Abstract
All cells possess surveillance and homeostatic mechanisms to adjust protein biogenesis to the demands of growth, differentiation, ageing and environmental stress. However, under certain circumstances, these mechanisms fail to adequately respond to proteotoxic imbalances and result in the accumulation of misfolded proteins. In humans, this can lead to neurodegeneration and other protein conformational diseases. To protect itself, the cell employs highly conserved stress responses and chaperone networks to maintain protein-folding homeostasis (proteostasis). Although the regulation of stress responses, such as the heat-shock response, and of proteostasis have been widely considered to be cell autonomous, recent studies using Caenorhabditis elegans have shown that these processes are regulated by neuronal signaling and endocrine pathways and integrated into other functions of the organism. The hierarchical control of the cellular proteostasis machinery affords insight into the organization of stress regulatory networks in multicellular organisms and offers novel targets for the treatment of human protein conformational diseases.
Protein misfolding and the stress response
Cells respond to rapid changes in their protein biogenesis requirements or exposure to proteotoxic environmental conditions, such as heat, oxidative stress or transition metals, by inducing a highly conserved program of gene regulation (the stress response) [1–4]. Studies on unicellular organisms such as bacteria and yeast and on isolated Drosophila and mammalian cells have shown that the stress response is triggered cell autonomously by changes in the intracellular flux of misfolded proteins that accompany these physiological or environmental challenges [1–4]. However, the genetic program implemented by isolated cultured cells in response to stress can differ from that of organs or whole organisms exposed to similar stress regimens [5,6]. The exposure of an organism to stress conditions can result in a selective and asynchronous activation of the stress response in different tissues [6,7]. Moreover, in human diseases of protein misfolding, such as Parkinson’s disease and Huntington’s disease, only certain cells and tissues are at risk. Despite the accumulation of damaged proteins, the stress response is not appreciably induced [3,8–10] in the affected cells, indicating that additional levels of control can exist at the organismal level. Recent advances in the understanding of stress responses and proteostasis in C. elegans show that the cell-autonomous stress responses of individual cells are regulated by neuronal and endocrine signaling to yield an integrated systemic response [11–13]. C. elegans has proven valuable as a model system to study this issue [4,14–22] (Box 1). Here, we summarize studies on the complex interplay between cell-autonomous and organismal regulation of the stress response and proteostasis in C. elegans.
Box 1. C. elegans as a model system to study stress responses.
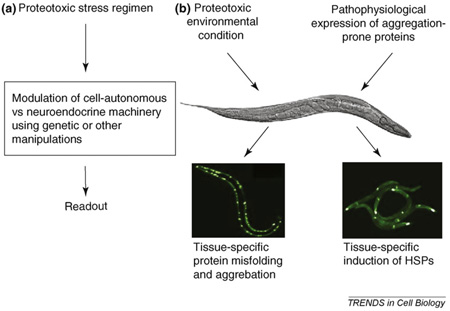
C. elegans is an ideal system to study the organismal integration of stress responses. Its short life cycle, optical transparency, amenability to genetic manipulation, fully sequenced genome and the extensive sequence homology between C. elegans and mammalian genes are complemented by its small size and simple method of cultivation [14]. This enables many regimens of stress imposed on the whole animal, such as a transient increase in temperature or exposure to oxidants, to penetrate all 959 cells of the adult animal. Tissue-specific differences in the stress response and hsp induction can be investigated to determine whether they are cell intrinsic or a result of cell non-autonomous regulation. In addition, protein misfolding and aggregation can be directly visualized and assayed within various tissues of the intact organism under different physiological or environmental conditions [15,16] (Figure I).
Environment regulates C. elegans growth and development in numerous ways. Temperature alone affects development, lifespan, brood size, defecation cycles, pharyngeal pumping rates, behavior and metabolism [17]. The life cycle of C. elegans larvae consists of four larval (L) stages (L1–L4). The life cycle is delayed at lower temperatures (15 °C) and animals have a larger body size; higher temperatures (25 °C) accelerate the life cycle and result in smaller body sizes [17]. This indicates that the growth and differentiation of all 959 cells is coordinately regulated by ambient temperature. Exposure to temperatures >27 °C or to other environmental stressors, such as starvation and high population densities, activates neuroendocrine pathways that induce larvae to enter an alternative long-lived, stress-resistant L3 state called ‘dauer’ [18]. The dauer decision is made, in part, by signaling through a pheromone called the dauer pheromone, which is detected by chemosensory neurons. Thermotaxis behavior is also regulated by neuroendocrine signaling and enables C. elegans to seek out temperatures associated with the availability of food and avoid temperatures associated with starvation [19]. Similar organismal responses occur in response to other environmental variables [20–22].
Figure I. C. elegans as a model system to study stress responses and protein misfolding. (a) Forward and reverse genetics and the wealth of molecular tools enable the modulation of the neuroendocrine system of C. elegans to study the organismal regulation of stress responses, (b) The effects of proteotoxic stress regimen imposed on the whole organism can be determined by assaying protein folding in specific tissue and the tissue-specific expression of transcriptional and translational reporters that are induced upon stress.
The role of heat-shock proteins in cytoprotection
The stress response and the molecular machinery for its implementation are conserved from archaebacteria to mammals. The upregulation of heat-shock proteins (HSPs) is central to the stress response. This is concomitant with the downregulation of genes for normal cellular function [1,2,4]. Elevated expression of HSPs is sufficient to protect cells from a wide range of cytotoxic conditions [1,2,4]. HSPs, as molecular chaperones, typically bind to non-native conformations of proteins that persist upon cell stress and these interactions protect against misfolding, aggregation or premature clearance and enable refolding and the restoration of native conformations [23–25] (Figure 1a). Thus, the interaction of chaperones with diverse substrates in stressed cells or upon increased protein biogenesis enhances the stability of the proteome and restores the activities of signaling and growth regulatory molecules re-establishing cellular homeostasis [1,2,23,25].
Figure 1.
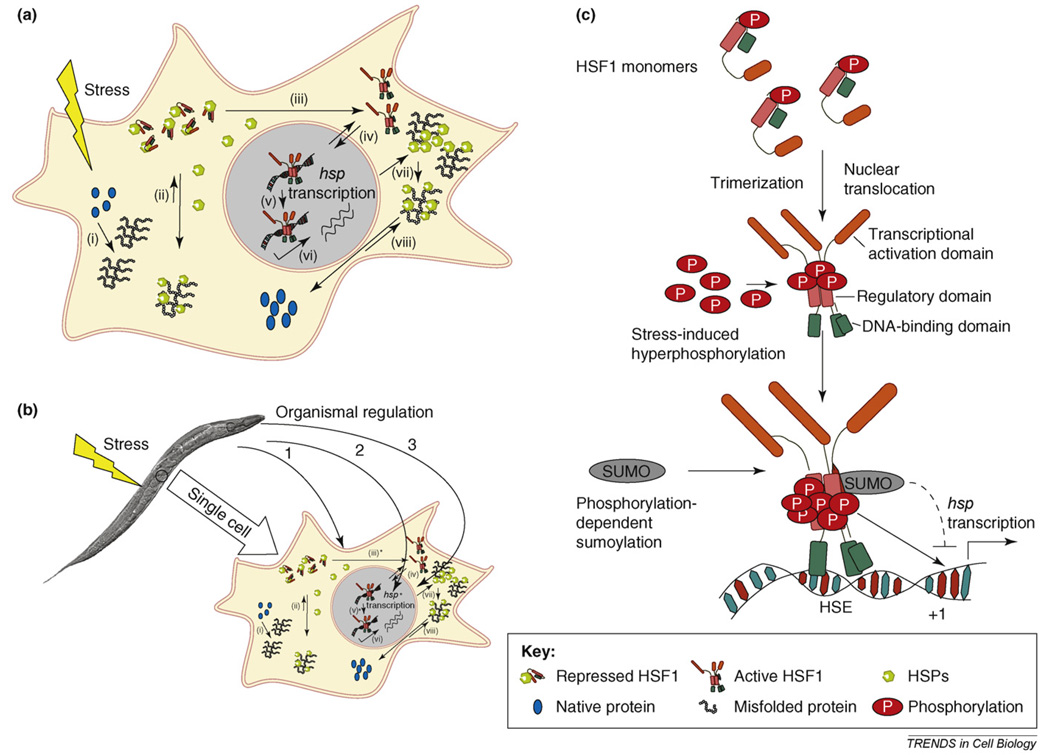
Cell-autonomous and organismal regulation of stress responses. (a) Model for the cell-autonomous regulation of the stress response. (i) The increased flux of damaged or misfolded proteins in response to proteotoxic environmental conditions (stress) is the trigger for the induction of the cellular stress response. (ii) The damaged or misfolded proteins titrate away HSPs that are bound to HSF1 and maintain it in a repressed state before stress, resulting in its activation. (iii) Numerous post-translational modifications influence the ability of HSF1 to trimerize, (iv) translocate into the nucleus, and bind DNA. (v) The binding of HSF1 to DNA alone is insufficient to initiate HSP transcription and requires at least one additional signal. (vi) HSF1-dependent HSP transcription upregulates the cellular levels of HSPs (vii), enabling cells to re-establish cellular protein homeostasis by various processes such as selective degradation, or (viii) refolding the misfolded proteins. (b) Organismal regulation of the cellular stress response via neuroendocrine signaling pathways can conceivably act at several points. Three possible levels of control of the stress response are depicted as: 3*, 5* and hsp transcription*. Adverse environmental conditions (stress) imposed on the animal activate neuroendocrine signaling pathways. 1. Neuroendocrine signaling can lead to changes in HSF1 post-translational modifications (iii*), regulating the ability of HSF1 to trimerize, translocate into the nucleus, or bind DNA. These effects on HSF1 and HSP induction can be activating or inhibitory. 2. Neuroendocrine signaling can also regulate the unknown step between DNA binding by HSF1 and HSP transcription (v*). This modulation can also be activating or inhibitory. 3. Alternative pathways that lead to the transcription of hsp genes by C. elegans DAF-16 or SKN-1 can independently upregulate HSP expression (hsp transcription*), leading to the higher tolerance of cells to misfolded protein species and re-setting the threshold for HSF1 activation. (c) Multi-step activation of HSF1 involves post-translational modifications. When exposed to stress, HSF1 is activated through a monomer to trimer transition that enables it to translocate into the nucleus and bind to heat-shock elements (HSEs) in the promoter regions of its target hsp genes. Activation involves hyperphosphorylation of HSF1 at multiple sites within its regulatory domain including on serine residues 303 and 307. Phosphorylation of HSF1 on Ser303 is required for the stress-induced addition of a SUMO residue on Lys298, which represses HSF1 activity. The interactions between the activating and repressing modifications are not well understood. +1 depicts the transcriptional start site.
The key regulator of HSP transcription in eukaryotes is heat-shock factor 1 (HSF1), which is highly conserved and ubiquitously expressed [24,26]. In the absence of stress, the DNA-binding and transcriptional activities of HSF1 are inhibited by HSPs, which associate weakly to maintain a repressed state (Figure 1a). An increase in the level of intracellular misfolded proteins thought to trigger the stress response [2,25,27,28] is proposed to titrate HSPs away from their association with HSF1, enabling HSF1 to trimerize and translocate into the nucleus and activate HSP gene transcription (Figure 1a). Thus, it seems that the cell has evolved an elegant and efficient mechanism to autonomously deploy resources proportional to protein biogenesis needs, or in response to damage incurred by the environmental insult. The basal levels of HSPs set the threshold of the stress response, whereas the autoregulation of HSF1-dependent HSP transcription ensures the re-establishment and maintenance of proteostasis [1–3,23,24].
Whereas the stress response describes the molecular events associated with damaged proteins in the cytoplasm and nucleus, the unfolded protein response (UPR) provides the same functionalities for protein misfolding in the endoplasmic reticulum and the mitochondria [29–32] (Box 2). Currently, there is limited understanding of why protein misfolding and aggregation cause cellular toxicity. Although cellular dysfunction caused by aggregation can, in part, be attributed to the loss of function of these proteins, protein misfolding seems to also have more general pleiotropic effects on cellular function by limiting essential factors for folding, transport and secretion [3,10,23]. In many cases, these deleterious effects seem to be a result of the ability of misfolded and aggregated proteins to engage and perturb the proteostasis machinery of the cell [3,33].
Box 2. Unfolded protein response.
ER stress response (UPRer)
The endoplasmic reticulum (ER) is the cellular compartment within which proteins destined for insertion into the plasma membrane or secretion are folded and post-translationally modified. An increase in the ER protein-folding demands, such as increased secretory function of the cell or exposure to environmental toxins that disrupt ER function, result in an accumulation of unprocessed ER client proteins. This triggers an ER stress response called the unfolded protein response (UPRer) [29,30]. As with cytoplasmic stress response, the UPRer is thought to be cell autonomously triggered by the increase in unfolded proteins within the ER. UPRer induction results in the transcriptional upregulation of ER-specific HSPs and proteins involved in processing, trafficking and degradation of the unprocessed ER client proteins and the re-establishment of ER folding homeostasis. The UPRer uses transcription factors and signaling molecules distinct from those that induce the cytoplasmic stress response. In yeast, the transcription factor responsible for the UPRer is Hac1. Hac1 upregulation requires the activity of a transmembrane protein kinase and endonuclease, Ire1, which modulates the post-transcriptional processing of the Hac1 mRNA to enable its accumulation and the transcription of its Hac1 genes. In addition to IRE1 homologues and a HAC1-like transcription factor, XBP-1, metazoans also possess two new pathways for UPRer induction. The pancreatic-enriched ER kinase (PERK) phosphorylates eukaryotic translation initiation factor 2 to attenuate protein synthesis during the UPRer, decreasing the protein folding load on the ER during stress. PERK also activates the expression of UPRer target genes [29,30]. The transcription factor ATF6 also directly activates UPRer target genes. Inactivation of UPR signaling impairs cell survival and the accumulation of misfolded proteins in the ER have an important role in human diseases [1–3].
Mitochondrial stress response (UPRmt)
The mitochondria consist of numerous multimeric protein complexes, which require the synthesis and assembly of subunits transcribed by both the nuclear and mitochondrial genomes. Disruption of the protein-folding environment in the mitochondria, either owing to the malfunction of transport into the mitochondria, a global compromise in expression of the mitochondrial genome (the rho− state), or to the expression of a aggregation-prone mitochondrial proteins, leads to accumulation of unassembled subunits in the mitochondria and elicits the mitochondrial unfolded protein response (UPRmt) [31]. The UPRmt upregulates mitochondrial chaperones and other factors that assist in the re-folding and degradation of the unassembled mitochondrial subunits and remodel the mitochondrial-folding environment [31]. The components of the UPRmt are only now being elucidated. Recent data from C. elegans reveal similarities between the machinery that activates the UPRmt and components of the bacterial stress response [31,32].
Neuroendocrine pathways regulate growth, development and stress tolerance in C. elegans
Although HSP expression is required to re-establish proteostasis after stress exposure, the prolonged overexpression of HSPs is detrimental to cell growth and division [2,34,35]. Because the autonomous triggering of the stress response within individual cells in multicellular organisms could disrupt the coordinated functions and interactions of their cells and tissue, metazoans must have evolved mechanisms to integrate their cellular stress response with other organismal processes.
The autoregulatory mechanism of HSP induction and the steps involved in HSF1 activation provide multiple opportunities for additional regulatory input (Figure 1b,c). HSP genes have cis-regulatory elements that can bind transcription factors other than HSF1. In C. elegans, two other stress-regulated transcription factors, the FOXO homologue DAF-16 [36,37] and the nrf-1 homologue SKN-1 [38] can also transcribe hsp genes. The upregulation of HSPs by these alternative pathways would increase the tolerance of the cells for misfolded proteins and reset a higher threshold for HSF1 activation upon stress. In addition, HSP induction by HSF1 is itself a multi-step process (Figure 1b,c): activation of HSF1 from an inert monomer to trimer and binding to DNA are insufficient for HSP transcription and at least one other signal is required [24,26]. HSF1 also undergoes numerous post-translational modifications (Figure 1c) including phosphorylation by growth-related kinases that affect DNA binding, trimerization and transcriptional activity, thereby providing possibilities for the regulation of the cellular stress response by signals other than the increase in misfolded protein species [24,26].
Every aspect of C. elegans biology is affected by environmental conditions [17,39] (Box 1). Conversely, the overall ability of the organism to withstand exposure to adverse environmental conditions is modulated by its developmental, physiological, metabolic and nutritional state [11,40,41]. Two kinds of stress response have been widely studied in C. elegans. The first is the switch in the developmental program of young (L1–L2; Box 1) larvae from continuous reproductive development to developmentally arrested, stress-resistant dauers [40] (Box 1). The entry into dauer is regulated by neuroendocrine signaling [40]. Thus, the ability of neuroendocrine pathways to regulate stress at the organismal level in C. elegans has been predominantly examined with respect to their ability to regulate dauer-specific gene expression. The second type of response, active in both larvae and adults, is the upregulation of hsps and other stress-inducible genes by transcription factors such as HSF1, DAF-16 and SKN-1 [13,42]. These responses are not necessarily distinct [43]: neuroendocrine pathways that modulate dauer development also regulate the transcription factors involved in stress-induced gene expression [38,44]. Likewise, DAF-16 and HSF1 are also involved in the regulation of the dauer program [44,45].
Three neuroendocrine signaling pathways modulate stress tolerance in C. elegans [11,46–48] (Figure 2). These pathways, insulin-like growth factor (IGF)/insulin-like signaling (ILS) pathway, the transforming growth factor-β (TGF-β) pathway and the nuclear hormone receptor (NR) pathway also regulate development, growth, body size, reproduction, fecundity, metabolism and behavior. The majority of studies have focused on DAF-16 as the downstream effector of neuroendocrine regulation [36,44,48–51]. SKN-1 is also regulated by some of the same neuroendocrine pathways [38]. DAF-16 transcribes specific hsps and genes that regulate growth and metabolism and normal and dauer development. Similarly, SKN-1 transcribes hsps and other genes induced by oxidative stress, in addition to genes required for the development of the intestine and mesendoderm [52]. HSF1, besides being the master regulator of stress-dependent hsp expression, also has a role in C. elegans growth and development: hsf1 deletion renders the organism inviable [45]. The regulation of cell growth, differentiation and stress tolerance by the same neuroendocrine signaling pathways and transcription factors indicates that these functions need to be coordinately regulated to yield a unified response at the organismal level. In yeast, cell growth induces HSF1 phosphorylation at specific sites repressing its activation [53]. An examination of the relationships between growth, development and the stress response within the different cells and tissues of C. elegans, and their regulation and roles in systemic function will provide an understanding of how stress responses and proteostasis are integrated within multicellular organisms.
Figure 2.
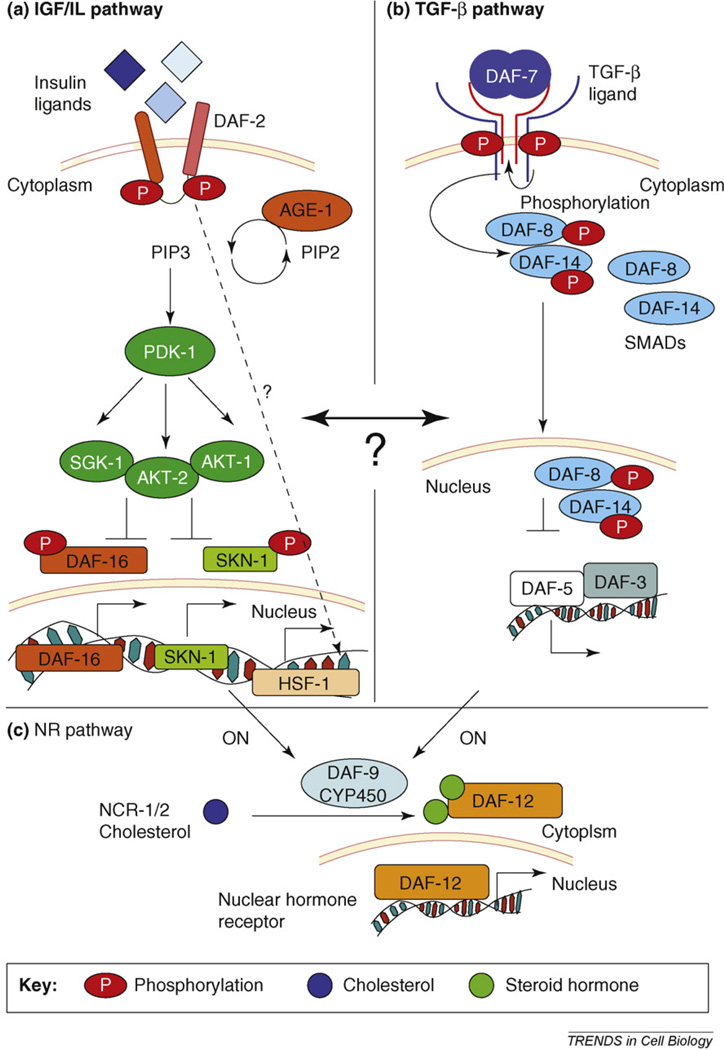
Neuroendocrine pathways regulate stress tolerance in C. elegans [13]. Schematic depiction of the three neuroendocrine pathways that regulate stress responses in C. elegans. (a) The IGF/IL signaling pathway. IL ligands bind the IL receptor DAF-2 to activate a phosphorylation cascade, which then activates the PI3 kinase AGE-1. This results in the phosphorylation of the ACG kinases, AKT-1 AKT-2 and SGK-1, which ultimately phosphorylate DAF-16 and SKN-1. Phosphorylated DAF-16 and SKN-1 are cytoplasmic and inactive. Unphosphorylated DAF-16 and SKN-1 are active and translocate to the nucleus to transcribe their target genes. IL signaling also modulates the activity of HSF1 in manner yet to be characterized, (b) The TGF-β signaling pathway in C. elegans has all the components of the canonical TGF-β signaling pathway. The TGF-β ligand DAF-7 binds the type I and type II receptors DAF-1 and DAF-4 to modulate the phosphorylation state of the downstream SMAD proteins DAF-8 and DAF-14. DAF-8 and DAF-14 inhibit the function of a DAF-3, a co-SMAD and DAF-5, a protein homologous to the Sno/Ski family of oncogenes. Phosphorylated DAF-8 and DAF-14 translocate to the nucleus where they transcribe its target genes involved in the development of dauer larvae. (c) The nuclear hormone receptor (NR) signaling pathway. Insulin/IGF-I and TGF-β peptide signals converge on the nuclear receptor branch of the dauer pathway. NCR-1/2, a Niemann-Pick C1 homolog, delivers cholesterol to DAF-9, presumably triggering the synthesis of steroid hormone. In the presence of hormone, DAF-12 directs expression of genes involved in reproductive development and the animals are not stress tolerant. In unfavorable environments, hormonal pathways are suppressed and unliganded DAF-12 specifies dauer development and stress resistance.
Insulin-like signaling (ILS) pathway
The fundamental principles of IGF/IL signaling seem to be the same in C. elegans and higher organisms [11,46,48,54]. The binding of insulin ligands to the insulin receptor signals the presence of abundant food and optimal growth conditions. Under these conditions, animals are less stress tolerant. Inhibition of IL signaling indicates stressful conditions and results in an increase in organismal stress tolerance. The C. elegans genome encodes one insulin receptor, DAF-2, and 38 insulin-like ligands, most of which remain to be characterized. Ligand binding to DAF-2 initiates a phosphorylation cascade (Figure 2a) whereby DAF-2 phosphorylation activates a P13 kinase (AGE-1 in C. elegans), which results in the phosphorylation of several ACG kinases (AKT-1 and AKT-2, homologous to human serine–threonine [Akt]/protein kinase B [PKB] kinase, and serum and glucocorticoid inducible kinase 1 [SGK-1], homologous to serum and corticoid-responsive kinase [SGK] in humans) and ultimately affects the phosphorylation status and localization of DAF-16. Under optimal growth conditions, DAF-16 is phosphorylated, inactivated and retained in the cytoplasm. High temperatures, the absence of food, oxidative stress or other suboptimal conditions disrupt the DAF-2 phosphorylation cascade: DAF-16 is not phosphorylated, translocates to the nucleus and induces the transcription of hsps and other target genes [11,46,48,54]. More recently, SKN-1 has also been shown to be regulated in a similar manner whereby the nuclear localization of SKN-1 and the transcription of its target genes upon oxidative stress depend on DAF-2 and ILS [38] (Figure 2a). ILS also modulates HSF1 function [55]. However, the molecular and cellular details of this regulation have yet to be worked out.
Many of the insulin ligands are expressed in the nervous system and intestine of C. elegans, indicating that these tissues could be the primary initiators of ILS and that the ILS-dependent regulation of stress tolerance is cell non-autonomous [54,56,57]. Indeed, neuronal pathways that regulate sensory perception affect longevity through ILS [58], which, in C. elegans, is inextricably linked to stress tolerance [59]. Changing DAF-16 levels in the intestine and neuronal cells changes lifespan [56,57,60] and stress tolerance. Overexpression of HSF1 or expression of a dominant-negative HSF1 in neuronal or muscle cells also increases or decreases longevity (and perhaps stress tolerance), respectively [55]. Both these effects depend on DAF-2/ILS. Similarly, whereas exposure to stress increases life span, the tissue in which the stress response is activated and the concomitant growth and metabolic changes within the organism can be decoupled depending on the cell type within which the IL signal is modulated [56,61,62]. Thus, although downregulation of DAF-2 signaling has similar effects on stress resistance, its down-regulation during early development induces larvae to develop into stress-resistant dauers (Box 1), whereas its downregulation at late developmental stages or in adults increases longevity [11,46]. Caloric restriction via ILS signaling activates SKN-1 within specific neurons, whereas the resultant increase in metabolic activity occurs in peripheral tissues [63]. A systematic understanding of the response of the different tissues to ILS and the resulting changes in their gene expression profiles upon stress will enable us to better comprehend the cell non-autonomous regulation of organismal stress responses.
Transforming growth factor β (TGF-β) signaling pathway
The TGF-β signaling pathway consists of a family of secreted peptides that include the activins and bone morphogenetic proteins (BMP) in vertebrates. Five TGF-β-related ligands, numerous receptors and SMAD homologues have been identified in C. elegans based on sequence homology [64] (Figure 2b). TGF-β signaling affects stress tolerance by inducing the dauer program in C. elegans [40,65], and DAF-7 is a prominent ligand regulating this process [40]. Under favorable conditions, DAF-7 is expressed in a pair of sensory neurons called ASI and promotes non-dauer development. The repression of DAF-7 under stressful environmental conditions leads to the development of dauer larvae [40,65]. The transcription factors that are downstream of DAF-7 signaling and implement daf-7-dependent dauer formation are distinct from the three stress-activated transcription factors. However, the nuclear localization of DAF-16 has been shown to be modulated by DAF-7 at the time of commitment between dauer and reproductive development [40,65]. In addition, the TGF-β signaling pathway interacts with the ILS pathway to regulate longevity. Because the dauer and/or longevity programs must be implemented by all cells, the effects of TGF-β signaling on organismal stress resistance must be cell non-autonomous. The stress resistance of dauer larvae is associated with elevated expression of certain hsp genes [66]. However, analyses of the tissues within which hsp expression is necessary and sufficient to confer the stress-resistant properties of dauer larvae have not been conducted.
Nuclear hormone signaling pathway
The nuclear hormone receptor (NR) pathway in metazoans consists of a family of transcription factors regulated by small lipophilic molecules including steroids, retinoids and bile and fatty acids, which mediate endocrine control [67]. The C. elegans genome encodes 284 NR receptors, compared to 48 for humans and 21 for flies [68]. Many of these receptors have homologs in other species and function in metabolism, nervous system development, sex determination, developmental timing, molting and entry into the stress-resistant dauer pathway. Two components of the NR pathway, DAF-12 and DAF-9, have been studied in C. elegans with respect to their involvement in organismal stress tolerance. DAF-12, the steroid hormone receptor, is thought to bind to hormones that are cholesterol derivatives (D4-dafachronic acid and D7-dafachronic acid [69]) [69–71] (Figure 2c). DAF-9, a P450 cytochrome related to fatty acid and steroidogenic hydroxylases is involved in the biogenesis of these hormones. Genetic studies place DAF-12 downstream of both the ILS and TGF-β pathways and it is, therefore, thought to have a crucial role in integrating neuroendocrine signaling to regulate the dauer program. DAF-12 is widely expressed throughout development and adulthood [71,72]. DAF-9 expression, however, is spatially restricted to a pair of anterior ganglion nuclei, hypodermis and spermatheca and depends on the developmental stage of the animal [70]. Although a strong loss-of-function mutation of DAF-9 also results in stress resistance and the arrest of larvae in dauer, expression in the hypodermis alone is sufficient to rescue this dauer phenotype [70]. Thus, dauer-specific gene expression is cell non-autonomously regulated by the steroid hormone signaling pathway. Cell non-autonomous regulation of the stress response by steroid hormone signaling is also evident in experiments in which removal of the germline by microsurgery increases the resistance of the animal to oxidative and heat stress and promotes the nuclear localization of DAF-16 in the intestine [60,73,74]. The effects of germline ablation are thought to be caused by the regulation of daf-9 expression [70,71] and seem to be conserved in other organisms such as Drosophila and the mouse [75].
In all the cases described here, it is unclear whether hsp expression within specific cells or tissues can suffice to protect the entire organism from stress-induced cellular protein damage. Neuroendocrine regulation of stress could indicate that proteotoxic damage within different cell types have different outcomes with regard to organismal health. It would be interesting to understand whether organisms can tolerate different levels of misfolded and damaged proteins under certain growth or metabolic conditions, even at the cost of cytotoxic damage to certain cells, and whether neuroendocrine signals can override cellular autonomy in the response of cells to environmental stress.
Neuronal signaling overrides cell-autonomous HSP induction and response to stress
The neuroendocrine pathways described earlier could regulate organismal stress tolerance by changing the basal levels of expressed HSPs and decreasing the overall cellular proteotoxicity that results from exposure to stress. However, there is also evidence that neuronal signaling is required for HSP induction upon the administration of temperature stress (heat shock), despite the probable increase in cellular misfolded and damaged proteins [13]. In C. elegans, two neurons called AFDs detect ambient temperature and regulate thermotaxis behavior [19]. Mutations affecting only these neurons inhibited heat-shock-dependent HSP induction throughout C. elegans. This included tissues such as the intestine and spermatheca, which were not directly innervated by these neurons, indicating that AFD regulation occurred through neuroendocrine pathways. The AFD-dependent upregulation of HSPs after heat shock depended on the metabolic status of the animal. Thus, neuronal regulation seemed to integrate the stress response with other organismal functions. A model proposed to explain these findings postulates that the cellular proteostasis machinery is negatively regulated by (at least) two mutually inhibitory pathways: a temperature-sensing pathway and a growth-regulated pathway. Disruption of either pathway results in the net inhibition of heat-shock-dependent HSP transcription, whereas the presence or absence of both pathways enables cells to express HSPs upon heat stress. The downstream target of the AFDs seems to be HSF1. Data from other studies on nutrient-dependent signaling in C. elegans indicate that the growth-related signal could act through DAF-16 [58]. Thus, as in mammalian tissue culture cells or yeast [24,26], organismal growth and HSF1-dependent HSP expression within C. elegans could also be mutually antagonistic.
C. elegans as a model system for neurodegenerative and other protein misfolding diseases
Neurodegenerative disease models in C. elegans created by expressing human disease-related proteins in C. elegans in some cases show cell-autonomous induction of HSPs. However, the ILS pathway also modulates the protein misfolding and aggregation of these disease-related proteins, indicating that hierarchical interactions exist between cell-autonomous and cell non-autonomous controls on the proteostasis machinery.
Huntington’s disease (HD)
PolyQ-containing proteins are implicated in HD [76–80] (Box 3) and other human age-related neurodegenerative diseases [81]. Expression of aggregation-prone polyQ-containing proteins within C. elegans muscle and neuronal tissues [82–84] recapitulates aspects of HD, including formation of Q-length-dependent intracellular aggregates that cause toxicity [81]. The aggregation-dependent toxicity phenotypes in C. elegans reflect the cell type in which the polyQ protein is expressed; in muscle cells, polyQ proteins cause muscle-cell dysfunction, whereas expression in neuronal cells causes neuronal dysfunction [82,83]. The toxicity associated with polyQ proteins seems to be caused by the global disruption of the cellular proteostasis machinery [77]. This disruption is cell autonomous: when temperature-sensitive metastable proteins were used as sensors of cellular protein folding capacity, polyQ expression in muscle cells destabilized temperature-sensitive proteins in muscle but not in neurons; likewise, polyQ aggregates in neurons destabilized temperature-sensitive proteins expressed only in neurons [77].
Box 3. Neurodegenerative diseases of protein misfolding and aggregation.
Huntington’s disease
Huntington’s disease (HD) is a genetic neurological disorder that affects up to seven in 100 000 people. It is characterized by uncoordinated body movements (chorea), a decline in cognitive abilities and, ultimately, a severe reduction in life expectancy. HD is one of many trinucleotide repeat diseases caused by an increase in the length of a repetitive DNA sequence, CAG, within the Huntintin gene (Htt) and is inherited in an autosomal dominant manner. CAG encodes the amino acid glutamine (Q). The number of Q repeats correlates with disease severity, age-dependent onset and the rate of progression of neurological symptoms. In the general population, the number of Q repeats within Htt rarely exceeds 27; sequences of 36 or more glutamines result in the selective neurodegeneration of striatal projection neurons and cortical pyramidal neurons in various regions of the brain [76].
The function of Htt and the reason why an increase in its Q length causes neurodegeneration is unclear. Numerous potential mechanisms for neurotoxicity have been proposed, including dysregulation of transcriptional pathways, disruption of mitochondrial transport, excitotoxicity, caspase activation and disruption of proteostasis [76,77]. The expression of the aggregation-prone polyQ stretch alone within animal models is sufficient to mimic toxic aspects of HD, indicating that its pathology is, at least in part, a consequence of misfolding and aggregation of mutant Htt in neurons [76,77]. Treatment for Huntington’s disease is at an early stage and currently the symptoms can be alleviated through the use of medication including antidepressants and antipsychotics, rehabilitation methods and nutrition management [76–78].
Alzheimer’s disease
Alzheimer’s disease (AD) is the most common form of dementia estimated to affect >30 million people worldwide. This neurodegenerative disease is terminal, age-related and typically diagnosed in people >65 years of age. The earliest observable symptoms include memory loss with subsequent symptoms including confusion, irritability, language breakdown and general social withdrawal. AD is associated with the appearance of plaques and tangles in affected brains. However, the cause of these deposits and their correlation with the progression of AD is poorly understood [79].
There are currently several hypotheses put forth to explain the cause of AD [79]. Currently available drug therapies are mainly based on the cholinergic hypothesis, which proposes that AD is caused by reduced synthesis of the neurotransmitter acetylcholine. Another theory is the amyloid hypothesis, which postulates that amyloid β (Aβ) deposits are the fundamental cause of AD. In support of this, people with an extra copy of the amyloid β precursor (APP) protein exhibit an earlier onset of AD symptoms, and a major genetic risk factor for AD is apolipoprotein E (APOE4), which causes excessive amyloid buildup. Transgenic mice that express a mutant form of the human APP gene develop fibrillar amyloid plaques and Alzheimer’s-like brain pathology and neurological symptoms. The appearance of damaged proteins is furthered by the tau hypothesis, which indicates that hyperphosphorylation and aggregation of the microtubule-associated protein tau causes AD. Both the amyloid and the tau hypotheses have in common the cellular misfolding and aggregation of proteins, indicating that the general dysfunction in proteostasis could have a central role in AD pathogenesis [79,80].
However, although some features of the response of C. elegans to polyQ misfolding are decidedly cell autonomous, the onset of aggregation and toxicity in the animal, as in the human disease, is age-dependent and regulated by the ILS pathway [55,85]. Downregulation of ILS by mutations in phosphoinsitide-3-kinase, AGE-1 or the insulin receptor DAF-2 causes the constitutive activation of DAF-16 (Figure 2) and this delays and/or suppresses polyQ aggregation and toxicity. ILS modulation of polyQ aggregation requires HSF1 [55,85] (Figure 2). The effects of ILS are thought to be caused by the cellular upregulation of chaperones and other transcriptional targets of DAF-16 and HSF1 [3].
PolyQ aggregation within C. elegans muscle cells is also regulated cell non-autonomously by neuronal gamma aminobutyric acid (GABA)-ergic and/or cholinergic signaling pathways [12]. Mutations in these pathways and the modulation of neuronal signaling by small molecules altered polyQ aggregation and increasing amounts of GABA even rescued polyQ aggregation and toxicity. GABA-ergic signaling also modulated the misfolding and accumulation of endogenous temperature-sensitive proteins in C. elegans muscle. Although the effects of GABA-ergic signaling are thought to be caused by increased excitotoxicity in muscle cells, in C. elegans, GABA-ergic neurons also regulate numerous motor functions [12] and are themselves targets of stress-induced signaling pathways [86]. This indicates that the observed cell non-autonomous regulation of polyQ proteins by these neurons could be part of a larger neuroendocrine network that modulates organismal function.
Alzheimer’s disease (AD)
The A(1–42 peptide, a prevalent component of plaques in the brains of patients with AD (Box 3), also forms aggregates within C. elegans muscle cells [16]. These aggregates interact with and induce specific HSPs that are downstream targets of both HSF1 and DAF-16 in the tissue in which they are expressed, again revealing the cell-autonomous nature of the regulation of proteostasis [87,88]. In addition, as in the polyQ models, Aβ1–42 aggregation-toxicity is also regulated by the ILS pathway [89] through DAF-16 and HSF1. Downregulation of DAF-16 and HSF1, although having similar effects on Aβ1–42-dependent toxicity, has opposing effects on intracellular A1–42 accumulation and misfolding. Aβ1–42 aggregation and misassembly within cells is increased upon lowering HSF1 levels but is decreased upon lowering DAF-16 [89].
Thus, both the polyQ and Aβ1–42 models of protein misfolding are regulated by neuroendocrine signaling and specifically the ILS pathway, through DAF-16 and HSF1. Whereas the neuroendocrine pathway that affects proteostasis is common to both models, the effects at the cellular level, implemented through the two stress-regulated transcription factors, differ depending on the misfolded or disease-causing protein species. One perplexing feature of protein misfolding in C. elegans, as has been noted for various human protein conformational diseases, is that the intracellular accumulation of misfolded proteins such as polyQ only sporadically activates HSP expression [84] and requires the downregulation of ILS signaling for its clearance and modulation [55,89]. This indicates that the cell-autonomous response to protein misfolding can be overridden by organismal regulation.
Concluding remarks
The cell non-autonomous regulation of stress responses and proteostasis by neurohormonal signaling in C. elegans indicates that similar forms of control are likely to occur in other eukaryotes. Although a systematic study of the hierarchical layers of organismal stress regulation has not been conducted in other animal models, ILS signaling is known to control lifespan in Drosophila melanogaster and mice [50,90–92]. Furthermore, the machinery for cell non-autonomous neurohormonal control of HSF1 and HSPs has been observed in specific tissues of mammals: restraint stress [7,93] results in the upregulation of specific HSPs in the adrenal cortex and the endothelial cells of the thoracic aorta of rats. This requires the hypothalamicpituitary-adrenal (HPA) axis and is adrenocorticotropic hormone-dependent. Likewise, the cell non-autonomous control of HSPs by HSF1 is dependent on organismal physiology and declines with age [7]. The ability of multi-cellular organisms to regulate cellular HSP expression, the stress response and proteostasis machinery by neurohormonal signaling could be influenced further by the ecology and life history of the organism [94].
Neurohormonal cell non-autonomous regulation might account, in part, for the pathophysiological situations that accompany protein misfolding diseases whereby the intracellular accumulation of misfolded and aggregated protein fails to adequately upregulate protective HSPs [3,8–10]. Similarly, diseases such as Hashimoto’s thyroiditis, Graves disease, arthritis and cancer [95,96] have increased levels of HSP expression within affected cells, indicating that these cells might have lost their responsiveness to the neurohormonal regulation imposed by the organism. An understanding of the systemic stress regulation in organisms would enable an examination of whether neurohormonal perturbations indeed correlate with disease conditions. Small molecules that can modulate the components of ILS or other neuroendocrine pathways to result in the upregulation of specific cytoprotective HSPs in the tissue harboring disease-related protein aggregates and re-establish cellular proteostasis could provide novel drug targets for combating diseases of protein conformation.
Acknowledgments
We thank the members of the Morimoto laboratory for discussions and comments on the manuscript and Sue Fox and Kai Orton for help with the figures. These studies were supported by grants to R.I.M from the NIH (NIGMS and NIA), the Huntington’s Disease Society of America Coalition for the Cure and the Daniel F. and Ada L. Rice Foundation.
References
- 1.Lindquist S, Craig EA. The heat-shock proteins. Annu. Rev. Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 2.Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 3.Balch WE, et al. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 4.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pardue ML. The heat shock response in biology and human disease: a meeting review. Genes Dev. 1988;2:783–785. doi: 10.1101/gad.2.7.783. [DOI] [PubMed] [Google Scholar]
- 6.Blake MJ, et al. Discordant expression of heat shock protein mRNAs in tissues of heat-stressed rats. J. Biol. Chem. 1990;265:15275–15279. [PubMed] [Google Scholar]
- 7.Fawcett TW, et al. Effects of neurohormonal stress and aging on the activation of mammalian heat shock factor 1. J. Biol. Chem. 1994;269:32272–32278. [PubMed] [Google Scholar]
- 8.Hay DG, et al. Progressive decrease in chaperone protein levels in a mouse model of Huntington’s disease and induction of stress proteins as a therapeutic approach. Hum. Mol. Genet. 2004;13:1389–1405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- 9.Zourlidou A, et al. Hsp27 overexpression in the R6/2 mouse model of Huntington’s disease: chronic neurodegeneration does not induce Hsp27 activation. Hum. Mol. Genet. 2007;16:1078–1090. doi: 10.1093/hmg/ddm057. [DOI] [PubMed] [Google Scholar]
- 10.Barral JM, et al. Roles of molecular chaperones in protein misfolding diseases. Semin. Cell Dev. Biol. 2004;15:17–29. doi: 10.1016/j.semcdb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 11.Baumeister R, et al. Endocrine signaling in Caenorhabditis elegans controls stress response and longevity. J. Endocrinol. 2006;190:191–202. doi: 10.1677/joe.1.06856. [DOI] [PubMed] [Google Scholar]
- 12.Garcia SM, et al. Neuronal signaling modulates protein homeostasis in Caenorhabditis elegans post-synaptic muscle cells. Genes Dev. 2007;21:3006–3016. doi: 10.1101/gad.1575307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prahlad V, et al. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 2008;320:811–814. doi: 10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brignull HR, et al. Modeling polyglutamine pathogenesis in C. elegans. Methods Enzymol. 2006;412:256–282. doi: 10.1016/S0076-6879(06)12016-9. [DOI] [PubMed] [Google Scholar]
- 16.Link CD, et al. Visualization of fibrillar amyloid deposits in living, transgenic Caenorhabditis elegans animals using the sensitive amyloid dye, X-34. Neurobiol. Aging. 2001;22:217–226. doi: 10.1016/s0197-4580(00)00237-2. [DOI] [PubMed] [Google Scholar]
- 17.Devaney E. Thermoregulation in the life cycle of nematodes. Int. J. Parasitol. 2006;36:641–649. doi: 10.1016/j.ijpara.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Hu PJ. WormBook, ed. The C. elegans Research Community, WormBook. 2007. Dauer. [Google Scholar]
- 19.Mori I, et al. Worm thermotaxis: a model system for analyzing thermosensation and neural plasticity. Curr. Opin. Neurobiol. 2007;17:712–719. doi: 10.1016/j.conb.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 20.de Bono M, et al. Social feeding in Caenorhabditis elegans is induced by neurons that detect aversive stimuli. Nature. 2002;419:899–903. doi: 10.1038/nature01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray JM, et al. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. 2004;430:317–322. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- 22.Wang DY, Wang Y. Phenotypic and behavioral defects caused by barium exposure in nematode Caenorhabditis elegans. Arch. Environ. Contam. Toxicol. 2008;54:447–453. doi: 10.1007/s00244-007-9050-0. [DOI] [PubMed] [Google Scholar]
- 23.Hartl FU, et al. Molecular chaperones in protein folding: the art of avoiding sticky situations. Trends Biochem. Sci. 1994;19:20–25. doi: 10.1016/0968-0004(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 24.Morano KA, Thiele DJ. Heat shock factor function and regulation in response to cellular stress, growth, and differentiation signals. Gene Expr. 1999;7:271–282. [PMC free article] [PubMed] [Google Scholar]
- 25.Bukau B, et al. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 26.Anckar J, Sistonen L. Heat shock factor 1 as a coordinator of stress and developmental pathways. Adv. Exp. Med. Biol. 2007;594:78–88. doi: 10.1007/978-0-387-39975-1_8. [DOI] [PubMed] [Google Scholar]
- 27.Hightower LE. Heat shock, stress proteins, chaperones, and proteotoxicity. Cell. 1991;66:191–197. doi: 10.1016/0092-8674(91)90611-2. [DOI] [PubMed] [Google Scholar]
- 28.Goldberg AL. Degradation of abnormal proteins in Escherichia coli (protein breakdown-protein structure-mistranslation-amino acid analogs-puromycin) Proc. Natl. Acad. Sci. U. S. A. 1972;69:422–426. doi: 10.1073/pnas.69.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 30.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 31.Broadley SA, Hartl FU. Mitochondrial stress signaling: a pathway unfolds. Trends Cell Biol. 2008;18:1–4. doi: 10.1016/j.tcb.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 32.Haynes CM, et al. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev. Cell. 2007;13:467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 34.Abravaya K, et al. Attenuation of the heat shock response in HeLa cells is mediated by the release of bound heat shock transcription factor and is modulated by changes in growth and in heat shock temperatures. Genes Dev. 1991;5:2117–2127. doi: 10.1101/gad.5.11.2117. [DOI] [PubMed] [Google Scholar]
- 35.Feder JH, et al. The consequences of expressing hsp70 in Drosophila cells at normal temperatures. Genes Dev. 1992;6:1402–1413. doi: 10.1101/gad.6.8.1402. [DOI] [PubMed] [Google Scholar]
- 36.Huang H, Tindall DJ. Dynamic FoxO transcription factors. J. Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 37.Murphy CT. The search for DAF-16/FOXO transcriptional targets: approaches and discoveries. Exp. Gerontol. 2006;41:910–921. doi: 10.1016/j.exger.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 38.Tullet JM, et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutteling EW, et al. Environmental influence on the genetic correlations between life-history traits in Caenorhabditis elegans. Heredity. 2007;98:206–213. doi: 10.1038/sj.hdy.6800929. [DOI] [PubMed] [Google Scholar]
- 40.Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen D, et al. Longevity determined by developmental arrest genes in Caenorhabditis elegans. Aging Cell. 2007;6:525–533. doi: 10.1111/j.1474-9726.2007.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rea SL, et al. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat. Genet. 2005;37:894–898. doi: 10.1038/ng1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harvey SC, et al. Quantitative genetic analysis of life history traits of Caenorhabditis elegans in stressful environments. BMC Evol. Biol. 2008;8:15. doi: 10.1186/1471-2148-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- 45.Walker GA, et al. Heat shock factor functions at the convergence of the stress response and developmental pathways in Caenorhabditis elegans. FASEB J. 2003;17:1960–1962. doi: 10.1096/fj.03-0164fje. [DOI] [PubMed] [Google Scholar]
- 46.Tatar M, et al. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- 47.Antebi A. Nuclear hormone receptors in C. elegans. WormBook. 2006:1–13. doi: 10.1895/wormbook.1.64.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braeckman BP, et al. Insulin-like signaling, metabolism, stress resistance and aging in Caenorhabditis elegans. Mech. Ageing Dev. 2001;122:673–693. doi: 10.1016/s0047-6374(01)00222-6. [DOI] [PubMed] [Google Scholar]
- 49.Antebi A. Tipping the balance toward longevity. Dev. Cell. 2004;6:315–316. doi: 10.1016/s1534-5807(04)00068-1. [DOI] [PubMed] [Google Scholar]
- 50.Finch CE, Ruvkun G. The genetics of aging. Annu. Rev. Genomics Hum. Genet. 2001;2:435–462. doi: 10.1146/annurev.genom.2.1.435. [DOI] [PubMed] [Google Scholar]
- 51.Lee SS, et al. DAF-16 target genes that control C. elegans lifespan and metabolism. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- 52.An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee P, et al. Yeast Yak1 kinase, a bridge between PKA and stress-responsive transcription factors, Hsf1 and Msn2/Msn4. Mol. Microbiol. 2008;70:882–895. doi: 10.1111/j.1365-2958.2008.06450.x. [DOI] [PubMed] [Google Scholar]
- 54.Samuelson AV, et al. Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants. Genes Dev. 2007;21:2976–2994. doi: 10.1101/gad.1588907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolkow CA, et al. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- 57.Libina N, et al. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 58.Alcedo J, Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2004;41:45–55. doi: 10.1016/s0896-6273(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 59.Cypser J, Johnson TE. Hormesis extends the correlation between stress resistance and life span in long-lived mutants of Caenorhabditis elegans . Hum. Exp. Toxicol. 2001;20:295–296. doi: 10.1191/096032701701548070. [DOI] [PubMed] [Google Scholar]
- 60.Lin K, et al. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat. Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 61.Apfeld J, Kenyon C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell. 1998;95:199–210. doi: 10.1016/s0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]
- 62.Iser WB, et al. Insulin signaling in Caenorhabditis elegans regulates both endocrine-like and cell-autonomous outputs. Dev. Biol. 2007;303:434–447. doi: 10.1016/j.ydbio.2006.04.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bishop NA, Guarente L. Two neurons mediate dietrestriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 64.en Dijke, P. and Hill, C.S. New insights into TGF-β-Smad signalling. Trends Biochem. Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 65.Patterson GI, Padgett RW. TGF β-related pathways. Roles in Caenorhabditis elegans development. Trends Genet. 2000;16:27–33. doi: 10.1016/s0168-9525(99)01916-2. [DOI] [PubMed] [Google Scholar]
- 66.Wang J, Kim SK. Global analysis of dauer gene expression in Caenorhabditis elegans. Development. 2003;130:1621–1634. doi: 10.1242/dev.00363. [DOI] [PubMed] [Google Scholar]
- 67.Mangelsdorf DJ, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sluder AE, Maina CV. Nuclear receptors in nematodes: themes and variations. Trends Genet. 2001;17:206–213. doi: 10.1016/s0168-9525(01)02242-9. [DOI] [PubMed] [Google Scholar]
- 69.Motola DL, et al. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 70.Gerisch B, Antebi A. Hormonal signals produced by DAF-9/ cytochrome P450 regulate C. elegans dauer diapause in response to environmental cues. Development. 2004;131:1765–1776. doi: 10.1242/dev.01068. [DOI] [PubMed] [Google Scholar]
- 71.Gerisch B, et al. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev. Cell. 2001;1:841–851. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- 72.Antebi A, et al. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 2000;14:1512–1527. [PMC free article] [PubMed] [Google Scholar]
- 73.Arantes-Oliveira N, et al. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- 74.Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- 75.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 76.Bates G, et al. Huntington’s Disease. 3rd. Oxford University Press; 2002. [Google Scholar]
- 77.Gidalevitz T, et al. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 78.Roze E, et al. Pathophysiology of Huntington’s disease: from huntingtin functions to potential treatments. Curr. Opin. Neurol. 2008;21:497–503. doi: 10.1097/WCO.0b013e328304b692. [DOI] [PubMed] [Google Scholar]
- 79.Selkoe DJ. Developing preventive therapies for chronic diseases: lessons learned from Alzheimer’s disease. Nutr. Rev. 2007;65:S239–S243. doi: 10.1111/j.1753-4887.2007.tb00370.x. [DOI] [PubMed] [Google Scholar]
- 80.Skovronsky DM, et al. Neurodegenerative diseases: new concepts of pathogenesis and their therapeutic implications. Annu. Rev. Pathol. 2006;1:151–170. doi: 10.1146/annurev.pathol.1.110304.100113. [DOI] [PubMed] [Google Scholar]
- 81.Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 2003;4:49–60. doi: 10.1038/nrn1007. [DOI] [PubMed] [Google Scholar]
- 82.Brignull HR, et al. Polyglutamine proteins at the pathogenic threshold display neuron-specific aggregation in a pan-neuronal Caenorhabditis elegans model. J. Neurosci. 2006;26:7597–7606. doi: 10.1523/JNEUROSCI.0990-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morley JF, et al. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Satyal SH, et al. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hsu AL, et al. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 86.Jorgensen EM. WormBook, ed. The C. elegans Research Community, Wormbook. 2005. Gaba. [Google Scholar]
- 87.Fonte V, et al. Interaction of intracellular β amyloid peptide with chaperone proteins. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9439–9444. doi: 10.1073/pnas.152313999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fonte V, et al. Suppression of in vivo β-amyloid peptide toxicity by overexpression of the HSP-16.2 small chaperone protein. J. Biol. Chem. 2008;283:784–791. doi: 10.1074/jbc.M703339200. [DOI] [PubMed] [Google Scholar]
- 89.Cohen E, et al. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 90.Holzenberger M, et al. IGF-1 signaling and aging. Exp. Gerontol. 2004;39:1761–1764. doi: 10.1016/j.exger.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 91.Gems D, Partridge L. Insulin/IGF signalling and ageing: seeing the bigger picture. Curr. Opin. Genet. Dev. 2001;11:287–292. doi: 10.1016/s0959-437x(00)00192-1. [DOI] [PubMed] [Google Scholar]
- 92.Suh Y, et al. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3438–3442. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Blake MJ, et al. Stress-induced heat shock protein 70 expression in adrenal cortex: an adrenocorticotropic hormone-sensitive, age-dependent response. Proc. Natl. Acad. Sci. U. S. A. 1991;88:9873–9877. doi: 10.1073/pnas.88.21.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- 95.Van Eden W, et al. Stress, heat shock proteins, and autoimmunity: how immune responses to heat shock proteins are to be used for the control of chronic inflammatory diseases. Ann. N. Y. Acad. Sci. 2007;1113:217–237. doi: 10.1196/annals.1391.020. [DOI] [PubMed] [Google Scholar]
- 96.Calderwood SK, et al. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem. Sci. 2006;31:164–172. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]