Significance
Neutrophils are the major effectors of acute inflammation responding to tissue injury or infection. The clearance of apoptotic neutrophils by inflammatory macrophages also provides a powerful proresolution signal. Apoptotic or necrotic neutrophils also release abundant amounts of the antimicrobial peptides alpha defensins. In this report, we show that the most abundant of these peptides, HNP1 (Human Neutrophil Peptide 1), profoundly inhibits protein translation. It achieves this without affecting mRNA stability or preventing mRNA polysomal association. This is, to our knowledge, the first demonstration of a peptide released from one cell, a leukocyte, entering and directly modulating the translatome of another cell. It alludes to a previously unidentified mechanism, driven by dying neutrophils, that ensures the timely resolution of macrophage-driven inflammation, without compromising antimicrobial function.
Keywords: macrophages, α-defensins, mRNA translation, inflammation, cytokines
Abstract
Neutrophils are the first and most numerous cells to arrive at the site of an inflammatory insult and are among the first to die. We previously reported that alpha defensins, released from apoptotic human neutrophils, augmented the antimicrobial capacity of macrophages while also inhibiting the biosynthesis of proinflammatory cytokines. In vivo, alpha defensin administration protected mice from inflammation, induced by thioglychollate-induced peritonitis or following infection with Salmonella enterica serovar Typhimurium. We have now dissected the antiinflammatory mechanism of action of the most abundant neutrophil alpha defensin, Human Neutrophil Peptide 1 (HNP1). Herein we show that HNP1 enters macrophages and inhibits protein translation without inducing the unfolded-protein response or affecting mRNA stability. In a cell-free in vitro translation system, HNP1 powerfully inhibited both cap-dependent and cap-independent mRNA translation while maintaining mRNA polysomal association. This is, to our knowledge, the first demonstration of a peptide released from one cell type (neutrophils) directly regulating mRNA translation in another (macrophages). By preventing protein translation, HNP1 functions as a “molecular brake” on macrophage-driven inflammation, ensuring both pathogen clearance and the resolution of inflammation with minimal bystander tissue damage.
Neutrophils, via the release of key inflammatory mediators, convey signals to practically all other immune cells, orchestrating both the innate inflammatory and subsequent adaptive immune responses (1). Through the de novo generation of lipid mediators, they are also key players in the resolution of inflammation (reviewed in ref. 2). Following neutrophil apoptosis, their subsequent uptake by human monocyte-derived macrophages (HMDMs) induces complex phenotypic changes, including the release of the immunosuppressive cytokines IL-10 and TGF-β (reviewed in ref. 3). We previously reported that the human antimicrobial peptides α-defensins [which are released following apoptosis, necrosis, or NET-osis (4) of neutrophils] also inhibited the secretion of multiple cytokines from activated HMDMs for up to 72 h, with full recovery thereafter and no effect on cell viability (5). In vivo, in mice, neutrophil derived α-defensins, given at the time of inducing peritonitis, led to a diminished inflammatory exudate (5). In addition, mice infected with pathogenic Salmonella enterica serovar Typhimurium showed a reduced bacterial load and serum TNFα levels upon administration of exogenous α-defensin. Hence, neutrophil-derived α-defensins were able to affect profound changes in the inflammatory environment while also serving as effective antimicrobial peptides.
Alpha defensins are small (3–4 kDa) cationic peptides that form part of a larger family of defensins (that also includes beta and theta peptides). Four structurally related peptides (HNP1–4) exist within the azurophil granules of neutrophils, of which HNP1 is the most abundant (6–9). They share a similar triple-stranded β-sheet structure, which is critically held together by three intramolecular disulphide bridges. Once the azurophil granules fuse with phagosomes, they release high concentrations of α-defensins close to the pathogen surface, where their amphipathic nature allows them to rapidly gain entry to the cell’s membrane (10). The permeabilization of membranes by α-defensins is believed to be crucial for their ability to kill microbes and host cells, elicited by membrane disruption and leakage of cellular contents (9, 11). Importantly, however, α-defensins only kill proliferating Escherichia coli and a simple model of “death by pore formation” is inadequate to explain all their antibacterial properties (12). They have also been noted to inhibit bulk bacterial protein synthesis in E. coli, although this is thought to be a consequence of membrane disruption and is temporally associated with cell death (11, 12). Additionally, following HIV-1 infection, α-defensins play a crucial role in inhibiting their life cycle (13, 14), suggesting that they have at their disposal a number of different mechanisms to kill diverse pathogens (7, 15). In favor of this hypothesis is the observation that α-defensin dimerization (which requires a tryptophan residue at position 26) is vital for its ability to kill Staphylococcus aureus (16) but has little effect on its ability to kill E. coli (17).
We wished to understand how α-defensins could simultaneously function as an effective antimicrobial antibiotic while also inducing profound changes in HMDM gene expression. We report here that HNP1 enters HMDMs, where it profoundly inhibits protein translation in both resting and activated macrophages, without affecting mRNA stability or turnover. Instead it abrogates mRNA translation without affecting mRNA polysomal association.
Results
HNP1 Inhibits the Synthesis of Proteins, Which Is Dependent on HNP1 Tertiary Structure.
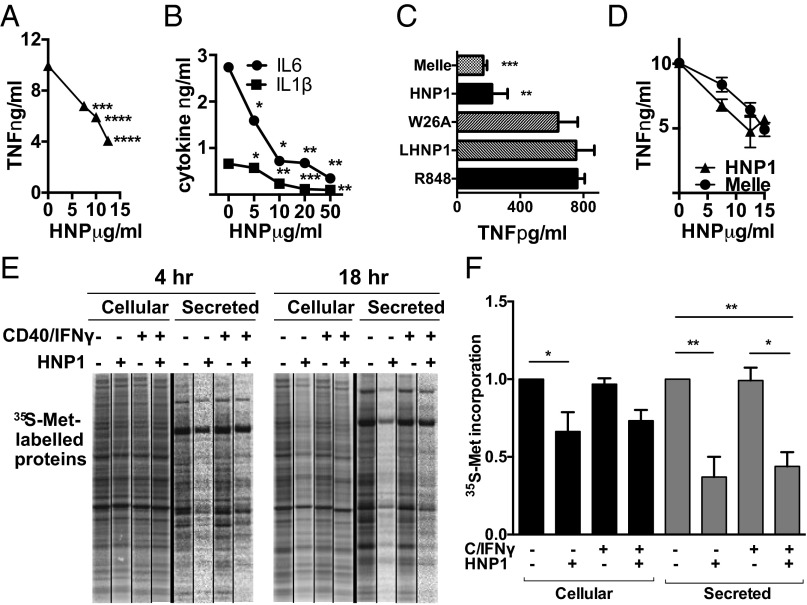
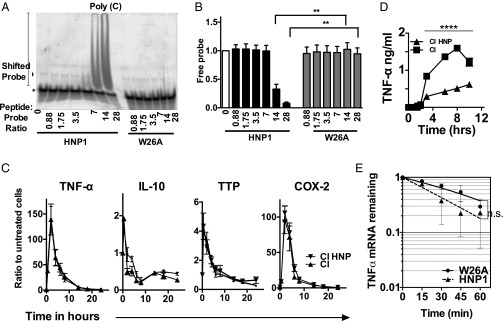
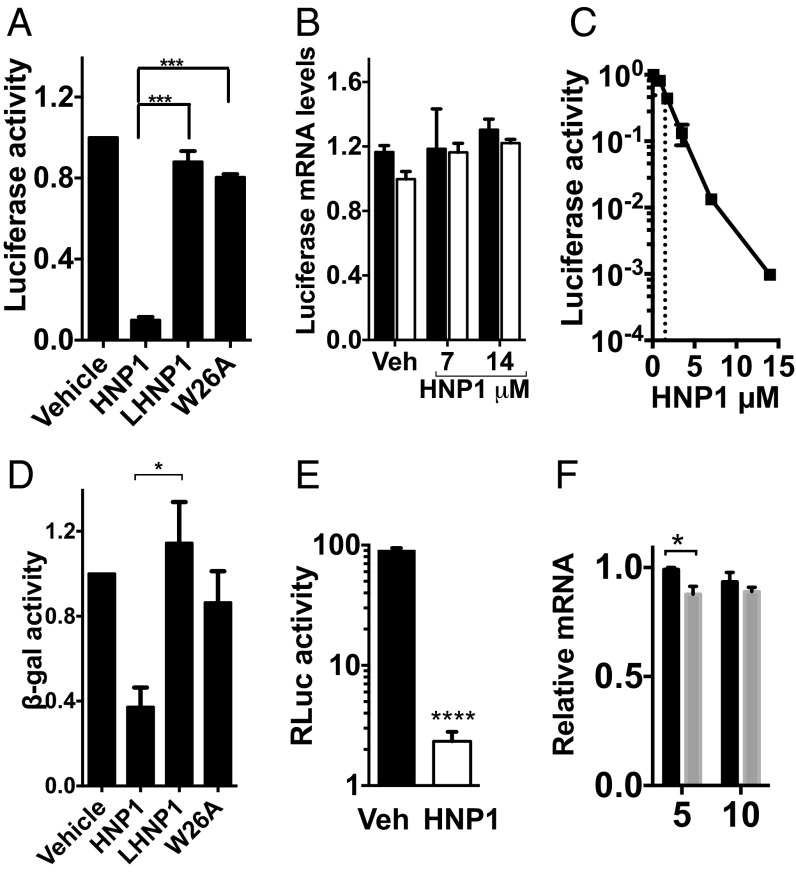
We have previously shown that although alpha defensins augmented the macrophage’s ability to kill intracellular Pseudomonas aeruginosa, these peptides simultaneously inhibited the production of multiple cytokines (TNFα, IL-6, IL-8, and IL-1β) (5). HNP1 also inhibited TNFα biosynthesis from HMDMs stimulated with the toll-like receptor 7/8 (TLR7/8) agonist R848 (Fig. 1A). The biosynthesis of IL-6 and IL-1β induced via the T-cell surrogate stimulus CD40L/IFNγ was also reduced (Fig. 1B), confirming that disparate stimuli and multiple secreted proteins were susceptible to HNP1-mediated inhibition. The structure of HNP1 was crucial for its cytokine inhibitory potential. When the intramolecular disulphide bonds that stabilize the triple-stranded beta-sheet structure of HNP1 were disrupted (linearized HNP1, l-HNP) or when dimerization was prevented by replacing the tryptophan residue at position 26 with the nonpolar amino acid alanine (W26A) (16), a complete loss of cytokine inhibitory potential was seen (Fig. 1C and ref. 5). In contrast N-methylation of Ile20 (Melle), which also prevents dimerization, had a minimal effect on the ability of HNP1 to inhibit R848-induced TNFα production by HMDMs (Fig. 1 C and D).
Fig. 1.
HNP1 inhibits bulk protein synthesis, which is dependent on HNP1 tertiary structure. (A and B) HNP1-treated HMDMs were stimulated with the TLR7 ligand R848 (1 μg/mL) (A) or with 3 μg/mL CD40L + 5 ng/mL IFNγ (B) for 18 h. TNFα (A) and IL-6 and IL-1β (B) were assayed by ELISA. (C and D) HMDMs were stimulated as for A and treated with 12.5 μg/mL of HNP1 or the mutant peptides LHNP, W26A, or Melle at the same (C) or variable concentrations (D). TNFα assayed by ELISA after 18 h. Results are representative of five independent experiments. One-way ANOVA with Dunnett’s multiple comparison tests; **P < 0.01, *P < 0.05. (E) Methionine-starved HMDMs were then cultured with 10 μCi/mL [35S]methionine ± activation (with 3 μg/mL CD40L and 5 ng/mL IFNγ) and ± addition of HNP1 (25 μg/mL) for 4 or 18 h. Secreted and intracellular proteins were resolved by SDS/PAGE. Phosphorimages of radiolabeled cellular and secreted protein gels show de novo protein synthesis. (F) De novo protein synthesis of 35S-Methionine–labeled proteins following 18 h of culture, quantified by scintillation counting and normalized to untreated controls. n = 3. Error bars represent mean ± SEM; **P < 0.01, *P < 0.05 (Tukey’s post hoc test following a one-way ANOVA).
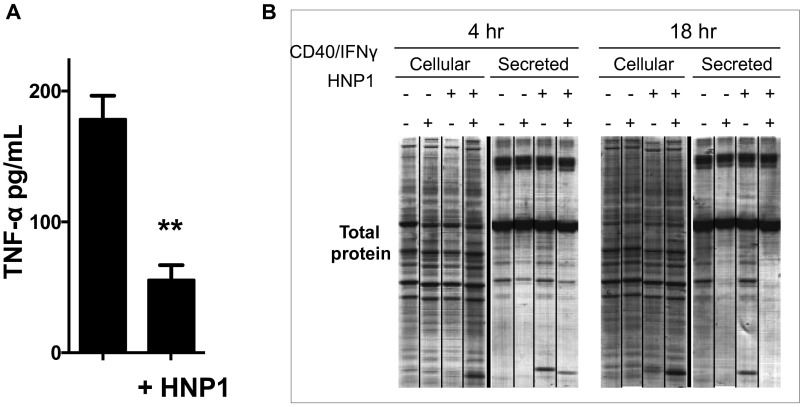
To test if HNP1 might inhibit protein synthesis per se, stimulated HMDMs were labeled with [35S]methionine in the presence of HNP1. [35S]methionine incorporation into proteins within cellular lysates (i.e., cellular proteins) and the culture media (i.e., secreted proteins) was visualized (Fig. 1E) and quantified following 18 h of culture (Fig. 1F). Strikingly, HNP1 treatment significantly reduced the quantity of both 35S-labeled cellular and secreted proteins in unstimulated HMDMs and robustly inhibited the labeling of secreted proteins in CD40L/IFNγ-stimulated HMDMs, possibly reflecting the highly secretory phenotype of the stimulated macrophage. As expected, secreted TNFα was significantly reduced by HNP1 (Fig. S1A). However, the overall cellular protein levels were unchanged during the time course of the experiment (Fig. S1B), consistent with a lack of increased global protein turnover and with maintenance of cell number and viability, as previously reported (5). Taken together, neutrophil-derived HNP1 profoundly inhibits global protein synthesis within the resting or activated macrophage.
Fig. S1.
Methionine-starved HMDMs were cultured with 10 μCi/mL [35S]methionine ± activation (with 3 μg/mL CD40L and 5 ng/mL IFNγ) and ± addition of HNP1 (25 μg/mL) for 4 or 18 h. (A) TNFα in the culture supernatants at 18 h. (B) Total intracellular and secreted proteins are shown in these Gelcode blue stained gels. Results are representative of experiments repeated three times. Tukey’s post hoc test following a one-way ANOVA. **P < 0.005.
Exogenous HNP1 Accumulates in the Macrophage.
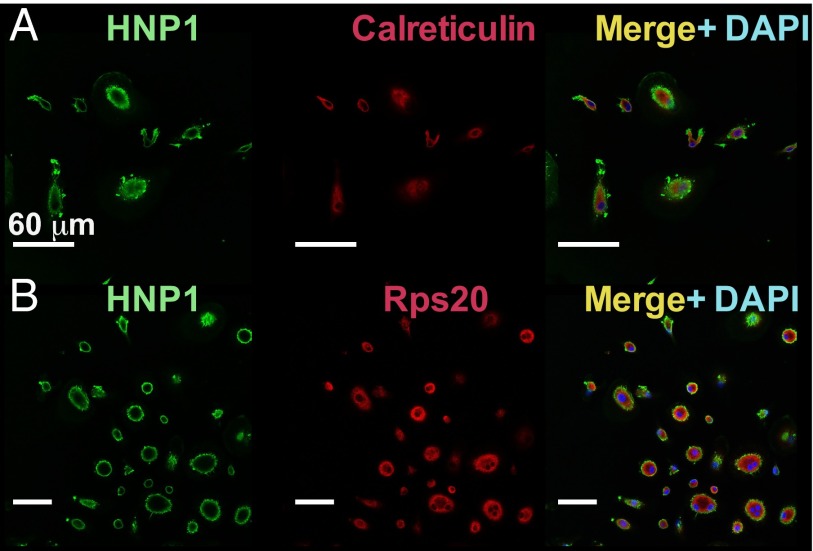
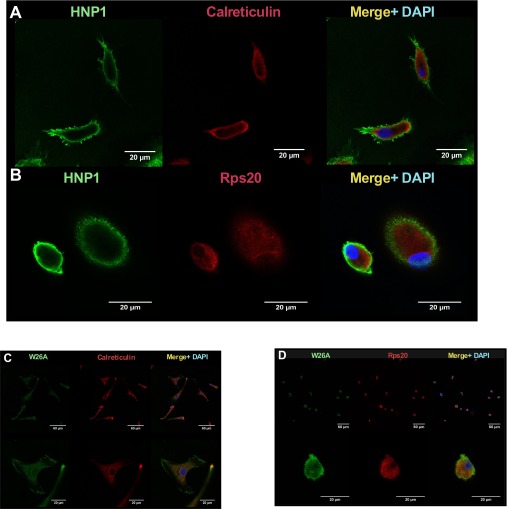
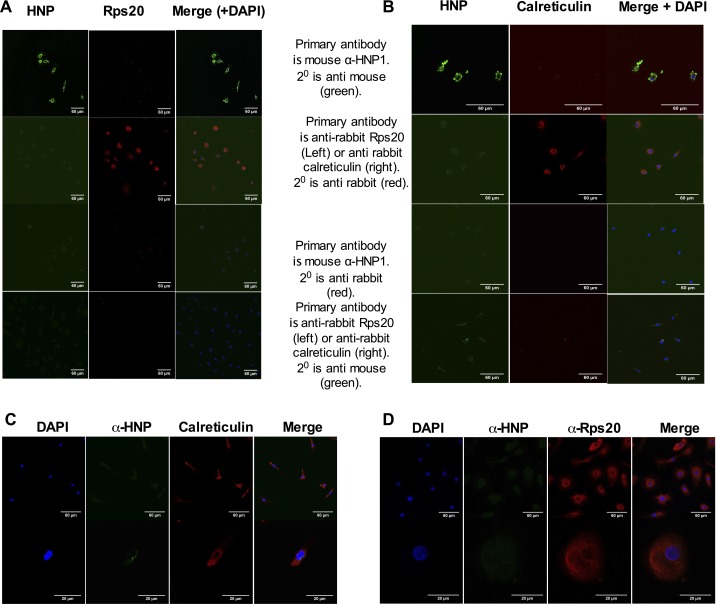
HNP1 gained entry to macrophages and was found within the membrane and cytoplasm. However, there was no clear colocalization of HNP1 (or the control peptide W26A) with the ER marker calreticulin (Fig. 2A and Fig. S2 A and C) or with ribosomes (stained with anti-Rps20; Fig. 2B and Fig. S2 B and D). Control experiments also showed no nonspecific staining or cross-reactivity between HNP1 and the ER or ribosomal secondary antibodies (Fig. S3).
Fig. 2.
HNP1 enters HMDMs. Confocal microscopy images of HNP1-treated HMDMs before visualization of anti-HNP1 (green) and DAPI (blue) seen on the merged images. In addition, red secondary staining indicates calreticulin (specific for the ER) in A and the ribosomal-associated protein Rps20 in B. Representative images are from one of six independent experiments. (White scale bars, 60 μm.)
Fig. S2.
(A and B) HNP1 localizes to the cytoplasm of HMDMs. Confocal microscopy images of HNP1-treated HMDMs before visualization of anti-HNP1 (green) and DAPI (blue) seen on the merged images. In addition, red secondary staining indicates calreticulin (specific for ER) in A and the ribosomal-associated protein Rps20 in B. Representative images are from one of six independent experiments. (C and D) W26A staining of HMDMs and confocal microscopy images of W26A-treated HMDMs before visualization with anti-HNP1–3 (green) and DAPI (blue) seen on the merged images. In addition, red secondary staining indicates calreticulin (specific for ER) in C and the ribosomal-associated protein Rps20 in D. Representative images are from one of six independent experiments.
Fig. S3.
(A and B) Confocal microscopy images of HNP1-treated HMDMs. As staining controls, anti-HNP1–3, anti-Rps20, or anti-calreticulin primary antibodies were followed by the “wrong” secondary antibodies (either anti-rabbit or anti-mouse secondary antibodies, respectively). DAPI (blue) is seen on the merged images. HMDMs that had not been treated with HNP1 were stained with primary and secondary antibodies to HNP1–3, (C) Rps20, or (D) calreticulin as indicated. Nonspecific staining of HNP1 is not seen. n = 3.
HNP1 Binds Nonspecifically to RNA but Does Not Alter mRNA Transcription or Stability.
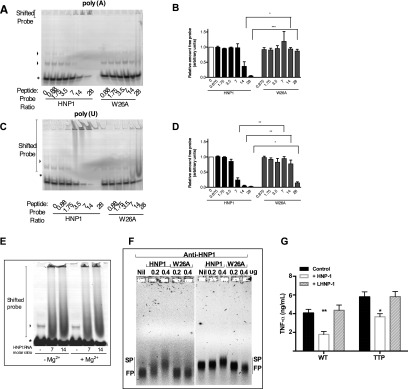
As HNP1 enters the macrophage, it may, by reason of its positive charge and amphipathic nature (10, 18), bind to mRNA, so altering its turnover and inhibiting protein synthesis. This was tested using electrophoretic mobility shift assays (EMSAs) with 25-mer homopolymeric RNA oligonucleotides. In contrast to W26A, HNP1 showed concentration-dependent shifts of poly(C) (Fig. 3 A and B), poly(A) (Fig. S4 A and B), and poly(U) RNA (Fig. S4 C and D), which were observed in both the presence or absence of Mg2+ (Fig. S4E), a cation often required for nucleic acid binding by proteins. An antibody supershift EMSA also confirmed that HNP1 could bind to mRNA [coding for the firefly luciferase (fLuc) or β-galactosidase (β-gal) reporters] (Fig. S4F).
Fig. 3.
HNP1 binds to mRNA but does not affect mRNA stability. (A) EMSA. Shown are the poly(C)25 RNA oligonucleotide probe (10 pmoles) incubated with molar ratios of HNP1 or W26A and RNA:peptide complexes resolved by nondenaturing acrylamide gel electrophoresis. Asterisk, free poly(C) probe; arrowhead, nonspecific complex. Error bars represent mean ± SD. (B) Binding of HNP1 and W26A to poly(C)25 RNA relative to total input RNA (where the relative amount of free probe is given in arbitrary units). (C) RNA was extracted from CD40L/IFNγ-stimulated HMDMs, and mRNA of TNF-α, IL-10, TTP, and Cox-2 was quantified by quantitative real time-PCR (qRT-PCR) and expressed as the ratio of mRNA from treated to untreated HMDMs. (D) Supernatants were collected for the first 10 h from cells treated as in C and TNFα protein assayed by ELISA. (E) TNFα mRNA levels were quantified from HMDMs that had been treated with 12.5 μg/mL of HNP1 or W26A and then stimulated with R848 (1 μg/mL) for 1 h before adding actinomycin D (5 μg/mL). TNFα is expressed relative to T = 0 min. Error bars are mean ± SEM for each time point, and a line represents a nonlinear two-phase decay fit with R2 values of 0.8667 and 0.8351 for W26A and HNP1, respectively. Results are derived from three separate experiments. A–D represent experiments repeated three times. (A and B) Tukey’s post hoc test following a one-way ANOVA; ****P < 0.0001, *P < 0.03. (C and D) Tukey’s post hoc test following a two-way ANOVA. n.s., not significant; P = 0.094.
Fig. S4.
(A and C) EMSA. (A) Poly(A)25 or Poly(U)25 RNA oligonucleotide probe (1 μM) incubated with molar ratios of HNP1 or W26A and RNA:peptide complexes resolved by nondenaturing acrylamide gel electrophoresis. Asterisk, free poly(C) probe; arrowhead, nonspecific complex. Error bars represent mean ± SD. (B and D) Binding of HNP1 and W26A to poly(A)25 or poly(U)25 RNA relative to total input RNA. (E) EMSA. A poly(C)25 RNA oligonucleotide probe (10 pmol; 1 μM final concentration) was incubated with the indicated molar ratios of HNP1 in the presence or absence of Mg2+ ions, and RNA:peptide complexes were resolved by nondenaturing acrylamide gel electrophoresis. (F) Electrophoretic mobility antibody supershift assay. HNP1 and W26A (0–0.4 μg) binding to m7G-fLuc-A0 (Left) or m7G-β-Gal-A0 (Right) reporter mRNAs (1 μg) were resolved following incubation with anti-HNP1–3 antibody. FP, free probe; SP, shifted probe. (G) Wild-type (WT) or TTP−/− (TTP)-deficient BMDMs were stimulated with R848 (1 μg/mL) for 18 h along with either HNP1 or the control peptide L-HNP1. TNFα protein secretion was analyzed by ELISA.
To ask if HNP1 affected mRNA transcription, we quantified the steady-state mRNA levels generated by CD40L/IFNγ-stimulated HMDMs. The mRNA levels of TNFα, IL-10, cyclooxygenase (Cox2), and tristetraprolin (TTP) were unaffected by HNP1 treatment of HMDMs over a 24-h time course (Fig. 3C), despite a clear reduction in TNFα protein production (Fig. 3D). To assess mRNA decay, HNP1- or W26A-treated HMDMs were stimulated (with R848) for 1 h, resulting in maximal TNF-α mRNA levels, before the addition of actinomycin D to arrest further transcription. The decay rate of TNF-α mRNA was not significantly modulated in HNP1 versus W26A-treated HMDMs over a further 1-h time course (Fig. 3E). As TNF-α mRNA stability is mediated in part by the zinc-finger protein TTP, which binds AU-rich sequences, we also assessed TNF-α protein secretion from activated mouse bone marrow-derived macrophages (BMDMs) isolated from TTP-deficient (TTP−/−) mice or wild-type littermate controls. Again, HNP1 (but not l-HNP1) was still able to significantly inhibit the secretion of TNF-α from TTP−/− BMDMs (Fig. S4G). Taken together, these data show that HNP1 can bind to RNA, likely in a sequence-independent manner, but does not affect mRNA stability or turnover.
HNP1 Does Not Induce ER Stress.
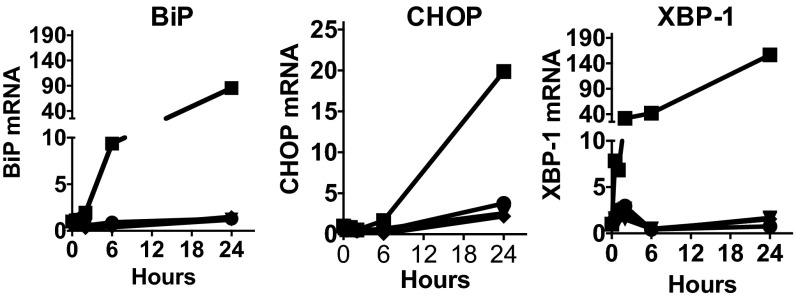
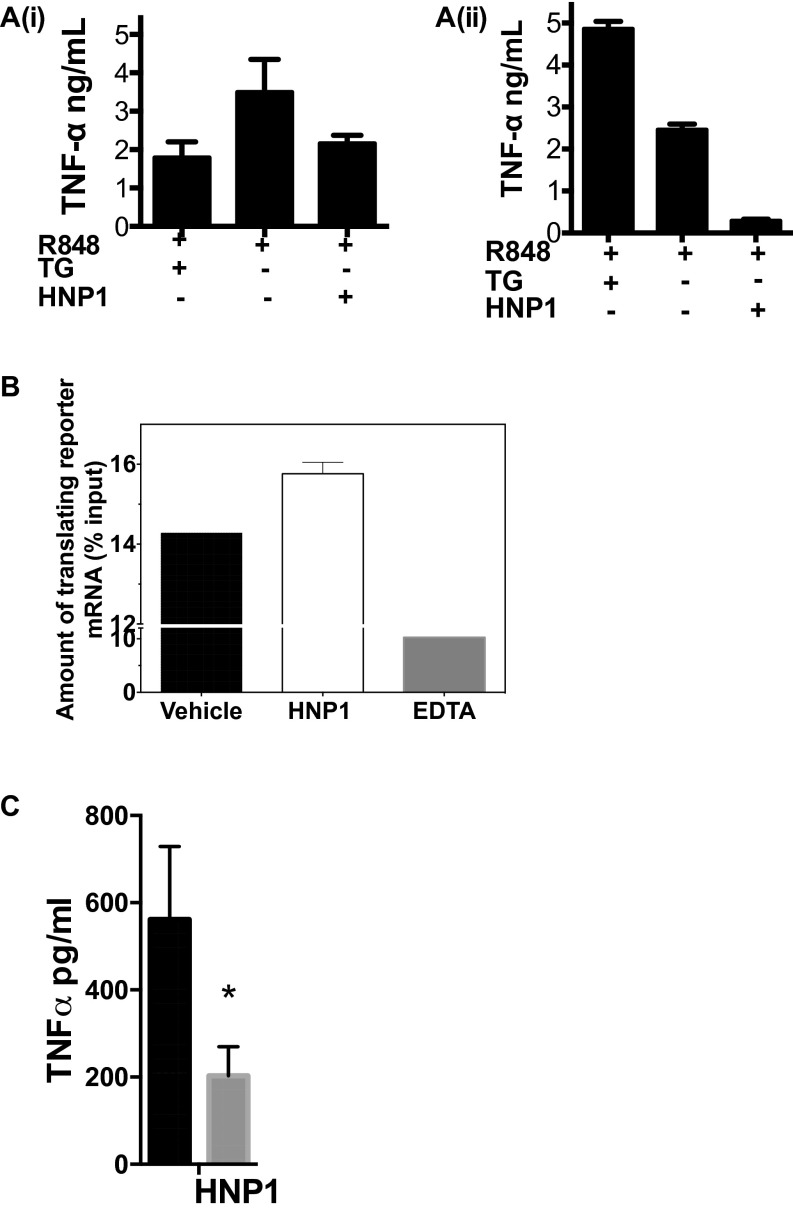
We have previously shown that HNP1 does not inhibit the exocytosis of TNFα from HMDMs (5). We also wished to confirm that it did not prevent protein synthesis by inducing the unfolded protein response (UPR) (reviewed in ref. 19). In contrast to the positive control thapsigargin (TG), we did not detect an increase in the synthesis of glucose-regulated protein 78 (Grp78), X-box-binding protein (XBP1), or CCAAT/enhancer-binding protein homologous protein (CHOP) in HNP1-treated and -stimulated HMDMs (Fig. 4), despite a clear inhibition of R848-induced TNFα production at 6 and 24 h (Fig. S5A). Hence the profound inhibition of protein synthesis by HNP1 was not the result of an induced UPR.
Fig. 4.
HNP1 does not cause ER stress. R848 (1 μg/mL) stimulated (filled diamond) HMDMs with either 12.5 µg/mL HNP1 (filled inverted triangle), l-HNP1 (dot), or 1 µM TG (filled square). Macrophage mRNAs for CHOP, spliced XBP1, and BiP were quantified by qRT-PCR and expressed relative to the same mRNAs in untreated control HMDMs. Hours represent time following stimulation. n = 3. Error bars represent mean ± SD.
Fig. S5.
(A, i) R848 (1 μg/mL)-stimulated HMDMs were treated with either 12.5 µg/mL HNP1 or 1 µM TG. Shown is TNFα in culture supernatants after 6 h. (A, ii) TNFα in culture supernatants after 24 h. n = 3. Error bars represent mean ± SD. (B) mRNA determined from the total mRNA content of fractions cosedimenting with one or more 80S ribosomes. Error bars represent mean ± SD (n = 2). (C) TNFα protein production (control, black bars; HNP1-treated, gray bar). Error bars represent mean ± SD (n = 4). *P < 0.05 (unpaired t test).
HNP1 Does Not Block Translation Initiation.
To ask if HNP1 affected translation directly and to avoid the confounding effects of mRNA transcription, processing, or nuclear export, we used the cell-free rabbit reticulocyte lysate (RRL) in vitro translation system. Translation of the canonical fLuc reporter mRNA was profoundly inhibited in the presence of HNP1 but not by the mutant control peptides l-HNP or W26A (Fig. 5A). As with TNFα mRNA, HNP1 did not destabilize the reporter mRNA because input mRNA levels were maintained (Fig. 5B). The IC50 value for this effect was ∼1.6 μM (or 5.5 μg/mL) (Fig. 5C), a concentration that significantly reduces the production of proinflammatory cytokines from stimulated HMDMs in vitro (Fig. 1).
Fig. 5.
HNP1 inhibits protein synthesis downstream of translation initiation. (A) 1 ng m7G-fLuc-A0 reporter mRNA, translated in vitro using the RRL with 25 μg/mL [7.3μM] HNP1, LHNP1, W26A, or vehicle control (0.01% acetic acid). Translational output was quantified as relative fLuc activity (normalized to vehicle control-treated samples). Error bars represent mean ± SEM (n = 3). (B) Similar to A but relative m7G-luciferase-A0 reporter mRNA levels were quantified by qRT-PCR. Black bars represent pretranslation levels and white bars the posttranslation levels. Shown are the results of three experiments. (C) Similar to A, with 400 pg m7G-fLuc-A0 reporter mRNA translated in the presence of increasing concentrations of HNP1. The IC50 (shown by the dotted line) is 1.6 ± 0.02 μM. Mean ± SEM from two independent experiments. (D) 1 ng CSFV IRES-β-gal-A0 reporter mRNA was in vitro-translated as for A. Values were plotted relative to vehicle control (n = 3). Error bars represent mean ± SEM (n = 3). For A and D, ***P < 0.001, *P < 0.05 (analyzed by Tukey’s multiple comparison post hoc test following one-way ANOVA). (E) 1 ng CrPV IRES-β-gal-A0 reporter mRNA translated as in A. ****P < 0.0001, analyzed by unpaired t test. Values are plotted relative to vehicle control. (F) RRL was pretreated with 150 μg/mL cycloheximide and either 25 μg/mL HNP1 or vehicle control. We then added 1 ng 32P-labeled m7G-fLuc-A0 reporter mRNA for the indicated times (shown in minutes) before 15–30% sucrose density gradient fractionation. The graph depicts the relative amounts of mRNA sedimenting with initiating ribosomes, normalized to the amount recruited at 5 min in vehicle control-treated RRL. Black bars represent the control group, and gray bars represent the HNP1-treated group. Error bars represent mean ± SEM (n = 3). *P < 0.05 (unpaired t test).
Eukaryotic mRNA has a 5′ monomethylated cap structure (m7G) that is crucial for canonical translation initiation, the rate-limiting and primary node of translation regulation (reviewed in ref. 20). To interrogate the role of translation initiation in HNP1-mediated inhibition, we used reporter mRNAs that contained a viral internal ribosome entry site (IRES) in their 5′ untranslated regions (5′ UTRs), bypassing some or all of the eukaryotic translation initiation factor (eIF) requirements and initiating translation cap-independently (reviewed in ref. 21). The Classical Swine Fever Virus (CSFV) IRES mRNA reporter initiates translation independently of the majority of eIFs but is dependent on the ternary complex (eIF2, GTP, and tRNAi), whereas the Cricket Paralysis Virus (CrPV) IRES allows the direct assembly of the 80S ribosome at the start codon, bypassing all canonical initiation factor requirements (22). Remarkably, despite their diverse mechanisms of translation initiation, HNP1 was also able to prevent the synthesis of both the CSFV-driven translation of β-Gal (Fig. 5D) and the CrPV-driven translation of Renilla luciferase (RLuc) (Fig. 5E). As HNP1 is able to prevent the translation of mRNAs using diverse mechanisms of translation initiation, it is most likely that it is acting downstream of this point. To confirm this empirically, ribosomal recruitment onto a radiolabeled m7G-capped fLuc reporter mRNA was quantified in the presence of cycloheximide to halt the 80S ribosome at the start codon, preventing translation elongation. Although HNP1 weakly inhibited translation initiation at 5 min following mRNA addition, by 10 min similar maximal 80S recruitment to that seen in vehicle control-treated extracts was observed (Fig. 5F), indicating only a small reduction in the rate of 80S recruitment in the presence of HNP1 and supporting the conclusion that HNP1 predominantly inhibits mRNA translation postinitiation.
HNP1 Does Not Affect Ribosomal Association with mRNA.
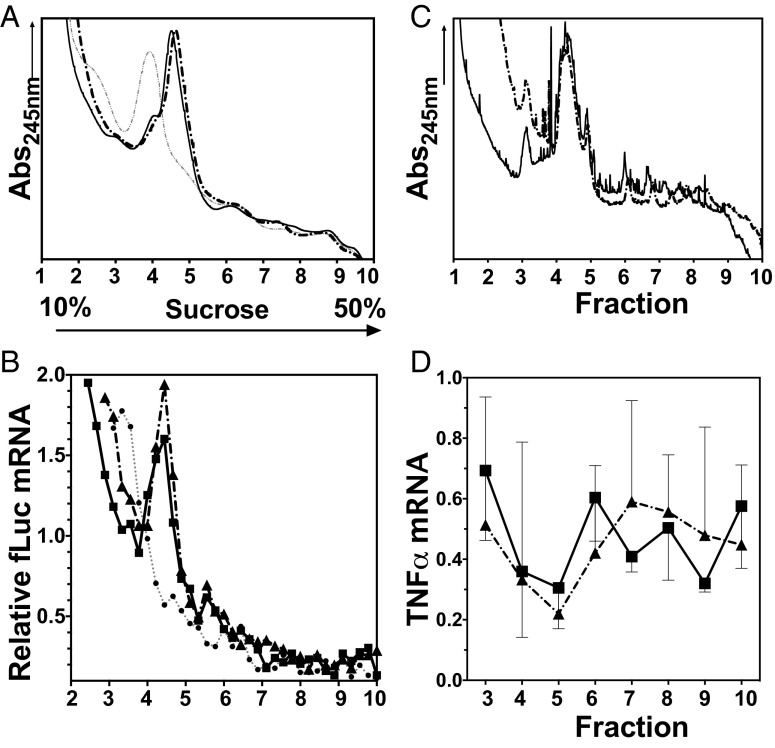
Finally to ask if luciferase mRNA was maintained on polysomes despite its significantly reduced translation, we assessed the steady-state ribosomal association of m7G-fLuc mRNA in the presence or absence of HNP1. Despite using a concentration of HNP1 that profoundly inhibited reporter protein synthesis (Fig. 1E), we observed no change in the polysomal profile (Fig. 6A) or the distribution of m7G-fLuc mRNA across the polysomal region of the density gradient (fractions 4–10) (Fig. 6B). In contrast, the presence of EDTA resulted in polysomal dissociation and depletion of the reporter mRNA from the fractions containing translating mRNA (Fig. 6B and Fig. S5B). We also wished to confirm if a similar mode of action was seen in HMDMs that had been treated with HNP1 or vehicle control (for 18 h). HMDMs so treated were then stimulated with R848 for 2 h to up-regulate the synthesis of TNFα. Again, the bulk polysome profile for HNP1-treated HMDMs was similar to that of control-stimulated cells (Fig. 6C). Importantly, the polysomal association of TNFα mRNA in untreated or HNP1-treated stimulated HMDMs was not significantly altered (Fig. 6D), despite the significant inhibition of TNFα protein synthesis (Fig. S5C). These data confirm that although HNP1 profoundly alters protein translation at a point after translation initiation, it does not prevent mRNA polysomal association.
Fig. 6.
HNP1 has no effect on polysome profile. (A) RRL pretreated with 25 μg/mL HNP1 or vehicle control. Shown is 2 ng 32P-labeled m7G-fLuc-A0 reporter mRNA translated for 30 min before addition of 150 μg/mL cycloheximide or 25 mM EDTA and 10–50% sucrose density gradient fractionation. Solid black line, vehicle control-treated; broken black line, HNP1-treated; dotted gray line, EDTA-treated. (B) Relative reporter mRNA content of gradient fractions expressed as a percentage of the total input mRNA. Solid black line with squares, vehicle control-treated; broken black line with triangles, HNP1-treated; dotted gray line with filled circles, EDTA-treated. (C) HMDMs treated with 25 μg/mL HNP1 or vehicle control before R848 stimulation for 2 h. We added 150 μg/mL cycloheximide for 10 min before lysis and 10–50% sucrose density gradient fractionation. An Abs254 nm trace was used to determine sedimentation of 80S ribosomes and polysomes. Solid black line, vehicle control-treated; dotted line, HNP1-treated. (D) TNFα mRNA content of gradient fractions expressed relative to maximal TNFα mRNA detected in fractions 3–10 (43S/60S to polysomal). Solid black line, vehicle control-treated; broken black line, HNP1-treated. Error bars represent mean ± SD (n = 4); paired t test, no significant differences detected.
Discussion
Cells of the immune system have developed tightly regulated systems to ensure the timely resolution of inflammation. The control of mRNA translation is emerging as a major mechanism that regulates the levels of proteins within leukocytes (reviewed in refs. 23, 24). We have now identified a previously unidentified mechanism in which the most abundant neutrophil α-defensin, HNP1, which is readily released as these cells die (5), inhibits bulk protein translation within macrophages. Although the characteristic hydrophobic, amphipathic nature of α-defensins allows them to partition into the membrane lipid layer (25), it also ensures ready access to the cell’s interior. Confocal imaging showed that HNP1 entered macrophages (Fig. 2) without inducing a UPR (Fig. 4) or affecting mRNA stability (Fig. 3). To our knowledge, this is the first description of an eobiotic peptide released by one cell profoundly affecting the translational capacity of another, in the absence of a requirement for de novo transcription and without compromising antimicrobial function.
HNP1 was able to inhibit translation initiation via diverse mechanisms. Both canonical cap-dependent (Fig. 5) and noncanonical, cap-independent translation (driven by either a CSFV or CrPV IRES) were profoundly inhibited in vitro. However, the small inhibitory effect of HNP1 on translation initiation (Fig. 5F) was insufficient to explain the magnitude of the effects seen in vitro and within macrophages. Rather, the dramatic inhibition of CrPV IRES-driven translation, which dispenses with the initiation event, implicates an HNP1-mediated inhibition downstream of translation initiation. HNP1 could inhibit translation by binding nonspecifically to mRNA, or equally it could sequester factors essential for translation, such as tRNA or ribosomal protein and/or rRNA components. Previous reports point to several RNA-binding proteins that require a net positive charge and arginine side chains (18). Alpha defensins also possess four positively charged arginines, which might allow it to interact with RNA (Fig. 3). These side chains are important for its function, as the substitution of these amino acids for similarly charged lysine significantly reduces its bactericidal activity (17, 26) (reviewed in ref. 10). Considering the ability of HNP1 to kill a diverse array of bacterial and viral pathogens, it will be of interest to determine whether HNP1 can similarly prevent prokaryotic protein translation.
Because HNP1 binds nonspecifically to RNA, we asked if it could inhibit translation by modulating ribosome engagement with mRNA. However, both reporter and cellular mRNAs remained polysome-associated (Figs. 5 and 6), and the polysomal distribution of these mRNAs was similar in control and HNP1-treated RRL and HMDMs. Translational repression could be occurring via either elongation and/or termination (27), and we would speculate that HNP1 prevents translation elongation (22), which has recently been established as a major control point for protein synthesis (28).
Previous studies also allude to the greater importance of protein synthesis rate over degradation rate in determining overall protein levels (29, 30). However, the lack of a significant change in overall HMDM cellular protein level (Fig. S1B) argues against an HNP1-mediated increase in nonspecific cellular protein degradation. Further, HNP1 profoundly inhibits reporter protein synthesis in cell-free assays in which protein turnover pathways are fundamentally compromised and HNP1 itself has no known protease activity. Taken altogether, we believe these data indicate that HNP1 affects de novo protein synthesis.
The tertiary structure of monomeric HNP1 is also clearly important for translational inhibition, as highlighted by the loss of efficacy observed for l-HNP1 or W26A (Fig. 1C). However, the N-methylation of HNP1 Ile-20 (Melle), which prevents dimerization, does not alter the ability of Melle to inhibit TNF-α production, confirming that HNP1 dimerization is not required to inhibit macrophage protein translation (Fig. 1D). The concentration of HNP1–3 in the synovial fluid of patients with rheumatoid arthritis is between 3 and 25 μg/mL, with an average of 12.4 μg/mL, suggesting that the concentration reached in tissues is similar to that used in our assays (5). Our previous studies have shown that HMDMs fully recover their proinflammatory potential within 72 h following exposure to α-defensins. So although they clearly disable the macrophage protein translation machinery, they do not induce macrophage apoptosis (5). A previous study reported that α-defensins reduced the release of IL-1β from activated monocytes, while not affecting the transcription of IL-1β mRNA (28). Based on our findings, these observations can likely be explained by the translation of pro–IL-1β being impaired.
In summary, we have uncovered that neutrophil α-defensins abrogate the bulk mRNA translation of proteins within HMDMs, without affecting mRNA transcription or stability. In this way they prevent an excessive proinflammatory response that would create its own collateral damage while still acting as powerful antimicrobial peptides. This is the first demonstration, to our knowledge, of an antimicrobial peptide that also has a translation-based antiinflammatory role, acting as a “molecular brake.” It opens the way for developing similar peptide-based therapeutics that would act as effective combined antiinflammatory and antimicrobial agents.
Materials and Methods
All experiments on mice were covered by a project license granted by the Home Office under the Animal (Scientific Procedures) Act of 1986. Locally, this license was approved by the University of Edinburgh's Ethical Review Committee. All materials and the following protocols are fully described in SI Materials and Methods. Briefly, synthetic HNP1 and mutant derivatives were prepared by solid-phase synthesis as previously described (31). Template plasmids pCSFV-lacZ (32) and pT7-Luc (33) for reporter mRNA transcription were previously described, and pSL200-CrPV-RLuc, RLuc downstream of a CrPV IRES, was a kind gift from Matthias Hentze, European Molecular Biology Laboratory, Heidelberg, Germany. Healthy donor peripheral blood mononuclear cells (PBMCs) were purified from whole blood as previously described (5). Stimuli included 1 μg/mL R848 (Invivogen), 3 μg/mL CD40L (Peprotech), and 5 ng/mL IFNγ (Peprotech). Cytokines were quantified by sandwich ELISA (R&D Systems). For assessment of protein synthesis, HMDMs were incubated in l-Methionine–free DMEM (MP Biomedicals) for 2 h at 37 °C, followed by 10 μCi/mL 35S-Methionine (Perkin-Elmer) and stimulation with CD40L and IFNγ and defensin peptides. In vitro transcription was assessed by m7G- or ApG-capped, nonadenylated, 32P-UTP–labeled or nonlabeled reporter mRNAs, and the mRNAs were synthesized as previously described (34). In vitro translation was assessed using the nuclease-treated RRL in vitro translation kit (Promega) according to the manufacturer’s recommendations. For EMSAs, 10 pmoles 5′-Cy5–labeled 25-mer oligonucleotides (poly-Adenine, poly-Cytosine, or poly-Uracil) (Eurogentec) were incubated with HNP1 or W26A peptide in 10 μL binding and then resolved by electrophoresis. For immunocytochemistry, HMDMs were grown on glass coverslips and stained with mouse monoclonal anti-human HNP1–3 antibody together with polyclonal rabbit anti-human ribosomal protein Rps20 (Abcam, dilution 1:250) or polyclonal rabbit anti-human calreticulin (Abcam, dilution 1:250). ER stress and the UPR and mRNA stability assay along with RNA quantitation and polysome analysis are fully explained in SI Materials and Methods.
SI Materials and Methods
Materials.
Synthetic HNP1 and mutant derivatives were prepared by solid-phase synthesis as previously described (31). Mutant derivatives included LHNP1 (structurally l-HNP1 by the substitution of cysteine amino acids with alanine), W26A-HNP1 (tryptophan substitution with alanine at residue position 26), and Me-Ile20-HNP1 (N-methylation of Isoleucine at position 20). Anti-HNP1–3 antibody was from Hycult Biotech. Template plasmids pCSFV-lacZ (32) and pT7-Luc (33) for reporter mRNA transcription were previously described, and pSL200-CrPV-RLuc, RLuc downstream of a CrPV IRES, was a kind gift from Matthias Hentze, European Molecular Biology Laboratory, Heidelberg, Germany.
Mice.
Mice lacking TTP−/− on a mixed 129/C57BL6 background were generated by Perry Blackshear as described (35). BMDMs (4 × 106 cells per mL) were cultured in Iscove's Modified Dulbecco's Medium (IMDM) (supplemented with 10% FCS, penicillin/streptomycin, 50 μM 2-ME, and 10% L929 conditioned media).
HMDM Cell Culture.
Healthy donor PBMCs were purified from whole blood as previously described (5), cultured in IMDM (Gibco) (supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, 10% autologous serum), and used from day 7–10. Stimuli included 1 μg/mL R848 (Invivogen), 3 μg/mL CD40L (Peprotech), and 5 ng/mL IFNγ (Peprotech). Cytokines were quantified by sandwich ELISA (R&D Systems).
Quantification of Protein Synthesis in HMDMs.
HMDMs were incubated in l-Methionine–free DMEM (MP Biomedicals) for 2 h at 37 °C, 5% CO2, followed by 10 μCi/mL 35S-Methionine (Perkin-Elmer) and stimulation with CD40L and IFNγ and defensin peptides. Secreted proteins in culture supernatants and cellular proteins in lysates were recovered by precipitation using a ProteoExtract Protein Precipitation Kit (Millipore) before scintillation analysis or SDS/PAGE analysis to visualize the total (SyproRuby stain; Life Technologies) and radiolabeled proteomes (phosphorimaging; Fuji FLA5100).
In Vitro Transcription.
m7G- or ApG-capped, nonadenylated, 32P-UTP–labeled or nonlabeled reporter mRNAs were synthesized as previously described (34).
In Vitro Translation.
In vitro translation was performed using the nuclease-treated RRL in vitro translation kit (Promega) according to the manufacturer’s recommendations. We used 1–2 ng of each reporter mRNA in all assays (90 min at 30 °C), except for HNP1 titration experiments, where 400 pg mRNA was used. Firefly and RLuc and β-gal activities were quantified using the Dual Luciferase Assay system (Promega) and Tropix Galacton Plus (Applied Biosystems) system, respectively. Luminescence was quantified using the Lumat LB 9507 (Berthold) luminometer. Total RNA was purified using TRI reagent (ThermoFisher), and 1 μg was reverse-transcribed using the High Capacity cDNA RT-PCR kit (Applied Biosystems) according to the manufacturer’s instructions. Luciferase cDNA levels were quantified by qPCR [Applied Biosystems 7900HT Fast Real-Time PCR system using SDS software (v2.4); forward primer, 5′-GGCGCGGTCGGTAAAGTT-3′; reverse primer, 5′-AGCGTTTTCCCGGTATCCA-3′].
EMSAs.
We incubated 10 pmoles 5′-Cy5–labeled 25-mer oligonucleotide (poly-Adenine, poly-Cytosine, or poly-Uracil) (Eurogentec) with HNP1 or W26A peptide in 10 μL binding buffer [20 mM Tris·HCl, pH 8.0, 20% (vol/vol) glycerol, 0.1% (vol/vol) Igepal CA-630, 2 mM DTT, 140 mM KCl, 200 ng/mL BSA, 3 mM MgCl2) and then resolved it by electrophoresis on a nondenaturing 7% (vol/vol) polyacrylamide gel. The probe was quantified using a FLA5100 image reader (FujiFilm), and the free probe was quantitated using AIDA Image Analyzer (RAYTEST). Supershift experiments were performed similarly, except 1 μg of mRNA was used as the probe and all binding reactions were supplemented by subsequent addition of anti-human HNP1–3 antibody (Hycult, 1 ng/μL) and complexes resolved by electrophoresis on a 1% TBE agarose gel and visualized using GelRed nucleic acid stain (Biotium).
Immunocytochemistry.
Day 6 HMDMs were grown on glass coverslips in serum-free IMDM and treated with HNP1 or W26A (12.5 μg/mL). HMDMs were washed, fixed (in 3% paraformaldehyde for 20 min), quenched with 50 mM glycine, permeabilized with 0.1% triton-X 100, and blocked (5% goat serum). Antibody staining was performed with mouse monoclonal anti-human HNP1–3 antibody (Hycult, dilution 1:100 dilution) together with polyclonal rabbit anti-human ribosomal protein Rps20 (Abcam, dilution 1:250) or polyclonal rabbit anti-human calreticulin (Abcam, dilution 1:250). Secondary antibodies were goat anti-mouse IgG Alexa Fluor 488 and goat anti-rabbit IgG Alexa Fluor 647 (Molecular Probes, Invitrogen) (1:400 dilution). Nuclei were stained with DAPI (Sigma). Images were acquired on a Leica TCS SP5 (Leica Microsystems) confocal laser scanning microscope.
ER Stress and UPR.
HMDMs in culture were treated with 12.5 μg/mL HNP1 or with 1 μM TG (Sigma) as a positive control for UPR induction, before stimulation with R848. BiP, XBP1, and CHOP cDNAs were quantified according to previously published methods (36).
mRNA Stability Assay.
HMDMs were treated with 25 μg/mL HNP1 for 18 h before R848 stimulation for 2 h. Transcription was arrested by addition of actinomycin D (5 μg/mL) for various times. Total RNA was extracted using a NucleoSpin RNA II kit (Macherey-Nagel).
RNA Quantitation.
Total RNA was reverse-transcribed (High Capacity cDNA reverse transcriptase kit; Applied Biosystems), and TNFα (Hs99999043_m1), IL-10 (Hs99999035_m1), TTP (Hs00185658_m1), and Cox2 (Hs00153133_m1) cDNA levels were quantitated by Taqman qPCR assay using the Taqman Gene Expression Assay predesigned primers for human with intrasample expression normalized to Eukaryotic 18S rRNA Endogenous control (FAM/MGB probe) and run on an Applied Biosystems 7900HT Fast-Real Time System using SDS software (v2.4). Data were analyzed using the comparative CT method (∆/∆CT), where fold differences in gene expression between control and test samples (ΔCT) were normalized to CT values of the 18S rRNA reference gene.
Polysome Analysis.
Analysis of mRNA polysomal recruitment in RRL was performed according to the manufacturer’s instructions with the following modifications. For analysis of translation initiation, RRL was pretreated with 25 μg/mL HNP1 and 150 μg/mL cycloheximide for 30 min before the addition of 2 ng 32P-labeled m7G-fLuc-A0 mRNA for various times. Polysomes were resolved by sucrose density gradient centrifugation and fractionation as previously described (37). Reporter mRNA content in each fraction was quantified as direct disintegration per minute on a scintillation counter. For analysis of steady-state polysome association, RRL was pretreated with 25 μg/mL HNP1 for 30 min before the addition of 2 ng 32P-labeled m7G-fLuc-A0 mRNA for 30 min. Translation elongation was arrested by addition of 150 μg/mL cycloheximide, or ribosomes were dissociated by addition of 25 mM EDTA. Polysomes were resolved and mRNA distribution quantified as above. For the quantification of TNFα mRNA polysomal association in HMDMs, cells were treated with vehicle control (0.01% acetic acid) or 25 μg/mL HNP1 for 18 h in serum-free conditions and then stimulated with R848 for 2 h. Cycloheximide at 150 μg/mL was added 10 min before lysis and polysome fractionation as previously described (36). Fractions were collected directly into TRIzol-LS (ThermoFisher) and spiked with 1 μg mouse total RNA before RNA purification using DirectZol spin columns (Zymo Research). Relative human TNFα mRNA levels in each fraction were quantitated as described above, and mouse 18S rRNA was simultaneously quantitated to control for variability in RNA recovery between fractions.
Acknowledgments
This work was supported by Arthritis Research UK Grant 18722 and Medical Research Council (MRC) Grant MR/J009555/1 (to M.G.) and by MRC Program Grant MR/J003069/1 (to N.K.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.B.I. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601831113/-/DCSupplemental.
References
- 1.Nathan C. Points of control in inflammation. Nature. 2002;420(6917):846–852. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- 2.Serhan CN, Savill J. Resolution of inflammation: The beginning programs the end. Nat Immunol. 2005;6(12):1191–1197. doi: 10.1038/ni1276. [DOI] [PubMed] [Google Scholar]
- 3.Savill J, Dransfield I, Gregory C, Haslett C. A blast from the past: Clearance of apoptotic cells regulates immune responses. Nat Rev Immunol. 2002;2(12):965–975. doi: 10.1038/nri957. [DOI] [PubMed] [Google Scholar]
- 4.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 5.Miles K, et al. Dying and necrotic neutrophils are anti-inflammatory secondary to the release of alpha-defensins. J Immunol. 2009;183(3):2122–2132. doi: 10.4049/jimmunol.0804187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganz T, Lehrer RI. Antimicrobial peptides of leukocytes. Curr Opin Hematol. 1997;4(1):53–58. doi: 10.1097/00062752-199704010-00009. [DOI] [PubMed] [Google Scholar]
- 7.Lehrer RI. Primate defensins. Nat Rev Microbiol. 2004;2(9):727–738. doi: 10.1038/nrmicro976. [DOI] [PubMed] [Google Scholar]
- 8.Lehrer RI, Ganz T. Cathelicidins: A family of endogenous antimicrobial peptides. Curr Opin Hematol. 2002;9(1):18–22. doi: 10.1097/00062752-200201000-00004. [DOI] [PubMed] [Google Scholar]
- 9.Lehrer RI, Lichtenstein AK, Ganz T. Defensins: Antimicrobial and cytotoxic peptides of mammalian cells. Annu Rev Immunol. 1993;11:105–128. doi: 10.1146/annurev.iy.11.040193.000541. [DOI] [PubMed] [Google Scholar]
- 10.Lehrer RI, Lu W. α-Defensins in human innate immunity. Immunol Rev. 2012;245(1):84–112. doi: 10.1111/j.1600-065X.2011.01082.x. [DOI] [PubMed] [Google Scholar]
- 11.Kagan BL, Selsted ME, Ganz T, Lehrer RI. Antimicrobial defensin peptides form voltage-dependent ion-permeable channels in planar lipid bilayer membranes. Proc Natl Acad Sci USA. 1990;87(1):210–214. doi: 10.1073/pnas.87.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehrer RI, et al. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J Clin Invest. 1989;84(2):553–561. doi: 10.1172/JCI114198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furci L, Sironi F, Tolazzi M, Vassena L, Lusso P. Alpha-defensins block the early steps of HIV-1 infection: Interference with the binding of gp120 to CD4. Blood. 2007;109(7):2928–2935. doi: 10.1182/blood-2006-05-024489. [DOI] [PubMed] [Google Scholar]
- 14.Gallo SA, et al. Theta-defensins prevent HIV-1 Env-mediated fusion by binding gp41 and blocking 6-helix bundle formation. J Biol Chem. 2006;281(27):18787–18792. doi: 10.1074/jbc.M602422200. [DOI] [PubMed] [Google Scholar]
- 15.Ganz T. Defensins: Antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3(9):710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 16.Wei G, et al. Trp-26 imparts functional versatility to human alpha-defensin HNP1. J Biol Chem. 2010;285(21):16275–16285. doi: 10.1074/jbc.M110.102749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou G, et al. Toward understanding the cationicity of defensins. Arg and Lys versus their noncoded analogs. J Biol Chem. 2007;282(27):19653–19665. doi: 10.1074/jbc.M611003200. [DOI] [PubMed] [Google Scholar]
- 18.Tan R, Frankel AD. Structural variety of arginine-rich RNA-binding peptides. Proc Natl Acad Sci USA. 1995;92(12):5282–5286. doi: 10.1073/pnas.92.12.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutkowski DT, Kaufman RJ. A trip to the ER: Coping with stress. Trends Cell Biol. 2004;14(1):20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 21.Firth AE, Brierley I. Non-canonical translation in RNA viruses. J Gen Virol. 2012;93(Pt 7):1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernández IS, Bai XC, Murshudov G, Scheres SH, Ramakrishnan V. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell. 2014;157(4):823–831. doi: 10.1016/j.cell.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piccirillo CA, Bjur E, Topisirovic I, Sonenberg N, Larsson O. Translational control of immune responses: From transcripts to translatomes. Nat Immunol. 2014;15(6):503–511. doi: 10.1038/ni.2891. [DOI] [PubMed] [Google Scholar]
- 24.Carpenter S, Ricci EP, Mercier BC, Moore MJ, Fitzgerald KA. Post-transcriptional regulation of gene expression in innate immunity. Nat Rev Immunol. 2014;14(6):361–376. doi: 10.1038/nri3682. [DOI] [PubMed] [Google Scholar]
- 25.Brogden KA. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3(3):238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 26.de Leeuw E, Rajabi M, Zou G, Pazgier M, Lu W. Selective arginines are important for the antibacterial activity and host cell interaction of human alpha-defensin 5. FEBS Lett. 2009;583(15):2507–2512. doi: 10.1016/j.febslet.2009.06.051. [DOI] [PubMed] [Google Scholar]
- 27.Richter JD, Coller J. Pausing on polyribosomes: Make way for elongation in translational control. Cell. 2015;163(2):292–300. doi: 10.1016/j.cell.2015.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi J, et al. A novel role for defensins in intestinal homeostasis: Regulation of IL-1beta secretion. J Immunol. 2007;179(2):1245–1253. doi: 10.4049/jimmunol.179.2.1245. [DOI] [PubMed] [Google Scholar]
- 29.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 30.Kristensen AR, Gsponer J, Foster LJ. Protein synthesis rate is the predominant regulator of protein expression during differentiation. Mol Syst Biol. 2013;9:689. doi: 10.1038/msb.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Z, Ericksen B, Tucker K, Lubkowski J, Lu W. Synthesis and characterization of human alpha-defensins 4-6. J Pept Res. 2004;64(3):118–125. doi: 10.1111/j.1399-3011.2004.00179.x. [DOI] [PubMed] [Google Scholar]
- 32.Smith RW, et al. DAZAP1, an RNA-binding protein required for development and spermatogenesis, can regulate mRNA translation. RNA. 2011;17(7):1282–1295. doi: 10.1261/rna.2717711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5(11):2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 34.Gray NK. Translational control by repressor proteins binding to the 5'UTR of mRNAs. Methods Mol Biol. 1998;77:379–397. doi: 10.1385/0-89603-397-X:379. [DOI] [PubMed] [Google Scholar]
- 35.Taylor GA, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4(5):445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 36.van Schadewijk A, van’t Wout EF, Stolk J, Hiemstra PS. A quantitative method for detection of spliced X-box binding protein-1 (XBP1) mRNA as a measure of endoplasmic reticulum (ER) stress. Cell Stress Chaperones. 2012;17(2):275–279. doi: 10.1007/s12192-011-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burgess HM, Gray NK. An integrated model for the nucleo-cytoplasmic transport of cytoplasmic poly(A)-binding proteins. Commun Integr Biol. 2012;5(3):243–247. doi: 10.4161/cib.19347. [DOI] [PMC free article] [PubMed] [Google Scholar]