Significance
Impairment of global genome nucleotide excision repair (NER) leads to extreme sun sensitivity and predisposition to cancers. The xeroderma pigmentosum C (XPC) complex senses diverse environmentally induced DNA lesions from predominantly normal DNA, and initiates NER by recruiting downstream factors. Using unique fluorescent approaches, this study unveils previously unresolved DNA dynamics during lesion recognition by radiation-sensitive 4 (Rad4; yeast XPC ortholog) and demonstrates that Rad4 nonspecifically deforms (“twists”) the DNA before specifically recognizing (“opening”) target lesions. These results mark the first observation, to our knowledge, of DNA distortional dynamics that reflect a nonspecific search/interrogation process by a DNA repair protein that relies entirely on DNA deformability to recognize its lesions, and provides keys to understanding the protein’s ability to search rapidly and yet also reliably recognize diverse lesions.
Keywords: DNA damage recognition, time-resolved fluorescence spectroscopy, temperature-jump perturbation, DNA unwinding dynamics, xeroderma pigmentosum
Abstract
DNA damage repair starts with the recognition of damaged sites from predominantly normal DNA. In eukaryotes, diverse DNA lesions from environmental sources are recognized by the xeroderma pigmentosum C (XPC) nucleotide excision repair complex. Studies of Rad4 (radiation-sensitive 4; yeast XPC ortholog) showed that Rad4 “opens” up damaged DNA by inserting a β-hairpin into the duplex and flipping out two damage-containing nucleotide pairs. However, this DNA lesion “opening” is slow (˜5–10 ms) compared with typical submillisecond residence times per base pair site reported for various DNA-binding proteins during 1D diffusion on DNA. To address the mystery as to how Rad4 pauses to recognize lesions during diffusional search, we examine conformational dynamics along the lesion recognition trajectory using temperature-jump spectroscopy. Besides identifying the ˜10-ms step as the rate-limiting bottleneck towards opening specific DNA site, we uncover an earlier ˜100- to 500-μs step that we assign to nonspecific deformation (unwinding/“twisting”) of DNA by Rad4. The β-hairpin is not required to unwind or to overcome the bottleneck but is essential for full nucleotide-flipping. We propose that Rad4 recognizes lesions in a step-wise “twist-open” mechanism, in which preliminary twisting represents Rad4 interconverting between search and interrogation modes. Through such conformational switches compatible with rapid diffusion on DNA, Rad4 may stall preferentially at a lesion site, offering time to open DNA. This study represents the first direct observation, to our knowledge, of dynamical DNA distortions during search/interrogation beyond base pair breathing. Submillisecond interrogation with preferential stalling at cognate sites may be common to various DNA-binding proteins.
Essential genetic mechanisms, such as replication, transcription, recombination, and repair, all require recognition of specific DNA sequences or structures by specialized proteins. How DNA-binding proteins search for and identify their target sites embedded in a vast excess of nontarget sites, especially if using only thermal energy, is a fundamental question in biology. Several lines of evidence indicate that proteins use some combination of 3D diffusion in the bulk solution and 1D diffusion while nonspecifically bound to DNA, and use this “facilitated diffusion” as a means to search efficiently in genomic DNA for their targets (1–7). Direct observations of proteins diffusing on nonspecific DNA have revealed residence times per base pair site ranging from 50 ns to 300 μs (7–13). On the other end, high-resolution structures and thermodynamic studies on a wide range of specific protein–DNA complexes have revealed significant distortions in otherwise B-form DNA duplex structures and concerted rearrangements in the bound protein to accommodate the deformed DNA; this “induced-fit” mechanism has emerged as a general principle of target recognition (14, 15). In many cases, the proteins discriminate between specific and nonspecific sites primarily by sensing differences in local DNA deformability (“indirect readout”), rather than by relying on direct interactions with target nucleotides (16). However, how rapidly the deformations occur over the course of target recognition and how they compare with the protein’s residence time on a given DNA site before it diffuses away remain largely unknown, obscuring our understanding of target recognition mechanisms.
Previous measurements of DNA conformational dynamics have revealed millisecond time-scale motions during specific recognition (17–22), which are slower compared with the submillisecond 1D residence times of various DNA-binding proteins when diffusing nonspecifically on DNA, as discussed above. It has been proposed that a conformational switch between a rapidly diffusing “search” mode and a more tightly bound “recognition” mode is needed for a protein to identify a potential target site without losing overall speed (23–26). Consistent with these arguments, alternative binding modes of various proteins interacting with nonspecific DNA have been observed (27–37), and have also been inferred from single-molecule studies of proteins diffusing on DNA (13, 38–40). Microsecond-to-millisecond conformational fluctuations have been reported in proteins nonspecifically bound to DNA in a few systems amenable to NMR (28, 32, 36).
For systems that are too large for NMR, the submillisecond conformational dynamics are however difficult to capture, either by stopped-flow or single-molecule techniques: Stopped-flow can only directly detect motions slower than a few milliseconds, and single-molecule fluorescence is limited to measurements with high-quantum-yield fluorescence labels, and can typically detect only large conformational changes. A laser temperature-jump (T-jump) approach in combination with time-resolved optical spectroscopy enables kinetics measurements with high (submicrosecond) temporal resolution [reviewed by Kubelka (41)]. The T-jump approach, together with probes such as tryptophans and fluorescent nucleotide analogs, has led to the characterization of otherwise unresolved conformational dynamics during protein folding (42–45), RNA folding (46), and protein–DNA (47–49) and protein–RNA (50) interactions.
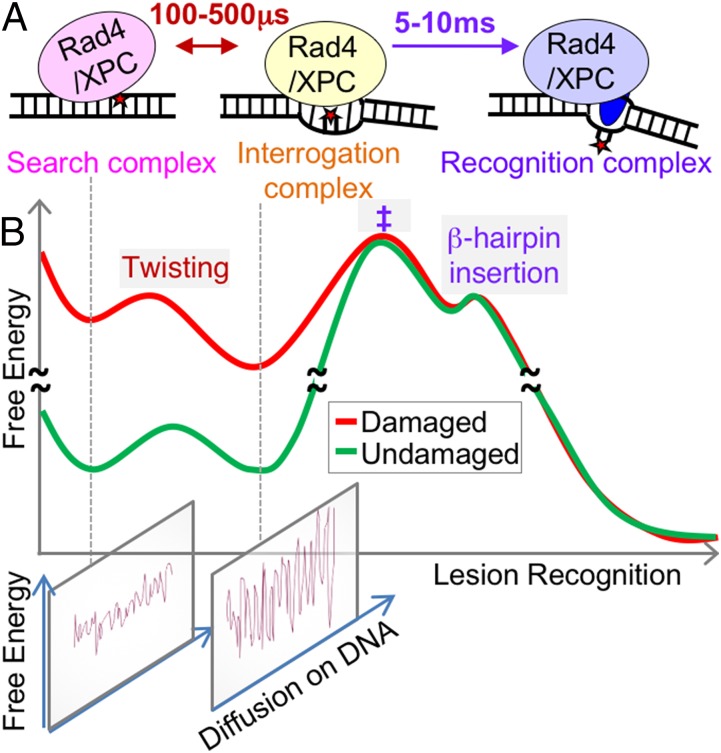
In this study, we applied T-jump to detect transient intermediates in a system that relies exclusively on indirect readout, the radiation-sensitive 4 (Rad4)–radiation-sensitive 23 (Rad23) protein complex (hereafter Rad4). Rad4 is a yeast homolog of the xeroderma pigmentosum C (XPC)–RAD23B complex (hereafter XPC), a key initiator of the global genome nucleotide excision repair (NER) pathway in eukaryotes [reviewed by Schärer (51)]. Using only thermal energy, Rad4/XPC surveils the genome and specifically recognizes lesions, initiating the recruitment of other NER factors that ultimately excise and repair the damaged portion of DNA. The lesions recognized by Rad4/XPC include photoproducts induced by UV light or bulky base adducts and intrastrand adducts induced by environmental genotoxins (52). These lesions are structurally diverse, but the damage is confined on one strand of the duplex DNA and commonly accompanies local helix destabilization/distortion. In vitro, Rad4/XPC can also specifically bind to 2-bp or 3-bp mismatch DNA (49, 53). The crystal structures of Rad4 bound to UV-induced model lesions showed that Rad4 inserts a β-hairpin into the lesion site and flips out two damage-containing nucleotide pairs; the duplex around the lesion is also partially unwound and bent (54) (Fig. 1A). In this “open” recognition complex, Rad4 does not directly contact damaged nucleotides but selectively accommodates the undamaged nucleotides on the complementary strand; such indirect binding enables Rad4/XPC to recognize extraordinarily diverse lesions. Intriguingly, we have recently observed that the same open structure is also formed when Rad4 is covalently tethered to undamaged DNA (49); thus, the lesion recognition must rely not on the structural differences in the thermodynamically most stable states of the bound complexes but on the differences in the kinetics of recognition.
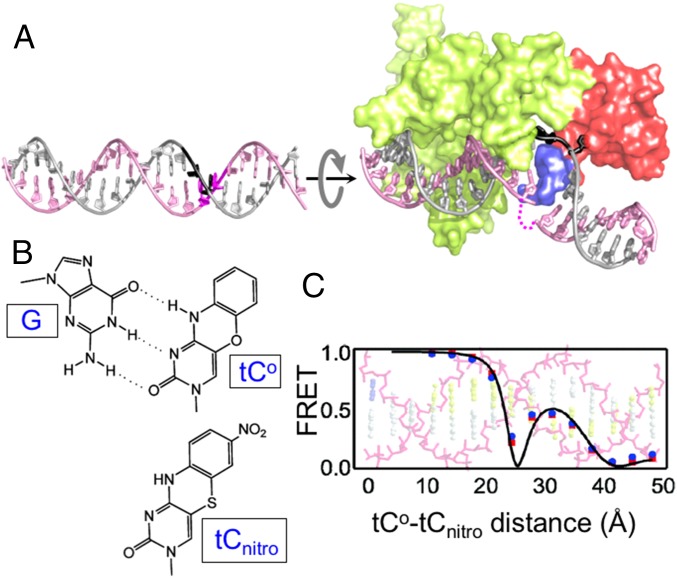
Fig. 1.
DNA conformational rearrangements during lesion recognition by Rad4 probed by tCo/tCnitro. (A) Structures of an ideal B-form DNA (Left) and Rad4-bound–specific lesion recognition (open) complex (Right; Protein Data Bank ID code 2QSH) (54). The gray rotation arrow indicates the direction of DNA unwinding upon opening. The DNA-binding domains of Rad4 are shown: TGD-BHD1-BHD2 (lime green), BHD3 (red), and the β-hairpin within BHD3 (blue). In the open complex, the flipped-out nucleotides on the undamaged strand (black) directly contact Rad4, whereas the flipped-out nucleotides on the damaged strand (magenta) do not and become disordered (dotted magenta line). (B) Chemical structures of tCo and tCnitro and Watson–Crick type base pairing of tCo with a guanine (G). (C) FRET efficiency between the tCo and tCnitro incorporated within normal B-DNA is plotted as a function of the distance between the two probes. FRET decreases as the distance increases, but additionally depends on the relative orientations of the absorption and emission dipoles of the fluorophores. B and C are adapted with permission from ref. 56; copyright (2009) American Chemical Society.
As a first step to characterize these recognition dynamics, we measured the Rad4-induced “opening” of mismatch DNA using the T-jump perturbation approach and showed that it occurred on time scales of ∼5–10 ms (49). The opening times for more stable, undamaged/matched DNA are expected to be much longer than for mismatch DNA (55). Although the 1D diffusion constants of Rad4/XPC on DNA are yet to be reported, studies have revealed microsecond-regime residence times per DNA site for a wide range of DNA-binding proteins (7–13). Thus, the observed opening time of Rad4 is many orders of magnitude longer than the typical residence times per DNA site. This result, we proposed, explains how Rad4/XPC avoids flipping open every undamaged site when searching DNA for a lesion, although having selectively higher probabilities to open helix-destabilizing NER lesion sites. However, this “kinetic gating” mechanism also posed a dilemma as to how Rad4 reliably recognizes damaged DNA if the recognition (∼5–10 ms) is much slower than the typical diffusion of proteins on DNA during a nonspecific search.
Here, we report on capturing kinetic intermediates that could bridge the gap between the fast, nonspecific search and the slow recognition of the specific damaged site. Taking advantage of the T-jump approach with the unique sensitivity of a FRET pair [1,3-diaza-2-oxophenoxazine (tCo) as the donor and 7-nitro-1,3-diaza-2-oxophenothiazine (tCnitro) as the acceptor] to DNA helicity (56, 57), we uncovered previously unidentified conformational dynamics occurring on the time scales of ∼100–500 μs when Rad4 is nonspecifically bound to DNA. We interpret these dynamics as rapid unwinding (“twisting”) of DNA by Rad4 resulting from the conformational interconversion between a rapidly scanning search mode and a momentarily stalled “interrogation” mode. We also demonstrated that the β-hairpin that stabilizes the open conformation is inserted after a rate-limiting (bottleneck) step for lesion-specific opening. The progressive, multistep nature of target recognition uncovered for this DNA repair protein provides critical insights into the speed-and-stability paradox of specific DNA recognition.
Results
Laser T-Jump Spectroscopy to Probe Conformational Dynamics.
We used ∼10-ns IR laser pulses to increase rapidly the temperature of a small volume of the sample by 5–10 °C within the duration of each pulse. We then monitored the temporal response of the ensemble of molecules as it reequilibrated (relaxed) from the initial temperature population to the population at the higher temperature, by recording the intensities of conformation-sensing fluorescent probes (further described below). The temperature of the heated volume stays approximately constant until about 50 ms and then decays (“recovers”) back to the initial temperature (i.e., the temperature of the surrounding bath), with a characteristic “T-jump recovery” time of ∼200 ms (58). Thus, our T-jump apparatus enables us to monitor the dynamics of protein–DNA interactions in a time window of ∼20 μs (the time resolution of our spectrometer) to ∼50 ms (before the T-jump starts to recover) (20, 47). The microsecond time resolution of the T-jump was critical in this study so as to overlap better with the time scales for fast, 1D diffusion of proteins along DNA. The application of the tCo/tCnitro FRET pair in characterizing rapid DNA dynamics, such as partial unwinding or twisting, also opens the door for uncovering new dynamics for a wide range of DNA-binding proteins.
tCo and tCnitro FRET Pair as Probes for Sensing Changes in DNA Helical Structure.
We have previously reported T-jump measurements of Rad4-induced DNA opening kinetics, probed specifically at mismatch model lesion sites using fluorescent 2-aminopurine (2AP) as one of the mismatched nucleotides (49). Here, we aimed at probing motions that affect regions beyond the mismatched nucleotides such as local unwinding around the lesion, as observed in the open structures (Figs. 1A and 2A). To monitor these motions, we adopted a pair of recently developed FRET probes, tCo (as the donor) and tCnitro (as the acceptor) (56, 59) (Fig. 1B). These FRET probes are tricyclic cytosine analogs that retain Watson–Crick-type pairing with guanines in duplex DNA; they also do not compromise the overall DNA duplex stability nor affect the binding affinities of DNA duplexes to Rad4 (56) (discussed in the next section). Importantly, the FRET efficiency of the probes incorporated within the DNA duplex was shown to exhibit an oscillatory trend as a function of separation within DNA, which reflects the helical periodicity of B-DNA (56) (Fig. 1C). Such unique characteristics of these probes make them highly suited to capture protein-induced DNA motions involving changes in helicity, such as local unwinding. Although it has been shown that conventional FRET probes, typically tethered to DNA through flexible linkers, can be responsive to DNA helicity when stacked at the ends of DNA duplexes (60), these probes are not readily incorporated as rigid internal labels within duplex DNA without disrupting the DNA backbone/structure (61), as is possible with the tCo/tCnitro probes (56, 59). Accordingly, we incorporated the tCo/tCnitro probes on either side of the mismatch site, spanning the region expected to undergo the most dramatic unwinding upon Rad4 binding (SI Appendix, Fig. S1).
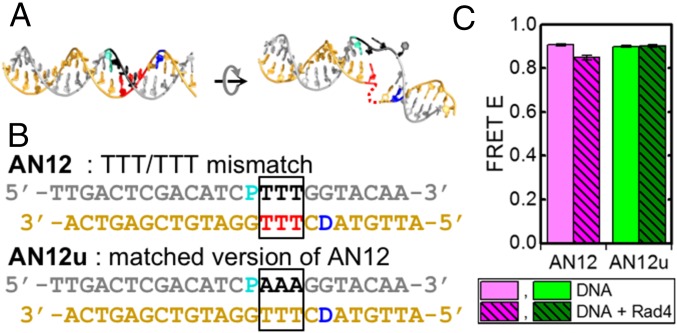
Fig. 2.
AN12 and AN12u DNA constructs. (A) Models of DNA duplexes when in B-DNA conformation (Left) and in the open complex (Right), in the same orientations as in Fig. 1A. The positions are marked for the mismatched nucleotides (black and red) and the FRET probes, tCo (donor, cyan) and tCnitro (acceptor, blue) of AN12. (B) DNA sequences of AN12 and AN12u. D, donor; P, acceptor. (C) FRET at 25 °C in free AN12 (pink, n = 11), AN12–Rad4 (magenta, n = 4), free AN12u (lime, n = 18), and AN12u–Rad4 (green, n = 9).
tCo/tCnitro-Labeled TTT/TTT-Mismatch DNA (AN12) as a Model Lesion for Specific Binding.
First, we tested a 24-bp DNA duplex sequence containing a TTT/TTT mismatch (AN12) as a model lesion for Rad4 and its undamaged/matched counterpart (AN12u). The TTT/TTT mismatch has been shown to form an open structure in crystals when bound to Rad4 (54) (Fig. 1A and SI Appendix, Fig. S1B). In these constructs, tCnitro was placed immediately abutting the mismatch site and tCo was placed 1 bp away from the other side of the mismatch bubble (Fig. 2 A and B). The melting temperatures (Tms) of the mismatch AN12 DNA were 8–11 °C lower than the Tms of the corresponding undamaged AN12u counterparts, as expected from the destabilization caused by the mismatched bases (SI Appendix, Fig. S2 A–D). Introducing FRET labels affected the Tm for each construct by less than 2 °C, compared with unlabeled DNA, reaffirming that the FRET probes do not significantly alter the thermodynamic stability of AN12 or AN12u (SI Appendix, SI Methods 1.3 and Fig. S2 B‒D).
Next, we examined the relative binding affinities of AN12 and AN12u to Rad4 using a competition EMSA, where the protein’s binding to 5 nM 32P-labeled substrate DNA is monitored in the presence of 1,000 nM unlabeled, undamaged “competitor” DNA (CH7_NX) (54) (SI Appendix, SI Methods 1.4 and Fig. S3). The benefit of using this competition assay over the conventional single-substrate EMSA is that one can directly observe any preferential binding over the nonspecific binding, including factors such as potential DNA end-binding while avoiding multiple proteins aggregating on a single DNA, as is the case when protein is in excess of total DNA (SI Appendix, Fig. S4).
The WT Rad4 construct we used in this study spans residues 101–632 and contains all four domains involved in DNA binding. This construct has previously been shown to exhibit the same DNA-binding characteristics as the full-length protein (54). The presence of tCo/tCnitro labels in the AN12/AN12u did not affect the Rad4-binding affinities of each DNA compared with unlabeled DNA. However, regardless of the labels, the mismatch-containing AN12 constructs showed about a fourfold lower apparent dissociation constant (Kd,app) than the corresponding undamaged AN12u or other undamaged DNA constructs. Although this specificity of AN12 is slightly lower than the approximately sevenfold specificity exhibited by another DNA construct containing CCC/CCC mismatches (49) (CH10_NX; SI Appendix, Fig. S3), the results indicate that the TTT/TTT mismatch in AN12 is specifically recognized by Rad4 (as also expected from the crystal structure), whereas AN12u is bound nonspecifically. The small differences in the apparent binding affinities between damaged and undamaged DNA are consistent with other studies for human XPC protein when using short duplex oligonucleotides as substrates (62, 63). These calculations are based on the assumption that there is only one protein-binding site per DNA substrate, even for nonspecific DNA; however, this assumption underestimates the real number of nonspecific sites available to the protein, because Rad4 can bind in more than one register and orientation even for these short DNA duplexes. Thus, it is possible that the actual differences between the specific and nonspecific binding are larger than those differences reflected in these Kd,app values.
We highlight here that all our equilibrium FRET and T-jump studies were carried out for equimolar protein–DNA concentrations higher than 4 μM, greatly exceeding the Kd,app for any of these complexes (discussed below). Native gel electrophoresis and dynamic light scattering independently confirmed that the Rad4–DNA samples under our conditions are indeed uniformly sized 1:1 complexes (SI Appendix, SI Methods 1.5 and 1.6 and Figs. S4 and S5).
Next, we measured the FRET efficiency for each DNA (10 μM) at equilibrium in the absence and presence of equimolar Rad4. We first acquired two donor fluorescence emission spectra (SI Appendix, Fig. S6), one from the DNA containing only the donor (denoted as AN12_D and AN12u_D) and the other from the DNA containing both the donor and acceptor probes (AN12_DA and AN12u_DA), and calculated the FRET efficiencies from the measured spectra (Fig. 2C and SI Appendix, SI Methods 1.7). The experimental FRET value for the undamaged AN12u in the absence of Rad4 was in reasonable agreement with predictions based on the relative orientations and distances between the probes in a canonical B-DNA structure (56, 64) (SI Appendix, Fig. S7A). Furthermore, the mismatched AN12 yielded FRET values very similar to AN12u, which indicates that the DNA largely retains the B-DNA conformation despite the helix-destabilizing mismatches, or at least that any changes in the average DNA conformation due to mismatches are not detected in these bulk FRET measurements.
In the presence of Rad4, the FRET of AN12 decreased in comparison to protein-free DNA, whereas the FRET of AN12u did not show a measurable difference (Fig. 2C and SI Appendix, Fig. S7A). As described above, AN12 is a specific substrate for Rad4 and is expected to undergo lesion-specific opening when bound to Rad4 under the experimental conditions. A decrease in FRET upon Rad4 binding is expected based on the changes in DNA conformation from B-DNA to the open conformation seen in the crystal structures (54). However, the observed FRET value (0.849 ± 0.011) differed significantly from the value calculated (0.04) based on the crystal structures (SI Appendix, Table summarizes the FRET values for all constructs in this study). Various factors may contribute to the deviations between the theoretical estimates and the observed FRET in the protein-bound samples (SI Appendix, SI Discussion 2.1), with perhaps the biggest contribution coming from enhanced dynamics of the tCnitro probe in the AN12 context, where the probe is placed right next to the mismatch site (Fig. 2B). It cannot be ruled out that the average orientation of this probe in the AN12–Rad4 solution structure is altered in comparison to the average orientation of the corresponding nucleotide in the crystal structure. Nevertheless, the distinct decrease in FRET in AN12, but not in AN12u, upon Rad4 binding indicates that the probes detect the change in average conformation in the mismatched AN12, induced upon specific binding to Rad4.
Increase in Temperature Amplifies Rad4-Induced Opening Sensed by the Probes in AN12.
We next monitored the extent to which temperature may alter the average DNA conformations probed by tCo/tCnitro by measuring equilibrium FRET at different temperatures. The donor emission intensities measured in AN12_D containing only the donor (IDs) decreased monotonically with increasing temperature from 10–40 °C for both free and Rad4-bound DNA (Fig. 3 A and B, black), corresponding to the quantum yield decrease of the donor with increasing temperature (65, 66). In contrast, the donor emission intensities in in AN12_DA containing both the donor and acceptor (IDAs) increased with increasing temperature for both free and Rad4-bound DNA (Fig. 3 A and B, magenta/pink). Although these measurements show similar trends for free and Rad4-bound AN12, the corresponding FRET values show a slightly larger change with increasing temperature for the AN12–Rad4 complex than for free AN12; in the 10–40 °C temperature range, the FRET in the AN12–Rad4 complex decreased by 0.084 ± 0.018, whereas for free AN12, it decreased by 0.051 ± 0.006 (SI Appendix, Fig. S7B). These results indicate that although a temperature increase alters the ensemble of conformations in free AN12, it also amplifies the extent and/or population of the tCo/tCnitro-sensed, Rad4-induced conformational changes (SI Appendix, Fig. S7B), similar to the previously observed conformational changes sensed by 2AP at mismatch sites (49) (SI Appendix, Fig. S8). Furthermore, T-jump perturbation kinetics measured on free and Rad4-bound AN12 samples are distinctly different (discussed below). AN12u, on the other hand, exhibited smaller changes in FRET with increasing temperature, which are almost the same for free DNA (0.028 ± 0.009) and Rad4-bound DNA (0.021 ± 0.008) (SI Appendix, Table). Thus, temperature increases result in smaller changes in the average conformations of the undamaged/matched AN12u than of mismatch AN12, regardless of the presence of the protein.
Fig. 3.
Equilibrium and T-jump measurements on AN12 bound to Rad4. (A and B) IDA as a function of temperature for AN12–Rad4 (magenta, n = 4) and AN12 (pink, n = 11); the corresponding ID is in black. ID and IDA have been normalized to match at the lowest temperature. The intensities at 25 °C measured before and after the heating/cooling cycle are both indicated. T-jump relaxation traces are shown for AN12_DA in the presence/absence of Rad4 (C and D) and for AN12_D in the presence/absence of Rad4 (E and F). Only AN12_DA–Rad4 in C exhibits kinetics in the T-jump time-window (7.7 ± 0.5 ms after a T-jump from 19–26 °C), whereas D–F exhibit only the slow T-jump recovery after a similar T-jump. (G) Relaxation rates of AN12–Rad4 (pink) vs. the inverse of the final temperature (after the T-jump). The different symbols indicate three independent sets of measurements. The continuous pink line is an Arrhenius fit to the relaxation rates, yielding an activation enthalpy of 18.5 ± 1.1 kcal/mol. The dashed black line is an Arrhenius fit to the relaxation rates on 2AP-labeled mismatch DNA (AN3) bound to Rad4, from a study by Chen et al. (49). A schematic representation of the mismatched AN12_DA construct design is also shown. a.u., arbitrary unit.
tCo/tCnitro-Sensed Dynamics in Rad4-Bound AN12 Are Concurrent with 2AP-Sensed, Rad4-Induced Lesion Opening Dynamics.
To measure the dynamics of conformational changes in AN12 upon binding to Rad4, we performed T-jump measurements on AN12 as free DNA (4–40 μM) or as 1:1 complexes with Rad4. The relaxation traces in response to an ∼5–10 °C T-jump perturbation were monitored by recording the donor fluorescence intensities (IDs or IDAs) over time, for a series of initial temperatures with final temperatures in the range of 16–41 °C. First, ID values from AN12_D showed a rapid initial drop (much faster than our earliest observable time of ∼20 μs), followed by a slow relaxation of 200 ± 60 ms, characteristic of T-jump recovery kinetics (Fig. 3F). The initial rapid drop in AN12_D is due to the decrease in the quantum yield of the donor probe in response to the T-jump to a higher temperature, as also observed in the equilibrium measurements (Fig. 3B, black). The presence of bound Rad4 did not change the observed kinetics of AN12_D (Fig. 3E). Thus, the donor-only AN12_D samples do not exhibit any detectable conformational relaxation kinetics for either free or Rad4-bound DNA.
On the other hand, IDA values from free AN12_DA revealed an immediate increase after the T-jump perturbation, on a time scale faster than the ∼20-μs resolution limit of our T-jump apparatus (Fig. 3D), followed by the slow T-jump recovery kinetics (SI Appendix, SI Methods 1.10 and Fig. S9 B and C). The rapid increase in AN12_DA is in a direction opposite of the rapid decrease seen in AN12_D; instead, it mirrors the increase in IDA seen in equilibrium measurements on the AN12_DA sample (Fig. 3B, pink) and is reminiscent of the rapid kinetics of mismatch unstacking (“premelting”) previously observed using 2AP-labeled DNA (49). We thus propose that the rapid increase in AN12’s IDA after the T-jump reflects similar premelting kinetics, as sensed by the rapid reorientation of tCnitro adjoining the mismatch bubble. The temperature-dependent FRET decrease in free AN12 DNA in the equilibrium measurements discussed above is also consistent with DNA distortions sensed by tCnitro placed adjacent to the mismatch as the DNA “premelts” at the higher temperatures.
Notably, AN12_DA, when bound to Rad4, showed additional relaxation kinetics in the T-jump time window of 20 μs to 32 ms, distinct from the rapid premelting (<20 μs) and slow T-jump recovery (∼200 ms) (Fig. 3C and SI Appendix, Figs. S9 and S10). Furthermore, the amplitude of the rapid (unresolved) kinetics was diminished in the complex (SI Appendix, Fig. S10), again reminiscent of our previous observations with 2AP constructs (49), indicating that mismatch premelting is partially suppressed in the presence of Rad4. The relaxation kinetics in the T-jump time window are detected only in the presence of the protein and require both the donor and the acceptor in AN12; they were not detected in the absence of the protein (Fig. 3D) or in AN12_D–Rad4 (Fig. 3E). The undamaged counterpart, AN12u_D and AN12u_DA, also did not show any relaxation kinetics in the same time window in either the presence or absence of Rad4 (SI Appendix, Fig. S11 C–F). Thus, the kinetics uniquely observed in Rad4-bound mismatch AN12_DA reflect DNA motions specifically induced by Rad4, because they alter the FRET between the tCo/tCnitro probes in the vicinity of the TTT/TTT mismatch. Considering the sensitivity of the probes to DNA helical conformations, our results also indicate that the mismatch premelting and the Rad4-induced open conformation sensed by the FRET probes in AN12 may share similar directionality of structural distortions (unwinding).
To determine the minimum number of relaxation phases needed to describe the kinetics observed within the T-jump time window in the AN12_DA–Rad4 complex, we analyzed the relaxation traces using maximum entropy analysis (SI Appendix, SI Methods 1.11). The analysis revealed monophasic kinetics with average relaxation times ranging from 15.9–1.6 ms (7.7 ± 0.8 ms at 25 °C) in the temperature range 16–41 °C (final temperature) (Fig. 3G and SI Appendix, Fig. S9). These ∼8-ms relaxation kinetics are on similar time scales as the previously reported Rad4-induced DNA opening dynamics probed by 2AP (49) (SI Appendix, Fig. S8). Because the 2AP probes were placed as one of the mismatched nucleotides that are flipped out when Rad4 binds specifically, 2AP likely senses the kinetics of nucleotide flipping. Our results suggest that the tCo/tCnitro-monitored DNA dynamics in the AN12_DA–Rad4 complex involve the same rate-limiting distortions as in the nucleotide-flipping during specific opening. However, 2AP in the previous study and tCo/tCnitro in AN12 do not necessarily probe identical motions during opening, as evidenced by the measurements with Rad4 mutants described below.
β-Hairpin Mutants Exhibit Previously Unobserved Submillisecond Kinetics and Help Reveal the Multistep Nature of Lesion Recognition by Rad4.
Previous studies have shown that the β-hairpin from the β-hairpin domain 3 (BHD3) plays an important role in Rad4’s ability to recognize lesions (49, 54, 67). In the open conformation, the β-hairpin is shown to be inserted into the damaged DNA site, filling the gap in the DNA duplex resulting from the flipped-out nucleotides (49, 54). The mutant Rad4 lacking either the β-hairpin in BHD3 (residues 599–605, blue in Fig. 1A, denoted as Δβ-hairpin3) or the entire BHD3 domain (residues 541–632, blue and red in Fig. 1A, denoted as ΔBHD3) retains nanomolar affinities to the undamaged DNA duplex but has significantly reduced specificity for DNA damage compared with the WT Rad4 in competition EMSA assays (49). Importantly, neither mutant revealed any Rad4-induced nucleotide-flipping kinetics detected by 2AP (49).
Here, we examine the role of the β-hairpin in inducing the conformational changes monitored in AN12. The FRET changes in AN12 upon binding to the mutants, Δβ-hairpin3 and ΔBHD3, were barely detectable in equilibrium measurements at 25 °C, consistent with the impaired specificity of the mutants (SI Appendix, Fig. S7A). The FRET measured on these samples as the temperatures were raised from 10–30 °C essentially overlapped with the FRET measured on free AN12 (Fig. 4 A and B and SI Appendix, Fig. S7B). Thus, the FRET in the AN12–mutant complexes in this temperature range predominantly indicate enhanced premelting of the mismatch region sensed by the tCnitro probe at the higher temperatures, as in free AN12. For Δβ-hairpin3, the temperature range was limited to up to ∼30 °C because the protein/DNA sample heated above ∼35 °C showed irreversible behavior in fluorescence measurements indicating early denaturation/aggregation of the protein (49). For ΔBHD3, the measurements were carried out up to 40 °C as with the WT Rad4. Although the FRET vs. temperature trends in AN12–ΔBHD3 resembled the temperature trends of the free DNA below 30 °C, small deviations were observed at temperatures above ∼30 °C (SI Appendix, Fig. S7B), indicating a contribution from the ΔBHD3-induced conformational changes at these higher temperatures. The FRET vs. temperature measurements on the undamaged AN12u with either of the two mutants essentially overlapped with the FRET vs. temperature measurements on free AN12u and on the AN12u–WT Rad4 complex (SI Appendix, Fig. S7C).
Fig. 4.
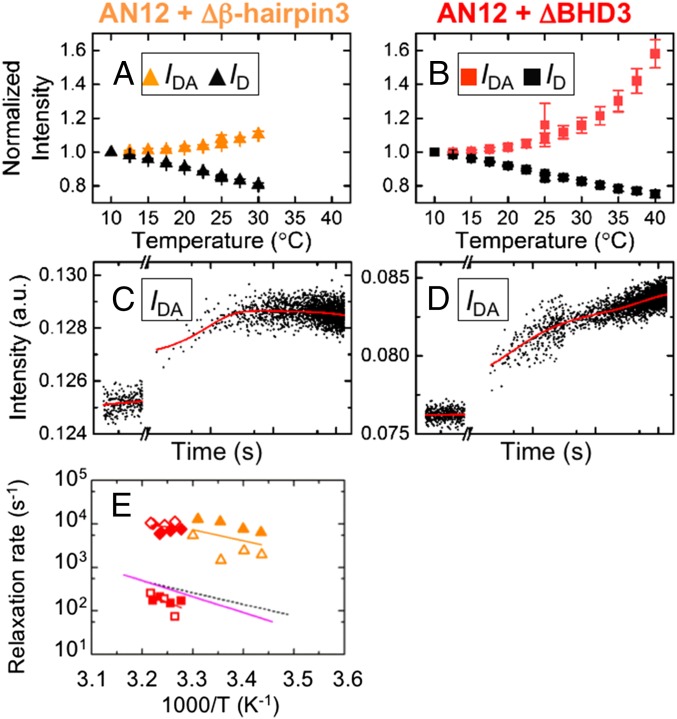
Equilibrium and T-jump measurements on AN12 bound to Rad4 β-hairpin mutants. (A and B) Equilibria IDA for AN12–Δβ-hairpin3 (orange, n = 4) and for AN12–ΔBHD3 (red, n = 4) are shown vs. temperature, with the corresponding ID in black. (C) T-jump relaxation traces with AN12_DA–Δβ-hairpin3 show a single phase: 89 ± 7.4 μs after a T-jump from 18–25 °C. (D) AN12_DA–ΔBHD3 shows two phases: 131 ± 12 μs and 5.8 ± 0.3 ms after a T-jump from 26–32 °C. (E) Relaxation rates of AN12–Δβ-hairpin3 (orange triangles) and AN12–ΔBHD3 (red diamonds for fast phase, red squares for slow phase) vs. inverse temperature from two independent sets of measurements (open/filled symbols). The continuous lines are Arrhenius fits to the rates, with activation enthalpies of 11.9 ± 0.3 kcal/mol for AN12–Δβ-hairpin3 and 3.3 ± 2.4 kcal/mol (fast phase) and 27.5 ± 32.4 kcal/mol (slow phase) for AN12–ΔBHD3. The pink and dashed black lines are from Fig. 3G.
T-jump measurements with the mutants detected no kinetics with the mismatched AN12 in the absence of the acceptor probe (ID from AN12_D) (SI Appendix, Figs. S12B and S13B) or with the undamaged AN12u, as was the case with WT Rad4. However, IDA in mutant-bound AN12_DA showed not only the rapid (<20 μs) premelting kinetics but also unique, protein-induced kinetics in the T-jump window of 20 μs to 32 ms: First, Δβ-hairpin3–bound AN12_DA revealed monophasic relaxation kinetics, 374.0–174.0 μs in the range from 18–29 °C, the highest temperature testable with this mutant (Fig. 4C and SI Appendix, Fig. S12 A and E). These kinetics (227.5 ± 18.2 μs at 25 °C) are about 30-fold faster than the 5- to 10-ms kinetics observed with the same DNA bound to the WT or the 2AP-monitored nucleotide-flipping kinetics. We note that these kinetics were observed despite the lack of detectible differences between the bound and free AN12_DA in equilibrium fluorescence measurements (SI Appendix, Fig. S7 A and B), underscoring the sensitivity of the tCo/tCnitro-based T-jump approach to detect relaxation kinetics with small (<2% change) amplitudes (SI Appendix, SI Methods 1.15 and Fig. S12F). Second, ΔBHD3-bound AN12_DA showed biphasic relaxation kinetics in the temperature range of 30–40 °C (final temperature) but no kinetics below ∼30 °C. The fast component of the biphasic behavior had 110.6- to 118.2-μs relaxation times, and overlapped the relaxation times of AN12–Δβ-hairpin3 (Table 1), whereas the slow component was 4.4–11.1 ms, similar to the 5- to 10-ms relaxation kinetics discussed above (Fig. 4 D and G and SI Appendix, Fig. S13).
Table 1.
Relaxation times for Rad4–DNA complexes at 25 °C and 32 °C
| Sample | T = 25 °C (fast), μs | T = 25 °C (slow), ms | T = 32 °C (fast), μs | T = 32 °C (slow), ms |
| AN12-Rad4 | — | 7.7 ± 0.8 | — | 4.1 ± 0.6 |
| AN12–ΔBHD3 | 113.1 ± 24.0 | nd | 118.2 ± 35.2 | 11.1 ± 7.1 |
| AN12–Δβ-hairpin3 | 227.5 ± 18.2 | — | 142.7 ± 115 | — |
| AN14–Rad4 | 448.4 ± 98.3 | — | 228.1 ± 66.7 | — |
| AN14u–Rad4 | 292.4 ± 173.9 | — | 136.0 ± 25.8 | — |
nd, not determined; T, temperature.
The submillisecond kinetics observed for both Δβ-hairpin3 and ΔBHD3 had similar relaxation times (113.1 μs and 227.5 μs at 25 °C) with closely overlapping Arrhenius trends, which are distinct from the relaxation times observed with the WT (Fig. 4E and Table 1). The low specificity of these mutants to the mismatch lesion in comparison to WT (49) raises the possibility that the predominant conformations in the AN12–mutant Rad4 complexes are of nonspecific modes, and that the fast phase observed in T-jump kinetics with these mutants reflects motions in these nonspecific modes (further discussed below).
It is also noteworthy that the ΔBHD3-bound AN12 showed the slow phase matching the kinetics of DNA opening induced by WT Rad4 in addition to the fast phase. The relative amplitudes of the fast and slow phases were similar (∼60% in the fast phase at 30 °C and ∼50% at 40 °C), whereas the overall amplitude in the two phases combined was similar to the overall amplitude with WT Rad4 (SI Appendix, Fig. S13F). We recall that neither ΔBHD3 nor Δβ-hairpin3 exhibited 2AP-based nucleotide-flipping kinetics (49). Thus, the results suggest that the slow phase in AN12–ΔBHD3 captures the rate-limiting structural distortion along the recognition trajectory, and that this distortion does not require the BHD3 domain, at least at temperatures higher than 30 °C. However, the formation of the fully flipped-out open conformation as monitored by 2AP must still require the insertion of the β-hairpin, which is lacking in these mutants.
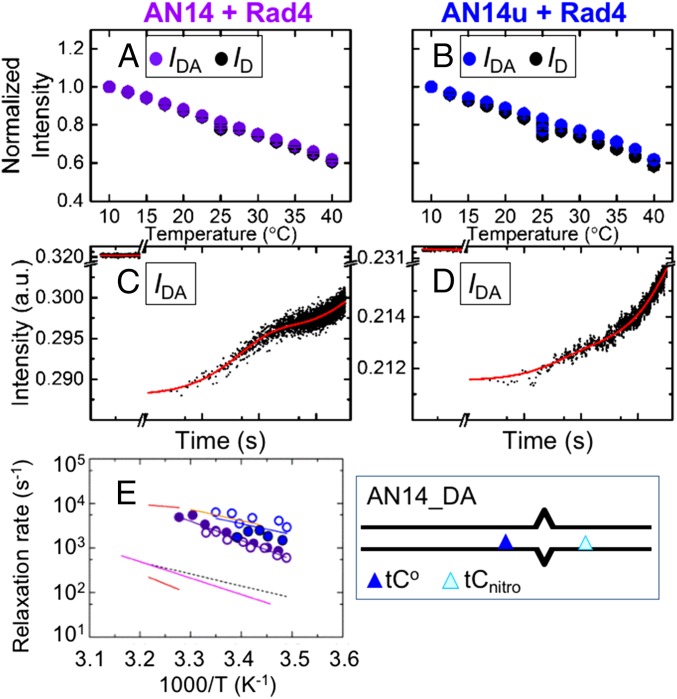
Fast Submillisecond Kinetics Are also Observed with WT Rad4 on Nonspecific DNA Substrates: AN14/AN14u.
As discussed above, the fast submillisecond kinetics observed with AN12 bound to β-hairpin mutants may indicate kinetics resulting from nonspecific binding modes of the mutants. We thus further investigated whether these nonspecific-mode kinetics could be captured with nonspecifically bound WT Rad4, particularly if the probes were positioned further away from the mismatch. Moving the probes further apart has two advantages: (i) the probes are less sensitive to locally enhanced dynamics in the immediate vicinity of the mismatch site, for example, from premelting of the mismatch as seen in AN12_DA, and (ii) nonspecific distortions may involve longer range conformational changes in DNA that are better picked up when the probes are positioned further apart. Thus, in the new DNA substrates, AN14 and AN14u, we placed both FRET probes 2 bp away from either side of the 3-bp mismatch (Fig. 5 A and B and SI Appendix, Fig. S1). In AN14, we also incorporated the TAT/TAT mismatch instead of the TTT/TTT mismatch in AN12. Although the TAT/TAT mismatch destabilizes the DNA duplexes by ∼7–8 °C (SI Appendix, Fig. S2 E–H) in a manner similar to the 8–11 °C destabilization caused by the TTT/TTT mismatch, AN14 (in contrast to AN12) does not show specific binding to Rad4 compared with other undamaged DNA constructs, including its own matched version, AN14u (SI Appendix, Fig. S14). We further confirm that AN14’s low specificity is due to its mismatch sequence rather than the presence of the FRET labels: tCo/tCnitro-labeled AN14/AN14u exhibited Rad4-binding affinities similar to the Rad4-binding affinities of unlabeled constructs (SI Appendix, Fig. S14), as was also the case with AN12/AN12u. We posit that TAT/TAT mismatch could perhaps mimic lesions such as cyclobutane pyrimidine dimer (CPD): A CPD lesion also lowers the melting temperature of the duplex DNA (68) but is not specifically recognized by XPC in vitro (53).
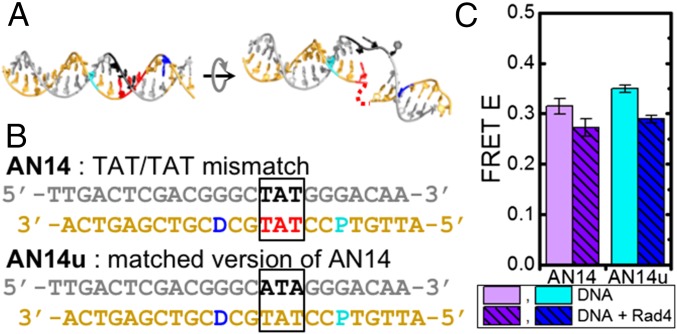
Fig. 5.
AN14 and AN14u DNA constructs. DNA models (A) and sequences (B) are shown for AN14 and AN14u, as in Fig. 2 A and B. (C) FRET at 25 °C in free AN14 (light purple, n = 12), AN14–Rad4 (purple, n = 6), free AN14u (cyan, n = 13), and AN14u–Rad4 (blue, n = 7).
The FRET values of free AN14 and AN14u in equilibrium were very close to the calculated values assuming B-DNA conformations, again indicating similar average B-DNA conformations for both constructs (Fig. 5C and SI Appendix, Fig. S15A). The FRET of AN14 decreased from 0.315 ± 0.015 in free DNA to 0.272 ± 0.018 upon Rad4 binding at 25 °C (Fig. 5C and SI Appendix, Fig. S15A). Interestingly, AN14u also showed a decrease in FRET from 0.350 ± 0.008 in free DNA to 0.289 ± 0.007 upon Rad4 binding, similar to the decrease in FRET seen with AN14, indicating that Rad4 induces similar conformational changes in AN14 and AN14u. Because Rad4 exhibits little specificity toward these DNA constructs, this decrease in FRET is primarily ascribed to DNA deformations induced by nonspecific interactions of the protein with DNA. The direction of these FRET changes is consistent with the direction calculated for conversion from the B-DNA conformation to the conformation of the specific open complex (SI Appendix, Fig. S15A); these results suggest that even in these nonspecific complexes, the bound DNA may be distorted in a manner that shares structural resemblances to the open conformation, for example, by unwinding and/or bending partway to the full open complex.
Equilibrium fluorescence measurements as a function of temperature showed a nearly identical decrease in both ID and IDA values in both AN14 and AN14u, with and without Rad4, as the temperature was increased from 10–40 °C (Fig. 6 A and B and SI Appendix, Fig. S16 A and B). These data correspond to a much smaller change in FRET as the temperature is raised, thus indicating little change in the equilibrium population distribution of DNA conformations sensed by the probes over this temperature range (SI Appendix, Fig. S15 B and C). However, the outstanding sensitivity of the T-jump approach, as also demonstrated with AN12 bound to Δβ-hairpin3, enabled us to observe the relaxation kinetics in both AN14 and AN14u when bound to Rad4.
Fig. 6.
Equilibrium and T-jump measurements on AN14 and AN14u bound to WT Rad4. (A and B) Equilibria IDA for AN14–Rad4 (purple, n = 5) and for AN14u–Rad4 (blue, n = 4) are shown vs. temperature with the corresponding ID in black. T-jump relaxation traces with AN14_DA–Rad4 (C) and AN14u_DA–Rad4 (D) show relaxation times of 603 ± 35 μs and 575 ± 114 μs, respectively, after a T-jump from 15–22 °C. (E) Relaxation rates of AN14–Rad4 (purple) and AN14u–Rad4 (blue) vs. inverse temperature from two independent sets of measurements (open/filled symbols). The continuous lines are Arrhenius fits to the rates, with activation enthalpies of 18.2 ± 2.2 kcal/mol (AN14–Rad4) and 11.6 ± 3.3 kcal/mol (AN14u–Rad4). The pink, orange, red, and dashed black lines are from Fig. 4E. A schematic representation of the nonspecific mismatched AN14_DA construct design is also shown.
In T-jump experiments with AN14, relaxation kinetics (besides the typical, initial rapid drop and the T-jump recovery) were observed only with AN14_DA bound to Rad4 (Fig. 6C and SI Appendix, Fig. S17A), but not in the absence of the acceptor (SI Appendix, Fig. S17 B and C) or the protein (SI Appendix, Fig. S16 C and E). These results therefore capture the conformational dynamics of AN14 induced by Rad4. Interestingly, the relaxation kinetics on AN14_DA bound to Rad4 occurred with a characteristic time constant of 448.4 ± 98.8 μs at 25 °C (Fig. 6E), similar to the submillisecond kinetic phase previously observed with AN12 bound to Δβ-hairpin3 and ΔBHD3 (Fig. 4E). Notably, similar relaxation kinetics (292.4 ± 173.9 μs at 25 °C) were also observed with undamaged AN14u_DA bound to Rad4 (Fig. 6 D and G and SI Appendix, Fig. S18 A and G), although the amplitudes for AN14u–Rad4 were smaller than for AN14–Rad4 (SI Appendix, Figs. S17H and S18H). Again, no relaxation kinetics other than the initial rapid drop and slow T-jump recovery were observed in the absence of the acceptor (SI Appendix, Fig. S18 B and C) or the protein (SI Appendix, Fig. S16 D and F). These results suggest that the 292- to 448-μs kinetics observed with WT Rad4 on the AN14/AN14u DNA substrates (Table 1) reflect the same nonspecific mode dynamics that underlie the 113- to 228-μs kinetics observed when the low-specificity Rad4 mutants were bound to a specific substrate, AN12.
Finally, we note that no relaxation kinetics with Δβ-hairpin3 or ΔBHD3 were observed on AN14 or AN14u. The amplitudes of the relaxation kinetics on AN14/AN14u–Rad4 were already significantly smaller than the amplitudes on AN12–Rad4, and at the limit of detection of the current setup (AN12, SI Appendix, Fig. S9H; AN14/AN14u, SI Appendix, Figs. S17H and S18H). Any conformational changes induced by the mutants in AN14/AN14u were thus likely to be below the detection limit. Similar reasons can be attributed to the absence of kinetics on AN12u–Rad4 (SI Appendix, SI Discussion 2.2).
Discussion
Although previous studies have shown that Rad4/XPC must sense diverse NER lesions in a uniquely indirect manner, the sequence and dynamics of the structural changes in Rad4 and DNA that lead to recognition have remained elusive. By combining the tCo/tCnitro FRET pair with microsecond-resolved T-jump spectroscopy, we have uncovered novel conformational dynamics in DNA during the damage search/recognition process: (i) a slow (∼5–10 ms) phase representing the rate-limiting step of the full opening kinetics and (ii) a fast (∼100–500 μs) phase reflecting collective DNA deformations during nonspecific interactions, which we interpret as DNA twisting (discussed below). The relative amplitudes of the two phases vary depending on the presence/absence of the β-hairpin or the BHD3 domain of Rad4 and the location of the probes in DNA as well as DNA sequences. Altogether, these results provide keys to constructing the lesion recognition trajectory of Rad4.
I. Slow Phase: The “Bottleneck” for Forming the Open Conformation Does Not Require β-Hairpin Insertion.
The mismatched AN12 that exhibited specific binding to Rad4 showed ∼8-ms kinetics when bound to WT Rad4. This slow phase is concurrent with the previously reported ∼5- to 10-ms kinetics of the Rad4-induced DNA opening, probed at specific mismatch sites by 2AP (49). Thus, we propose that the slow phase reflects the kinetics of overcoming the rate-limiting transition state ensemble (bottleneck) during the DNA opening process. However, despite a common bottleneck, the structural distortions captured by tCo/tCnitro in AN12 are not necessarily the same as those structural distortions reported by the 2AP probe: Whereas the slow phase probed by tCo/tCnitro (AN12) could be observed at least with ΔBHD3 mutant at higher temperatures, no such kinetics were observed in either of the β-hairpin mutants by 2AP (49). We positioned the 2AP as one of the flipped-out damaged/mismatched nucleotides (49), and thus assigned the 2AP-probed motions to full flipping of the nucleotides for which the β-hairpin is required (49). On the other hand, the tCo/tCnitro FRET probes, especially in AN12, where the acceptor flanks the mismatch site, may be sensing distortions/unwinding around that site even with the mismatched nucleotides still intrahelical. The ΔBHD3 mutant, despite the lack of the entire β-hairpin domain, could induce the rate-limiting distortions sensed by tCo/tCnitro probes at >30 °C, although incapable of the 2AP-sensed, full flipping at the same temperatures. (Whether Δβ-hairpin3 could also induce the bottleneck distortions at these higher temperatures could not be tested because of instability of this mutant above 35 °C.) We conclude that full nucleotide flipping that engages the β-hairpin occurs after the rate-limiting distortion or partial opening on the path to the fully open conformation. We envision this partially open conformation as one that entails a significantly unwound DNA structure with disrupted hydrogen bonding and unstacking, but with the nucleotides at the lesion site still mostly intrahelical. The late involvement of the β-hairpin during Rad4-induced DNA opening is also consistent with our observation that the preliminary interrogation step also does not require the β-hairpin (discussed below).
II. Fast Phase: Rapid DNA Deformations, Such as Unwinding (Twisting), Are Induced by Nonspecific Interrogation of DNA by Rad4.
The ∼100- to 500-μs fast phase is more than 10-fold faster than the slow phase. It was observed both with β-hairpin deletion mutants bound to AN12 and with WT protein bound to AN14 and AN14u, but not in free DNA. These protein–DNA complexes share nonspecific binding as a common feature, either due to a low-specificity mutant protein or a low-specificity DNA construct. We therefore interpret the rapid dynamics as arising from conformational fluctuations during nonspecific interactions with the protein. The nonspecific complexes are inherently heterogeneous in their structures and conformations because Rad4 can bind to DNA in multiple registers or orientations. Despite this inherent heterogeneity and the differences in the location or the sequence context of the probes in AN12/AN14/AN14u, we saw a single relaxation phase for the fast dynamics with approximately the same Arrhenius behavior. The results thus indicate that (i) the intrinsic energetic differences in distorting this DNA by the protein must have been removed upon formation of the complex, apparently by using the binding energy, and (ii) these dynamics must involve motions of comparable free-energy barriers that are smaller than the distortions during the slower rate-limiting step (Fig. 6E). Based on the sensitivity of the FRET probes to DNA helicity and a comparison of the Rad4-bound DNA and B-form structures (54), we attribute the collective nonspecific mode dynamics primarily to partial unwinding/rewinding fluctuations (twisting) of DNA during nonspecific search and interrogation interactions (discussed in next section), although we cannot exclude the possibility that other motions, such as DNA bending, may also contribute. In addition, the fast dynamics did not require the β-hairpin or BHD3 in contrast to the 2AP-monitored full nucleotide-flipping kinetics. Thus, the nonspecifically “twisted” structures may retain intrahelical conformations of nucleotides in DNA. Our results mark the first direct observation, to our knowledge, of DNA conformational fluctuations captured during nonspecific interrogation by a large protein complex among DNA repair proteins or among “indirect-readout” DNA-binding proteins.
Rapid Twisting May Represent Conformational Switching Between Search and Interrogation Modes, en Route to Damage Recognition.
How site-specific DNA-binding proteins are able to scan the genome rapidly while reliably locating and recognizing their respective binding sites has been dubbed as the “search-speed stability” paradox (23). Previous studies to resolve the problem have invoked two modes of protein–DNA binding: the search mode and the recognition mode (23–26). The search mode is presumed to have few stabilizing protein–DNA interactions, allowing rapid 1D diffusion of protein on DNA in a relatively smooth energy landscape and/or dissociation from DNA. However, the lack of specific interactions in the search mode comes at a price in that the protein has limited ability to sense/probe the presence of the target site. In the recognition mode, on the other hand, the protein assumes a conformation that resembles its conformation in the final recognition complex; in this mode, it can make more extensive protein–DNA contacts, allowing the protein to test the targets with increased accuracy/specificity. It has been proposed that an efficient, stochastic interchange between the two modes enables the resolution of the speed-stability paradox in searching and recognition.
Consistent with these notions, various studies have shown evidence of multiple binding modes in nonspecific protein–DNA complexes (13, 27–39, 69, 70). The structures of DNA repair glycosylases captured on nonspecific DNA showed severely kinked conformations of DNA even on those nontarget sites (27, 29–31, 34, 35). Although enabling specific probing of a potential target lesion, such distortions seem incompatible with fast sliding/scanning of the protein on DNA, as anticipated in the search mode. These complexes have been termed interrogation complexes distinct from a search complex but lying along the conformational trajectory toward forming the lesion-specific, recognition complex (25). NMR analyses of lac repressor and uracil DNA glycosylase bound to nonspecific DNA have revealed large-scale, submillisecond conformational fluctuations in the proteins that are potential candidates for such search/interrogation conformational switches (28, 36). However, direct measurements of corresponding DNA dynamics to account for such switches in the submillisecond regime have been limited to a few NMR imino–proton exchange experiments that detect base pair “breathing” (37, 71, 72). Although previous stopped-flow studies on DNA glycosylases have indicated multiple kinetic steps for damage recognition and repair (21, 22), these observed kinetics are too slow to capture interrogation compatible with 1D diffusion and are likely probing additional steps leading to recognition and catalysis.
Our study directly revealed that there are at least two distinct kinetic phases: a fast nonspecific mode that we ascribe to twisting and a slow opening. The fast phase, we propose, reflects conformational interconversions between a rapidly diffusing search and a slowly diffusing or momentarily stalled interrogation mode, whereas the slow phase reflects the recognition step (i.e., the conversion from the search/interrogation to the final open conformation). The fast twisting may represent “search/interrogation” not only for proteins bound to nonspecific sites but also for those proteins on specific sites, and thus lie along the specific multistep lesion recognition trajectory (Fig. 7). The following observations support this interpretation: (i) Our previous crystal structure of undamaged DNA covalently tethered to Rad4 showed the same open conformation as damaged DNA specifically bound by Rad4 (49), indicating that the general process of opening may not be structurally different for the damaged and undamaged DNA; (ii) the FRET changes induced by nonspecific Rad4 binding to AN14/AN14u are in the same direction as the FRET changes expected in a conformational change from “straight” B-DNA to the open conformation; and (iii) both the fast and the slow phases are observable simultaneously in AN12–ΔBHD3. The fact that we observe the fast phase only in nonspecific or low specificity complexes and not in the specific AN12–Rad4 complex is not inconsistent with our proposed scheme. A T-jump perturbation of a specific complex may not populate these nonspecific modes sufficiently to detect the fast phase (SI Appendix, SI Discussion 2.3).
Fig. 7.
Conformational trajectory of lesion recognition by Rad4. (A) Three distinct binding modes for Rad4/XPC as it searches for, interrogates, and recognizes a damaged site and the time scales for fluctuations between these modes are shown. (B, Top) A free-energy profile that may underlie the observed kinetics along the recognition trajectory. Damaged DNA in nonspecific binding modes is shown with inherently higher free energy than undamaged DNA due to the damaged/induced destabilization. The 100- to 500-μs twisting step is depicted with a smaller energetic barrier than the 5- to 10-ms rate-limiting distortion step (‡), which is followed by rapid β-hairpin insertion and full-nucleotide flipping. (B, Bottom) Putative diffusional landscapes of the protein along DNA are illustrated along coordinates orthogonal to the conformational trajectory. As the recognition proceeds, the diffusional landscape gets progressively rough; once the β-hairpin is inserted, the protein is practically obstructed from diffusing away, and thus recognition.
Interrogation May Promote Kinetic Gating by Stalling the Protein Preferentially at Damaged Sites.
Previously, we have proposed a kinetic gating model for damage recognition by XPC/Rad4 (49). In this model, discrimination between normal and damaged DNA sites is determined by the differential probabilities of the Rad4-induced DNA opening of a DNA site before it diffuses away from the site. Residence times reported for damage repair proteins on normal, nonspecific DNA range from 0.1–300 μs (7, 9, 10, 13). Thus, as previously argued, it has not been clear how Rad4 can efficiently open to recognize damaged DNA, which takes ∼5–10 ms, if Rad4 should indeed exhibit the microsecond-regime residence times per DNA site during the nonspecific search. The interrogation mode of Rad4 revealed in this study provides critical insights into bridging this gap between fast diffusion and slow recognition: The increased interactions between the protein and the DNA in the interrogation mode may work to slow down the diffusion of XPC/Rad4 on DNA, thus increasing the probability that it can open a given site, according to the kinetic gating mechanism (Fig. 7). Rapidly diffusing proteins on DNA switching to slow diffusers have been reported in single-molecule studies (40). It is tempting to speculate that abnormal DNA structures and dynamics induced by NER lesions may promote the conversion from search to interrogation in comparison to normal DNA, and also further stall XPC/Rad4 when in the interrogation mode, ultimately to allow residence times in the millisecond regime. Not inconsistent with this notion, the nonspecific twisting rates for undamaged AN14u were slightly faster than the nonspecific twisting rates for mismatched AN14 (Fig. 6E), indicating that Rad4 may engage more extensively with the AN14 mismatch site than normal sites. These results suggest that the free-energy barriers that a nonspecifically engaged Rad4 experiences may indeed differ depending on the nature of the DNA sequences and structures, which may ultimately contribute to its specific ability to recognize NER lesions. Additionally, it is also conceivable that some DNA-stabilizing lesions may present a significantly high barrier even for twisting, leading to a perpetual search mode at the lesion site that impairs recognition, and thus repair.
In summary, we conclude that Rad4 interrogates DNA by twisting DNA on 100- to 500-μs time scales. Such a mechanism provides a compelling account for how Rad4 may examine DNA while also diffusing on DNA, and thus reconciles rapid searching with reliable recognition. Our T-jump approach that also took advantage of the FRET probes uniquely sensitive to unwinding (twisting) of DNA was critical in unveiling these submillisecond dynamics. A rapid (∼100 μs) nonspecific interrogation preceding a slow (∼10 ms) target recognition has previously also been observed in T-jump studies of the integration host factor, a DNA-bending bacterial architectural protein (58, 73). Thus, interrogation on time scales of hundreds of microseconds compatible with rapid diffusion of proteins on DNA may well be a common feature during target recognition by diverse DNA-binding proteins. In general, the multistep recognition process through dynamic conformational changes would allow proteins to span a wide range of residence times on DNA that can be fine-tuned at each successive step. The actual diffusion landscapes of XPC/Rad4 on DNA in each binding mode, and the structural and kinetic parameters that control each step along the recognition trajectory, remain as fascinating problems to be explored.
Materials and Methods
Materials.
HPLC-purified DNA oligonucleotides were purchased from TriLink Biotechnologies and were annealed as described in SI Appendix, SI Methods 1.1. Uniformly sized, homogeneous Rad4–Rad23 complexes were prepared as described (49, 54) (SI Appendix, SI Methods 1.2 and Fig. S19). Rad4–Rad23–DNA complexes were prepared by combining purified Rad4–Rad23 and annealed duplex DNA substrates in a 1:1 molar ratio in PBS buffer containing 1 mM DTT.
Equilibrium FRET Measurements.
Fluorescence emission spectra of samples with 10 μM tCo- or tCo/tCnitro-labeled DNA duplexes in the absence and presence of equimolar proteins were obtained in the range 375–550 nm, with excitation at 365 nm, using a FluoroMax4 spectrofluorometer (JobinYvon, Inc.). The donor fluorescence intensities (ID or IDA) were computed as described in SI Appendix, SI Methods 1.7; the FRET efficiency (E) between the tCo/tCnitro probes was calculated as E .
Laser T-Jump Spectroscopy.
The home-built laser T-jump spectrometer (47, 74) uses 10-ns laser pulses at 1,550 nm, generated by Raman-shifting the 1,064-nm pulses from the output of a neodymium/yttrium aluminium garnet laser, which are focused to an ∼1-mm spot size onto a 2-mm-wide sample cuvette with a path length of 0.5 mm; each laser pulse yields ∼5–10 °C of T-jump at the center of the heated volume. The probe source for excitation of tCo was a 200-W Hg-Xe lamp, with the excitation wavelengths (335–375 nm) selected by a broadband filter and focused to an ∼300-μm spot in the middle of the heated volume. The fluorescence emission was monitored perpendicular to the excitation direction using a photomultiplier tube and a 500-MHz transient digitizer. For each measurement, ∼500 kinetics traces were acquired and averaged. Further details are provided in SI Appendix, SI Methods 1.8–1.15. Protein–DNA 1:1 complex concentrations were 20 μM or 40 μM for tCo/tCnitro-labeled samples and 4 μM or 8 μM for tCo-only samples.
Supplementary Material
Acknowledgments
We dedicate this article to the memory of Kamal Shukla, a staunch advocate for basic research in molecular biophysics. A major part of the research in this article was supported by his program at National Science Foundation (NSF), and his encouragement over the years has been invaluable. We thank Marcus Wilhelmsson and Søren Preus for their help in designing the tCo/tCnitro-labeled DNA and in calculating the expected FRET values, and Peter J. Steinbach for his maximum entropy analysis algorithm. We thank Suse Broyde, Bennett Van Houten, and L. James Maher III, for their careful reading of the manuscript and helpful comments. This work was funded by NSF Grants MCB-0721937 and MCB-1158217 (to A.A.) and MCB-1412692 (to J.-H.M.), the Chancellor’s Discovery Fund (to A.A. and J.-H.M.), and a startup fund from the University of Illinois at Chicago (to J.-H.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1514666113/-/DCSupplemental.
References
- 1.Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970;53(3):401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- 2.Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor--operator interaction: Kinetic measurements and conclusions. Biochemistry. 1981;20(24):6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 3.Stanford NP, Szczelkun MD, Marko JF, Halford SE. One- and three-dimensional pathways for proteins to reach specific DNA sites. EMBO J. 2000;19(23):6546–6557. doi: 10.1093/emboj/19.23.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32(10):3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gowers DM, Wilson GG, Halford SE. Measurement of the contributions of 1D and 3D pathways to the translocation of a protein along DNA. Proc Natl Acad Sci USA. 2005;102(44):15883–15888. doi: 10.1073/pnas.0505378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Widom J. Target site localization by site-specific, DNA-binding proteins. Proc Natl Acad Sci USA. 2005;102(47):16909–16910. doi: 10.1073/pnas.0508686102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schonhoft JD, Stivers JT. Timing facilitated site transfer of an enzyme on DNA. Nat Chem Biol. 2012;8(2):205–210. doi: 10.1038/nchembio.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang YM, Austin RH, Cox EC. Single molecule measurements of repressor protein 1D diffusion on DNA. Phys Rev Lett. 2006;97(4):048302. doi: 10.1103/PhysRevLett.97.048302. [DOI] [PubMed] [Google Scholar]
- 9.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci USA. 2006;103(15):5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorman J, et al. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Mol Cell. 2007;28(3):359–370. doi: 10.1016/j.molcel.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tafvizi A, et al. Tumor suppressor p53 slides on DNA with low friction and high stability. Biophys J. 2008;95(1):L01–L03. doi: 10.1529/biophysj.108.134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonnet I, et al. Sliding and jumping of single EcoRV restriction enzymes on non-cognate DNA. Nucleic Acids Res. 2008;36(12):4118–4127. doi: 10.1093/nar/gkn376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson SR, Dunn AR, Kathe SD, Warshaw DM, Wallace SS. Two glycosylase families diffusively scan DNA using a wedge residue to probe for and identify oxidatively damaged bases. Proc Natl Acad Sci USA. 2014;111(20):E2091–E2099. doi: 10.1073/pnas.1400386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263(5148):777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 15.Garvie CW, Wolberger C. Recognition of specific DNA sequences. Mol Cell. 2001;8(5):937–946. doi: 10.1016/s1097-2765(01)00392-6. [DOI] [PubMed] [Google Scholar]
- 16.Lawson CL, Berman HM. Indirect readout of DNA sequence by proteins. In: Rice PA, Correll CC, editors. Protein-Nucleic Acid Interactions. Royal Society of Chemistry; Cambridge, UK: 2008. [Google Scholar]
- 17.Stivers JT, Pankiewicz KW, Watanabe KA. Kinetic mechanism of damage site recognition and uracil flipping by Escherichia coli uracil DNA glycosylase. Biochemistry. 1999;38(3):952–963. doi: 10.1021/bi9818669. [DOI] [PubMed] [Google Scholar]
- 18.van den Broek B, Noom MC, Wuite GJ. DNA-tension dependence of restriction enzyme activity reveals mechanochemical properties of the reaction pathway. Nucleic Acids Res. 2005;33(8):2676–2684. doi: 10.1093/nar/gki565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugimura S, Crothers DM. Stepwise binding and bending of DNA by Escherichia coli integration host factor. Proc Natl Acad Sci USA. 2006;103(49):18510–18514. doi: 10.1073/pnas.0608337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuznetsov SV, Sugimura S, Vivas P, Crothers DM, Ansari A. Direct observation of DNA bending/unbending kinetics in complex with DNA-bending protein IHF. Proc Natl Acad Sci USA. 2006;103(49):18515–18520. doi: 10.1073/pnas.0608394103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koval VV, Kuznetsov NA, Ishchenko AA, Saparbaev MK, Fedorova OS. Real-time studies of conformational dynamics of the repair enzyme E. coli formamidopyrimidine-DNA glycosylase and its DNA complexes during catalytic cycle. Mutat Res. 2010;685(1-2):3–10. doi: 10.1016/j.mrfmmm.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 22.Kuznetsov NA, et al. Conformational Dynamics of DNA Repair by Escherichia coli Endonuclease III. J Biol Chem. 2015;290(23):14338–14349. doi: 10.1074/jbc.M114.621128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slutsky M, Mirny LA. Kinetics of protein-DNA interaction: Facilitated target location in sequence-dependent potential. Biophys J. 2004;87(6):4021–4035. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savir Y, Tlusty T. Conformational proofreading: the impact of conformational changes on the specificity of molecular recognition. PLoS One. 2007;2(5):e468. doi: 10.1371/journal.pone.0000468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman JI, Stivers JT. Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry. 2010;49(24):4957–4967. doi: 10.1021/bi100593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou HX. Rapid search for specific sites on DNA through conformational switch of nonspecifically bound proteins. Proc Natl Acad Sci USA. 2011;108(21):8651–8656. doi: 10.1073/pnas.1101555108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wibley JE, Waters TR, Haushalter K, Verdine GL, Pearl LH. Structure and specificity of the vertebrate anti-mutator uracil-DNA glycosylase SMUG1. Mol Cell. 2003;11(6):1647–1659. doi: 10.1016/s1097-2765(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 28.Kalodimos CG, et al. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305(5682):386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 29.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434(7033):612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 30.Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science. 2006;311(5764):1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 31.Banerjee A, Verdine GL. A nucleobase lesion remodels the interaction of its normal neighbor in a DNA glycosylase complex. Proc Natl Acad Sci USA. 2006;103(41):15020–15025. doi: 10.1073/pnas.0603644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwahara J, Zweckstetter M, Clore GM. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc Natl Acad Sci USA. 2006;103(41):15062–15067. doi: 10.1073/pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440(7088):1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 34.Maiti A, Morgan MT, Pozharski E, Drohat AC. Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition. Proc Natl Acad Sci USA. 2008;105(26):8890–8895. doi: 10.1073/pnas.0711061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi Y, et al. Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature. 2009;462(7274):762–766. doi: 10.1038/nature08561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedman JI, Majumdar A, Stivers JT. Nontarget DNA binding shapes the dynamic landscape for enzymatic recognition of DNA damage. Nucleic Acids Res. 2009;37(11):3493–3500. doi: 10.1093/nar/gkp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker JB, et al. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature. 2007;449(7161):433–437. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leith JS, et al. Sequence-dependent sliding kinetics of p53. Proc Natl Acad Sci USA. 2012;109(41):16552–16557. doi: 10.1073/pnas.1120452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghodke H, et al. Single-molecule analysis reveals human UV-damaged DNA-binding protein (UV-DDB) dimerizes on DNA via multiple kinetic intermediates. Proc Natl Acad Sci USA. 2014;111(18):E1862–E1871. doi: 10.1073/pnas.1323856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cuculis L, Abil Z, Zhao H, Schroeder CM. Direct observation of TALE protein dynamics reveals a two-state search mechanism. Nat Commun. 2015;6:7277. doi: 10.1038/ncomms8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kubelka J. Time-resolved methods in biophysics. 9. Laser temperature-jump methods for investigating biomolecular dynamics. Photochem Photobiol Sci. 2009;8(4):499–512. doi: 10.1039/b819929a. [DOI] [PubMed] [Google Scholar]
- 42.Thompson PA, Eaton WA, Hofrichter J. Laser temperature jump study of the helix<==>coil kinetics of an alanine peptide interpreted with a ‘kinetic zipper’ model. Biochemistry. 1997;36(30):9200–9210. doi: 10.1021/bi9704764. [DOI] [PubMed] [Google Scholar]
- 43.Qiu L, Pabit SA, Roitberg AE, Hagen SJ. Smaller and faster: The 20-residue Trp-cage protein folds in 4 micros. J Am Chem Soc. 2002;124(44):12952–12953. doi: 10.1021/ja0279141. [DOI] [PubMed] [Google Scholar]
- 44.Yang WY, Gruebele M. Folding at the speed limit. Nature. 2003;423(6936):193–197. doi: 10.1038/nature01609. [DOI] [PubMed] [Google Scholar]
- 45.Kubelka J, Henry ER, Cellmer T, Hofrichter J, Eaton WA. Chemical, physical, and theoretical kinetics of an ultrafast folding protein. Proc Natl Acad Sci USA. 2008;105(48):18655–18662. doi: 10.1073/pnas.0808600105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narayanan R, Velmurugu Y, Kuznetsov SV, Ansari A. Fast folding of RNA pseudoknots initiated by laser temperature-jump. J Am Chem Soc. 2011;133(46):18767–18774. doi: 10.1021/ja205737v. [DOI] [PubMed] [Google Scholar]
- 47.Kuznetsov SV, Kozlov AG, Lohman TM, Ansari A. Microsecond dynamics of protein-DNA interactions: Direct observation of the wrapping/unwrapping kinetics of single-stranded DNA around the E. coli SSB tetramer. J Mol Biol. 2006;359(1):55–65. doi: 10.1016/j.jmb.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 48.Vivas P, Velmurugu Y, Kuznetsov SV, Rice PA, Ansari A. Mapping the transition state for DNA bending by IHF. J Mol Biol. 2012;418(5):300–315. doi: 10.1016/j.jmb.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 49.Chen X, et al. Kinetic gating mechanism of DNA damage recognition by Rad4/XPC. Nat Commun. 2015;6:5849. doi: 10.1038/ncomms6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anunciado D, Dhar A, Gruebele M, Baranger AM. Multistep kinetics of the U1A-SL2 RNA complex dissociation. J Mol Biol. 2011;408(5):896–908. doi: 10.1016/j.jmb.2011.02.054. [DOI] [PubMed] [Google Scholar]
- 51.Schärer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5(10):a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gillet LC, Schärer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev. 2006;106(2):253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 53.Sugasawa K, et al. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001;15(5):507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449(7162):570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 55.Zheng H, et al. Base flipping free energy profiles for damaged and undamaged DNA. Chem Res Toxicol. 2010;23(12):1868–1870. doi: 10.1021/tx1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Börjesson K, et al. Nucleic acid base analog FRET-pair facilitating detailed structural measurements in nucleic acid containing systems. J Am Chem Soc. 2009;131(12):4288–4293. doi: 10.1021/ja806944w. [DOI] [PubMed] [Google Scholar]
- 57.Shi Y, et al. Mammalian transcription factor A is a core component of the mitochondrial transcription machinery. Proc Natl Acad Sci USA. 2012;109(41):16510–16515. doi: 10.1073/pnas.1119738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vivas P. Mechanism of Integration Host Factor, a DNA-Bending Protein, Probed with Laser Temperature-Jump. University of Illinois at Chicago; Chicago: 2009. [Google Scholar]
- 59.Preus S, Börjesson K, Kilså K, Albinsson B, Wilhelmsson LM. Characterization of nucleobase analogue FRET acceptor tCnitro. J Phys Chem B. 2010;114(2):1050–1056. doi: 10.1021/jp909471b. [DOI] [PubMed] [Google Scholar]
- 60.Iqbal A, et al. Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proc Natl Acad Sci USA. 2008;105(32):11176–11181. doi: 10.1073/pnas.0801707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ranjit S, Gurunathan K, Levitus M. Photophysics of backbone fluorescent DNA modifications: Reducing uncertainties in FRET. J Phys Chem B. 2009;113(22):7861–7866. doi: 10.1021/jp810842u. [DOI] [PubMed] [Google Scholar]
- 62.Hey T, et al. The XPC-HR23B complex displays high affinity and specificity for damaged DNA in a true-equilibrium fluorescence assay. Biochemistry. 2002;41(21):6583–6587. doi: 10.1021/bi012202t. [DOI] [PubMed] [Google Scholar]
- 63.Trego KS, Turchi JJ. Pre-steady-state binding of damaged DNA by XPC-hHR23B reveals a kinetic mechanism for damage discrimination. Biochemistry. 2006;45(6):1961–1969. doi: 10.1021/bi05196t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Preus S, Kilså K, Miannay FA, Albinsson B, Wilhelmsson LM. FRETmatrix: A general methodology for the simulation and analysis of FRET in nucleic acids. Nucleic Acids Res. 2013;41(1):e18. doi: 10.1093/nar/gks856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sandin P, et al. Characterization and use of an unprecedentedly bright and structurally non-perturbing fluorescent DNA base analogue. Nucleic Acids Res. 2008;36(1):157–167. doi: 10.1093/nar/gkm1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Preus S, Kilså K, Wilhelmsson LM, Albinsson B. Photophysical and structural properties of the fluorescent nucleobase analogues of the tricyclic cytosine (tC) family. Phys Chem Chem Phys. 2010;12(31):8881–8892. doi: 10.1039/c000625d. [DOI] [PubMed] [Google Scholar]
- 67.Fei J, et al. Regulation of nucleotide excision repair by UV-DDB: Prioritization of damage recognition to internucleosomal DNA. PLoS Biol. 2011;9(10):e1001183. doi: 10.1371/journal.pbio.1001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jing Y, Kao JF, Taylor JS. Thermodynamic and base-pairing studies of matched and mismatched DNA dodecamer duplexes containing cis-syn, (6-4) and Dewar photoproducts of TT. Nucleic Acids Res. 1998;26(16):3845–3853. doi: 10.1093/nar/26.16.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Townson SA, Samuelson JC, Bao Y, Xu SY, Aggarwal AK. BstYI bound to noncognate DNA reveals a “hemispecific” complex: Implications for DNA scanning. Structure. 2007;15(4):449–459. doi: 10.1016/j.str.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 70.Melero R, et al. Electron microscopy studies on the quaternary structure of p53 reveal different binding modes for p53 tetramers in complex with DNA. Proc Natl Acad Sci USA. 2011;108(2):557–562. doi: 10.1073/pnas.1015520107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao C, Jiang YL, Stivers JT, Song F. Dynamic opening of DNA during the enzymatic search for a damaged base. Nat Struct Mol Biol. 2004;11(12):1230–1236. doi: 10.1038/nsmb864. [DOI] [PubMed] [Google Scholar]
- 72.Cao C, Jiang YL, Krosky DJ, Stivers JT. The catalytic power of uracil DNA glycosylase in the opening of thymine base pairs. J Am Chem Soc. 2006;128(40):13034–13035. doi: 10.1021/ja062978n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ansari A, Kuznetsov SV. Dynamics and mechanism of DNA-bending proteins in binding site recognition. In: Williams MC, Maher LJ, editors. Biophysics of DNA-Protein Interactions. Springer; New York: 2010. pp. 107–142. [Google Scholar]
- 74.Vivas P, Kuznetsov SV, Ansari A. New insights into the transition pathway from nonspecific to specific complex of DNA with Escherichia coli integration host factor. J Phys Chem B. 2008;112(19):5997–6007. doi: 10.1021/jp076042s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.