Significance
DNA epigenetic modifications in the forms of 5-methylcytosine (5mC) and 5-hydroxylmethylcytosine (5hmC) play crucial regulatory functions in the mammalian genome. Here we developed an ultrasensitive single-molecule epigenetic imaging technology for detecting and quantifying 5hmC and 5mC. By conducting single-molecule fluorescence resonance energy transfer experiments, we discovered high levels of adjacent and opposing methylated and hydroxymethylated CpG sites in the mouse genome, a previously unappreciated structure which may play an important role in gene regulation.
Keywords: single-molecule imaging, epigenetics, 5-hydroxymethylcytosine, 5-methylcytosine
Abstract
The modifications 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) are the two major DNA epigenetic modifications in mammalian genomes and play crucial roles in development and pathogenesis. Little is known about the colocalization or potential correlation of these two modifications. Here we present an ultrasensitive single-molecule imaging technology capable of detecting and quantifying 5hmC and 5mC from trace amounts of DNA. We used this approach to perform single-molecule fluorescence resonance energy transfer (smFRET) experiments which measure the proximity between 5mC and 5hmC in the same DNA molecule. Our results reveal high levels of adjacent and opposing methylated and hydroxymethylated CpG sites (5hmC/5mCpGs) in mouse genomic DNA across multiple tissues. This identifies the previously undetectable and unappreciated 5hmC/5mCpGs as one of the major states for 5hmC in the mammalian genome and suggest that they could function in promoting gene expression.
Epigenetic modifications of DNA contribute critical regulatory functions to the underlining genetic sequence. The two major DNA modifications in the mammalian genome are 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC), which are often referred to as the “fifth base” and “sixth base,” respectively; 5mC is generated by DNA methyltransferase (DNMTs) mainly at CpG dinucleotides and generally results in gene silencing (1, 2), and 5hmC is oxidized from 5mC by ten-eleven translocation (TET) family dioxygenases and mostly enriched in brain (3, 4). The modification 5hmC is generally believed to be a gene activation mark for two reasons. First, it is enriched in active genes in brain and other tissues (5–9). Second, 5hmC is the key intermediate in the mammalian active DNA demethylation pathway in which 5hmC is further oxidized by TET to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) followed by removal of 5fC and 5caC through base excision repair (10–12).
Intensive research on 5hmC in recent years indicated the TET-mediated oxidation process plays important roles in diverse biological processes ranging from embryonic development to carcinogenesis; however, how 5hmC exerts its biological role is largely unclear (13–15). One important piece of information that has been missing is the interplay between 5hmC and its precursor 5mC. Despite many techniques that have been developed to detect and sequence 5mC and 5hmC, including recent advances in base-resolution mapping of 5hmC (16), no method to date can simultaneously reveal 5mC and 5hmC sites in the same DNA molecule. Here we present an ultrasensitive single-molecule imaging technology capable of detecting and quantifying 5mC and 5hmC from trace samples, which we used to study the distance relationship between 5mC and 5hmC with single-molecule fluorescence resonance energy transfer (smFRET).
Results and Discussion
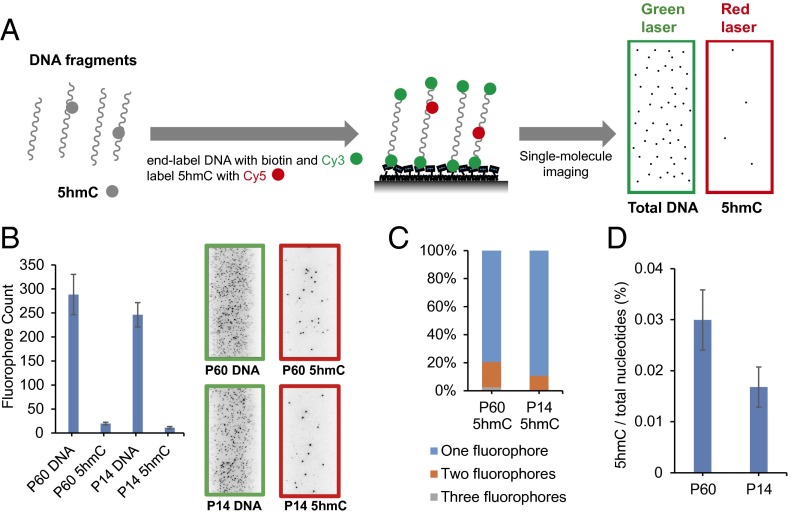
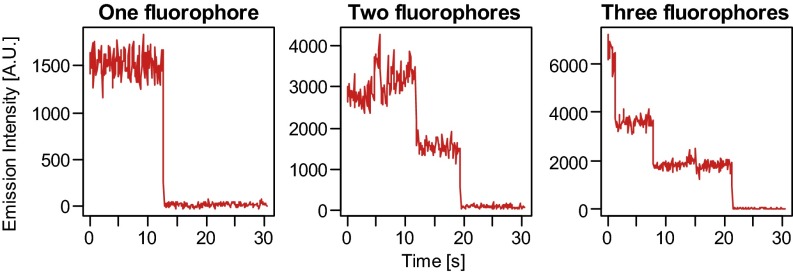
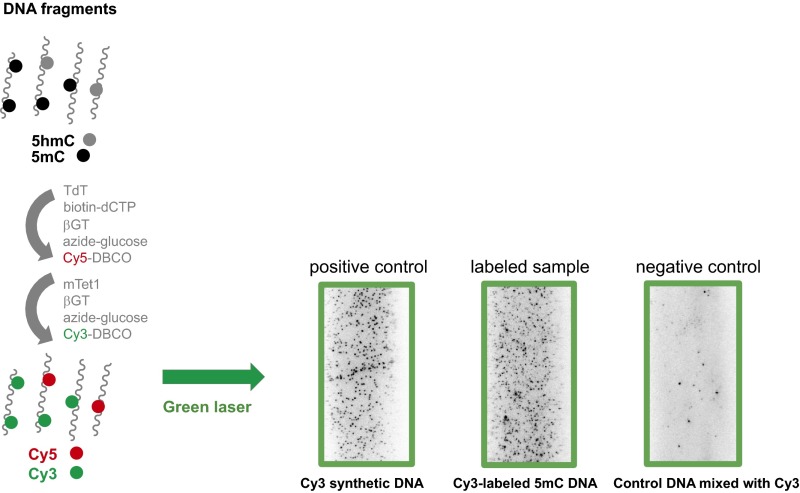
Our imaging approach uses a selective chemical labeling strategy (17) to label DNA base modifications with specific fluorophores, followed by single-molecule imaging fluorescent assays (18) (Fig. 1A and Fig. S1). This method is highly modular and can be used to image just one modification or multiple modifications simultaneously. To image 5hmC, the DNA fragments are first end-labeled with biotin and Cy3 by using Terminal Transferase (TdT) and modified dCTP. The biotin is used to immobilize DNA molecules to the microscope slide, and the Cy3 serves as a counter for total amount of DNA. Next, β-glucosytransferase (βGT) is used to label 5hmC with Cy5 via an azide-modified glucose. The dye-labeled biotinylated DNA is then captured by surface-tethered neutravidin on a passivated microscope slide and imaged with single-molecule total internal reflection fluorescence (TIRF) microscopy (Fig. 1A and Fig. S1). The number of 5hmC containing molecules and total amount of DNA can be determined by counting the fluorophores in the red channel (Cy5) and green channel (Cy3), respectively. Using synthetic DNA constructs we confirmed that the stepwise labeling is highly efficient and shows minimum background (Fig. S1). We then applied the method to postnatal day 60 (P60) and postnatal day 14 (P14) mouse cerebellum genomic DNA (Fig. 1B). In addition to counting the fluorophores by direct emission, we also used photobleaching to detect multiple fluorophores in individual DNA molecules (Fig. 1C and Fig. S2). The 5hmC level can then be calculated based on the fluorophore counts, multiple fluorophore correction, and the average length of the DNA fragments (Fig. 1D). Our results for 5hmC are comparable to what has been found in bulk samples by previous HPLC-MS techniques (19) and also reveal the age-dependent increase of 5hmC in mouse cerebellum from P14 to P60 as previously reported (17).
Fig. 1.
Single-molecule imaging of 5hmC. (A) General procedure for single-molecule imaging of 5hmC. DNA fragments are end-labeled with biotin and Cy3, and 5hmC is labeled with Cy5. The labeled DNA is immobilized to the microscope slide and imaged with single-molecule TIRF microscopy. (B) Fluorophore counts of labeled P60 and P14 mouse cerebellum genomic DNA with example images shown on the right. (C) Distribution of multiple fluorophores on DNA fragment of mouse cerebellum when detecting 5hmC. (D) The 5hmC level of mouse cerebellum DNA. Error bars, mean ± SD (n = 15 counting regions).
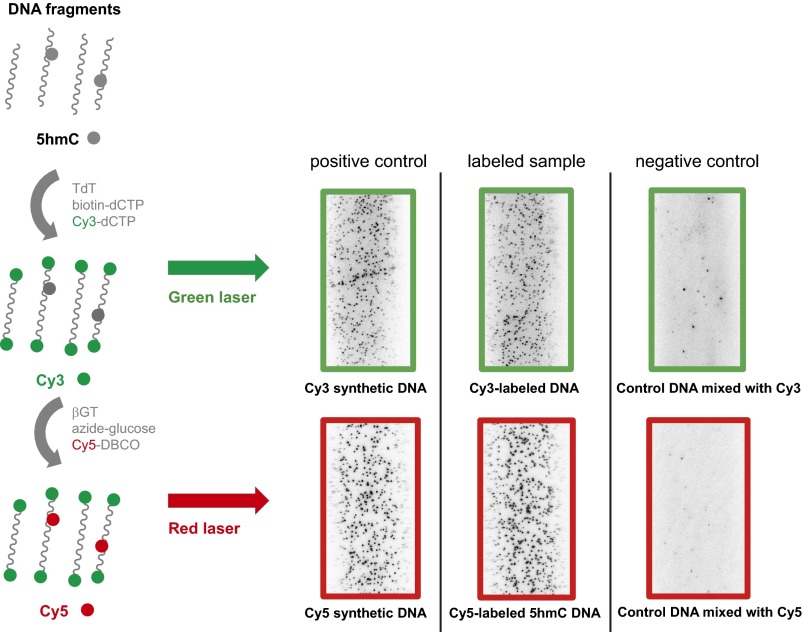
Fig. S1.
Example images of stepwise labeling reactions using model DNA. Positive controls are synthetic DNA bearing biotin and fluorophore. Top row labeled sample is biotin and Cy3 end-labeled genomic DNA fragments. Bottom row labeled sample is synthetic DNA bearing 5hmC and biotin after labeling reaction. Negative controls are synthetic DNA bearing biotin incubated with fluorophore without the enzymes. All DNA concentrations are 30 pM.
Fig. S2.
Example photobleaching traces of single and multiple fluorophores.
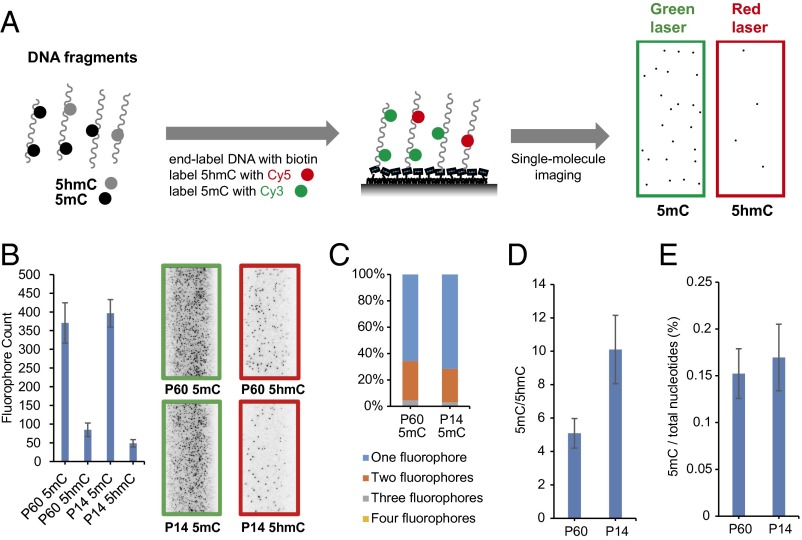
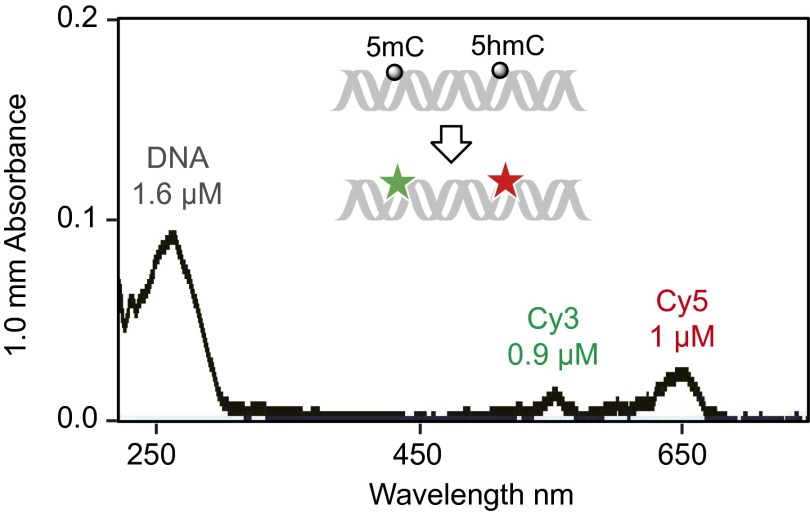
To image 5mC and 5hmC simultaneously, we developed a dual-labeling strategy (Fig. 2A and Fig. S3). We first end-labeled DNA fragments with biotin and labeled 5hmC with Cy5 as described above. Then we used βGT and Tet1 in a one-pot procedure to label 5mC with Cy3 (20) (Fig. 2A and Fig. S3). The number of 5hmC and 5mC modifications can be determined by counting the fluorophores in the red channel (Cy5) and green channel (Cy3), respectively. The 5mC labeling was highly efficient as indicated by experiments with synthetic DNA (Fig. S3). The labeling efficiencies of both 5hmC and 5mC are about 60% as estimated by the absorption spectrum of the dual-labeled DNA (Fig. S4). The dual-labeling strategy was also validated on mouse cerebellum genomic DNA. By counting Cy3 on 5mC and Cy5 on 5hmC we obtained the ratio between 5mC and 5hmC occurrence (Fig. 2B). As expected, in addition to higher counts of 5mC than that of 5hmC, we observed more multiple fluorophores events on 5mC than on 5hmC (Fig. 2C), which was factored into the final 5mC to 5hmC ratio (Fig. 2D). We calculated the absolute 5mC level from the previous 5hmC measurement (Fig. 2E). Unlike 5hmC, the 5mC level did not change significantly between P14 and P60.
Fig. 2.
Dual-labeling of 5mC and 5hmC for simultaneous imaging. (A) General procedure for dual-labeling and simultaneous imaging of 5mC and 5hmC. DNA fragments are end-labeled with biotin, 5hmC is labeled with Cy5, and 5mC is labeled with Cy3. The labeled DNA is immobilized to the microscope slide and imaged with single-molecule TIRF microscopy. (B) Fluorophore counts of dual-labeled P60 and P14 mouse cerebellum genomic DNA with example images shown on the right. (C) Distribution of multiple fluorophores on DNA fragment of mouse cerebellum when detecting 5mC. (D) The 5mC to 5hmC ratio in mouse cerebellum genomic DNA after adjusting multiple fluorophores. (E) The 5mC level of mouse cerebellum DNA. Error bars, mean ± SD (n = 15 counting regions).
Fig. S3.
Example images of 5mC labeled model DNA. Positive control is synthetic DNA bearing biotin and fluorophore. Labeled sample is synthetic DNA bearing 5mC and biotin after labeling reaction. Negative controls are synthetic DNA bearing biotin incubated with fluorophore without the enzymes. All synthetic DNA concentrations are 30 pM.
Fig. S4.
An absorbance spectrum of a 36mer synthetic DNA bearing one 5mC and one 5hmC after the dual labeling reaction in which 5mC is labeled with Cy3 and 5hmC is labeled with Cy5. The absorption peaks at 260, 555, and 647 nm correspond to the absorption maxima of DNA, Cy3, and Cy5, respectively. The concentrations are calculated using the extinction coefficients of the synthetic DNA, Cy3, and Cy5.
Thanks to the ultrahigh sensitivity of single-molecule imaging, this method only requires 50 pg of DNA or less for each measurement, representing orders of magnitude less DNA than is required by previous quantification methods such as the HPLC-MS or other fluorescence-based methods (19, 21–23). Besides being a general detection and quantification method, single-molecule imaging provides a unique opportunity to study the colocalization states of 5mC and 5hmC, which has been unknown because no previous method could perform an integrated analysis of 5mC and 5hmC in the same DNA molecule. With the dual-labeling scheme described above, we were able for the first time to measure the proximity between 5mC and 5hmC in the same DNA molecule.
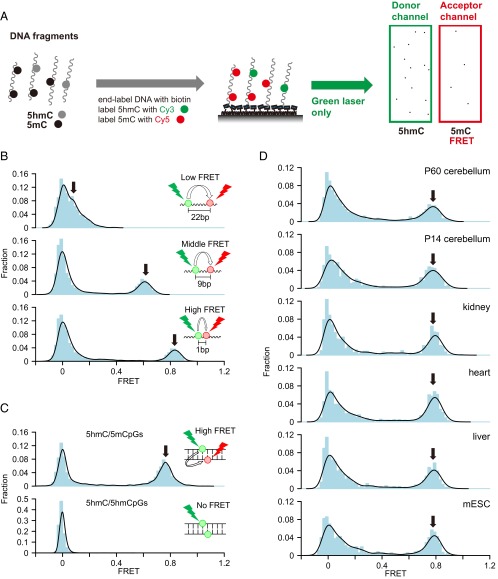
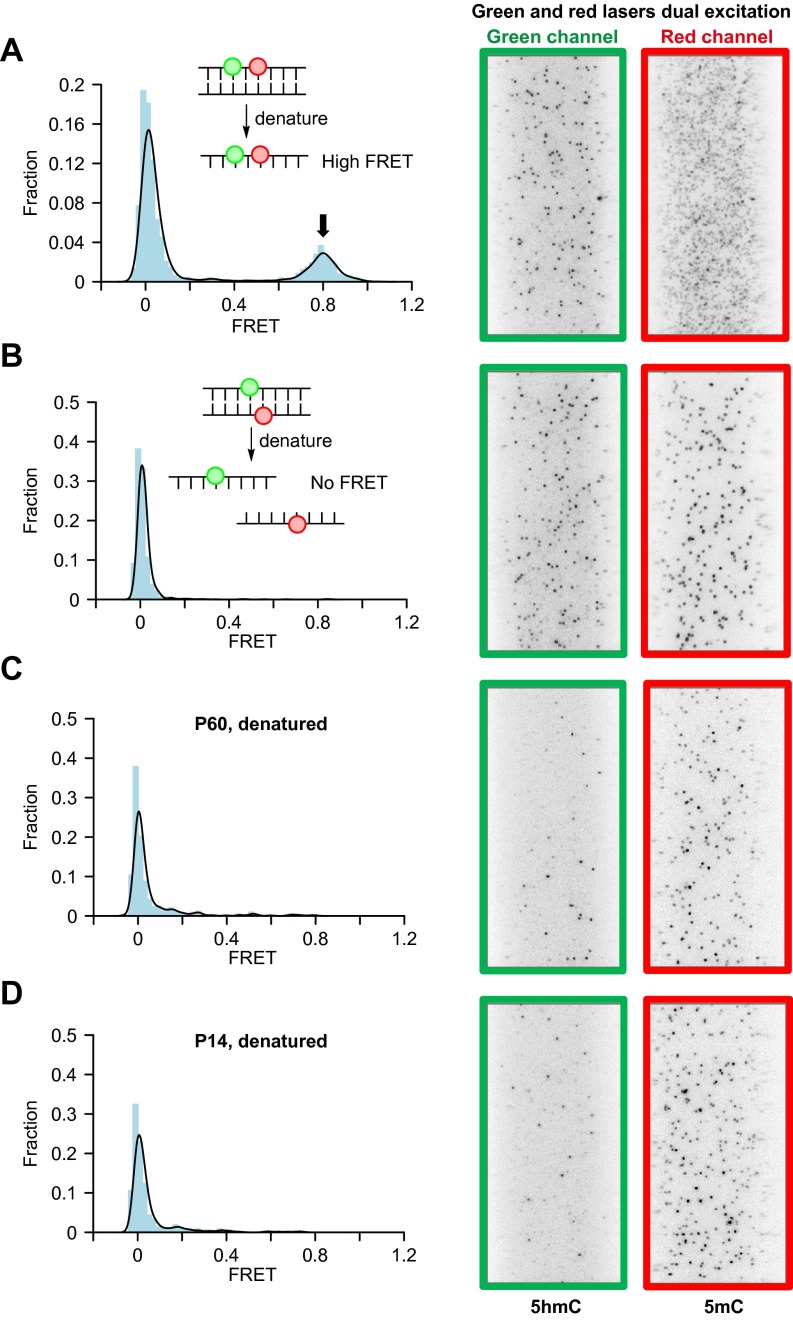
Because 5mC is much more abundant than 5hmC, we switched the fluorescence labels on the modification for the smFRET experiment so that there are more acceptors (5mC-Cy5) than donors (5hmC-Cy3) (Fig. 3A). We first used synthetic DNA with 5hmC and 5mC separated by defined distances for smFRET measurement. We observed low FRET (∼0.1), middle-FRET (∼0.6), and high-FRET (∼0.82) states when 5hmC and 5mC are 22, 9, and 1 bp apart, respectively (Fig. 3B). We also constructed a synthetic DNA with adjacent and opposing hemihydroxymethylated/hemimethylated CpG sites (5hmC/5mCpGs) and observed a high-FRET (∼0.78) state (Fig. 3C). Surprisingly, when performing the smFRET measurement on mouse cerebellum DNA, we observed a distinct high-FRET peak (∼0.78) with no middle-FRET peaks in between in both P60 and P14 samples (Fig. 3D). To determine whether this high-FRET state was from 5hmC and 5mC on the same strand or from 5hmC and 5mC on the CpG site, we denatured the DNA before smFRET experiment. We verified that synthetic DNA with 5hmC and 5mC on the same strand retained the FRET signal, whereas synthetic DNA with 5hmC/5mCpGs lost the FRET signal after denaturing (Fig. S5 A and B). Both P60 and P14 mouse cerebellum DNA lost the FRET signal after denaturing, showing that the high FRET state was from adjacent and opposing CpG sites (Fig. S5 C and D).
Fig. 3.
smFRET analysis between dual-labeled 5mC and 5hmC. (A) General procedure for smFRET analysis between 5mC and 5hmC. DNA fragments are end-labeled with biotin, 5hmC is labeled with Cy3, and 5mC is labeled with Cy5. The labeled DNA is immobilized to the microscope slide and imaged with single-molecule TIRF microscopy for smFRET analysis. (B) smFRET distributions of dual-labeled synthetic DNA with 5mC and 5hmC separated with different lengths in the same DNA strand. (C) smFRET distributions of dual-labeled synthetic DNA with 5hmC/5mCpG or 5hmC/5hmCpG sites. (D) smFRET distributions of dual-labeled mouse genomic DNA from various tissues and mESC. In B–D, solid lines indicate the Gaussian kernel density estimation. Arrows indicate FRET peaks. The zero FRET peak is mainly from DNAs with donor only.
Fig. S5.
smFRET distributions and example images of denatured samples. (A) Denatured dual-labeled synthetic DNA with 5mC and 5hmC separated with 1 bp in the same strand. (B) Denatured dual-labeled synthetic DNA with 5hmC/5mCpG site. (C) Denatured dual-labeled mouse P60 cerebellum genomic DNA. (D) Denatured dual-labeled mouse P14 cerebellum genomic DNA. Each panel has smFRET distribution on the left and dual excitation example images on the right showing the fluorophore labeling remains after denaturing. Arrows indicate FRET peak.
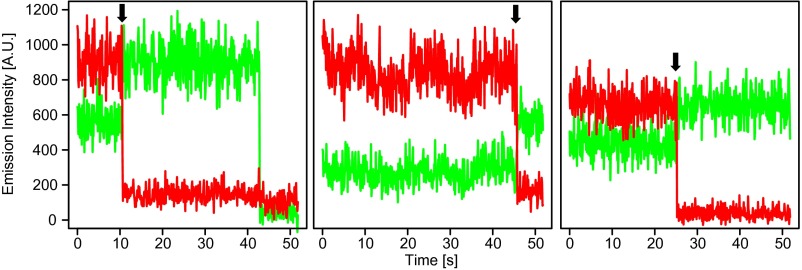
We also constructed synthetic DNAs with fully hydroxymethylated (5hmC/5hmCpGs) or fully methylated CpG sites (5mC/5mCpGs) and verified the smFRET signals were not from these CpG sites (Fig. 3C and Fig. S6). Additionally, we used mouse cerebellum genomic DNA to confirm that the high-FRET events can only be observed in 5mC and 5hmC dual-labeled samples but not in donor-only or acceptor-only single-labeled samples (Fig. S7). Moreover, we observed anticorrelated intensity changes of the donor and acceptor due to accepter bleaching in mouse cerebellum genomic DNA, a validation of the smFRET events (Fig. S8). These results confirm the existence of high levels of 5hmC/5mCpGs in the mouse cerebellum. We further conducted smFRET measurements in genomic DNA from different mouse tissues with different 5hmC levels and cell proliferation rates (10, 24) and from mouse embryonic stem cell (mESC) as well. We observed consistently high levels of 5hmC/5mCpGs in all of the tissues and the cell line, indicating that such hybrid CpG sites are a universal phenomenon across the mammalian genome independent of cell proliferation and 5hmC level (Fig. 3D). After accounting for the efficiency of the labeling and detection on synthetic DNA, we estimated that 5hmC/5mCpGs roughly account for 60% of 5hmC. Our results revise the current understanding that 5hmC mainly exists as fully hydroxymethylated form (5hmC/5hmCpGs) (15, 25) and point to the previously indiscernible 5hmC/5mCpGs as a major state for 5hmC in vivo.
Fig. S6.
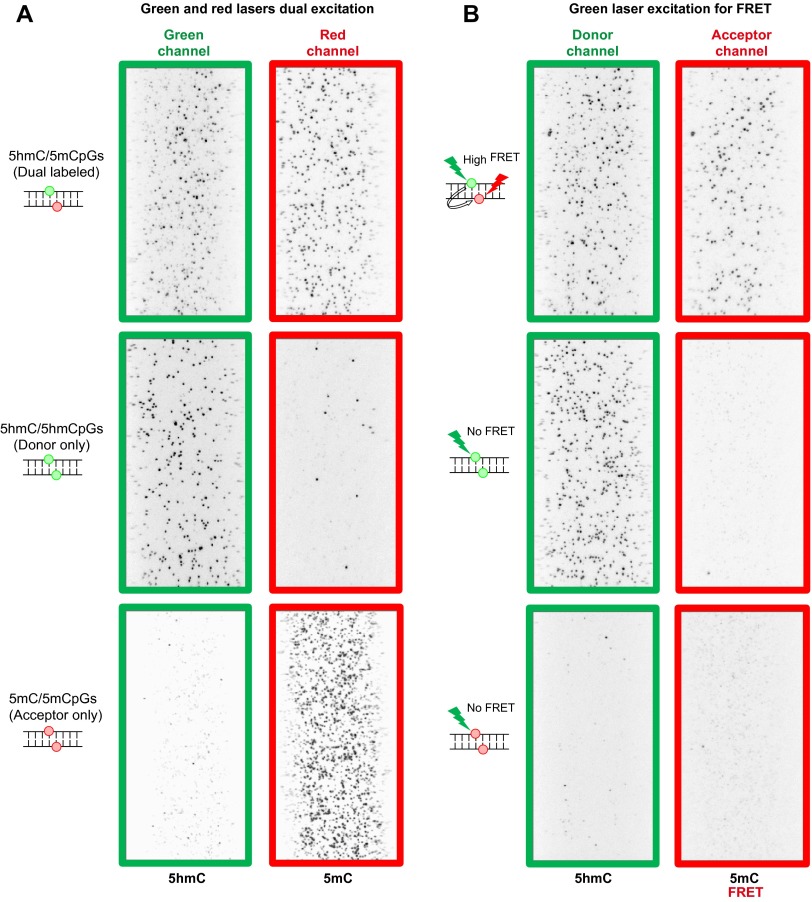
Example images of model DNA with different CpG sites. (A) Green and red lasers dual excitation and (B) green laser excitation-only smFRET experiments of model DNA with different CpG sites. (Top) Example image of model DNA containing a 5hmC/5mCpG site. (Middle) Example image of model DNA containing a 5hmC/5hmCpG site. (Bottom) Example image of model DNA containing a 5mC/5mCpG site.
Fig. S7.
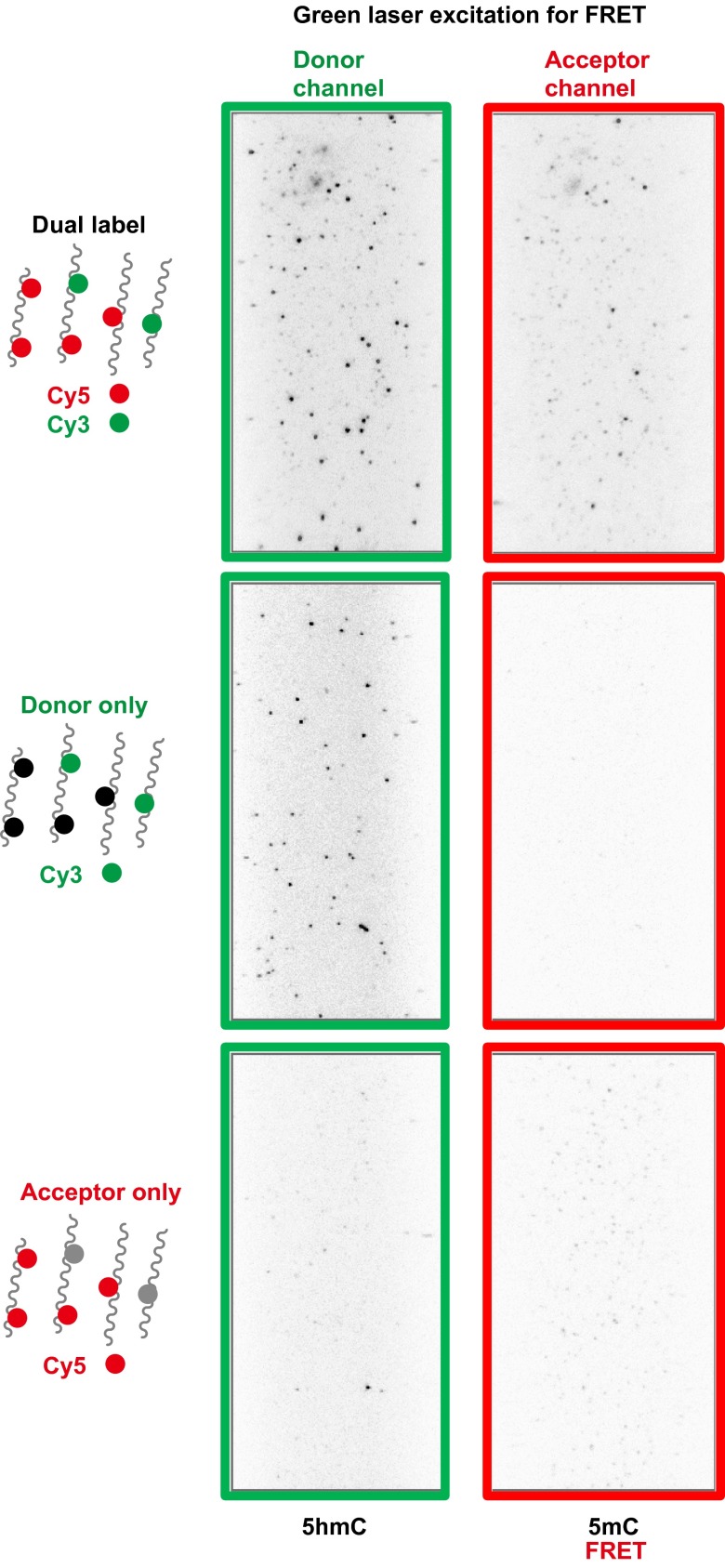
Example images of the smFRET experiment of P60 mouse cerebellum DNA. (Top) Example image of the dual labeled DNA. (Middle) Example image of the donor-only labeled DNA. (Bottom) Example image of the acceptor-only labeled DNA.
Fig. S8.
Example of anticorrelated time traces of smFRET experiment of P60 mouse cerebellum DNA. Arrows indicate anticorrelated intensity changes.
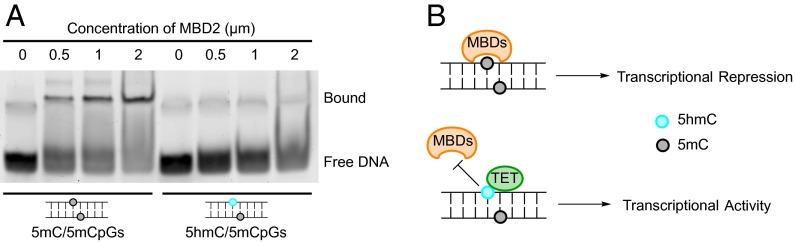
The widespread and highly abundant 5hmC/5mCpGs could potentially serve important functions such as gene activation and protein binding. One general mechanism of DNA methylation mediated gene repression is through the recruitment of methyl-CpG binding domain (MBD)-containing proteins to fully methylated CpG sites (1). Being the first intermediate state of TET oxidation of 5mC/5mCpGs, one immediate consequence of 5hmC/5mCpGs could be to inhibit such interactions. Others have investigated MBD binding to asymmetrically methylated sites in the context of oxidative DNA damage (26) and genome replication (25) and found reduced binding affinity. We tested the protein MBD2 using an electrophoretic mobility shift assay (EMSA) on synthetic DNA and found that 5hmC/5mCpGs significantly inhibited its binding compared with 5mC/5mCpGs (Fig. 4A). This result is consistent with previous investigation using other methods (25), and it implies the potential function of 5hmC/5mCpGs may be to induce transcriptional activation through the inhibition of MBD protein binding (Fig. 4B). Future studies are needed to elucidate the functional significance of 5hmC/5mCpGs.
Fig. 4.
Reduced binding to MBD proteins is shown in 5hmC/5mCpGs compared with 5mC/5mCpGs. (A) Binding of 5mC/5mCpGs and 5hmC/5mCpGs containing DNA to varying concentrations of the methyl-CpG binding domain of human MBD2 protein assayed via EMSA. (B) Model for potential function of 5hmC/5mCpGs in promoting gene expression.
Here we described a versatile single-molecule technology to image 5mC and 5hmC from trace amounts of DNA. The ultralow input requirement enables this approach to be applied to limited and sensitive samples, such as cell-free circulating DNA (27). It is also the first measurement technology to our knowledge that can be used to study the colocalization status between 5mC and 5hmC with smFRET. The modification 5mC occurs almost exclusively in the form of 5mC/5mCpGs and is maintained by maintenance methyltransferase DNMT1 following DNA replication (1, 2). Recent base-resolution sequencing suggested 5hmC to be less symmetric in CpG than 5mC (28). Our results show that a large proportion of 5hmC exists in the form of 5hmC/5mCpGs, a previously undetectable and unappreciated state. TET proteins can convert 5mC in all contexts to 5hmC efficiently in vitro (20, 28). The widespread occurrence of 5hmC/5mCpGs in the mammalian genome suggests TET proteins may be regulated by yet unidentified mechanisms to preferentially oxidize only one strand of 5mC/5mCpGs in vivo. Alternatively, 5hmC/5mCpGs can be generated by the de novo methyltransferase DNMT3 following DNA replication (25). Also, 5hmC/5mCpGs may play important functions such as gene activation (Fig. 4B) and protein binding, which warrant further investigation. Currently, to our knowledge, our method is the only way to detect such hybrid CpG sites. It highlights the importance of developing new methods that can detect multiple DNA modifications in the same DNA context. Future development and application of this and other single-molecule technologies such as the direct detection of DNA modifications by single-molecule, real-time sequencing (29) would enable the study of a large set of epigenetic modifications.
Materials and Methods
Preparation of Genomic DNA.
All animal procedures were performed in accordance with approved Institutional Animal Care and Use Committee protocols by Stanford University. Mouse tissue genomic DNA was extracted using Wizard Genomic DNA Purification Kit (Promega) following the manufacturer's recommendation. Genomic DNA was digested with dsDNA Fragmentase (NEB) to 50–200 bp following the manufacturer's recommendation and analyzed by agarose gel electrophoresis to determine the average fragment length.
Preparation of Synthetic DNA.
Oligonucleotides containing 5mC, 5hmC, biotin, Cy3, Cy5, and FAM were ordered from Integrated DNA Technologies. Equal molar of two complementary strands are slowly annealed to form duplex DNA. Sequences of synthetic DNA are as follows:
5hmC_5mC_22bp, X = 5mC, Y = 5hmC:
5′ CCCGAXGCATGATCTGTACTTGATCGACYGTGCAAC 3′
3′ GGGCTGCGTACTAGACATGAACTAGCTGGCACGTTG-biotin 5′
5hmC_5mC_9bp, X = 5mC, Y = 5hmC:
5′ CCCGACGCATGATCTGTAXTTGATCGACYGTGCAAC 3′
3′ GGGCTGCGTACTAGACATGAACTAGCTGGCACGTTG-biotin 5′
5hmC_5mC_1bp, X = 5mC, Y = 5hmC:
5′ CCCGAXGYATGATCTGTACTTGATCGACCGTGCAAC 3′
3′ GGGCTGCGTACTAGACATGAACTAGCTGGCACGTTG-biotin 5′
5hmC/5mC_CpG, X, Y= 5mC or 5hmC:
5′ TCGATGTAGTGCGTCACYGGATGATAGCTGTACTCA 3′
3′ AGCTACATCACGCAGTGGXCTACTATCGACATGAGT-biotin 5′
5hmC/5mC_CpG_FAM, X = 5mC, Y = 5mC or 5hmC:
5′ TCGATGTAGTGCGTCACYGGATGATAGCTGTACTCA 3′
3′ AGCTACATCACGCAGTGGXCTACTATCGACATGAGT-FAM 5′
End-Labeling of Genomic DNA Fragments with Biotin and Cy3.
To end-label genomic DNA fragment with biotin and Cy3, 50 ng genomic DNA fragment was incubated in a 10-µL solution containing 1X Terminal Transferase Reaction Buffer (NEB), 0.25 mM CoCl2, 50 µM biotin-16-Aminoallyl-2'-dCTP (Trilink), 50 µM Cy3-dCTP (GE Healthcare), and 10 U Terminal Transferase (NEB) for 0.5 h at 37 °C. To end-label genomic DNA fragment with biotin only, 50 ng genomic DNA fragment was incubated with Terminal Transferase as described in the presence of 250 µM biotin-16-Aminoallyl-2'-dCTP. The end-labeled DNA was purified by DNA Clean & Concentrator-5 (Zymo) following the manufacturer’s recommendation and eluted in 8 µL H2O.
Single-Labeling of 5hmC with Cy5.
Biotin and Cy3 end-labeled DNA was incubated in a 10-µL solution containing 50 mM Hepes buffer (pH 8), 25 mM MgCl2, 75 μM UDP-6-N3-Glc (Active Motif), and 1 U βGT (5-hmC glucosyltransferase; Zymo) for 1 h at 37 °C. Then 2.5 μL Cy5 DBCO (10 mM stock in DMSO; Click Chemistry Tools) was directly added to the reaction mixture and incubated overnight at 37 °C. The 5hmC-labeled DNA was purified by DNA Clean & Concentrator-5 (Zymo) following the manufacturer’s recommendation and eluted in EB buffer (Qiagen). DNA concentration was determined by Qubit Fluorometer (Life Technologies).
Dual-Labeling of 5hmC and 5mC with Cy5 and Cy3.
Biotin end-labeling of DNA and 5hmC labeling were performed as described above, and the 5hmC-labeled DNA was purified into 8 µL H2O. Next, Tet1 oxidation was carried out by incubating the 5hmC-labeled DNA in a 10-µL solution containing 50 mM Hepes buffer (pH 8), 10 mM MgCl2, 75 mM ammonium iron (II) sulfate, 2 mM ascorbic acid, 1 mM α-ketoglutarate, 90 μM UDP-6-N3-Glc, 1 mM DTT, 2 U βGT, and 2 μM Tet1 (Wisegene) for 1 h at 37 °C. The DNA was purified and incubated with 2 mM Cy3 DBCO (Click Chemistry Tools) overnight at 37 °C in presence of 50 mM Hepes buffer (pH 8) and 10 mM MgCl2. The dual-labeled DNA was purified by DNA Clean & Concentrator-5 into EB buffer. DNA concentration was determined by Qubit Fluorometer.
Dual-Labeling of 5hmC and 5mC with Cy3 and Cy5 for smFRET.
The biotin end-labeled DNA was incubated in a 10-µL solution containing 50 mM Hepes buffer (pH 8), 25 mM MgCl2, 150 μM UDP-6-N3-Glc (Active Motif), and 1 U βGT for 1 h at 37 °C. Then 1 U fresh βGT and 200 μM unmodified UDP-Glc (NEB) was added to the reaction and incubated for 1 h at 37 °C. The DNA was purified and incubated with 2 mM Cy3 DBCO (Click Chemistry Tools) overnight at 37 °C in presence of 50 mM Hepes buffer (pH 8) and 10 mM MgCl2. The 5hmC-labeled DNA was purified and oxidized with Tet1 and labeled with Cy5 DBCO as described above. The dual-labeled DNA was purified by DNA Clean & Concentrator-5 into EB buffer. DNA concentration was determined by Qubit Fluorometer.
Denaturing Dual-Labeled Sample for smFRET.
Dual-labeled synthetic DNA was first end-labeled with biotin as described above. Fifty microliters of 50 pM dual-labeled synthetic DNA or genomic DNA samples in EB buffer were heated for 10 min at 100 °C and immediately put on ice for 10 min before smFRET experiments.
Single-Molecule Imaging Through Total Internal Reflection Fluorescence Microscope.
A quartz slide was first coated with polyethylene glycol (PEG) molecules [99:1 (mol/mol) mPEG-SVA:biotin-PEG-SVA (Laysan Bio)] to eliminate nonspecific binding of DNA (30). The slide was then assembled into a flow chamber and coated with neutravidin by injecting 0.2 mg/mL neutravidin solution. Through the specific interaction between biotin and neutravidin, the dye-labeled DNAs conjugated with biotin were immobilized on the PEG-coated surface by an incubation at a concentration of 30∼100 pM for 15 min. After washing out the free DNAs, the FRET measurements by a TIRF microscope were performed with an oxygen scavenger system (0.1 mg/mL glucose oxidase and 0.02 mg/mL catalase) and Trolox to eliminate single-molecule blinking events (31). Details of the wide-field TIRF microscope have been reported (30). Briefly, the excitation beam was focused into a pellin broca prism (CVI Laser), which was placed on top of a quartz slide with a thin layer of immersion oil in between to match the index of refraction. Cy3 (donor) and Cy5 (acceptor) dyes were excited through the dual-laser excitation system (532 and 633 nm) via TIRF. The fluorescence signals from Cy3 and Cy5 that were collected by a water immersion objective lens (60×, 1.2 N.A. Nikon) and then passed through a notch filter to block out excitation beams. The emission signals of Cy5 dyes were separated by a 630-nm dichroic mirror (630DCXR; Chroma Technology) and detected by the electron-multiplying charge-coupled device camera (iXon 897; Andor Technology) with a time resolution of 100 ms. The fluorescence signal, recorded in real time by using software written in Visual C++ (Microsoft), was amplified before camera readout, which produced an arbitrary unit for the recorded fluorescence intensity. The single-molecule data analysis was carried out by programs written in Visual C++. The FRET efficiency, E, was approximately calculated as the intensity of the acceptor channel divided by the total intensity, which is the sum of donor and acceptor channel intensities. Leakages from the donor channel to the acceptor channel and vice versa were corrected.
Electrophoretic Mobility Shift Assay.
The MBD domain of MBD2 (from MethylMiner Methylated DNA Enrichment Kit, Life Technologies) at varying concentrations was incubated with 10 nM 36_5hmC/5mC_CpG_FAM duplex DNA, 50 ng/µL of poly(dA-dT)/poly(dA-dT) (Sigma) in 20 mM Hepes (pH 8), 1 mM EDTA, 0.05% Triton X-100, and 30 mM KCl for 15 min at room temperature in a 10 µL reaction volume, before the addition of 2.5 µL Hi-Density TBE Sample Buffer (Life Technologies). The binding reactions were then loaded onto 6% DNA retardation gel (Life Technologies) and visualized with a Typhoon 9410 imager by using standard Blue FAM filter set (λex = 488 nm, λem = 520 nm).
Acknowledgments
We thank Dr. Chuan He for providing mECS genomic DNA. We thank Dr. Ozgun Gokce for providing mouse tissues. This study was supported by National Institutes of Health (U01 CA154209 to S.R.Q.) and Department of Defense (W81XWH1110287 to S.R.Q.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600223113/-/DCSupplemental.
References
- 1.Klose RJ, Bird AP. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci. 2006;31(2):89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 3.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szulwach KE, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14(12):1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151(7):1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colquitt BM, Allen WE, Barnea G, Lomvardas S. Alteration of genic 5-hydroxymethylcytosine patterning in olfactory neurons correlates with changes in gene expression and cell identity. Proc Natl Acad Sci USA. 2013;110(36):14682–14687. doi: 10.1073/pnas.1302759110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomson JP, et al. Non-genotoxic carcinogen exposure induces defined changes in the 5-hydroxymethylome. Genome Biol. 2012;13(10):R93. doi: 10.1186/gb-2012-13-10-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor SE, et al. Stable 5-hydroxymethylcytosine (5hmC) acquisition marks gene activation during chondrogenic differentiation. J Bone Miner Res. 2016;31(3):524–534. doi: 10.1002/jbmr.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286(41):35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen L, Song CX, He C, Zhang Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu Rev Biochem. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y, Rao A. Connections between TET proteins and aberrant DNA modification in cancer. Trends Genet. 2014;30(10):464–474. doi: 10.1016/j.tig.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pastor WA, Aravind L, Rao A. TETonic shift: Biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14(6):341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H, Zhang Y. Charting oxidized methylcytosines at base resolution. Nat Struct Mol Biol. 2015;22(9):656–661. doi: 10.1038/nsmb.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song CX, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29(1):68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain A, et al. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473(7348):484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Globisch D, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5(12):e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, et al. Tet-mediated covalent labelling of 5-methylcytosine for its genome-wide detection and sequencing. Nat Commun. 2013;4:1517. doi: 10.1038/ncomms2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michaeli Y, et al. Optical detection of epigenetic marks: Sensitive quantification and direct imaging of individual hydroxymethylcytosine bases. Chem Commun (Camb) 2013;49(77):8599–8601. doi: 10.1039/c3cc42543f. [DOI] [PubMed] [Google Scholar]
- 22.Shahal T, et al. Spectroscopic quantification of 5-hydroxymethylcytosine in genomic DNA. Anal Chem. 2014;86(16):8231–8237. doi: 10.1021/ac501609d. [DOI] [PubMed] [Google Scholar]
- 23.Nifker G, et al. One-pot chemoenzymatic cascade for labeling of the epigenetic marker 5-hydroxymethylcytosine. ChemBioChem. 2015;16(13):1857–1860. doi: 10.1002/cbic.201500329. [DOI] [PubMed] [Google Scholar]
- 24.Bachman M, et al. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat Chem. 2014;6(12):1049–1055. doi: 10.1038/nchem.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashimoto H, et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40(11):4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valinluck V, et al. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32(14):4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan KC, et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci USA. 2013;110(47):18761–18768. doi: 10.1073/pnas.1313995110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu M, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149(6):1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flusberg BA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7(6):461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joo C, et al. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126(3):515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 31.Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Methods. 2006;3(11):891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]