The molecular nature of the interallelic interaction requires further elucidation. Because the interaction requires the presence of Gal4 on both alleles (Fig. 4A), but not the WT GAL1pr transcription (Fig. 4D), the simplest possibility is that the Gal4 dimers can interact and bring the two alleles together, but to our knowledge there is no evidence for Gal4 oligomerization. Alternatively, the interaction may be mediated by some downstream factors recruited by Gal4 before transcription initiation.
Second, among the integration sites we have chosen, the interaction was only detected between the allelic reporters. We can think of two explanations. The first possibility is that the GAL1 promoter does not explore the whole nuclear space for binding partners, and the contacts it can make are restricted to a nuclear subvolume. Similar observations have been made in mammalian cells (4), and it was proposed that this constraint is imposed by the limited motion of the chromosomal context. The second possibility is that when the two reporters are at nonallelic sites, one or both of them can find better binding partners, preventing them from interacting with each other. In either case, our results indicate that the allelic reporters occupy similar 3D space before galactose induction and, therefore, have higher probability to form contact.
Third, in all our experimental conditions where the interaction between the two reporters was lost, the acceleration effect also disappeared (e.g., in Figs. 1E and 4A). In contrast, there is a case where the interaction remained, but we could not detect any acceleration (Fig. 4B). These data suggest that the interallelic interaction is necessary but not sufficient for trans regulation.
How does the interallelic interaction lead to change in the Δ1/Δ2pr expression? One possibility is that the UAS on the WT GAL1pr can directly recruit and deposit transcription machinery onto the paired allele. However, this model could not explain why the WT GAL1pr selectively enhances the activation rate but not the steady-state level of the Δ1/Δ2pr expression. In addition, this model predicts a competitive relation between the cis and trans core promoters, and we would expect the trans-regulatory effect to be enhanced after the removal of the cis TATA element (an observation made for transvection in Drosophila; refs. 23 and 35). Instead, we found that the deletion of TATA in the WT GAL1pr eliminated the trans regulation. We therefore suspect that the acceleration of the Δ1/Δ2pr expression is not due to direct activation in trans. This result also argues that the mechanism used here is different from that of transvection (or at least some classes of transvection).
Another component in the GAL regulatory pathway is long noncoding RNA (lncRNA). Two lncRNAs from the GAL1-10 and GAL7 locus can regulate endogenous GAL1 expression both in cis and in trans (36–38). These lncRNAs are not expressed from our reporter constructs because they lack the 3′ end GAL10 ORF sequence that is required for the initiation of these lncRNAs. Our reporters also do not contain the 5′ end of the GAL1 ORF that is critical for the trans regulation of the lncRNA (38). The lncRNAs disappear after GAL activation, but the allelic Δ1pr-GFP and wt-gfp remain in contact (Fig. 3D). Δ1pr-GFP/- and Δ1pr-GFP/wt-gfp strains have the same amount of GAL10 lncRNAs from the native site, but Δ1pr-GFP has faster activation in the latter strain. Overall, the evidence argues against the idea that the allelic interaction and trans regulation is mediated by the GAL10 lncRNAs.
We propose an indirect mechanism where the WT GAL1pr stimulates the Δ1/Δ2pr expression by generating a more transcriptionally engaging environment. Recent studies indicate that pol II tends to form concentrated foci in the nucleus, and such pol II “clustering” may present a rate-limiting step for transcription activation (39). Because the Δ1/Δ2pr has less Gal4 binding capacity, we speculate that they are not as effective as the WT promoter in the initial recruitment of general transcription factors and pol II, causing the delayed activation. By physical association, actively transcribed WT GAL1pr may provide higher local pol II concentration, allowing Δ1/Δ2pr to have higher transcription rate at the early stage of induction when pol II is still limiting. In contrast, the steady-state expression level of the Δ1/Δ2pr may be limited by the maximum rate of the mediator/pol II recruitment at saturating concentration. Therefore, further increase in the pol II concentration will not result in an increase in the expression level.
Methods
Strain and Plasmids Construction.
All plasmids used in this study were derived from pRS yeast shuttle vectors, and the GAL1pr reporters were inserted into the multiple cloning sites. The Δ1, Δ2, and Δ4 promoters were constructed by deletion of the −395 to −335 bp, −312 to −277 bp, and −395 to −277 bp fragments from the WT GAL1pr, respectively (relative to the start of the ORF). For the ΔTATA promoter, the original TATA box “ATATAA” was mutated to “gcgTAA.” GFP frame shift mutation was generated by inserting a 4-bp sequence “ATCC” between 104 and 105 bp of GFP ORF. This mutated GFP shows no green fluorescence signal. All plasmids were confirmed by Sanger sequencing.
All of the yeast strains are W303-congenic (see Table S1 for strain list). We first generated two background haploid strains, MMY116 (W303a, ADE2) and W303-TRP (W303α, TRP1), and integrated the reporters into these strains (in most cases with the URA3 marker). To direct the reporters into ChrII, we inserted a homologous region (ChrII 664401–664682) into the plasmid, linearized by KpnI, and transformed into the haploid yeast following the standard protocol. Similarly, we used the segments ChrV 394711–394317 and ChrXVI 67026–67427 for integration into ChrV and ChrXVI.
Table S1.
Strain list
| Ploidy | Strains | Genotype |
| Haploids | MMY116 | MATa, ADE2 |
| W303-Trp | MATalpha, TRP1 | |
| yDZ01 | MATa, ADE2, ChrII:: GALpr-GFP-URA3 | |
| yDZ02 | MATa, ADE2, ChrII:: GALpr-gfp-URA3 | |
| yDZ03 | MATa, ADE2, ChrII:: Δ4pr-GFP-URA3 | |
| yDZ05 | MATa, ADE2, ChrII:: Δ2pr-GFP-URA3 | |
| yDZ06 | MATa, ADE2, ChrII:: Δ1pr-GFP-URA3 | |
| yDZ07 | MATalpha, TRP1, ChrII:: GALpr-GFP-URA3 | |
| yDZ08 | MATalpha, TRP1, ChrII:: GALpr-gfp-URA3 | |
| yDZ09 | MATalpha, TRP1, ChrII:: Δ4pr-GFP-URA3 | |
| yDZ11 | MATalpha, TRP1, ChrII:: Δ2pr-GFP-URA3 | |
| yDZ12 | MATalpha, TRP1, ChrII:: Δ1pr-GFP-URA3 | |
| yDZ13 | MATa, ADE2, ChrV:: GALpr-GFP-URA3 | |
| yDZ14 | MATa, ADE2, ChrV:: GALpr-gfp-URA3 | |
| yDZ15 | MATa, ADE2, ChrV:: Δ2pr-GFP-URA3 | |
| yDZ16 | MATa, ADE2, ChrV:: Δ1pr-GFP-URA3 | |
| yDZ21 | MATalpha, TRP1, ChrII:: ΔTATApr-gfp-URA3 | |
| yDZ22 | MATa, ADE2, ChrII:: XhoI-Δ1pr-GFP-XhoI-URA3 | |
| yDZ23 | MATalpha, TRP1, ChrII:: XhoI-GALpr-gfp-XhoI-URA3 | |
| yDZ24 | MATalpha, TRP1, ChrII:: XhoI-Δ4pr-GFP-XhoI-URA3 | |
| yDZ25 | MATalpha, TRP1, ChrII:: XhoI-ΔTATApr-GFP-XhoI-URA3 | |
| yDZ26 | MATalpha, TRP1, ChrV:: XhoI-GALpr-gfp-XhoI-URA3 | |
| yDZ41 | MATa, ADE2, Myo1::Myo1-mCherry-HIS3, ChrII:: Δ1pr-Venus-TRP1 | |
| yDZ42 | MATalpha, TRP1, ChrII:: GALpr-GFP-URA3 | |
| yDZ43 | MATalpha, TRP1, ChrXVI:: GALpr-GFP-URA3 | |
| Diploids | yDZ01 X W303-Trp | MATa/α, ADE2, TRP1, ChrII:: GALpr-GFP-URA3+/− |
| yDZ05 X W303-Trp | MATa/α, ADE2, TRP1, ChrII:: Δ2pr-GFP-URA3+/− | |
| yDZ06 X W303-Trp | MATa/α, ADE2, TRP1, ChrII:: Δ1pr-GFP-URA3+/− | |
| yDZ02 X yDZ11 | MATa/α, ADE2, TRP1, ChrII:: Δ2pr-GFP-URA3/GALpr-gfp-URA3 | |
| yDZ02 X yDZ12 | MATa/α, ADE2, TRP1, ChrII:: Δ1pr-GFP-URA3/GALpr-gfp-URA3 | |
| yDZ14 X yDZ12 | MATa/α, ADE2, TRP1, ChrII:: Δ1pr-GFP-URA3+/−, ChrV:: GALpr-gfp-URA3+/− | |
| yDZ06 X yDZ21 | MATa/α, ADE2, TRP1, ChrII:: Δ1pr-GFP-URA3/ΔTATApr-gfp-URA3 | |
| yDZ06 X yDZ09 | MATa/α, ADE2, TRP1, ChrII:: Δ1pr-GFP-URA3/Δ4pr-GFP-URA3 | |
| yDZ22 X yDZ23 | MATa/α, ADE2, TRP1, ChrII:: XhoI-Δ1pr-GFP-XhoI-URA3/XhoI-GALpr-gfp-XhoI-URA3 | |
| yDZ22 X yDZ26 | MATa/α, ADE2, TRP1, ChrII:: XhoI-Δ1pr-GFP-XhoI-URA3+/−, ChrV:: XhoI-GALpr-gfp-XhoI-URA3+/− | |
| yDZ22 X yDZ24 | MATa/α, ADE2, TRP1, ChrII:: XhoI-Δ1pr-GFP-XhoI-URA3/XhoI-Δ4pr-GFP-XhoI-URA3 | |
| yDZ22 X yDZ25 | MATa/α, ADE2, TRP1, ChrII:: XhoI-Δ1pr-GFP-XhoI-URA3/XhoI-ΔTATApr-GFP-XhoI-URA3 | |
| yDZ41 X yDZ42 | MATa/α, ADE2, TRP1, Myo1::Myo1-mCherry-HIS3, ChrII:: Δ1pr-Venus-TRP1/ GALpr-GFP-URA3 | |
| yDZ41 X yDZ43 | MATa/α, ADE2, TRP1, Myo1::Myo1-mCherry-HIS3, ChrII:: Δ1pr-Venus-TRP1+/−, ChrXVI:: GALpr-GFP-URA3+/− |
After each transformation, we picked six to eight yeast colonies from the selection plate and performed three PCR tests. (i) To confirm the location of integration, we carried out PCRs between primer pairs flanking the intended integration site, one in the genomic DNA and the other inside the plasmid. The presence of the PCR product confirms that the reporter is inserted into the intended locus. (ii) To make sure that the reporter is inserted as a single copy, we performed PCRs between two primers facing toward each other in the plasmid sequence flanking the homologous region. Because we cut the homologous region in the middle for transformation, this PCR should not produce any product; the presence of the product indicates multiintegration. For some strains, we also used the GFP fluorescence intensity to detect multiintegration. (iii) To confirm that the integrated Δ1, Δ2, and Δ4 promoters carry the intended deletions, we performed PCRs flanking the deleted sequence and we expected to see two products with different lengths, one from the endogenous GAL1pr, and the other from the reporter. After validation of the haploid strains, we mated them into diploids (in most cases selected with SCD-Ade-Trp). All of the experiments were repeated with at least two different diploid colonies.
FACS.
Overnight culture in glucose (OD 0.5–0.7) was washed and diluted to OD 0.1 in synthetic media containing 3% (wt/wt) galactose, 2% (wt/wt) raffinose, and 0.1% glucose. We diluted the culture by using fresh media after each hour of induction to replenish glucose and maintain the OD between 0.1 and 0.2. The samples were taken at different time points between 0–8 h of induction, kept on ice, sonicated, and subject to FACS measurement by using The BD LSRFortessa cell analyzer in a 96-plate format.
Time-Lapse Fluorescence Microscopy.
The time-lapse assay was performed as described (28). Briefly, live yeast cells were placed into CellAsic flow-cells, pregrown in 2% glucose for 4 h, and induced in 3% galactose, 2% raffinose, and 0.05% glucose for 8 h. In the meantime, we took GFP and YFP fluorescent images every 5 min. The movies were analyzed by using semiautomated data analysis software written in Matlab (27) to obtain the fluorescent intensity of individual cells at each movie frame. We then eliminated the cross-talk between the GFP and YFP signals through linear subtraction (28), which resulted in the single cell activation curves shown in Fig. 2C. The curves were then smoothed, and the activation time was quantified as the point where the second derivative reached the maximum.
3C Assay.
We cloned two distinct 100-bp fragments from KanMX ORF, 462–561 and 362–461 (KanMX1 and KanMX2 in Fig. 3A), and inserted them upstream of the GAL1 reporters. We introduced two XhoI sites into the plasmids, one upstream of the KanMX fragments and the other downstream of GFP transcript. After integrating the plasmids into yeast, we followed standard 3C protocol described (40). Cells were incubated overnight from a freshly growing colony and induced by switching from 2% glucose to 3% galactose and 2% raffinose for 8 h to an OD ∼0.65–0.8. Cells were cross-linked by formaldehyde (final concentration 1%) for 20 min and quenched by glycine for 5 min followed by centrifugation. Cell pellets were washed with TBS twice and frozen by liquid nitrogen. Five hundred microliters of FA-lysis buffer was added to resuspend the cells, and an equal volume of glass beads was added to disrupt the cells. The cell extracts were collected by centrifugation, washed with FA-lysis buffer, and resuspended in 500 μL of 10 mM Tris⋅HCl (pH 7.5). We digested 50 μL of the cross-linked chromatin sample with XhoI overnight before heating the sample at 65 °C to inactivate XhoI. After resuspension of the digested chromatin in 100 μL of 10 mM Tris⋅HCl, we added 400 U of T4 ligase and ligated overnight. We then digested RNA with RNase, and reversed cross-linking with SDS and proteinase K at 65 °C overnight. The DNA was pheno-extracted, resuspended in 50 μL of Tris-EDTA (pH 8.0), and subject to PCR/qPCR analysis. The two primers used here are “ATTCGTGATTGCGCCTGAGC” and “CGCCTGAGAATGGCAAAAGC”. The interaction between two cis regions on the ChrXV (near 878307 and 891189) was measured as a positive control and for normalization of the qPCR signal. The two primers used for the control are “CGGTAACCGAATTCTTCTCTCATG” and “TTCTTTGTCACTGTCGTGCGTG”.
Acknowledgments
We acknowledge Qian Zhang for technical support and all the members in L.B. laboratory for insightful comments on the manuscript. This work was supported by the Penn State start-up funding (to L.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601003113/-/DCSupplemental.
References
- 1.Hebenstreit D. Are gene loops the cause of transcriptional noise? Trends Genet. 2013;29(6):333–338. doi: 10.1016/j.tig.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Dean A. In the loop: Long range chromatin interactions and gene regulation. Brief Funct Genomics. 2011;10(1):3–10. doi: 10.1093/bfgp/elq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng W, et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149(6):1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noordermeer D, et al. Variegated gene expression caused by cell-specific long-range DNA interactions. Nat Cell Biol. 2011;13(8):944–951. doi: 10.1038/ncb2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Sullivan JM, et al. Gene loops juxtapose promoters and terminators in yeast. Nat Genet. 2004;36(9):1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 6.Singh BN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Mol Cell. 2007;27(5):806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 7.Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2(11):e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brickner DG, et al. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5(4):e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casolari JM, et al. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117(4):427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 10.Duncan IW. Transvection effects in Drosophila. Annu Rev Genet. 2002;36:521–556. doi: 10.1146/annurev.genet.36.060402.100441. [DOI] [PubMed] [Google Scholar]
- 11.Wu CT, Morris JR. Transvection and other homology effects. Curr Opin Genet Dev. 1999;9(2):237–246. doi: 10.1016/S0959-437X(99)80035-5. [DOI] [PubMed] [Google Scholar]
- 12.Burgess SM, Kleckner N, Weiner BM. Somatic pairing of homologs in budding yeast: Existence and modulation. Genes Dev. 1999;13(12):1627–1641. doi: 10.1101/gad.13.12.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keeney S, Kleckner N. Communication between homologous chromosomes: Genetic alterations at a nuclease-hypersensitive site can alter mitotic chromatin structure at that site both in cis and in trans. Genes Cells. 1996;1(5):475–489. doi: 10.1046/j.1365-2443.1996.d01-257.x. [DOI] [PubMed] [Google Scholar]
- 14.Lorenz A, Fuchs J, Bürger R, Loidl J. Chromosome pairing does not contribute to nuclear architecture in vegetative yeast cells. Eukaryot Cell. 2003;2(5):856–866. doi: 10.1128/EC.2.5.856-866.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aragón-Alcaide L, Strunnikov AV. Functional dissection of in vivo interchromosome association in Saccharomyces cerevisiae. Nat Cell Biol. 2000;2(11):812–818. doi: 10.1038/35041055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2(4):292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 17.Lewis JP, Tanke HJ, Raap AK, Beverstock GC, Kluin-Nelemans HC. Somatic pairing of centromeres and short arms of chromosome 15 in the hematopoietic and lymphoid system. Hum Genet. 1993;92(6):577–582. doi: 10.1007/BF00420942. [DOI] [PubMed] [Google Scholar]
- 18.Arnoldus EP, Peters AC, Bots GT, Raap AK, van der Ploeg M. Somatic pairing of chromosome 1 centromeres in interphase nuclei of human cerebellum. Hum Genet. 1989;83(3):231–234. doi: 10.1007/BF00285162. [DOI] [PubMed] [Google Scholar]
- 19.LaSalle JM, Lalande M. Homologous association of oppositely imprinted chromosomal domains. Science. 1996;272(5262):725–728. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]
- 20.Marahrens Y. X-inactivation by chromosomal pairing events. Genes Dev. 1999;13(20):2624–2632. doi: 10.1101/gad.13.20.2624. [DOI] [PubMed] [Google Scholar]
- 21.Homouz D, Kudlicki AS. The 3D organization of the yeast genome correlates with co-expression and reflects functional relations between genes. PLoS One. 2013;8(1):e54699. doi: 10.1371/journal.pone.0054699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirkin EV, Chang FS, Kleckner N. Dynamic trans interactions in yeast chromosomes. PLoS One. 2013;8(9):e75895. doi: 10.1371/journal.pone.0075895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellert DJ, Truman JW. Transvection is common throughout the Drosophila genome. Genetics. 2012;191(4):1129–1141. doi: 10.1534/genetics.112.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Floer M, et al. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010;141(3):407–418. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Acar M, Pando BF, Arnold FH, Elowitz MB, van Oudenaarden A. A general mechanism for network-dosage compensation in gene circuits. Science. 2010;329(5999):1656–1660. doi: 10.1126/science.1190544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biggar SR, Crabtree GR. Cell signaling can direct either binary or graded transcriptional responses. EMBO J. 2001;20(12):3167–3176. doi: 10.1093/emboj/20.12.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charvin G, Cross FR, Siggia ED. A microfluidic device for temporally controlled gene expression and long-term fluorescent imaging in unperturbed dividing yeast cells. PLoS One. 2008;3(1):e1468. doi: 10.1371/journal.pone.0001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q, et al. Stochastic expression and epigenetic memory at the yeast HO promoter. Proc Natl Acad Sci USA. 2013;110(34):14012–14017. doi: 10.1073/pnas.1306113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.West RW, Jr, Yocum RR, Ptashne M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: Location and function of the upstream activating sequence UASG. Mol Cell Biol. 1984;4(11):2467–2478. doi: 10.1128/mcb.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duan Z, et al. A three-dimensional model of the yeast genome. Nature. 2010;465(7296):363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gehlen LR, et al. Chromosome positioning and the clustering of functionally related loci in yeast is driven by chromosomal interactions. Nucleus. 2012;3(4):370–383. doi: 10.4161/nucl.20971. [DOI] [PubMed] [Google Scholar]
- 32.Ben-Elazar S, Yakhini Z, Yanai I. Spatial localization of co-regulated genes exceeds genomic gene clustering in the Saccharomyces cerevisiae genome. Nucleic Acids Res. 2013;41(4):2191–2201. doi: 10.1093/nar/gks1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fanucchi S, Shibayama Y, Burd S, Weinberg MS, Mhlanga MM. Chromosomal contact permits transcription between coregulated genes. Cell. 2013;155(3):606–620. doi: 10.1016/j.cell.2013.09.051. [DOI] [PubMed] [Google Scholar]
- 34.Schoenfelder S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42(1):53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee AM, Wu CT. Enhancer-promoter communication at the yellow gene of Drosophila melanogaster: Diverse promoters participate in and regulate trans interactions. Genetics. 2006;174(4):1867–1880. doi: 10.1534/genetics.106.064121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cloutier SC, Wang S, Ma WK, Petell CJ, Tran EJ. Long noncoding RNAs promote transcriptional poising of inducible genes. PLoS Biol. 2013;11(11):e1001715. doi: 10.1371/journal.pbio.1001715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32(5):685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cloutier SC, et al. Regulated formation of lncRNA-DNA hybrids enables faster transcriptional induction and environmental adaptation. Mol Cell. 2016;61(3):393–404. doi: 10.1016/j.molcel.2015.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cisse II, et al. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 2013;341(6146):664–667. doi: 10.1126/science.1239053. [DOI] [PubMed] [Google Scholar]
- 40.Singh BN, Hampsey M. Detection of short-range chromatin interactions by chromosome conformation capture (3C) in yeast. Methods Mol Biol. 2014;1205:209–218. doi: 10.1007/978-1-4939-1363-3_13. [DOI] [PubMed] [Google Scholar]