Significance
Reef corals in the Persian/Arabian Gulf (PAG) withstand exceptionally high salinity and regular summer temperatures of ∼35 °C that kill conspecifics elsewhere. These thermotolerant communities established themselves within only ∼6,000 y under the pressure of rapid climate change and can therefore inform how other coral reefs may respond to global warming. One key to the thermotolerance of PAG corals is their symbiosis with Symbiodinium thermophilum. Phylogeographic evidence indicates that this symbiont represents a stress-tolerant subpopulation of an ancestral taxonomic group with surprising genetic diversity that exists at barely detectable levels outside the PAG. Our results highlight the critical importance of present-day biodiversity for future adaptation to climate change for coral reefs and ecosystems in general.
Keywords: Persian/Arabian Gulf, adaptation, coral, Symbiodinium, climate change
Abstract
Coral communities in the Persian/Arabian Gulf (PAG) withstand unusually high salinity levels and regular summer temperature maxima of up to ∼35 °C that kill conspecifics elsewhere. Due to the recent formation of the PAG and its subsequent shift to a hot climate, these corals have had only <6,000 y to adapt to these extreme conditions and can therefore inform on how coral reefs may respond to global warming. One key to coral survival in the world’s warmest reefs are symbioses with a newly discovered alga, Symbiodinium thermophilum. Currently, it is unknown whether this symbiont originated elsewhere or emerged from unexpectedly fast evolution catalyzed by the extreme environment. Analyzing genetic diversity of symbiotic algae across >5,000 km of the PAG, the Gulf of Oman, and the Red Sea coastline, we show that S. thermophilum is a member of a highly diverse, ancient group of symbionts cryptically distributed outside the PAG. We argue that the adjustment to temperature extremes by PAG corals was facilitated by the positive selection of preadapted symbionts. Our findings suggest that maintaining the largest possible pool of potentially stress-tolerant genotypes by protecting existing biodiversity is crucial to promote rapid adaptation to present-day climate change, not only for coral reefs, but for ecosystems in general.
Episodes of heat stress cause coral bleaching, the breakdown of the obligate symbiosis between the coral host and its algal partner that contributes to the global decline of coral reefs (1). Despite their capacity to acclimate to rising seawater temperatures (2), corals will suffer from high-frequency bleaching episodes by the end of this century, threatening their survival in the warmer oceans of the future (3). Changing to symbiotic associations with more thermally tolerant types like Symbiodinium trenchii (synonym type D1a/D1–4) may increase the heat stress tolerance of corals rapidly (4–6). However, due to tradeoffs such as reduced calcification rates that can be associated with hosting alternative symbionts, reef ecosystems may ultimately fail to benefit from the increased thermal tolerance of their most important habitat-forming species (5, 6). Hence, it is unclear at present whether alternative symbiont associations will rescue reefs from their expected demise in response to global warming.
We are using the Persian/Arabian Gulf (PAG), the world’s hottest sea, as a natural laboratory where coral communities endure regular summer temperatures of up to ∼35 °C to address the question of how coral reefs relying on heat-tolerant Symbiodinium may respond to rapid climate change over a prolonged period. Due to rising sea levels associated with the last glacial retreat, the modern PAG started to form ∼12,500 y before present (BP) by ingression of the Indian Ocean into the previously dry basin, extending to present-day shorelines ∼6,000 y BP (7). By this time, the climate in the Middle East began to change from cooler and moister to warmer and more arid, reaching today’s conditions only ∼4,000 y ago (8–10). Hence, the coral communities of the PAG, composed mostly of a subset of Indian Ocean species (11), have had to adjust rapidly to temperatures not expected to occur in other parts of the world’s oceans before the next century (12). We recently discovered that corals of the southern, hottest region of the PAG form a prevalent, year-round association with a thermally tolerant symbiont species, Symbiodinium thermophilum (13). Although the symbiosis with S. thermophilum might not be the sole cause for the heat tolerance, and others factors including the host physiology (14) and environmental conditions (such as the exceptionally high salinity in the relevant PAG regions) (15) may contribute to the resilience of the holobiont, the striking dominance of this symbiont in the southern PAG strongly suggests that it represents a key component to the success of corals in this extreme environment.
S. thermophilum can be identified by specific intragenomic variants of the nuclear ribosomal second internal transcribed spacer region (ITS2), which carry an 8-bp duplication indel named “S. thermo.-indel” (13). In close to pure S. thermophilum populations, these variants represent an average ∼16% of the total ITS2 sequences, however this proportion can vary considerably (Fig. S1). Additionally, this symbiont can be distinguished by its genetically disparate resolution from other closely related ITS2 C3 types using the noncoding region of the chloroplastic psbA gene (psbAncr) among other markers (13). As yet undetected in other parts of the world, our previous work suggested that symbionts characterized by the S. thermo.-indel and a disparate psbAncr resolution (hereafter S. thermophilum group) may not be endemic to the PAG (15). Because the understanding of heat tolerance in coral–symbiont associations is crucial in gauging the adaptation potential of coral reefs to global warming, we investigated the origin of S. thermophilum to assess whether this thermotolerant symbiont emerged as the result of an unexpectedly rapid evolution under the pressure of the extreme environmental conditions in the PAG or whether this species originated elsewhere.
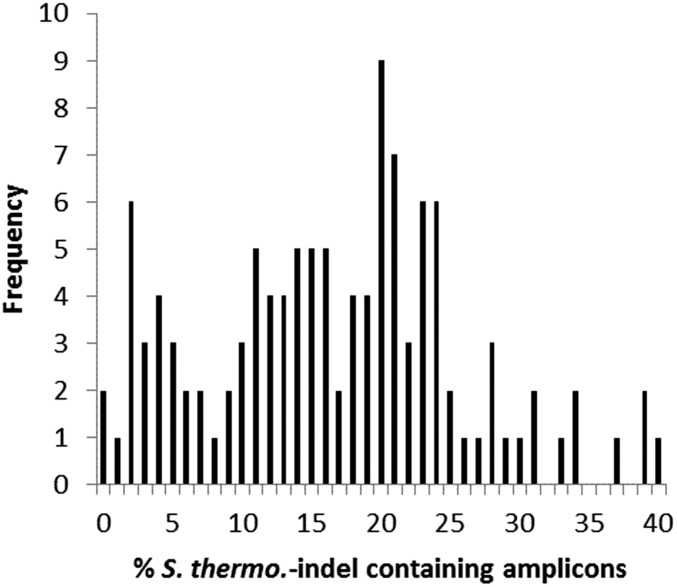
Fig. S1.
Proportion of S. thermo.-indel–containing variants in total ITS2 sequences from C3 predominated cnidarians (n = 116) from the PAG. Cnidarians included in the analysis contained >50% clade C Symbiodinium, of which >95% were C3 or closely related variants. Average % S. thermo.-indel–containing amplicons: 16.6%.
Results and Discussion
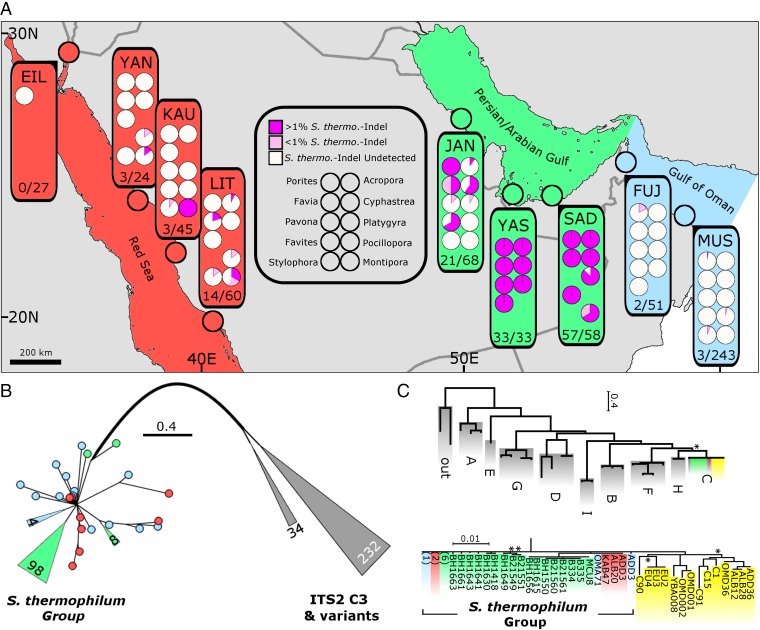
We screened for S. thermophilum at 23 sites across >5,000 km of coastline from the northwestern PAG to the Gulf of Eilat/Aqaba in the Red Sea (Fig. 1A). The symbiont was detected by the presence of the S. thermo.-indel in libraries containing 2.8 × 106 ITS2 amplicons obtained by 454 and MiSeq sequencing (except Eilat; see Methods) of >900 symbiotic hexa- and octocorallians from 46 genera (Tables S1 and S2). The results for the 10 most sampled taxa are presented in Fig. 1A, with details being given for all samples in Fig. 2 and Tables S1 and S2.
Fig. 1.
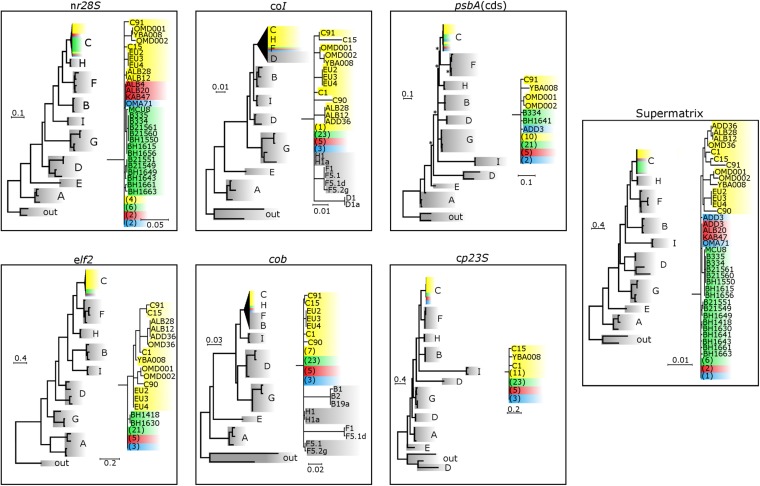
Identification and phylogenetic resolution of the S. thermophilum group. Colors denote sampling locations according to map coloration with yellow denoting non-S. thermophilum group clade C symbionts. (A) Sampling sites (three letter abbreviations; e.g., JAN) are detailed in Table S1. The presence of S. thermophilum group symbionts within the 10 most sampled genera are presented categorically (undetected, <1% and >1% S. thermo.-indel). Regional summaries (*)/(*) represent the proportion of samples in which S. thermophilum group symbionts were detected (Tables S1 and S2). The map was created as detailed in Hume et al., 2015 (13). (B) PsbAncr estimated phylogeny of S. thermophilum group samples created through the addition of samples collected in the Gulf of Oman and the Red Sea (sequenced as part of this study) to psbAncr alignments previously constructed (15). Symbionts within the gray branches are ITS2 type C3 and closely related ITS2 types from the Atlantic and Indo-Pacific (17). Wedges and circles represent collapsed groups and individual samples, respectively. Lengths of collapsed branches are drawn to scale with number of contained sequences indicated. (C) Supermatrix Bayesian estimation of phylogeny using six additional genetic markers (Fig. S3 and Table S5) on a selection of cnidarian-harbored Symbiodinium samples collected in the PAG, the Gulf of Oman, the Red Sea, as well as several reference locations (see Methods for further details). The full collapsed tree with clade C subtree is expanded below. Numbers in parentheses represent multiple sequences resolving at the same position and are colored according to collection site and sequence type (Gulf of Oman, blue; PAG, green; Red Sea, red; reference sequences, yellow). Gray groupings contain reference Symbiodinium from clades A–I as well as two outgroups (G. simplex and P. glacialis) (Table S5). Nodes supported with posterior probability (PP) above 0.8 are not displayed. Nodes with supports below 0.8 are marked with an asterisk (*).
Table S1.
Detection of S. thermophilum symbionts in the PAG, the Gulf of Oman, and the Red Sea through screening for the rDNA ITS2 S. thermo.-indel
| Water body | Region (abbreviation) | Reef | Number of samples | % hosts with S. thermo.-indel | Hosts >1%* S. thermo.-indel | Hosts <1%* S. thermo.-indel | Genera with S. thermo.-indel |
| PAG | Jana (JAN) | Subri | 31 | 29 | 6 | 3 | 5 of 13 |
| Subri (Jana) | 41 | 27 | 8 | 3 | 10 of 18 | ||
| Chandelier | 23 | 74 | 13 | 4 | 7 of 9 | ||
| Yassat (YAS) | Al Yassat | 33 | 100 | 33 | 0 | 7 of 7 | |
| Saadiyat (SAD) | Saadiyat | 42 | 98 | 40 | 1 | 11 of 12 | |
| Ras Ghanada | 26 | 96 | 22 | 3 | 7 of 7 | ||
| Gulf of Oman | Fujairah (FUJ) | Al Aqah | 34 | 15 | 1 | 4 | 4 of 14 |
| Dibba Rock | 37 | 0 | 0 | 0 | 0 of 14 | ||
| Muscat (MUS) | Bandar Khayran Islet | 90 | 0 | 0 | 0 | 0 of 21 | |
| Damanyat 3 sisters | 35 | 0 | 0 | 0 | 0 of 17 | ||
| Damanyat legends | 36 | 0 | 0 | 0 | 0 of 16 | ||
| Damanyat Noodle | 58 | 0 | 0 | 0 | 0 of 15 | ||
| Fahal Island Site I | 34 | 6 | 1 | 1 | 2 of 14 | ||
| Fahal Island Site II | 28 | 7 | 1 | 1 | 2 of 12 | ||
| Saifat Ash Shiekh | 87 | 1 | 1 | 0 | 1 of 23 | ||
| Red Sea | Eilat (EIL) | Eilat | 27 | 0 | 0 | 0 | 0 of 1 |
| Yanbu (YAN) | Yanbu 23 | 31 | 3 | 1 | 0 | 3 of 13 | |
| Yanbu Ayona | 36 | 0 | 0 | 0 | 0 of 17 | ||
| KAUST (KAU) | KAUST Al Fahal | 42 | 0 | 0 | 0 | 0 of 16 | |
| KAUST Inner Fsar | 31 | 3 | 1 | 0 | 2 of 13 | ||
| KAUST Shib Nazaar | 43 | 0 | 0 | 0 | 0 of 16 | ||
| Al Lith (LIT) | Al Lith Abu Lath | 40 | 15 | 3 | 3 | 7 of 17 | |
| Al Lith South Reef | 33 | 0 | 0 | 0 | 0 of 18 |
This is representative of total ITS2 amplicons detected (all clades).
Table S2.
Proportion of taxa in which S. thermophilum symbionts were detected through screening for the ribosomal DNA ITS2 S. thermo.-indel
| Taxa | Detected | Undetected | ||
| Subtaxa | Proportion | Subtaxa | Proportion | |
| Actiniaria | Unidentified | 1/3 | ||
| Entacmea | 1/1 | |||
| Alcyonacea | Unidentified | 0/4 | ||
| Alcyoniidae | Sarcophyton | 1/11 | Lobophytum | 0/3 |
| Sinularia | 1/4 | |||
| Nephtheidae | Nephthea | 0/1 | ||
| Xeniidae | Xenia | 0/23 | ||
| Scleractinia | ||||
| Acroporidae | Acropora | 12/107 | Astreopora | 0/14 |
| Montipora | 10/56 | |||
| Agariciidae | Pavona | 3/48 | Gardineroseris | 0/5 |
| Astrocoeniidae | Madracis | 1/2 | Stephanocoenia | 0/0 |
| Stylocoeniella | 0/1 | |||
| Dendriphyllidae | Turbinaria | 7/14 | ||
| Diploastreidae | Diploastrea | 0/5 | ||
| Caryophylliidae | Cladocora | 0/1 | ||
| Coscinaraeidae | Coscinaraea | 1/4 | ||
| Fungiidae | Fungia | 0/7 | ||
| Lobophyllidae | Acantastrea | 2/9 | Echinophyllia | 0/12 |
| Merulinidae | Cyphastrea | 26/42 | Echinopora | 0/20 |
| Favites | 9/40 | Hydnophora | 0/6 | |
| Goniastrea | 1/15 | Oulophyllia | 0/1 | |
| Leptoria | 2/4 | |||
| Platygyra | 16/53 | |||
| Montastraeidae | Montastraea | 0/1 | ||
| Mussidae | Favia | 20/60 | Symphyllia | 0/9 |
| Leptastrea | 2/26 | |||
| Unidentified | 1/1 | |||
| Oculinidae | Galaxea | 1/8 | ||
| Pectiniidae | Mycedium | 0/1 | ||
| Pocilloporidae | Pocillopora | 3/75 | ||
| Seriatopora | 1/6 | |||
| Stylophora | 3/43 | |||
| Poritidae | Goniopora | 1/18 | ||
| Porites | 27/98 | |||
| Siderastreidae | Psammocora | 9/26 | ||
| Zoantharia | ||||
| Sphenopidae | Palythoa | 1/1 | ||
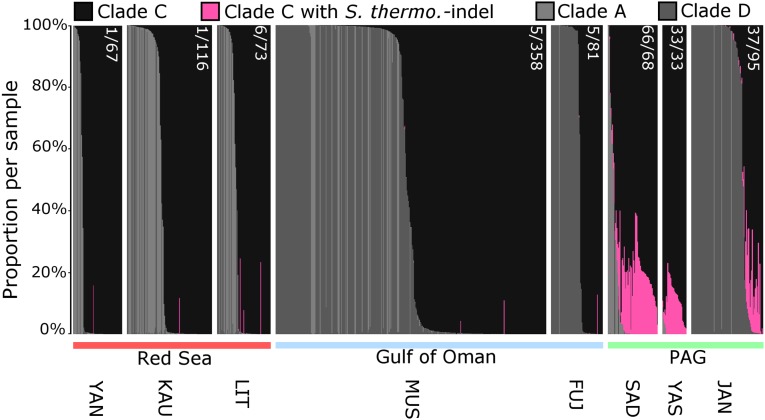
Fig. 2.
Identification of clade C sequences containing the S. thermo.-indel as a proxy for the presence of S. thermophilum and classification of the symbiont complements in all sampled individuals. Each vertical bar represents a single sample. The contribution of sequences representing different clades/species to the sequence complement of each sample is color-coded as defined in the legend. Sample origins are shown below the chart (Table S1). Numbers of samples containing the S. thermo.-indel as a proportion of the total samples are shown in white in the format (*)/(*) (Tables S1 and S2).
Members of the S. thermophilum group were found in 10 out of the 11 sampled genera in the southern PAG but were less frequent in the western PAG and could only be detected at low levels in ∼4% of samples in the Gulf of Oman and the Red Sea (Figs. 1 and 2 and Tables S1 and S2). Additionally, when screening public databases for ITS2 sequences containing the S. thermo.-indel, we retrieved a close match with a sequence originating from Kaneohe Bay, Hawaii, which was recently entered in GenBank (accession no. EF428343). This hit may indicate the presence of a S. thermophilum group member in the Indo-Pacific, implying an even wider, cryptic distribution.
PsbAncr sequences of samples containing the S. thermo.-indel from the Gulf of Oman and Red Sea are resolved within the strongly supported (PP = 1) monophyletic S. thermophilum group (Fig. 1B and Table S3) but as genetically separate from the majority of PAG sequences and with larger average within-group genetic distances (0.065 vs. 0.039; Table S4). The average genetic distance of the five most divergent sequence pairs within the S. thermophilum group was larger than that between the considerably divergent C27 and C40 ITS2 types (0.163 vs. 0.154; Fig. S2 and Table S4), demonstrating a genetic diversity in the group greater than that found among considerably divergent clade C lineages. This highlights the existence of a wide range of previously unidentified and possibly stress-tolerant genotypes within the group.
Table S3.
Accession numbers for the seven genetic markers (amplicons) used in this study
Samples were collected as part of this study.
Locations were as follows: MP, Musandam Peninsular; UAQ, Umm Al Quwain; SAS, Saifat Ash Shiekh.
The group is S. thermophilum.
Samples were collected and psbAncr sequences were amplified as part of D’Angelo et al., 2015 (15).
Table S4.
Pairwise and average between- and within-group genetic distances based on the psbAncr alignment from this study*
| Sequence/group 1 | Sequence/group 2 | Genetic distance |
| S. thermophilum | Average, 0.163 | |
| ALB30† (Al Lith) | BH1428 (Umm Al Quwain) | 0.155 |
| BH1660 (Umm Al Quwain) | BH1693 (Dalma) | 0.156 |
| B335 (Muscat) | BH1633 (Ras Ghanada) | 0.160 |
| B21557 (Musandam) | B315 (Saadiyat) | 0.167 |
| B21561 (Fujairah) | BH1642 (Saadiyat) | 0.178 |
| C40 and C27 | Average, 0.154 | |
| C27J043670 | C40K572369 | 0.147 |
| C27J043674 | C40K572361 | 0.147 |
| C27J043669 | C40K572360 | 0.157 |
| C27J043671 | C40K572365 | 0.158 |
| C27J043668 | C40K572367 | 0.163 |
| Between group | ||
| C3SID | C40 | 0.164 |
| C3SID | C7a | 0.126 |
| C3SID | C7 | 0.115 |
| C7 | C7a | 0.060 |
| S. thermophilum (PAG) | 0.039 | |
| S. thermophilum (external) | 0.065 |
Pairwise genetic distances of the five most disparate pairs within the S. thermophilum group and between the ITS2 type C40 and C27 groups are shown under the first two corresponding subheadings. Between-group genetic distance of three ITS2 groups (C40, C7, and C7a) and a monophyletic group of Symbiodinium ITS2 type C3 sequences originating from host S. siderea samples (C3SID) as highlighted in yellow in figure 8A from Thornhill et al., 2014 (17) are shown under the subheading ”Between group.” Finally, average within-group genetic distances of S. thermophilum group samples from the PAG and external to the PAG are shown. A partial graphical representation of the data is shown in Fig. S2. Sampling locations are detailed in parentheses.
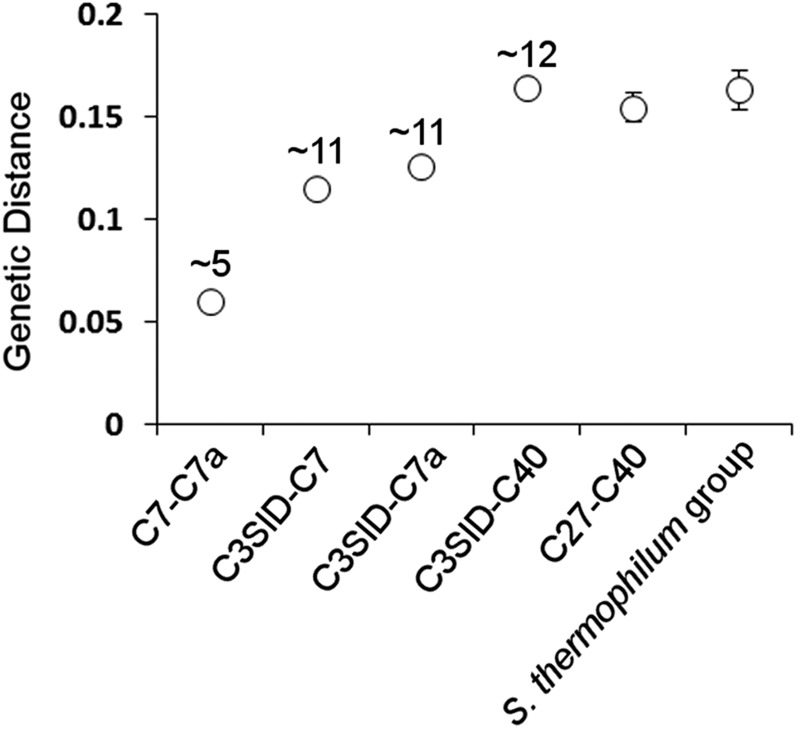
Fig. S2.
Estimated age of diversity in the S. thermophilum group. C7-C7a, C3SID-C7, C3SID-C7a, and C3SID-C40 represent between-group genetic distance for associated ITS2 groups, with C3SID being defined as the grouping of Symbiodinium ITS2 type C3 sequences originating from host Siderastrea siderea samples as highlighted in yellow in figure 8A from Thornhill et al., 2014 (17). Numbers above genetic distances represent corresponding lineage diversification ages (Mya) extracted from figure 8A of Thornhill et al., 2014 (17). C27–C40 and S. thermophilum group represent average pairwise genetic distances between the five most divergent pairs between ITS2 groups C27 and C40 and within the S. thermophilum group. Error bars represent 1 SD above and below the average (comparisons of five most divergent pairs only). Values for all genetic distances as well as details of the five most divergent pairs are given in Table S4.
To establish the age and taxonomic position of the S. thermophilum group within clade C, we estimated molecular phylogenies of this group with representatives from Symbiodinium clades A–I, including subclade C representatives, using a suite of single gene markers (Tables S3 and S5) through Bayesian analysis. These markers varied in their ability to resolve between clade C types (Fig. S3 and Table S6). Nine of the 31 S. thermophilum group samples showed identical resolutions across all markers and resolved on the ancestral node in a supermatrix phylogeny (Fig. 1C). All non-S. thermophilum group samples resolved as a strongly supported monophyletic group derived from this ancestral node, whereas those S. thermophilum group samples not resolved on the ancestral node separated as sister groups or individual samples. Given the molecular dating of the radiation of clade C placed in the mid-Miocene, these results provide strong evidence that the S. thermophilum group represents one of the oldest (∼13 Mya) and most genetically diverse groups of extant clade C symbionts (16). Furthermore, estimation of divergences between lineages within the ITS2 type C3 and closely related ITS2 variants according to the psbAncr as being between 2 and 12 My old (17), would suggest that within-group genetic distances of the S. thermophilum group represent evolutionary divergences occurring over at least several million years, a span considerably longer than the age of the PAG (Fig. S2).
Table S5.
Reference sequences used in the single-gene and supermatrix estimations of phylogeny
Fig. S3.
Rooted single-gene and supermatrix estimated phylogenies for Symbiodinium spp. Shown are single-gene and supermatrix estimated phylogenies (Bayesian analysis with nodes support assessed through posterior probabilities) for a suite of A–I clade sequences (gray), including subcladal C sequences (C3, C1, C41, and C39; yellow) and S. thermophilum sequences from the PAG (green), Gulf of Oman (blue), and Red Sea (red). For each phylogeny, a full tree is illustrated on the left, in which the clade C subtree is collapsed. The resolution of this collapsed subtree is shown in detail to the right. Numbers in parentheses represent multiple sequences resolving at the same position and are color coded according to collection site and sequence type (Gulf of Oman, blue; PAG, green; Red Sea, red; reference sequence, yellow). Nodes supported with PPs above 0.8 are not displayed. Nodes with supports below 0.8 are marked with an asterisk.
Table S6.
Phylogenetic differentiation of taxa from the clade C ancestral nodes in single-gene phylogenies
| Sample | ITS2 type | nr28S | elf2 | coi | psbAcds | cob | cp23S | Supermatrix | Total |
| C91 | C91 | 1* | 1 | 1 | 1 | 1 | 0† | 1 | 5 |
| OMD001 | C3 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 4 |
| OMD002 | C3 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 4 |
| C15 | C15 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 4 |
| YBA008 | C3 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 4 |
| EU2 | C3 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 4 |
| EU3 | C3 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 4 |
| EU4 | C3 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 4 |
| C1 | C1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 3 |
| C90 | C90 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 3 |
| ALB028 | C41 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 3 |
| ALB012 | C41 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 3 |
| ADD036 | C39 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 2 |
| B334 | S. thermophilum | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 2 |
| OMD036 | C39 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 |
| BH1418 | S. thermophilum | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 |
| BH1630 | S. thermophilum | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 |
| BH1641 | S. thermophilum | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 |
| ADD003 | S. thermophilum | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 |
| ALB004 | S. thermophilum | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| ALB020 | S. thermophilum | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| KAB047 | S. thermophilum | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| OMA071 | S. thermophilum | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| BH1659 | S. thermophilum | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| B335 | S. thermophilum | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| B21561 | S. thermophilum | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| B21560 | S. thermophilum | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| BH1550 | S. thermophilum | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| BH1615 | S. thermophilum | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| BH1656 | S. thermophilum | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| B21551 | S. thermophilum | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| BH21549 | S. thermophilum | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| BH1649 | S. thermophilum | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| BH1643 | S. thermophilum | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| BH1661 | S. thermophilum | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| BH1663 | S. thermophilum | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| OMD054 | S. thermophilum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| YBA016 | S. thermophilum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ALB030 | S. thermophilum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| BH1660 | S. thermophilum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| BH1660 | S. thermophilum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| BH1699 | S. thermophilum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| BH1693 | S. thermophilum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| BH1694 | S. thermophilum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| B359 | S. thermophilum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
For each sample, a “1” in a single-gene column denotes the sample was not resolved on the clade C ancestral node of that phylogeny.
“0” denotes resolution on the ancestral node.
The diversity of S. thermophilum–psbAncr was assessed by analyzing sequences from independent samples (Table S7) representing six sites in the PAG and nine sites in the Gulf of Oman and the Red Sea. The significant difference (two-sample t test, P value < 0.01) obtained for group comparisons between within-group genetic distances of PAG sequences (0.039; 112 samples) and Gulf of Oman/Red Sea sequences (0.065; 22 samples) (Table S4) provides evidence that the genetic diversity of S. thermophilum in the PAG is strongly reduced compared with the sites outside of this water body. Similarly, in the southern PAG, the overall Symbiodinium diversity at the species/clade level is lower compared with the western PAG and the Strait of Hormuz, which connects the PAG with the Gulf of Oman (15, 18) (Figs. 1A and 2).
Table S7.
Sequences included in the calculation of within-group (PAG vs. Gulf of Oman/Red Sea) genetic distance
| ID | Collection site | Accession number | Host taxon | ID | Collection site* | Accession number | Host taxon |
| Persian/Arabian Gulf sequences | Persian/Arabian Gulf sequences | ||||||
| BH1415 | Dalma | KM458276† | Porites harrisoni | BH1630 | Umm Al Quwain | KP280257 | P. lutea |
| BH1686 | Dalma | KP280207 | P. harrisoni | BH1668 | Umm Al Quwain | KP280258 | P. lutea |
| BH1448 | Dalma | KM458286 | P. harrisoni | BH1456 | Umm Al Quwain | KM458291 | P. lutea |
| BH1413 | Dalma | KM458274 | P. harrisoni | BH1626 | Umm Al Quwain | KP280259 | P. lutea |
| BH1684 | Dalma | KP280208 | P. harrisoni | BH1669 | Umm Al Quwain | KP280260 | P. lutea |
| BH1414 | Dalma | KM458275 | P. harrisoni | BH1457 | Umm Al Quwain | KP280261 | P. lutea |
| BH1685 | Dalma | KP280209 | P. harrisoni | BH1660 | Umm Al Quwain | KP280262 | P. lutea |
| BH1450 | Dalma | KM458288 | Porites lutea | BH1392 | Umm Al Quwain | KM458294 | P. lutea |
| BH1449 | Dalma | KM458287 | P. lutea | BH1661 | Umm Al Quwain | KP280263 | P. lutea |
| BH1689 | Dalma | KP280210 | P. lutea | BH1458 | Umm Al Quwain | KM458292 | P. lutea |
| BH1692 | Dalma | KP280211 | P. lutea | BH1427 | Umm Al Quwain | KM458284 | P. lutea |
| BH1690 | Dalma | KP280212 | P. harrisoni | BH1663 | Umm Al Quwain | KP280264 | P. lutea |
| BH1643 | Dalma | KP280213 | P. harrisoni | BH1679 | Umm Al Quwain | KP280265 | P. lutea |
| BH1411 | Dalma | KM458273 | P. harrisoni | BH1688 | Umm Al Quwain | KP280266 | P. lutea |
| BH1647 | Dalma | KP280215 | P. harrisoni | BH1675 | Ras Al Kaimah | KP280267 | Porites sp. |
| BH1691 | Dalma | KP280216 | P. harrisoni | BH1621 | Ras Al Kaimah | KP280268 | Porites sp. |
| BH1447 | Dalma | KP280217 | P. harrisoni | BH1671 | Ras Al Kaimah | KP280269 | Porites sp. |
| BH1693 | Dalma | KP280218 | P. lutea | BH1672 | Ras Al Kaimah | KP280270 | Porites sp. |
| BH1416 | Dalma | KP280219 | P. harrisoni | BH1676 | Ras Al Kaimah | KP280271 | Porites sp. |
| BH1702 | Saadiyat | KP280220 | Porites lobata | BH1623 | Ras Al Kaimah | KP280272 | Porites sp. |
| BH1704 | Saadiyat | KP280221 | P. lobata | BH1620 | Ras Al Kaimah | KP280273 | Porites sp. |
| BH1708 | Saadiyat | KP280222 | P. lobata | BH1624 | Ras Al Kaimah | KP280275 | Porites sp. |
| BH1356 | Saadiyat | KM458293 | P. lutea | BH1670 | Ras Al Kaimah | KP280276 | Porites sp. |
| B359 | Saadiyat | KP280223 | P. lutea | BH1673 | Ras Al Kaimah | KP280277 | Porites sp. |
| BH1422 | Saadiyat | KM458279 | P. lutea | BH1678 | Ras Al Kaimah | KP280278 | Porites sp. |
| BH1694 | Saadiyat | KP280224 | P. lutea | BH1680 | Ras Al Kaimah | KP280279 | Porites sp. |
| BH1451 | Saadiyat | KM458289 | P. lobata | BH1677 | Ras Al Kaimah | KP280280 | Porites sp. |
| BH1695 | Saadiyat | KP280225 | P. lobata | BH1683 | Ras Al Kaimah | KP280281 | Porites sp. |
| BH1417 | Saadiyat | KM458277 | P. lutea | BH1497 | Ras Al Kaimah | KP280282 | Porites sp. |
| BH1419 | Saadiyat | KM458278 | P. lutea | BH1625 | Ras Al Kaimah | KP280283 | Porites sp. |
| BH1640 | Saadiyat | KP280226 | P. lobata | BH1682 | Ras Al Kaimah | KP280284 | Porites sp. |
| BH1699 | Saadiyat | KP280227 | P. lutea | BH1681 | Ras Al Kaimah | KP280285 | Porites sp. |
| BH1701 | Saadiyat | KP280228 | P. lutea | BH1674 | Ras Al Kaimah | KP280286 | Porites sp. |
| BH1705 | Saadiyat | KP280229 | P. lutea | B21549 | Musandam‡ | KP280287 | P. lobata |
| BH1706 | Saadiyat | KP280230 | P. lutea | B21550 | Musandam‡ | KP280288 | P. lobata |
| B315 | Saadiyat | KP280231 | P. harrisoni | BH1652 | Musandam‡ | KP280289 | P. lobata |
| BH1696 | Saadiyat | KP280232 | P. harrisoni | BH1616 | Musandam‡ | KP280290 | P. lutea |
| BH1641 | Saadiyat | KP280233 | P. lobata | BH1617 | Musandam‡ | KP280291 | P. lobata |
| BH1703 | Saadiyat | KP280234 | P. harrisoni | B21551 | Musandam‡ | KP280292 | P. lutea |
| BH1642 | Saadiyat | KP280235 | P. harrisoni | B21552 | Musandam‡ | KP280293 | P. lutea |
| BH1698 | Saadiyat | KP280236 | P. lutea | B21553 | Musandam‡ | KP280294 | P. lutea |
| BH1487 | Saadiyat | KP280237 | P. lobata | B21554 | Musandam‡ | KP280295 | P. lutea |
| BH1418 | Saadiyat | KP280238 | P. lutea | BH1649 | Musandam‡ | KP280296 | P. lobata |
| BH1639 | Saadiyat | KP280239 | P. lutea | BH1653 | Musandam‡ | KP280297 | P. lutea |
| BH1700 | Saadiyat | KP280240 | P. harrisoni | BH1655 | Musandam‡ | KP280298 | P. lutea |
| BH1453 | Saadiyat | KM458290 | P. lobata | ||||
| BH1697 | Saadiyat | KP280241 | P. lobata | Gulf of Oman and Red Sea sequences | |||
| BH1636 | Ras Ghanada | KP280242 | P. lutea | B341 | Musandam§ | KP280299 | P. lutea |
| BH1547 | Ras Ghanada | KP280243 | P. lobata | BH1659 | Musandam§ | KP280300 | P. lutea |
| BH1632 | Ras Ghanada | KP280244 | P. lobata | B21556 | Musandam§ | KP280301 | P. lobata |
| BH1634 | Ras Ghanada | KP280245 | P. harrisoni | BH1658 | Musandam§ | KP280302 | P. lobata |
| BH1635 | Ras Ghanada | KP280246 | P. harrisoni | B21557 | Musandam§ | KP280303 | P. lobata |
| BH1548 | Ras Ghanada | KP280247 | P. lutea | BH1656 | Musandam§ | KP280304 | P. lobata |
| BH1631 | Ras Ghanada | KP280248 | P. harrisoni | BH1614 | Musandam§ | KP280305 | P. lobata |
| BH1633 | Ras Ghanada | KP280249 | P. harrisoni | BH1615 | Musandam§ | KP280306 | P. lobata |
| BH1664 | Umm Al Quwain | KP280250 | P. lutea | B21561 | Fujairah | KP280307 | P. lobata |
| BH1425 | Umm Al Quwain | KM458282 | P. lutea | BH1550 | Fujairah | KP280308 | P. lobata |
| BH1665 | Umm Al Quwain | KP280251 | P. lutea | B21560 | Fujairah | KP280309 | P. lobata |
| BH1662 | Umm Al Quwain | KP280252 | P. lutea | B334 | Fujairah | KP280310 | P. lobata |
| BH1428 | Umm Al Quwain | KM458285 | P. lutea | B335 | Muscat | KP280311 | Porites sp. |
| BH1627 | Umm Al Quwain | KP280253 | P. lutea | ADd014 | Fujairah AA | KT156662 | Galxea |
| BH1424 | Umm Al Quwain | KM458281 | P. lutea | ADd003 | Fujairah AA | KT156654 | Leptastrea |
| BH1666 | Umm Al Quwain | KP280254 | P. lutea | ALb020 | Al Lith ALS | KT156656 | Montipora. |
| BH1628 | Umm Al Quwain | KP280255 | P. lutea | ALb030 | Al Lith ALS | KT156660 | Montipora |
| BH1426 | Umm Al Quwain | KM458283 | P. lutea | ALb004 | Al Lith ALS | KT156655 | Montipora |
| BH1629 | Umm Al Quwain | KP280256 | P. lutea | KAb047 | KAUST KIF | KT156657 | Montipora |
| BH142 | Umm Al Quwain | KM458280 | P. lutea | OMa071 | Muscat FIS2 | KT156661 | Palythoa |
| OMd054 | Muscat SAS | KT156658 | Goniastrea | ||||
| YBa016 | Yanbu Y23 | KT156659 | Montipora | ||||
Collection site abbreviations are as follows: AA, Al Aqah; ALS, Abu lath Shallow Reef; FIS2, Fahal Island Site 2; KIF, KAUST Inner Fasr; SAS, Saifat Ash Shiekh; Y23, Yanbu 23.
Sequence accessions beginning with KM are from Hume et al., 2015 (13), beginning with KP are from D’Angelo et al., 2015 (15), and beginning with KT are from this study.
From samples collected on the PAG side of the Musandam peninsula.
From samples collected on the Gulf of Oman side of the Musandam peninsula.
The low genetic diversity of the S. thermophilum population in the southern PAG could be indicative of a recent bottleneck event or a founder effect. However, the PAG is well connected to the Gulf of Oman via the dominant inflow of surface water through the Strait of Hormuz (18), and various symbiont types including S. trenchii (synonym type D1a/D1-4), some of which are known to be heat stress-tolerant and to associate with many different coral species (19), are already established in other regions of the PAG and the Strait of Hormuz (15). Together with the fact that Symbiodinium can disperse rapidly over long distances (6, 19, 20), these conditions should promote a rapid homogenization of the symbiont distribution. Hence, the distinct distribution patterns presented in this study render founder or bottleneck effects rather unlikely causes of the low genetic diversity of S. thermophilum in the southern PAG. More plausibly, the genetic uniformity of this symbiont in the hottest coral reef ecosystem in the world may have resulted from a strong positive selection of a few of the most thermally tolerant genotypes from an old lineage with a more widespread but cryptic distribution. This selection pressure may not have resulted only from temperature extremes in the southern PAG, but might also have been influenced by the exceptionally high salinity of this habitat (15). Moreover, it cannot be ruled out that characteristics of today’s S. thermophilum populations were recently influenced by evolutionary processes despite the relatively short time (∼thousands of years) that this symbiont has been exposed to the pressures of the PAG environment.
A contrasting scenario was recently presented for the Caribbean, where the lack of genetic diversity among thermotolerant S. trenchii was interpreted as an indicator of a recent, long-range introduction of coral symbiont species (6). There, the opportunistic/invasive nature of a small founder population promoted its rapid spread to coral communities across the Greater Caribbean.
Although productive coral ecosystems exist in the southern PAG, the diversity of its habitat-forming scleractinian corals (34 species) (21) is substantially lower compared with the adjacent Gulf of Oman (68 species) (11) and the central and northern Red Sea (289 species) (22). Furthermore, in contrast to many other parts of the world, where corals construct reefs by forming vertically growing platforms through calcium carbonate accretion, corals in the southern PAG form only a living veneer over suitable substrates (8). Applying the findings from the PAG to the future of coral reefs elsewhere, it appears that coral–dinoflagellate symbioses may respond rapidly to increasing water temperature by the spread of tolerant symbiont associations, which are normally ecologically rare. Nonetheless, the PAG ecosystem has had millennia to adapt, whereas the adaptation to global warming will need to take place over decades to centuries. As exemplified by the coral communities of the PAG and changes in their composition in response to short-term temperature anomalies, not all species will survive beyond certain changes in the environment (21, 23). Inevitably, the shift toward more extreme environmental conditions on the global scale will be accompanied by a substantial loss of diversity both at the species and within-species level.
Although the failure of corals to build reefs in the PAG has been previously attributed to high-frequency disturbances, high-level bioerosion, and the constant exposure of corals to temperature and salinity extremes (8, 15, 24), the dominance of thermally tolerant coral–dinoflagellate combinations might also contribute to reduced reef accretion as recently exemplified for coral communities elsewhere (4–6). Because modeling predicts that reef accretion will not easily keep up with projected rates of sea-level rise at present day’s growth rates (25), the question arises whether the thermal adaptation of reef corals might come at the cost of an increased risk of reefs drowning in the rising oceans of the future.
Despite the potential tradeoffs that might be associated with the thermal adaptation of coral communities, the example of the PAG suggests that protecting present-day biodiversity is of upmost importance to provide the largest possible genepool from which more stress-tolerant species and genotypes may emerge and become more common under severe natural selection (26, 27). For coral reef ecosystems, this implies that any loss of biodiversity by causes other than heat stress including habitat destruction, pollution, and eutrophication (28–31) will reduce their likelihood of adapting to climate change.
Methods
Collection and DNA Extraction of Samples.
Cnidarian samples from 46 genera were collected at 23 reefs in the PAG, the Gulf of Oman, and the Red Sea. Sampling locations were categorized as follows: water bodies (the PAG, the Gulf of Oman, and the Red Sea), sampling regions (made up of one or more reef), and reefs. A detailed list of the sampling locations and number of cnidarians collected at each site can be found in Table S1. All samples were collected by SCUBA (self-contained underwater breathing apparatus) diving with ∼1 cm2 of tissue sampled from the surface of each cnidarian. Samples collected at Eilat were placed into 5 mL of RNAlater, whereas all other samples were flash-frozen in liquid nitrogen before storage at –20 °C.
Genomic DNA (gDNA) was extracted as described for environmental samples in Arif et al., 2014 (32), except for samples collected at Eilat. For Eilat samples, host and symbiont gDNA was extracted using a cetyl trimethylammonium bromide (CTAB) extraction. Before extraction, each of the samples was washed with 96% (vol/vol) ethanol to remove the majority of RNAlater storage buffer to minimize coprecipitation of salts during the DNA precipitation step of the extraction. Samples were then frozen in liquid nitrogen before being added to 1 mL of CTAB extraction buffer [2% (wt/vol) CTAB; 1.4 M NaCl; 0.5% 2-β-mercaptoethanol; 2% (wt/vol) polyvinylpyrrolidone (PVP); 20 mM EDTA; 100 mM Tris⋅HCl, pH 8.0] and beaten using a 5-mm stainless steel ball in a Tissue Lyser II (Qiagen) at maximum speed until the sample was completely homogenized. Samples were incubated at 60 °C for 30 min before three extractions in 1 mL of chloroform:isoamyl alcohol (IAA) (24:1), phenol:chloroform:IAA (25:24:1), and chloroform:IAA (24:1) with centrifugation after each extraction. The supernatant was added to an equal volume of isopropanol before incubation at –20 °C for 2 h. The DNA was pelleted through centrifugation before being washed in 750 µL of 96% ethanol and centrifuged. Supernatant was removed and pellets were dried before suspension in 50 µL of ddH2O. All centrifugation steps were carried out at 20,000 × g for 5 min at 4 °C.
ITS2 Genotyping of Symbiodinium spp. Harbored by Coral Samples.
The Symbiodinium nuclear ribosomal ITS2 region of all samples except those collected at Eilat was sequenced by 454 and MiSeq sequencing as detailed (32). The Symbiodinium ITS2 region of the Eilat-collected samples was sequenced by direct PCR sequencing (services provided by Eurofins MWG) using the internal primer SYM_VAR5.8SII (13) on the 18S-ITS1-5.8S-ITS2-28S amplicon amplified by SYM_VAR-FWD and SYM_VAR_REV as detailed (33), with the exception of an annealing temperature of 56 °C. Results of this sequencing revealed a mix of C15-cluster ITS2 sequences. Given that S. thermophilum group symbionts are characterized by an ITS2 type C3, these samples were excluded from further analysis.
Screening of Corals for Associations with S. thermophilum Group.
To identify the presence of S. thermophilum group symbionts, all corals were screened for the characteristic 8-bp indel sequence described in Hume et al., 2015 (13). To incorporate the possibility of PCR error and the existence of possible genetic variants, sequences 1 bp different (substitution only) from the originally described 8-bp sequences were also included in the results (hereafter referred to as the S. thermo.-indel) as indicative of the S. thermophilum group. Symbiont complements identified as having at least one ITS2 amplicon containing the S. thermo.-indel (hereafter referred to as S. thermo.-indel amplicons) were further categorized according to whether such amplicons made up more than or less than 1% of the total Symbiodinium amplicon sequences found in that organism (Fig. 1A). S. thermo.-indel–containing amplicons made up between 2% and 41% of the C3 type ITS2 amplicons in S. thermophilum samples (i.e., a coral sample hosting 100% S. thermophilum would likely have between 2% and 41% ITS2 amplicons containing the S. thermo.-indel; Fig. S1). As such, the 1% cutoff used in this study could potentially represent a 50% complement of S. thermophilum group symbionts, whereas symbionts containing >15% have a high likelihood of containing a close to pure complement of an S. thermophilum group symbiont.
Verification of S. thermophilum Group by PCR Amplification, Sequencing, and Phylogenetic Analysis of the psbAncr.
To validate the successful identification of the S. thermophilum group by the presence of the S. thermo.-indel, samples from the Gulf of Oman and Red Sea that contained more than 25% clade C ITS2 sequences and in which S. thermo.-indel amplicons made up more than 1% of the clade C sequences had the psbAncr analyzed by direct PCR sequencing as detailed in Hume et al., 2015 (13). Chromatograms were checked manually for miscalls. Chromatograms with multiple peaks were first assessed to determine whether the multiple peaks could be explained by a reading frame shift caused by indels by calling secondary peaks using the software Geneious 5.1.7 (www.geneious.com) before attempting to resolve indels using Indelligent 1.2 (dmitriev.speciesfile.org/indel.asp). If the multiple peaks could be resolved in this way, the majority sequence was associated with that sample. If multiple peaks were not explained by such “indel analysis,” chromatograms were characterized by no more than two peaks at each nucleotide location, and multiple peak locations clearly showed a predominant and lesser abundance of called nucleotide, then these predominant and lesser calls were used to identify a primary and secondary sequence, respectively. In this case, the primary sequence was associated with the sample. In cases where multiple-peaked chromatograms could not be explained by indel analysis or by identifying primary and secondary sequences, the sample genotype was not used in further analysis (1 out of 10 samples).
Nine samples that successfully returned psbAncr sequences were aligned manually with additional sequences from the psbAncr alignment created by Hume et al., 2015 (13) that contained sequences from corals collected within the PAG and C3 radiation sequences (17) collected external to the PAG.
Phylogenetic analysis was conducted by Bayesian inference using Mr. Bayes 3.2.2 (mrbayes.sourceforge.net/). Phylogenies were estimated using the Jukes–Cantor (JC) model with a gamma-shaped distribution (+G) with invariable sites (I) (according to Akaike Information Criterion using MEGA6; www.megasoftware.net/). Markov chain Monte Carlo (MCMC) analyses were run for 2.0 × 106 generations (SDs of split frequencies < 0.05), sampling every 1,000 generations. A relative burn-in of 0.25 was used in calculating a 50% majority rule consensus tree.
To compare psbAncr genetic diversity between the S. thermophilum group and the two most divergent ITS2 types (C40 and C27) that resolve within the C3-radiation psbAncr phylogeny, as well as between S. thermophilum group sequences found within and external to the PAG, pairwise genetic distances and within-group genetic distance were calculated in MEGA6 using the JC model +G (Table S4). Variance was determined to be equal (F test) before conducting a two sample Student’s t test to compare the within-group genetic distances of the samples from sites internal and external to the PAG.
Taxonomic Position of S. thermophilum Group Within Symbiodinium Clade C.
To elucidate the taxonomic position of the S. thermophilum group within clade C, a selection of S. thermophilum group, ITS2 type C3, and C41 and C39 (two of the top four numerically common subclades) harboring corals (including some corals under culture at the Coral Reef Laboratory Experimental Mesocosm Facility) (34) had their symbiont complement genotyped with six additional genetic markers (35): the nuclear ribosomal large subunit (nr28S), the nuclear elongation factor 2 (elf2), the chloroplastic ribosomal large subunit (cp23S) domain V, the coding region of the plastid-encoded photosystem II protein D1 (psbAcds), the mitochondrial cytochrome oxidase I (coi), and the mitochondrial cytochrome b (cob). To assess fine-scale taxonomic resolution, the psbAncr was also amplified.
Specifically, the following coral samples underwent this additional genotyping: all samples in the Gulf of Oman and the Red Sea characterized as S. thermophilum by psbAncr genotype and containing >99% clade C ITS2 sequences (eight samples); a selection of 23 samples collected in the PAG as part of studies by Hume et al., 2015 (13) and D’Angelo et al., 2015 (15), representing samples collected over >400 km within the PAG and as resolving in different positions within the psbAncr phylogeny of D’Angelo et al., 2015 (15); samples collected in the Red Sea and Gulf of Oman harboring ITS2 type C3 as their numerically dominant ITS2 amplicon and containing no S. thermo.-indel amplicons (three samples; OMD001, OMD002, and YBA008; Table S3); three ITS2-type C3-harboring Euphyllia spp. corals currently in culture at the Coral Reef Laboratory Experimental Mesocosm Facility (34) but originating from Indo-Pacific waters in proximity to Bali (EU2, EU3, and EU4; Table S3); and finally, four corals containing predominant ITS2 variants for C41 and C39 (representing the first and fourth most common ITS2 variants sampled, respectively) collected in the Red Sea and the Gulf of Oman (Table S3).
All PCR conditions were as for the SYM_VAR_FWD/SYM_VAR_REV primer pair with cycles as detailed in Pochon et al., 2014 (35), except for the psbAncr region that was amplified according to Hume et al., 2015 (13). Sequences were attained through direct PCR sequencing, services provided by Eurofins, using forward (elf2, psbAcds, coi, and cob) and reverse (nr28S and cp23S) primers. Sequences from this study were added to a selection of reference sequences (Table S5) representing members from clades A–I, including subcladal C1, C15, C90, and C91, as well as sequence collections from Gymnodinium simplex and Polarella glacialis as outgroups.
Sequences returned from the six additional genetic markers (i.e., not psbAncr) were aligned in MEGA 6 with the ClustalW algorithm and checked by eye. Hypervariable regions that prevented robust alignment were removed from the cp23S alignment sensu Pochon et al., 2006 (16). Phylogenies were estimated both from individual markers and from a concatenated supermatrix of all six markers. Phylogenies were estimated by Bayesian Inference in Mr. Bayes 3.2.2 using the following nucleotide substitution models: nr28S, Kimura 2-Parameter (K2) +G; Elf2, K2 +G +I; cp23S, Hasegawa Kishino Yano (HKY) +G; psbAcds, Generalized Time Reversible +G; coi, HYK +G; cob, HKY +G. The supermatrix analysis was partitioned with the separate nucleotide models being used for each marker’s region. MCMC analyses were run for 1.0 × 106 generations, sampling every 500 generations. A relative burn-in of 0.25 was used in calculating a 50% majority rule consensus tree. Alignments of the psbAncr sequences returned from the ITS2 type C3 (non-S. thermo.-indel–containing), C41, and C39 samples were not possible between ITS2 types from this study and with previous alignments by Hume et al., 2015 (13) (C3/S. thermophilum radiation) and Thornhill et al., 2014 (17) (C1 radiation) due to the dissimilarity of the sequences.
Acknowledgments
We appreciate the help of Cornelia Roder, Sergey Dobretsov, Julia Schnetzer, Todd LaJeunesse, and Drew Wham with sample collection. A. Al-Hemeri (UAE Federal Environment Agency), A. Al-Cibahy (Environment Agency of Abu Dhabi), and the Oman Ministry of Environment & Climate Affairs kindly provided Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) export permits (no. 09FEA555) and collection permits. We acknowledge Tropical Marine Centre (London) and Tropic Marin (Wartenberg) for sponsoring the Coral Reef Laboratory at the University of Southampton. We thank the NYU Abu Dhabi Institute for supporting the 2012/2013 field workshops during which samples for this study were collected and the Interuniversity Institute for Marine Sciences in Eilat for field work support. The study was funded by Natural Environment Research Council Grant NE/K00641X/1 (to J.W.), the European Research Council under the European Union’s Seventh Framework Programme Grant FP7/2007-2013/ERC Grant Agreement 311179 (to J.W.), the King Abdullah University of Science and Technology (C.R.V.), and Israel Science Foundation Grant 341/12, United States Agency for International Development/Middle East Regional Cooperation (USAID/MERC) No. M32-037 (to Y.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KR996268–KR996464 and KT156647–KT156665).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601910113/-/DCSupplemental.
References
- 1.Hughes TP, et al. Climate change, human impacts, and the resilience of coral reefs. Science. 2003;301(5635):929–933. doi: 10.1126/science.1085046. [DOI] [PubMed] [Google Scholar]
- 2.Palumbi SR, Barshis DJ, Traylor-Knowles N, Bay RA. Mechanisms of reef coral resistance to future climate change. Science. 2014;344(6186):895–898. doi: 10.1126/science.1251336. [DOI] [PubMed] [Google Scholar]
- 3.Logan CA, Dunne JP, Eakin CM, Donner SD. Incorporating adaptive responses into future projections of coral bleaching. Glob Change Biol. 2014;20(1):125–139. doi: 10.1111/gcb.12390. [DOI] [PubMed] [Google Scholar]
- 4.Jones MJ, Berkelmans R. Tradeoffs to thermal acclimation: Energetics and reproduction of a reef coral with heat tolerant Symbiodinium type-D. J Mar Biol. 2011;2011:185890. [Google Scholar]
- 5.Ortiz JC, González-Rivero M, Mumby PJ. Can a thermally tolerant symbiont improve the future of Caribbean coral reefs? Glob Change Biol. 2013;19(1):273–281. doi: 10.1111/gcb.12027. [DOI] [PubMed] [Google Scholar]
- 6.Pettay DT, Wham DC, Smith RT, Iglesias-Prieto R, LaJeunesse TC. Microbial invasion of the Caribbean by an Indo-Pacific coral zooxanthella. Proc Natl Acad Sci USA. 2015;112(24):7513–7518. doi: 10.1073/pnas.1502283112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambeck K. Shoreline reconstructions for the Persian Gulf since the last glacial maximum. Earth Planet Sci Lett. 1996;142(1):43–57. [Google Scholar]
- 8.Riegl B, Purkis S. Adaptations to climatic extremes in the world’s hottest sea. In: Riegl B, Purkis SJ, editors. Coral Reefs of the Gulf. Springer; Dordrecht, The Netherlands: 2012. pp. 1–4. [Google Scholar]
- 9.Arz HW, Lamy F, Pätzold J, Muller PJ, Prins M. Mediterranean moisture source for an early-Holocene humid period in the northern Red Sea. Science. 2003;300(5616):118–121. doi: 10.1126/science.1080325. [DOI] [PubMed] [Google Scholar]
- 10.Parker AG, et al. Holocene vegetation dynamics in the northeastern Rub’ al-Khali desert, Arabian Peninsula: A phytolith, pollen and carbon isotope study. J Quat Sci. 2004;19(7):665–676. [Google Scholar]
- 11.Coles SL. Coral species diversity and environmental factors in the Arabian Gulf and the Gulf of Oman: A comparison to the Indo-Pacific region. Atoll Res Bull. 2003;507:1–19. [Google Scholar]
- 12.Intergovernmental Panel on Climate Change . Long-term climate change: Projections, commitments and irreversibility. In: Stocker TF, et al., editors. Climate Change 2013 the Physical Science Basis Working Group 1 Contribution to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge Univ Press; New York: 2013. pp. 1029–1136. [Google Scholar]
- 13.Hume BCC, et al. Symbiodinium thermophilum sp. nov., a thermotolerant symbiotic alga prevalent in corals of the world’s hottest sea, the Persian/Arabian Gulf. Sci Rep. 2015;5:8562. doi: 10.1038/srep08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baird AH, Bhagooli R, Ralph PJ, Takahashi S. Coral bleaching: The role of the host. Trends Ecol Evol. 2009;24(1):16–20. doi: 10.1016/j.tree.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 15.D’Angelo C, et al. Local adaptation constrains the distribution potential of heat-tolerant Symbiodinium from the Persian/Arabian Gulf. ISME J. 2015;9(12):2551–2560. doi: 10.1038/ismej.2015.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pochon X, Montoya-Burgos JI, Stadelmann B, Pawlowski J. Molecular phylogeny, evolutionary rates, and divergence timing of the symbiotic dinoflagellate genus Symbiodinium. Mol Phylogenet Evol. 2006;38(1):20–30. doi: 10.1016/j.ympev.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 17.Thornhill DJ, Lewis AM, Wham DC, LaJeunesse TC. Host-specialist lineages dominate the adaptive radiation of reef coral endosymbionts. Evolution. 2014;68(2):352–367. doi: 10.1111/evo.12270. [DOI] [PubMed] [Google Scholar]
- 18.Sheppard C, et al. The Gulf: A young sea in decline. Mar Pollut Bull. 2010;60(1):13–38. doi: 10.1016/j.marpolbul.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 19.LaJeunesse TC, et al. Ecologically differentiated stress-tolerant endosymbionts in the dinoflagellate genus Symbiodinium (Dinophyceae) Clade D are different species. Phycologia. 2014;53(4):305–319. [Google Scholar]
- 20.Baird AH, Guest JR, Willis BL. Systematic and biogeographical patterns in the reproductive biology of scleractinian corals. Annu Rev Ecol Evol Syst. 2009;40:551–571. [Google Scholar]
- 21.Riegl B. Corals in a non-reef setting in the southern Arabian Gulf (Dubai, UAE): Fauna and community structure in response to recurring mass mortality. Coral Reefs. 1999;18(1):63–73. [Google Scholar]
- 22.Veron JEN, et al. Delineating the coral triangle. Galaxea. J Coral Reef Stud. 2009;11(2):91–100. [Google Scholar]
- 23.Loya Y, Sakai K, Nakano Y, Van Woesik R. Coral bleaching: The winners and the losers. Ecol Lett. 2001;4(2):122–131. [Google Scholar]
- 24.Riegl B. Inhibition of reef framework by frequent disturbance examples from the Arabian Gulf, South Africa, and the Cayman Islands. Palaeogeogr Palaeoclimatol Palaeoecol. 2001;175(1):79–101. [Google Scholar]
- 25.Hamylton SM, Leon JX, Saunders MI, Woodroffe CD. Simulating reef response to sea-level rise at Lizard Island: A geospatial approach. Geomorphology. 2014;222:151–161. [Google Scholar]
- 26.Dixon GB, et al. Genomic determinants of coral heat tolerance across latitudes. Science. 2015;348(6242):1460–1462. doi: 10.1126/science.1261224. [DOI] [PubMed] [Google Scholar]
- 27.van Oppen MJH, Oliver JK, Putnam HM, Gates RD. Building coral reef resilience through assisted evolution. Proc Natl Acad Sci USA. 2015;112(8):2307–2313. doi: 10.1073/pnas.1422301112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Angelo C, Wiedenmann J. Impacts of nutrient enrichment on coral reefs: New perspectives and implications for coastal management and reef survival. Curr Opin Environ Sustain. 2014;7:82–93. [Google Scholar]
- 29.Edinger EN, Jompa J, Limmon GV, Widjatmoko W, Risk MJ. Reef degradation and coral biodiversity in indonesia: Effects of land-based pollution, destructive fishing practices and changes over time. Mar Pollut Bull. 1998;36(8):617–630. [Google Scholar]
- 30.Vega Thurber RL, et al. Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Glob Change Biol. 2014;20(2):544–554. doi: 10.1111/gcb.12450. [DOI] [PubMed] [Google Scholar]
- 31.Sale PF. Addressing the decline of coral reefs. Reef Encounter. 2014;29:15–17. [Google Scholar]
- 32.Arif C, et al. Assessing Symbiodinium diversity in scleractinian corals via next-generation sequencing-based genotyping of the ITS2 rDNA region. Mol Ecol. 2014;23(17):4418–4433. doi: 10.1111/mec.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hume B, et al. Corals from the Persian/Arabian Gulf as models for thermotolerant reef-builders: Prevalence of clade C3 Symbiodinium, host fluorescence and ex situ temperature tolerance. Mar Pollut Bull. 2013;72(2):313–322. doi: 10.1016/j.marpolbul.2012.11.032. [DOI] [PubMed] [Google Scholar]
- 34.D’Angelo C, Wiedenmann J. An experimental mesocosm for long-term studies of reef corals. J Mar Biol Assoc U K. 2012;92(04):769–775. [Google Scholar]
- 35.Pochon X, Putnam HM, Gates RD. Multi-gene analysis of Symbiodinium dinoflagellates: A perspective on rarity, symbiosis, and evolution. PeerJ. 2014;2:e394. doi: 10.7717/peerj.394. [DOI] [PMC free article] [PubMed] [Google Scholar]