Significance
Superresolution microscopy has made significant progress with molecule localization, stimulated emission depletion, and structured illumination techniques. Here we describe a simple method to achieve ∼20-nm axial resolution based on total internal reflection fluorescence (TIRF) microscopy. In this approach, serial fluorescent images at multiple depths within the normal ∼200-nm TIRF illumination zone are obtained by the serial imaging and photobleaching at multiple TIRF excitation angles. These images with ∼20-nm axial depth intervals are then simply stacked together to differentiate the relative distributions of multiple proteins in the axial direction. Moreover, images collected during the photobleaching step of the experiment can be analyzed using Bayesian analysis of fluorophore blinking and bleaching to generate superresolution images in the lateral direction for each depth.
Keywords: total internal reflection fluorescence, axial superresolution, photobleaching
Abstract
We report superresolution optical sectioning using a multiangle total internal reflection fluorescence (TIRF) microscope. TIRF images were constructed from several layers within a normal TIRF excitation zone by sequentially imaging and photobleaching the fluorescent molecules. The depth of the evanescent wave at different layers was altered by tuning the excitation light incident angle. The angle was tuned from the highest (the smallest TIRF depth) toward the critical angle (the largest TIRF depth) to preferentially photobleach fluorescence from the lower layers and allow straightforward observation of deeper structures without masking by the brighter signals closer to the coverglass. Reconstruction of the TIRF images enabled 3D imaging of biological samples with 20-nm axial resolution. Two-color imaging of epidermal growth factor (EGF) ligand and clathrin revealed the dynamics of EGF-activated clathrin-mediated endocytosis during internalization. Furthermore, Bayesian analysis of images collected during the photobleaching step of each plane enabled lateral superresolution (<100 nm) within each of the sections.
Fluorescence microscopy is an essential technique for cell biologists, providing 3D views of protein distributions with high contrast and specificity. For conventional microscopes, however, these views are limited by diffraction and have a lateral resolution of approximately one-half the wavelength of the excitation light. Numerous advances have enabled cellular imaging beyond the lateral diffraction limit, pushing the smallest structures that can be resolved with optical wavelengths from a few hundred nanometers to a few tens of nanometers. On the other hand, axial resolution is even more strongly affected by diffraction and can be threefold more coarse than the lateral resolution (1). Progress in the axial dimension has lagged behind lateral improvements, but various methods that offer vastly improved axial resolution are now available, including interference methods like standing wave microscopy (2), I5M (3), 4Pi-microscopy (4), and SAIM (5) and molecule localization techniques (6) like 3D-PALM (7–11), 3D-STORM (12–14), iPALM (15), and 3D-STED (16). However, these techniques often rely on special optical components or photoactivatable/photoswitchable fluorophore behaviors (17) and/or extensive data analysis to contend with a multitude of artifacts.
Total internal reflection fluorescence (TIRF) microscopy (18), one of the first optical methods to provide subdiffractive axial information, is often overlooked in the expansion of superresolution techniques. The imaging is limited to cellular structures close to the coverglass, but TIRF microscopy has been widely applied to investigations of processes at cell membranes and to techniques requiring single-molecule imaging. During TIRF excitation, the evanescent field produced by total internal reflection decays in intensity exponentially with the depth into the sample from the surface. As a result, only a thin layer of illumination above the interface is created, and only fluorophores within this thin layer in the sample are excited. This property is advantageous for imaging by reducing the background and improving the signal-to-noise ratio. Furthermore, depending on the optics, excitation wavelength, and refractive indices of the media involved, the penetration depth of the evanescent wave can be <100 nm (17), much smaller than the axial extent of a point spread function in conventional confocal microscopy (1).
Because much fluorophore distribution information resides even within a TIRF excitation zone, efforts have focused on the dependence of penetration depth on laser illumination angle to provide quantitative information about the distances between fluorophores and the surface. Based on such information and a special optical design with a varying incident beam angle, TIRF microscopy has worked well in mapping fluorophore distribution and reconstructing 3D information in TIRF excitation zones for cell membranes (19, 20), secretory granules (21), and microtubules (22, 23). These techniques rely extensively on the relationship between the fluorescent intensity and incident beam angle, which is complicated by numerous factors (17). The refractive index gradient and incident beam angle change the evanescent wave profile and amplitude, with subsequent effects on fluorescence intensity. Furthermore, near-field interactions of fluorophores near the coverglass produce an axial fluorescence emission profile differing from the fluorescence excitation profile. Thus, the image analyses must account for these complications in the differentiation of two or more fluorophores at same lateral location but different axial locations. Despite these challenges, 50- to 100-nm axial resolutions have been reported recently in ∼800-nm-thick 3D reconstructions of actin filaments (24).
In this work, we have developed an alternative and simple imaging protocol to improve the accessible content of TIRF imaging and further improve the axial resolution to ∼20 nm. In this protocol, a normal TIRF excitation zone (∼200 nm) is divided into distinct layers by varying the incident beam angle. The distance between each layer, representing the axial resolution, is determined by the difference in the imaging depths of the evanescent wave at the two adjacent angles of incidence. Images of these layers are obtained by sequentially imaging and photobleaching the fluorescent molecules at the angle of incidence from the highest allowable value (the smallest TIRF depth) toward the critical angle (the largest TIRF depth). Photobleaching at each angle reduces background resulting from brighter signals originating closer to the coverglass and unmasks signals deeper in the TIRF excitation zone. Stacking these images in different layers reconstructs a 3D image with axial superresolution. In addition, images collected during the photobleaching steps can be used in Bayesian analysis of fluorophore blinking and bleaching (3B) (25) molecule localization analysis to provide superresolution in the lateral dimension of each imaging plane.
Results
TIRF Calibration Using Fluorescently Labeled Beads.
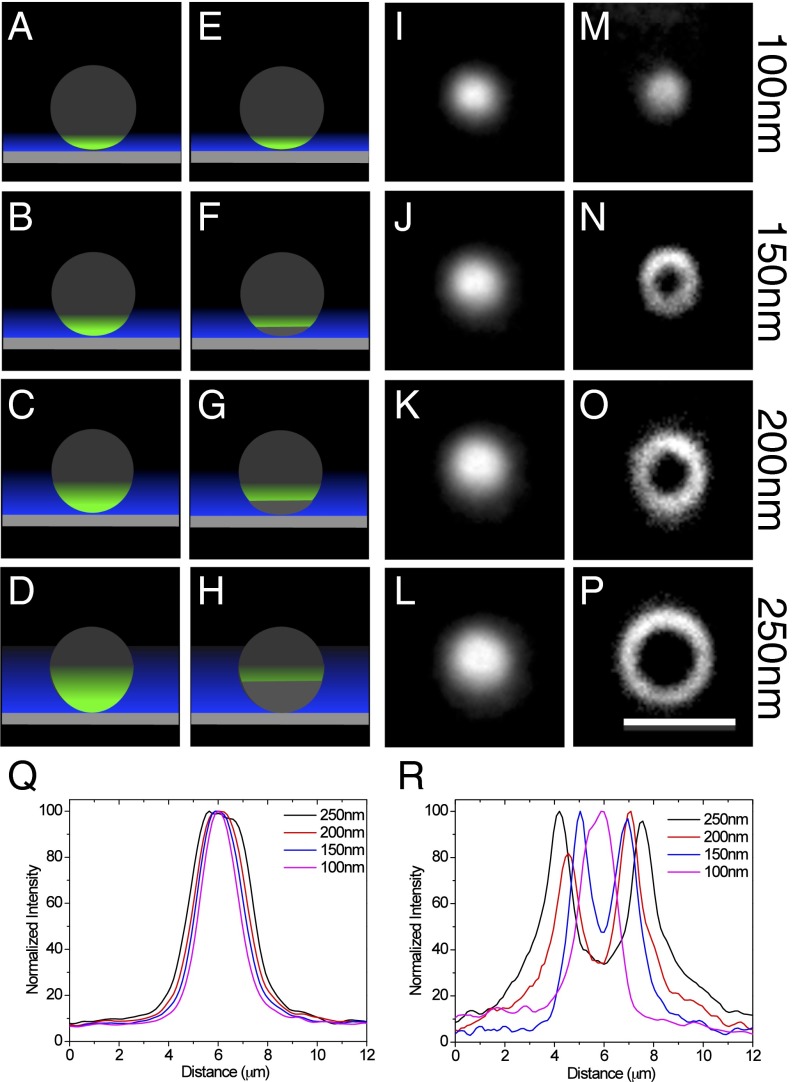
An imaging system was built with a two-axis galvanometer that moves a laser illumination beam around the periphery of the microscope objective lens rear aperture. This produced azimuthal rotation of the incident beam in the sample plane and provided a time-averaged homogeneous illumination at multiple TIRF incident angles (SI Appendix, Fig. S1). The TIRF depths were calculated and confirmed using calibration experiments (SI Appendix, Figs. S2 and S3), including the relationship between the galvanometer setting, TIRF angle of incidence, and TIRF depth. In addition, TIRF depth measurements were performed using a modified version of a previously published protocol (26) (Fig. 1). TIRF images of silica beads (7.27 µm diameter) labeled with purified mCherry were obtained at different incident beam angles labeled with the corresponding penetration depths (Fig. 1 I–L). Imaging was initiated at the largest incident beam angle (the shallowest penetration depth) and decreased toward the critical angle (the deepest TIRF depth) (Fig. 1 A–D). As the illumination field increased in depth, the bead was detected as a larger diameter spot (Fig. 1Q). Based on the geometry of the spherical beads (SI Appendix, Fig. S3 A and B), the calculated penetration depths were found to be similar to theoretical values (Fig. 1Q).
Fig. 1.
Multiangle TIRF images of beads with and without sequential imaging and photobleaching. (A–H) Schematic of the imaging protocols using multiangle TIRF illumination. (A–D) A fluorescent sphere placed on a coverglass (gray rectangle) is imaged using TIRF excitation (blue) at decreasing angles of incidence, which results in increasing evanescent field depths (blue). As the depth increases, more of the object is illuminated and the fluorescence signal is observed as a spot in the image that is increasing in diameter. (E–H) The fluorescent sphere is imaged at the highest angle of incidence (smallest TIRF depth) and photobleached at that angle before moving to the next angle. This is repeated for each angle. The signal is observed as a ring of fluorescence that is increasing in diameter with increasing TIRF depth. (I–P) Silica beads (diameter, 7.27 µm; refractive index, 1.42) were labeled with purified mCherry and imaged using different incident beam angles corresponding to the calculated penetration depths indicated (I–L) and imaged using the image-photobleach-image protocol at the calculated penetration depths indicated (M–P). (Q) Plot profiles of the beads in I–L were used to determine the dimensions of the bead fluorescence signals. Based on the geometry of the beads (SI Appendix, Fig. S3B), the measured penetration depths for the theoretically calculated values were 109.2 nm for 100 nm, 154.3 nm for 150 nm, 196.6 nm for 200 nm, and 278.9 nm for 250 nm. (R) Plot profiles of the beads in M–P were used to determine the dimensions of the bead fluorescence signals. Using the distance of two peaks as the diameters of imaged beads (SI Appendix, Fig. S3C), the measured penetration depths for the theoretically calculated values were 99.4 nm for 100 nm, 130.4 nm for 150 nm, 205.9 nm for 200 nm, and 385.1 nm for 250 nm. (Scale bar: 5 µm.)
The procedure for obtaining superresolution optical sections within the TIRF imaging zone requires collection of an image, referred to here as a “prebleach” image, at the steepest angle possible, which results in the lowest TIRF depth. Molecules in this region are then photobleached at the same incident beam angle, and a “postbleach” image is collected. After moving to a smaller angle and thus a deeper TIRF depth, a prebleach image is collected before photobleaching, and a postbleach image is collected (Fig. 1 E–H) after photobleaching. This process is repeated until TIRF depths of 200–300 nm are reached. Image processing simply involves subtraction of the postbleach image from the prebleach image.
Using this procedure, we performed another calibration using mCherry-stained silica beads starting at a calculated TIRF depth of ∼100 nm (Fig. 1M). As the TIRF depth increased, a ring of fluorescence of increasing diameter was observed (Fig. 1 N–P), instead of a round spot of fluorescence of increasing diameter. The ring was produced in each image because the molecules imaged at the previous TIRF depth were photobleached. As the depth increased and images were collected further from the coverslip, more of the bead was imaged, and the diameter of the ring increased. Normalized fluorescence intensities determined from plot profiles across the images showed a single peak at the lowest depth (Fig. 1M), but two peaks in subsequent images (Fig. 1 N–P).
Using the peak-to-peak distances to determine the diameter, we calculated the penetration depth in each layer based on the geometry of beads (SI Appendix, Fig. S3 A and C). The calculated penetration depth in each layer was similar to the theoretical value with the exception of the 250-nm layer (Fig. 1R).
Axial Superresolution of Images of Actin Filament Bundles.
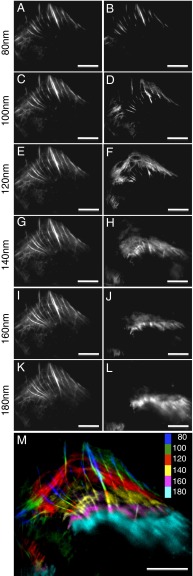
We tested the multiangle photobleaching approach in a biological specimen by imaging actin in a fixed U2OS cell. Actin filaments form a network near the plasma membrane. Near the cell periphery, actin filament bundles were observed at the shallowest TIRF depths to form numerous, almost parallel filaments extending from the edge of cell toward the center (Fig. 2A). As the penetration depth increased, the bundles elongated (Fig. 2 C, E, G, I, and K), suggesting that the bundles tilted away from the coverslip and were farther from the coverglass toward the center. In addition, an actin fiber network higher in the cell was observed as TIRF depth increased. However, because the TIR-induced evanescent field decays exponentially in the axial direction, the collected fluorescence signal originates from mainly the lowest layer even when the illumination field penetration depth is increased, and thus the fluorescence signals at the lower levels tend to mask or overwhelm the signals from deeper inside the specimen.
Fig. 2.
TIRF imaging of actin filaments using multiangle TIRF and multiangle TIRF with sequential imaging and photobleaching. A U2OS cell was fixed and labeled with phalloidin-Alexa Fluor 488. (A, C, E, G, I, and K) Images collected under TIRF illumination at penetration depths of 80–180 nm without photobleaching. (B, D, F, H, J, and L) Images of the same cell using the image-photobleach-image protocol at penetration depths of 80–180 nm. (M) A color-coded 3D image of actin filaments reconstructed using the images in B, D, F, H, J, and L. (Scale bars: 10 µm.)
Using the photobleaching method described above, actin structures deeper inside the cell were unmasked by removing the fluorescence signals below (SI Appendix, Fig. S4); for example, actin filament bundles along the cell edge at the increased depth were more evident and well defined in the 100-nm layer (Fig. 2D) compared with the 100-nm depth (Fig. 2C). At 120 nm (Fig. 2F), an actin network that appeared to be oriented parallel to the cell periphery became evident. Comparisons with depths of 120–180 nm (Fig. 2 E, G, I, and K) showed some evidence of this network, but this was not well defined. The actin network also appeared to be farther away from the coverglass toward the center of the cell compared with the periphery. Fig. 2M shows a color-coded image of the various depths of the actin structures. Several of the fibers can be seen to transverse multiple layers as they extend toward the center of the cell, and the network shown in red is ∼40–50 nm above the brightest and shallowest fibers located at the cell periphery.
When reconstructing a 3D image of the actin filaments, we report 20-nm sections between each layer based on the TIRF depths at which the imaging experiments were performed. To verify that the sections were indeed 20 nm apart, we imaged commercialized uniform 40-nm and 20-nm fluorescent beads embedded in a 0.4% agarose gel, which has a refractive index close to that of water (27). The results are shown in SI Appendix, Fig. S5 for images collected at different penetration depths 20 nm apart using the photobleaching approach. Whereas the 40-nm fluorescent beads were observed in two or three adjacent layers, the 20-nm fluorescent beads were observed in one or two adjacent layers. These results confirm that the images for each layer represent an axial depth of ∼20 nm.
Two-Color Images of Cellular Proteins.
A major benefit of this approach is its ability to differentiate the relative distributions of multiple proteins in the axial direction. Implementation is straightforward but requires calibration of both channels using silica beads labeled with mCherry (SI Appendix, Fig. S6 A–D) and EGFP (SI Appendix, Fig. S6 E–H). Our data show approximately the same peak-to-peak distances and indicate similar illumination depths at each layer using 561- and 488-nm laser beams (SI Appendix, Fig. S6 I–P). We further tested these in control experiments by labeling the same actin structures with two different color dyes, phalloidin-Alexa Fluor 488 (SI Appendix, Fig. S7 A–C) and phalloidin-Alexa Fluor 568 (SI Appendix, Fig. S7 D–F). Importantly, the results in SI Appendix, Fig. S7 G–I show similar actin filament structures at each layer in each channel. Intensity plot profiles from both channels of two fibers (SI Appendix, Fig. S7 J–L) show slightly different relative intensities but the same peak positions. Thus, these data showing the same structures at each layer in both channels confirm that the optical sectioning approach provides accurate information about the relative positions of two protein distributions.
We performed further tests using two different protein structures on U2OS cells immunolabeled for microtubules (SI Appendix, Fig. S8 A–C) and phalloidin-labeled for actin (SI Appendix, Fig. S8 D–F). Both actin filaments and microtubules were found in the shallowest depth images (∼80 nm), but sectioning through the TIRF excitation zone (SI Appendix, Fig. S8 H and I) revealed that most of the actin network was located closer to the cell periphery compared with microtubules, which showed a more pronounced network toward the center of the cell. In another example, we examined the locations of zyxin-Emerald in combination with actin filaments (SI Appendix, Fig. S8 J–M). Zyxin has been imaged by iPALM (28) as a component of focal adhesions and is located closer to the substrate (the coverglass) than actin. Consistent with those previous measurements, we found both proteins within the 80-nm layer (SI Appendix, Fig. S8 J and L), but also found that as the TIRF depth increased (SI Appendix, Fig. S8 K and M), the fluorescent signal from zyxin decreased, whereas a more extensive actin network became evident, indicating a deeper relative position.
Imaging of Clathrin-Mediated Endocytosis.
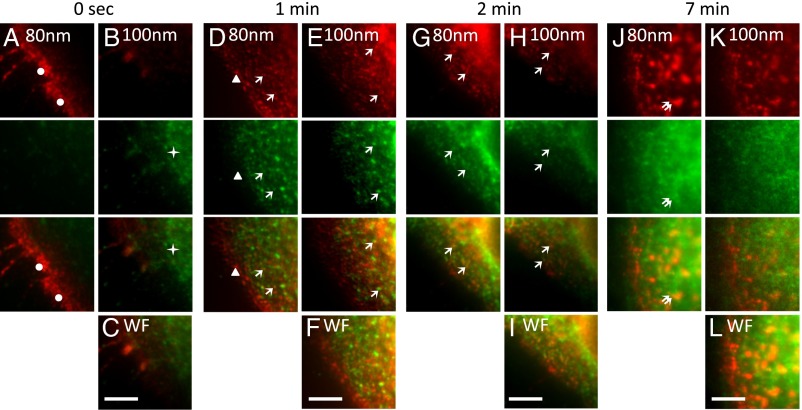
With the superresolution optical sectioning capability of our system, we revisited the dynamics of ligand-induced clathrin-mediated endocytosis in A431 cells. Clathrin-mediated endocytosis is essential for the internalization and transport to endosomal compartments of the epidermal growth factor (EGF) receptor (EGFR). EGFR endocytosis determines the longevity and biological response of EGF-activated signaling pathways (29, 30). To follow the fate of EGFR and its ligand EGF in association with clathrin over time, we first labeled cells with Alexa Fluor 555 conjugated to EGF for 1 h at 4 °C, a temperature that allows receptor-ligand binding but prevents their internalization. Cells were shifted to 37 °C to allow internalization and transport of the EGFR-ligand complex, via clathrin labeled with Alexa Fluor 488-coated pits and vesicles, to endosomal compartments located deeper within the cell.
The EGFR-ligand complex was found almost exclusively at the plasma membrane (Fig. 3A, dots below 80 nm depth) of cells fixed immediately after the receptor ligand incubation at 4 °C. In contrast, clathrin was located mainly in the cytoplasm near the plasma membrane (Fig. 3B, stars above 80 nm depth). Although a fraction of the EGFR-ligand complex was still present at the plasma membrane without clathrin after 1–2 min at 37 °C (Fig. 3D, arrowheads), some of the EGFR-ligand complex was also observed in clathrin-coated pits/vesicles (Fig. 3 D, E, G, and H, arrows) at both the 80- and 100-nm layers. At 7 min, the EGFR-ligand complex was found in endosomal compartments (double arrows in Fig. 3J) beneath the plasma membrane; there was less localization with clathrin at this time point. The axial superresolution optical sectioning capability of our system allowed us to observe the differential “association” between clathrin and the EGFR. At time 0 s, we observed little clathrin localized with the EGFR at the plasma membrane. After 1–2 min, we observed clathrin localized with EGFR in structures resembling clathrin-coated pits and vesicles. The localization between clathrin and the EGFR diminished at later time points, when the EGFR enters endosomal compartments. These observations are difficult to make with the wide-field images shown in Fig. 3 C, F, I, and L.
Fig. 3.
Multiangle-TIRF imaging of EGF-activated clathrin-mediated endocytosis. (A–C) A431 cells were incubated with Alexa Fluor 555-labeled EGF ligand (red) for 1 h at 4 °C. (D–L) Cells were shifted to 37 °C for 1 min (D–F), 2 min (G–I), and 7 min (J–L). Cells were fixed and stained with antibodies to clathrin and with secondary antibodies conjugated to Alexa Fluor 488 (green). Cells in A–C were fixed immediately after the 4 °C incubation with fluorescent EGF (0 s). Dots in A mark the EGFR-ligand complex located at the plasma membrane, seen at the 80-nm layer at time 0 s. The EGFR-ligand complex was found in clathrin-coated pits and vesicles at the plasma membrane and inside the cell at 100 nm and beneath after the 1- to 2-min incubation at 37 °C (D, E, G, and H, arrows). The EGFR-ligand complex was found in endosomes after 7 min (double arrows in J). Images were collected using the image-photobleach-image protocol at penetration depths of 80–100 nm. Wide-field (WF) images (C, F, I, and L) were collected as well. (Scale bars: 5 μm.)
Combining TIRF Sectioning with Molecule Localization Analysis.
The photobleaching procedure used in this technique offers the opportunity to achieve lateral superresolution through Bayesian analysis of the blinking and photobleaching (3B) of the fluorescent molecules (25). 3B analysis reveals superresolution information of structures not available in the diffraction-limited images by modeling an entire dataset of sequentially acquired images as arising from a number of overlapping fluorophores. 3B analysis models fluorophores in the dataset as occupying one of three states—“on,” “off,” or “bleached”—using a factorial hidden Markov model. Superresolution of spatially overlapping fluorophores is achieved in 3B by using a priori knowledge of the preceding frames of the dataset and estimation of both blinking and bleaching events from subsequent frames.
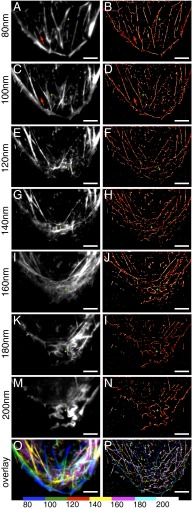
The images for 3B analysis are collected in multiangle TIRF mode during the photobleaching step of the sequential image and photobleach protocol. The image produced with 3B analysis is considered to display the structural information from the same layer as the final diffraction-limited image produced by subtracting the postbleach image from the prebleach image. In comparison, Fig. 4 shows actin fibers in different layers acquired using the image-photobleach-image protocol (Fig. 4 A, C, E, G, I, K, and M) and 3B analysis (Fig. 4 B, D, F, H, J, L, and N). Qualitatively, the images have similar observable structures in each layer as the TIRF depth is increased. Moreover, the images provide similar information as in Fig. 2, except that the 3B analysis improves the lateral resolution to ∼75 nm (SI Appendix, Fig. S9). This was determined from the full width at half-maximum of the smallest actin filament in the image. Although 75-nm resolution is insufficient to differentiate single fibers in each actin bundle, the relative locations of each bundle or fiber in the lateral dimension in each layer are more clearly resolved (stars in Fig. 4). Importantly, the images produced with 3B analysis still depicted the structures within each layer 20 nm apart; for example, the actin fiber indicated by the arrows in Fig. 4 B and D exists only within the 80-nm TIRF depth and with no actin fibers close to the cell edge oriented parallel to the cell periphery appears within the 80- to 100-nm layer (Fig. 4D). Therefore, combining multiangle TIRF sectioning with photobleaching and 3B analysis allows us to achieve 3D superresolution imaging (Fig. 4P). Moreover, 3B analysis allows observation of distinct fiber bundles in images collected at TIRF depths ≥160 nm (Fig. 4 J, L, and N), which are difficult to discern in the diffraction-limited images (Fig. 4 I, K, and M).
Fig. 4.
Comparison of multiangle TIRF images using the image-photobleach-image protocol and 3B analysis of actin fiber bundles. U2OS cells were fixed and stained with phalloidin-Alexa Fluor 488. (A, C, E, G, I, K, and M) Images collected using the image-photobleach-image protocol at penetration depths of 80–200 nm. (B, D, F, H, J, L, and N) 3B analysis of TIRF images acquired during the photobleaching steps at penetration depths of 80–200 nm. Stars indicate the improvement of x-y dimensional resolution in B, D, F, H, J, L, and N compared with A, C, E, G, I, K, and M, respectively. Arrows point to the structures appearing within the 80-nm layer (A and B), which are not observed within the 80- to 100-nm layer (C and D). (O) Color-coded 3D image of actin fibers reconstructed with the diffraction-limited images in A, C, E, G, I, K, and M. (P) Color-coded 3D image of actin fibers reconstructed with images from the 3B analysis in B, D, F, H, J, L, and N. (Scale bars: 5 μm.)
Discussion
TIRF microscopy exploits the unique properties of an induced evanescent field by total internal reflection in a thin-layer region of the sample immediately adjacent to the interface between coverslip and specimen. The evanescent wave eliminates the out-of-focus fluorescence background, improves the signal-to-noise ratio, and offers excellent axial spatial resolution. The method described here separates the fluorescence signal derived from conventional TIRF illumination into several layers through the dependence of evanescent field illumination on the incident beam angle. This allows preferentially induced photobleaching of fluorophores in shallow layers compared with deeper layers. The first layer’s depth is determined by the largest angle of the incident beam, which is limited by the numerical aperture of the objective. After the first and shallowest section, each section’s thickness is determined by the incident beam angle increment.
Limitations and potential artifacts of the technique are outlined in SI Appendix, Methods, but perhaps the most prominent is the need for careful control of the level of photobleaching at each depth. This in turn requires an evenly illuminated field or restriction of the field of view to an evenly illuminated region (SI Appendix, Fig. S10). In addition, because the evanescent wave decays exponentially at all angles, sharp edges between adjacent sections are not possible; thus, photobleaching in any section reduces the number of fluorescent molecules in that section as well as all subsequent sections, albeit to an exponentially decaying degree. The experimental consequence of this is that a section of insufficient thickness will not have sufficient fluorescent molecules to generate an image representative of the true structure. Another limitation is that the photobleaching event takes additional time and limits application in any studies of fast dynamics, and a redistribution of fluorophores during the photobleaching step could lead to misinterpretation.
Despite these limitations, we performed several single-color and two-color experiments using known and unknown structures in which we estimated the optimal photobleaching level based on uniform distributions (SI Appendix, Table S1). Using the decrease in fluorescence intensity to monitor for the proper level of photobleaching, our sequential imaging and photobleaching protocol produced images that encoded the relative axial positions of the actin filament network near the edge of a cell (Fig. 2) to a precision better than the axial diffraction limit found in conventional TIRF imaging. This method also provided axial ultrastructural information on EGF internalization dynamics that was not accessible in normal TIRF images. Moreover, combining this technique with 3B analysis improved the lateral resolution within each layer and produced <100-nm laterally resolved images at ∼20-nm axial intervals (Fig. 4).
Although this combination of techniques does not surpass the precision and resolution reported by other 3D molecule localization techniques (11, 14, 15), it does not require special fluorescent molecules, optical components, or optical configurations. No special fluorophore is required for this technique, although it must have the capability to be turned off by some mechanism, such as photobleaching or photoswitching. The photobleaching step can be performed in less than 1 min, depending on the incident illumination power, and 3B analysis can construct an image with ∼200 frames collected during this step. Moreover, the sequential photobleaching and imaging protocol provides a straightforward adaptation of 3B analysis in retrieving information in the lateral dimension.
Although the multidirection TIRF excitation used here has the advantages of averaging imperfections in the illumination field (SI Appendix, Fig. S11) and minimizing pattern artifacts in the images, it is not strictly necessary for the performance of this method, which should be readily adapted to a variable-angle TIRF microscope (SI Appendix, Fig. S12). The major experimental investment is the need to calibrate the illumination angles with the TIRF penetration depth. Image processing for the diffraction-limited images requires only a simple image subtraction. Both command line and ImageJ plugin (31) versions of the 3B molecule localization analysis software are available (www.coxphysics.com/3b/), as are the scripts and protocols for performing 3B analysis using cloud-based computation (32). In summary, the multiangle TIRF technique described here is straightforward to use, requires no special reagents, requires no special instrumentation, and needs straightforward image analysis to study protein structures at superresolution levels in axial as well as lateral dimensions.
Methods
Multiangle TIRF Imaging System.
A schematic representation of the multiangle TIRF system is shown in SI Appendix, Fig. S1. Details of the optical pathway and hardware components are provided in SI Appendix, Methods. Micro-Manager open source software (33) was used to control the microscope, and the images were processed and analyzed using ImageJ (34) or FIJI (35).
Fluorescent Beads and Fluorescent Monolayer Preparation.
Histidine6-tagged fluorescent proteins were produced and purified using Ni-NTA agarose in our laboratory as described previously (36). These were used to label uniform nonfunctionalized silica beads (mean diameter, 7.27 μm; refractive index, 1.42; Bangs Laboratories). The monolayer of His-EGFP for critical angle determination was prepared by sequentially coating a coverslip with monoclonal Histidine6-tag antibody (GenScript) and His-EGFP.
Cell Culture, Transfection, and Immunofluorescence Labeling.
U2OS cells were grown in standard DMEM-HG medium with 2 mM GlutaMAX, 1 mM sodium pyruvate (Invitrogen) and 10% (vol/vol) heat-inactivated FBS at 37 °C in a 5% CO2 incubator. A431 cells were cultured in DMEM supplemented with 10% (vol/vol) FCS, 2 mM GlutaMAX, and 1 mM sodium pyruvate.
For clathrin-mediated endocytosis, cells were serum starved in DMEM supplemented with 2 mM GlutaMAX and 1 mM sodium pyruvate for ∼10 h before the experiment. Cells were incubated in ice-cold Hepes-buffered DMEM for 1 h, followed by incubation on ice containing 2 μg/mL Alexa Fluor 555-labeled EGF ligand (Life Technologies) in Hepes-buffered DMEM. Cells were washed with ice-cold PBS and incubated at 37 °C in Hepes-buffered DMEM for the indicated times before fixation.
Fixation was performed with 4% (wt/vol) paraformaldehyde (Electron Microscopy Services) for 15 min at 37 °C. After fixation, the cells were washed in PBS twice, quenched with 100 mM glycine in PBS, and blocked in antibody-dilution buffer [150 mM NaCl, 20 mM Tris, 0.1% Triton X-100, 0.1% NaN3, and 2% (wt/vol) BSA, pH 7.4].
Immunolabeling was performed with anti-clathrin heavy-chain antibody (Thermo Fisher Scientific; MA1-065), α-tubulin monoclonal antibody (Invitrogen; 322500), and secondary antibodies goat anti-mouse IgG (H+L)-Alexa Fluor 568 (Invitrogen; A11004) and/or goat anti-mouse IgG (H+L)-Alexa Fluor 488 (Invitrogen; A11001). Actin filaments were stained with phalloidin-Alexa Fluor 488 (Invitrogen; A12379) and/or phalloidin-Alexa Fluor 568 (Invitrogen; A12380).
Transfections with plasmid DNA were performed with X-tremeGENE HP DNA transfection reagent. The transfected cells were incubated at 37 °C in 5% CO2 overnight before fixation and/or imaging. Detailed protocols are provided in SI Appendix, Methods.
Molecule Localization Analysis.
The 3B analysis (25) was performed using custom software designed for use on the National Institutes of Health’s Biowulf cluster parallel processing system (https://hpc.nih.gov). All custom software scripts needed to perform parallelized 3B analysis and detailed instructions on their use are available in SI Appendix.
Supplementary Material
Acknowledgments
We thank Hari Shroff for helpful discussions and a critical reading of the manuscript. This work was supported by the Intramural Research Program of the National Institutes of Health, including the National Institute of Biomedical Imaging and Bioengineering. This study was performed using the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (https://hpc.nih.gov).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516715113/-/DCSupplemental.
References
- 1.Inoué S. 2006. Foundations of confocal scanned imaging in light microscopy. Handbook of Biological Confocal Microscopy, ed Pawley JB (Springer, New York), 3rd Ed, pp 1–19.
- 2.Bailey B, Farkas DL, Taylor DL, Lanni F. Enhancement of axial resolution in fluorescence microscopy by standing-wave excitation. Nature. 1993;366(6450):44–48. doi: 10.1038/366044a0. [DOI] [PubMed] [Google Scholar]
- 3.Gustafsson MG, Agard DA, Sedat JW. I5M: 3D widefield light microscopy with better than 100 nm axial resolution. J Microsc. 1999;195(Pt 1):10–16. doi: 10.1046/j.1365-2818.1999.00576.x. [DOI] [PubMed] [Google Scholar]
- 4.Bewersdorf J, Schmidt R, Hell SW. Comparison of I5M and 4Pi-microscopy. J Microsc. 2006;222(Pt 2):105–117. doi: 10.1111/j.1365-2818.2006.01578.x. [DOI] [PubMed] [Google Scholar]
- 5.Paszek MJ, et al. Scanning angle interference microscopy reveals cell dynamics at the nanoscale. Nat Methods. 2012;9(8):825–827. doi: 10.1038/nmeth.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: Super-resolution imaging of cells. Cell. 2010;143(7):1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313(5793):1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 8.Vaziri A, Tang J, Shroff H, Shank CV. Multilayer three-dimensional superresolution imaging of thick biological samples. Proc Natl Acad Sci USA. 2008;105(51):20221–20226. doi: 10.1073/pnas.0810636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.York AG, Ghitani A, Vaziri A, Davidson MW, Shroff H. Confined activation and subdiffractive localization enables whole-cell PALM with genetically expressed probes. Nat Methods. 2011;8(4):327–333. doi: 10.1038/nmeth.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juette MF, et al. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat Methods. 2008;5(6):527–529. doi: 10.1038/nmeth.1211. [DOI] [PubMed] [Google Scholar]
- 11.Pavani SR, et al. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc Natl Acad Sci USA. 2009;106(9):2995–2999. doi: 10.1073/pnas.0900245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3(10):793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319(5864):810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu K, Babcock HP, Zhuang X. Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat Methods. 2012;9(2):185–188. doi: 10.1038/nmeth.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shtengel G, et al. Interferometric fluorescent superresolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci USA. 2009;106(9):3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt R, et al. Spherical nanosized focal spot unravels the interior of cells. Nat Methods. 2008;5(6):539–544. doi: 10.1038/nmeth.1214. [DOI] [PubMed] [Google Scholar]
- 17.Axelrod D. Total internal reflection fluorescence microscopy. Methods Cell Biol. 2008;89:169–221. doi: 10.1016/S0091-679X(08)00607-9. [DOI] [PubMed] [Google Scholar]
- 18.Axelrod D. Cell-substrate contacts illuminated by total internal reflection fluorescence. J Cell Biol. 1981;89(1):141–145. doi: 10.1083/jcb.89.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ölveczky BP, Periasamy N, Verkman AS. Mapping fluorophore distributions in three dimensions by quantitative multiple angle-total internal reflection fluorescence microscopy. Biophys J. 1997;73(5):2836–2847. doi: 10.1016/S0006-3495(97)78312-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stock K, et al. Variable-angle total internal reflection fluorescence microscopy (VA-TIRFM): Realization and application of a compact illumination device. J Microsc. 2003;211(Pt 1):19–29. doi: 10.1046/j.1365-2818.2003.01200.x. [DOI] [PubMed] [Google Scholar]
- 21.Rohrbach A. Observing secretory granules with a multiangle evanescent wave microscope. Biophys J. 2000;78(5):2641–2654. doi: 10.1016/S0006-3495(00)76808-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun W, Marchuk K, Wang G, Fang N. Autocalibrated scanning-angle prism-type total internal reflection fluorescence microscopy for nanometer-precision axial position determination. Anal Chem. 2010;82(6):2441–2447. doi: 10.1021/ac902789z. [DOI] [PubMed] [Google Scholar]
- 23.Yang Q, Karpikov A, Toomre D, Duncan JS. 3-D reconstruction of microtubules from multi-angle total internal reflection fluorescence microscopy using Bayesian framework. IEEE Trans Image Process. 2011;20(8):2248–2259. doi: 10.1109/TIP.2011.2114359. [DOI] [PubMed] [Google Scholar]
- 24.Boulanger J, et al. Fast high-resolution 3D total internal reflection fluorescence microscopy by incidence angle scanning and azimuthal averaging. Proc Natl Acad Sci USA. 2014;111(48):17164–17169. doi: 10.1073/pnas.1414106111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox S, et al. Bayesian localization microscopy reveals nanoscale podosome dynamics. Nat Methods. 2012;9(2):195–200. doi: 10.1038/nmeth.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattheyses AL, Axelrod D. Direct measurement of the evanescent field profile produced by objective-based total internal reflection fluorescence. J Biomed Opt. 2006;11(1):014006. doi: 10.1117/1.2161018. [DOI] [PubMed] [Google Scholar]
- 27.Byron ML, Variano EA. Refractive index-matched hydrogel materials for measuring flow-structure interactions. Exp Fluids. 2013;54:1456–1461. [Google Scholar]
- 28.Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468(7323):580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vieira AV, Lamaze C, Schmid SL. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274(5295):2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 30.Sigismund S, et al. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev Cell. 2008;15(2):209–219. doi: 10.1016/j.devcel.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 31.Rosten E, Jones GE, Cox S. ImageJ plug-in for Bayesian analysis of blinking and bleaching. Nat Methods. 2013;10(2):97–98. doi: 10.1038/nmeth.2342. [DOI] [PubMed] [Google Scholar]
- 32.Hu YS, Nan X, Sengupta P, Lippincott-Schwartz J, Cang H. Accelerating 3B single-molecule super-resolution microscopy with cloud computing. Nat Methods. 2013;10(2):96–97. doi: 10.1038/nmeth.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using microManager. Curr Protoc Mol Biol. 2010;92(14.20):14.20.1–14.20.17. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297(5588):1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.