Abstract
In muscle, the Sarco(Endo)plasmic Reticulum Calcium ATPase (SERCA) activity is regulated by two distinct proteins, PLB and SLN, which are highly conserved throughout vertebrate evolution. PLB is predominantly expressed in the cardiac muscle, while SLN is abundant in skeletal muscle. SLN is also found in the cardiac atria and to a lesser extent in the ventricle. PLB regulation of SERCA is central to cardiac function, both at rest and during extreme physiological demand. Compared to PLB, the physiological relevance of SLN remained a mystery until recently and some even thought it was redundant in function. Studies on SLN suggest that it is an uncoupler of the SERCA pump activity and can increase ATP hydrolysis resulting in heat production. Using genetically engineered mouse models for SLN and PLB, we showed that SLN, not PLB, is required for muscle-based thermogenesis. However, the mechanism of how SLN binding to SERCA results in uncoupling SERCA Ca2+ transport from its ATPase activity remains unclear. In this review, we discuss recent advances in understanding how PLB and SLN differ in their interaction with SERCA. We will also explore whether structural differences in the cytosolic domain of PLB and SLN are the basis for their unique function and physiological roles in cardiac and skeletal muscle.
Keywords: SERCA, Sarcolipin, Phospholamban, Ca2+ transport
1. Introduction
In cardiac and skeletal muscle, Ca2+ plays a central role in contractile physiology and is efficiently compartmentalized in the sarcoplasmic reticulum (SR). The SR membrane and T-tubular network are closely associated with the contractile machinery and coordinate the release and removal of Ca2+ during the contraction-relaxation cycle. In muscle at rest, the cytosolic Ca2+ concentration is in the nanomolar level; it rises to several micro molar during muscle contraction (1, 2). Ca2+ release from the SR lumen through the ryanodine receptor is responsible for muscle contraction, and the subsequent reuptake of Ca2+ by the Sarco(Endo)plasmic Reticulum Calcium ATPase (SERCA) pump leads to relaxation of the muscle (3). The rate and amount of Ca2+ taken up by the SR in cardiac vs. skeletal and fast vs. slow muscle types are regulated by the presence of specific SERCA isoforms (4). In addition, SERCA activity in muscle is regulated by Phospholamban (PLB- 52 aa) and Sarcolipin (SLN-31 aa) (Fig. 1), two small transmembrane proteins whose interaction with SERCA can alter the dynamics of Ca2+ cycling and SR Ca2+ load (4–6).
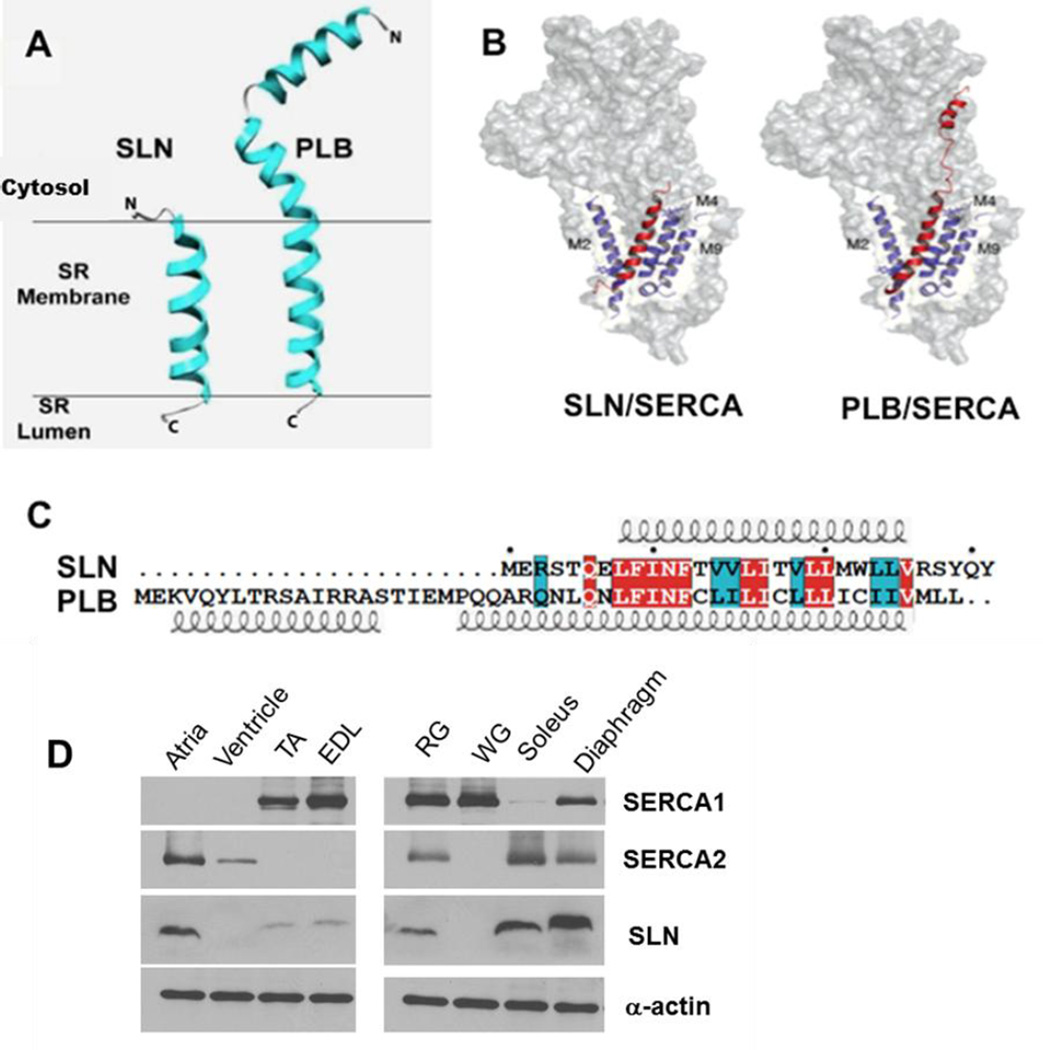
Figure 1. Comparison of SLN and PLB structure.
A, PLB has an extended N-terminal domain, whereas SLN has a shorter N-terminus. B, Both PLB and SLN bind to the same TM groove on SERCA (adapted from Traaseth et. al. (6)) C, SLN and PLB amino acid sequence from mouse, identical residues are highlighted in red and conserved residues in cyan. Both the N-terminal and C-terminal residues are distinct in SLN and PLB. D, Western blot analysis of SLN and SERCA expression in mouse cardiac and skeletal muscle tissues. α-actin was used as a loading control. SLN, sarcolipin; TA, tibialis anterior; EDL, extensor digitorum longus; RG, red gastrocnemius; WG, white gastrocnemius.
PLB is highly expressed in cardiac muscle, whereas SLN is quite abundant in skeletal muscle. SLN is also expressed in cardiac atria and to a lesser extent in the ventricle (5, 7–10). The physiological relevance of PLB is well known as a mediator of β-adrenergic response of the heart and it is an important regulator of cardiac ionotrophy and chronotrophy. The function of SLN was less clear until recently; some even thought that it was redundant in function. However, in vitro Ca2+ transport studies suggested that SLN binding to SERCA promotes uncoupling of Ca2+ transport from ATP hydrolysis. By this mechanism SLN can promote futile cycling of SERCA and increase heat production (11–13). Using genetically engineered mouse models for SLN, we recently showed that SLN is an important regulator of muscle-based thermogenesis and metabolism (14, 15). Interestingly, SLN-KO mice were able to maintain their core body temperature at thermoneutrality, but when exposed to acute cold (4°C) they were unable to maintain their body temperature as compared to their WT and PLB-KO counterparts. Furthermore, loss of SLN predisposed mice to develop diet-induced obesity whereas increased SLN expression in muscle offered resistance to obesity, suggesting that SLN can be recruited to increase oxidative metabolism (15, 16). These studies highlighted that SLN interaction with SERCA may serve an entirely different physiological role in muscle.
However, the mechanistic details of how SLN binding promotes uncoupling of SERCA Ca2+ transport and increases heat production remain unclear. Therefore, a major goal of this review is to discuss recent advances in our understanding of how PLB and SLN differ in their interaction with the SERCA pump. Another important focus of this review is to examine whether the unique cytosolic sequences in SLN and PLB might be responsible for their distinct functions. We tried to cite many relevant articles on SLN and PLB, but this review is not sufficient to cover all of the existing biological information on SLN and PLB. There are also excellent reviews on PLB that the reader may want to refer to for a greater understanding of PLB regulation of SERCA and its role in cardiac pathophysiology (5, 8, 9, 17).
2. SERCA is more than a calcium ion transport pump
The SERCA pump belongs to the P-type ATPase family and transports two Ca2+ ions into the SR lumen (in exchange of H+) (18–24). The SERCA pump is able to exploit chemical energy obtained from ATP hydrolysis to transport Ca2+ ions against a Ca2+ gradient across the SR membrane (osmotic energy); a part of the resulting chemical energy is released as heat (25). It comprises a single polypeptide chain that folds to form a transmembrane (TM) region and large cytoplasmic head. The TM region has 10 α-helices (M1–M10) of varying lengths, inclination and flexibility, that are connected to three large, highly mobile cytosolic domains. These domains are the nucleotide or ATP binding (N)-domain, the P-domain that gets phosphorylated by the γ-phosphate of ATP at a conserved Aspartate, and the actuator (A)-domain that coordinates the de-phosphorylation with the help of a conserved Glutamate (21). Two Ca2+ ions bind cooperatively at the high affinity sites I and II of the pump located within the TM region and open toward the cytosol (E1 state) (26). Large movements driven by ATP hydrolysis, phosphorylation, and dephosphorylation at the cytoplasmic domains are relayed to the TM region and cause re-arrangements in the TM helices, changing the affinity of the Ca2+ binding sites for directional transfer of Ca2+ into the SR lumen and for formation of the Ca2+-free (E2) state. The kinetic cycle of SERCA is represented in black, in Fig. 3. The vectorial transfer of Ca2+ ions leads to the formation of a Ca2+ ion gradient across the SR that in turn affects SERCA activity by back inhibition (27).
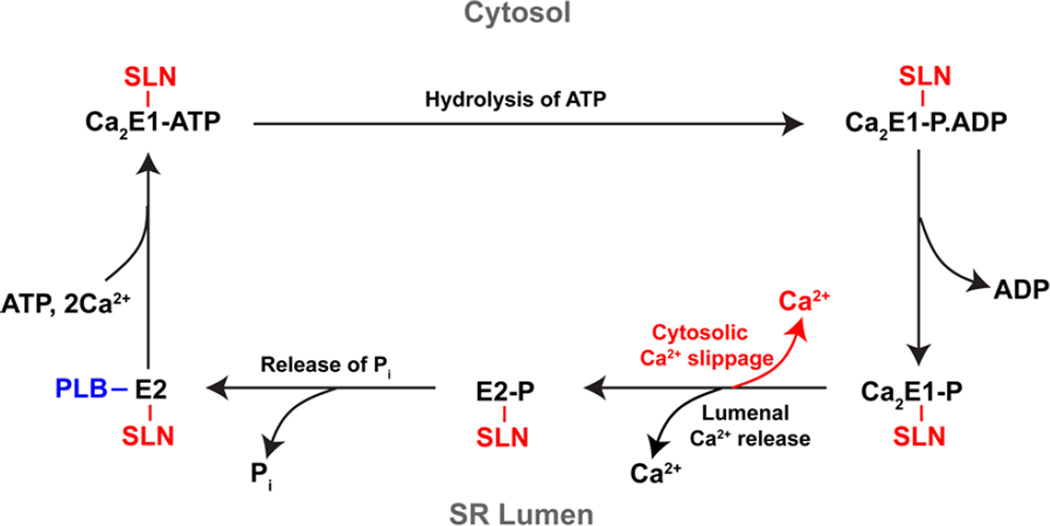
Figure 3. SLN binding to SERCA causes uncoupling of Ca2+ transport.
Upon binding of ATP and two Ca2+ ions in the E1 conformational state, SERCA becomes auto-phosphorylated by hydrolysis of the ATP and transitions to the high energy Ca2E1-P state, which releases the Ca2+ into the SR lumen in exchange for protons and converts to the low energy E2-P state. De-phosphorylation of the E2-P state results in the Ca2+-free, E2 conformation. E2 turns over to the E1 state in the presence of ATP and is able to undergo a subsequent cycle only on binding two Ca2+ ions from the cytosol. The interaction of PLB is represented in blue and that of SLN is shown in red. The functional interactions of PLB with SERCA and Ca2+ binding to SERCA are mutually exclusive. PLB binds to the Ca2+-free E2 state of SERCA pump and prolongs the pump in E2 state. On the other hand, SLN binds to SERCA in the E2 state but remains bound to SERCA during the kinetic cycle, which results in uncoupling of the pump and slippage of Ca2+ back into the cytosol.
The Ca2+ gradient across the SR membrane can influence the kinetic cycle of SERCA, resulting in different outcomes (28). When the SR luminal Ca2+ concentration is low, SERCA cycles most efficiently in the forward direction with a higher Ca2+/ATP coupling ratio, hydrolyzing one ATP to transport two Ca2+ ions into the lumen; however, this is not case when the luminal Ca2+ gradient is high. Studies by deMeis and others suggested that when the luminal Ca2+ concentration is high, SERCA ATP hydrolysis becomes uncoupled from Ca2+ transport and the uncoupled ATPase activity leads to more heat production (27). Further, when the ADP/ATP ratio is low, the Ca2+ transport is exothermic, and the formation of the gradient increases the amount of heat produced from the hydrolysis of each ATP molecule (29). However, when the ADP/ATP ratio is high, uncoupled ATPase activity is abolished, and the Ca2+ transport is endothermic. These studies were done in isolated SR vesicles, however, and the role of SERCA uncoupling in heat production in vivo remained unclear (27, 29–31).
3. PLB and SLN are integral SR membrane proteins that regulate SERCA function
3.1. PLB is an affinity modulator of SERCA
PLB is a 52 amino acid protein that has been extensively studied because of its role in cardiac muscle contractility. There are several excellent reviews focused on PLB and its role in cardiac contractile function, therefore we will limit our description on PLB. It was first identified as a phosphoprotein by Tada et. al., in 1975 (32, 33). Extensive work done in the MacLennan, Jones and Kranias laboratories has clearly demonstrated the regulation of SERCA by PLB (5, 7, 10, 17, 34–50). They showed that active PLB is a monomer and can easily associate into pentamers (43, 51–54). The phosphorylated monomers dissociate from SERCA and form oligomers to eventually pentamerize, forming a pool of inactive PLB unable to inhibit SERCA. These laboratories also showed that PLB pentamers are in a dynamic equilibrium and dissociate into active monomers on de-phosphorylation by protein phosphatase-1 (PP-1) when the cytosolic Ca2+ concentration is sufficiently low (55). Monomeric, unphosphorylated PLB binds to SERCA at low (< 1µM) Ca2+ concentrations and inhibits SERCA from cycling by affecting the rate of SERCA Ca2+ uptake as well as net ATP hydrolysis (48). However, at higher Ca2+ concentrations PLB dissociates from SERCA, allowing the pump to become fully activated (Fig. 2) (33, 56). Similarly, during adrenergic activation of the heart phosphorylation of PLB at Ser16 and/or Thr17 relieves inhibition of SERCA, accelerating SERCA mediated Ca2+ uptake and muscle relaxation. Further studies have identified that PLB interacts with a Ca2+-free, ATP bound, E2 conformation of SERCA, slowing the E2 to E1 transition of SERCA and reducing the apparent affinity of the pump for Ca2+ (5, 34–38, 40, 41, 50)(Fig. 2, 3). Thus PLB acts as a brake on SERCA until it is dissociated by either phosphorylation or by high Ca2+.
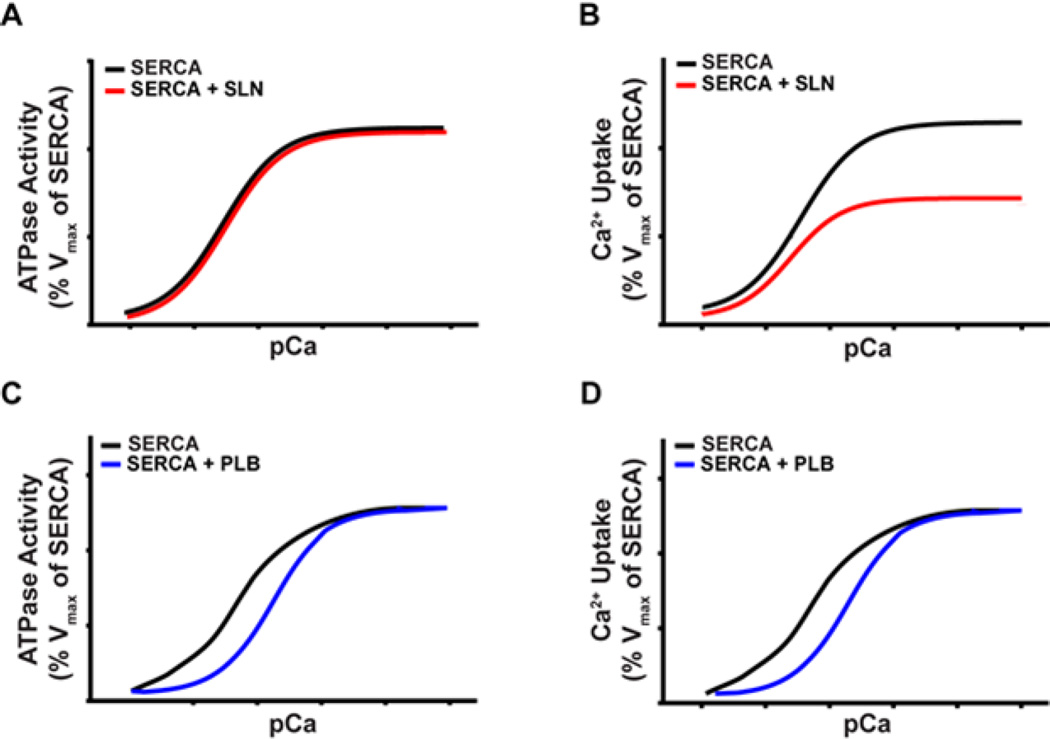
Figure 2. SLN and PLB differently affect SERCA ATPase activity and Ca2+ uptake.
Representative Ca2+ dependent ATPase and Ca2+ transport activity profiles of SERCA are shown. A and B, SLN does not inhibit SERCA ATPase activity but reduces the Vmax of SERCA Ca2+ transport. C and D, PLB inhibits SERCA ATPase activity and Ca2+ transport at low Ca2+ concentration but has no effect at high Ca2+ concentration.
Recent studies on human PLB have shown that mutations and deletions in the PLB gene can cause a detrimental effect on cardiac function. The mutation R9C or deletion of Arg14 in the cytosolic domain of human PLB has been reported to cause dilated cardiomyopathy (45, 57). These mutants fail to regulate the SERCA pump in a reversible manner. They cannot be phopsphorylated because amino acids Arg9 and Arg14 are required for protein kinase A (PKA) binding and recognition (58, 59). In addition, the L39stop mutation which truncates PLB is a loss of function mutation (60). Both the loss and/or alteration of PLB phosphoryaltion can cause heart failure because PLB is required for the increase in heart rate by two to three fold during flight-or-fight response (44). These studies further established that PLB regulation of SERCA is central to maintaining cardiac function at all times.
3.2. Identification of SLN as a regulator of SERCA
MacLennan (1974) first identified a ‘proteolipid’ in purified rabbit SR preparations that was soluble in acidic chloroform/methanol because it was highly hydrophobic and closely associated with the membrane phospholipids (61). The true function of this protein remained unknown for a long period of time with several conflicting ideas over the years. Initial studies suggested that this protein enhanced the coupling ratio of SERCA; later it was speculated that the protein acted as an ionophore in certain co-reconstitution studies (62). Few studies questioned its role in regulating SERCA activity and many suggested that it had no effect on SERCA Ca2+ uptake (63). In 1992, Wawrzynow et. al. determined the protein’s complete 31 amino acid sequence, confirmed its molecular weight as 3733Da, and named it Sarcolipin (SLN) with reference to its nature and origin (64). Early on, most of the characteristics of SLN were compared with and assumed to be similar to what was already known for PLB. Similar gene structure and homology of protein sequences of the TM of PLB and SLN led to the conclusion that SLN and PLB originated from the same family of proteins and, similar to PLB, the N-terminus of SLN is cytosolic and C-terminus, luminal (65, 66). There were conflicting views on the regulation of SERCA by SLN that emerged with further in vivo studies in skeletal and cardiac muscles. Nonetheless, recent in vivo studies involving SLN gene transfer in wild type as well as transgenic mice established that SLN by itself is capable of regulating SERCA and affecting muscle contractility (67–71). However, the mechanism of SERCA regulation by SLN was not clear until recently.
3.3. Regulation of SLN and PLB expression in development and disease states
PLB and SLN are expressed differentially in cardiac and skeletal muscle tissues across species. PLB expression is abundant in cardiac muscle and is present at low level in slow twitch soleus muscle. PLB expression and phosphorylation status is modified in cardiac hypertrophy and heart failure. The depressed contractility in experimental and human heart failure is partially attributed to increased inhibition by PLB due to: (a) increases in PLB/SERCA2 ratio; and (b) decreases in PLB phosphorylation. There is extensive literature on this and a number of reviews (5, 9, 72). More recent studies have shown that mutations or loss of PLB in humans can lead to dilated cardiomyopathy and death (57, 60).
SLN expression is relatively high in embryonic and neonatal skeletal muscle, but its expression is down regulated in adult fast twitch muscles of rodents (73). It is, however, expressed in adult slow/oxidative muscle fibers including the diaphragm and soleus (73, 74). Interestingly, SLN is also expressed at high levels in atria but very low levels in the ventricle of rodents (Fig 1D). SLN expression was shown to be increased in pacing induced heart failure in dogs but decreased in atria of dogs with ventricular fibrillation (75). SLN was found to be upregulated in ventricles of human patients with mitral regurgitation (76). Recent studies using SLN-KO mice showed that loss of SLN causes atrial remodeling and predisposes the mice to atrial arrhythmias upon aging. Further, loss of both PLB and SLN in the mouse heart can lead to excessive remodeling including hypertrophy, fibrosis and heart failure (77). These studies collectively suggest that SLN regulation of SERCA is critical for maintaining a fine Ca2+ gradient between cytosol and SR, and that the loss of SLN in atria leads to abnormal Ca2+ cycling and remodeling of the atria causing arrhythmias (75, 77–79).
4. SLN and PLB have different effects on SERCA pump activity
4.1. SLN affects the Vmax of SERCA Ca2+ uptake
It is well known that PLB binding to SERCA affects the apparent affinity for Ca2+ but does not affect the Vmax of Ca2+ uptake (Fig. 2) (34, 38–40, 48). The effect of SLN on SERCA has been studied extensively using in vitro functional assays. Although initial studies were less certain regarding the effect of SLN on SERCA, recent studies from our laboratory suggest that SLN does not affect pump affinity for Ca2+ but decreases the Vmax of SERCA Ca2+ uptake. Despite a reduction in SERCA Ca2+ uptake Vmax, the presence of SLN did not affect the total amount of ATP hydrolyzed (Fig. 2) (11, 12, 80). This suggests that, in the presence of SLN, the hydrolysis of ATP is not completely linked to the transport of Ca2+.
Using reconstituted synthetic SLN and SERCA, Lee et. al. first proposed the idea that SLN binding to SERCA could promote uncoupling of SERCA ATP hydrolysis from Ca2+ transport. They found that at saturating protein ratios of SLN/SERCA, SLN did not affect ATP hydrolysis and that the affinity of SERCA for Ca2+ remained unchanged (11, 12, 81). They further showed that Ca2+ accumulation in the vesicles decreased and that the heat released by SERCA increased in the presence of SLN. Based on this, it was suggested that SLN binding to SERCA prevents release of Ca2+ into the lumen and promotes slippage of Ca2+ back to the cytosol. The energy from the resulting ATP hydrolysis would thus be released as heat, without any Ca2+ transport. These initial in vitro studies predicted that SLN could play a role in muscle thermogenesis (14).
4.2. SLN plays a role in muscle thermogenesis
The role of SLN in muscle function remained unclear for quite some time and could not be addressed easily. Therefore, we decided to generate SLN-KO mice and study how SLN impacts muscle function. Initial studies did not clearly indicate that loss of SLN had any impact on muscle growth and/or function since the KO and WT animals could not be distinguished easily. To test if SLN could be an important player in muscle thermogenesis we decided to challenge the mice with acute cold (4°C) and found that the SLN-KO mice were unable to maintain their body temperature, developed hypothermia, and died if not removed from the cold (14). On the other hand, similarly cold challenged PLB-KO mice did not develop hypothermia (13). Further, acute cold induced hypothermia of SLN-KO mice was rescued by reintroduction of SLN. These cold exposure studies on WT, SLN-KO and PLB-KO mice indicated that SLN is indeed essential for muscle thermogenesis (13). These in vivo studies using SLN null mice validated the initial in vitro findings, which suggested that SLN interaction with SERCA promotes uncoupling of SERCA, resulting in increased ATP hydrolysis and heat production (13).
Furthermore, we recently showed that SLN is an important regulator of metabolism; loss of SLN resulted in diet-induced obesity compared to WT animals, which upregulated SLN expression in oxidative muscle (14, 15). We also found that SLN overexpression can affect basal metabolic rate and that higher SLN expression can be effective in providing resistance against diet induced obesity in mouse models (13). These studies suggested that uncoupling of SERCA not only contributes to heat production but also to increased energy expenditure, thereby affecting whole body metabolism. Further work is necessary to understand the detailed mechanism by which SLN regulates energy expenditure in muscle.
4.3. The interaction of SLN with SERCA
The mechanism of PLB interaction with SERCA has been known for some time. However, the role of SLN was unclear in the beginning, and some even questioned its relevance as a regulator of the SERCA pump. Studies using co-expression of PLB and SLN with SERCA appeared to have an additive effect on the inhibition of SERCA, and it was suggested that SLN and PLB bind together to SERCA and cause super-inhibition (82). Based on this data, it was proposed that SLN does not directly bind to SERCA but mediates its effect on SERCA through PLB. This binding of SLN could disrupt PLB pentamer formation and increase the interaction of PLB with SERCA, thereby inhibiting SERCA to a much greater extent. However, subsequent overexpression of SLN in the heart did not alter PLB expression, phosphorylation, or its monomer/pentamer ratio, suggesting independent regulation of SERCA by SLN (67–69). Functional studies by SLN overexpression in PLB-KO mouse hearts suggested that SLN can interact directly with SERCA and does not require PLB (67, 68, 74). Direct demonstration of SLN binding to SERCA was lacking, however initial studies probed SLN/SERCA interaction using co-immunoprecipitation assays, but these assays are not particularly suitable for membrane proteins since they require solubilization of membranes, disrupting native protein structure (82–86). Therefore we used a chemical cross-linking approach to determine the dynamic interaction of SLN with SERCA during Ca2+ transport (13, 14, 80). The results from these studies have shown that monomeric SLN directly interacts with SERCA (13, 14, 80). The cross-linking of specific residues at both cytosolic and luminal interfaces of the membrane suggested that SLN binds to the groove on the TM surface of SERCA, formed by helices M2, M4, M6 and M9; this groove is known to bind PLB as well (13, 14, 84). However, mutations introduced along this binding groove in SERCA had different effects on the binding characteristics of SLN and PLB to SERCA. Some mutations affected only PLB binding but not SLN, suggesting that SLN and PLB interact with a different set of residues within the TM groove (13).
4.4. SLN binds to Ca2+ bound SERCA and promotes uncoupling of SERCA
The mechanism of SLN interaction with SERCA remained a challenge for a long time; due to its small size and highly hydrophobic nature since it could not be easily purified in a manageable quantity. This was partially overcome using co-expression of SLN and SERCA in HEK cell culture, isolating the protein-expressing microsomes. Protein cross-linking studies using these microsomes showed that SLN binding to SERCA was maximal in the presence of ATP. Interestingly, SLN is able to bind to SERCA even at high Ca2+ and remains bound to SERCA throughout its kinetic cycle. PLB, on the other hand, only interacts with the Ca2+ free SERCA (E2 form of SERCA), and it is unable to bind to the Ca2+-bound phospho-intermediates of SERCA (Fig. 4) (13, 14, 34, 35, 38, 41).
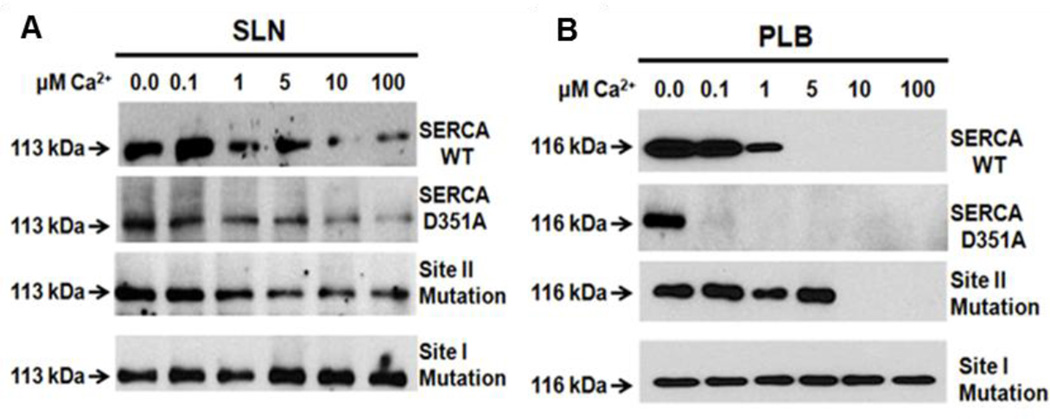
Figure 4. SLN interacts with SERCA in the presence of high Ca2+concentration.
SLN or PLB interaction with SERCA was studied using chemical cross-linking under increasing Ca2+ concentrations (Sahoo et.al. (13)). Only SLN cross-links with SERCA in the presence of high Ca2+-up to 100µm. Interestingly, PLB interacts with SERCA at nanomolar Ca2+ concentration and its binding is abolished at Ca2+ above 1µM (from Sahoo et.al. (13)). Cross-linking of A, SLN and B, PLB, with WT SERCA, phosphorylation–defective (D351A-SERCA), and Ca2+ binding site mutants. (Site II mutant-E309Q-SERCA and Site I mutant-E771Q-SERCA). Mutation of site I abolishes Ca2+ binding to both sites. These cross-linking studies were performed using N30C-PLB or E7C-SLN that cross-linked with C318 of SERCA.
During Ca2+ transport, Ca2+ binds to sites I and II of SERCA, sequentially, in a cooperative manner (87). Mutagenesis of the Ca2+ binding site I excludes the Ca2+ binding at site II and thus the SERCA pump is unable to hydrolyze ATP to form further phospho-intermediates, whereas mutagenesis of site II still allows Ca2+ binding to site I but does not allow cycling (88). We took advantage of these mutants (Fig. 4) to show that PLB and SLN bind equally well to the site I SERCA mutant (which cannot bind Ca2+) and increasing cytosolic Ca2+ fails to dissociate either PLB or SLN (13, 37, 80). On the other hand, when site II is mutated (SERCA can still bind one Ca2+ at site I) only SLN remains bound to SERCA while PLB easily dissociates from SERCA at high Ca2+ (10µM). In addition, the use of the phosphorylation site D351A mutant (which binds Ca2+ to both sites but cannot cycle) allowed us to demonstrate that only SLN (not PLB) can bind to Ca2+ bound SERCA and, further, showed simultaneous binding of SLN and Ca2+ to SERCA. This confirmed that Ca2+-induced conformational change precludes the functional interaction of PLB with SERCA, therefore PLB only inhibits Ca2+ free SERCA. In contrast, the ability of SLN to interact with Ca2+ bound SERCA during its kinetic cycle allows SLN to cause uncoupling of SERCA (Fig. 3, 4). When the SERCA pump is uncoupled, Ca2+ is released back to the cytosol at the expense of ATP hydrolysis. Thus SLN could promote futile cycling of SERCA pump, increasing energy cost and accelerating metabolism.
4.5. PLB and SLN bind to the same TM groove formed by helices M2, M6 and M9
Recent crystallographic studies of SLN bound SERCA provided an important breakthrough on SLN/SERCA interaction. The crystal structures of SLN/SERCA published by Toyoshima et al and Winther et. al. reaffirmed the protein cross-linking results and proved that monomeric SLN binds to a TM groove formed by helices M2, M6 and M9. These elegant studies suggested that SLN binds to a novel, Mg2+ bound SERCA intermediate state in the SERCA kinetic cycle; the studies provided valuable insights about the transition of SERCA from the Ca2+ free E2 state to the Ca2+ bound E1 state (89, 90). Toyoshima et. al. proposed that, similar to PLB, the binding of SLN would hinder the Actuator domain movement of SERCA and cause it to undergo a larger than normal rotation to enable Ca2+ entry into the Ca2+ binding sites within the pump. Furthermore, they suggested that SLN could inhibit Ca2+ transport by hindering the sliding of M1 and M2 TM helices that is required for transitioning of the pump to a phospho-intermediate state. Winther et. al. showed that the binding of SLN to SERCA promotes stabilization of the Ca2+ binding sites, and also mentioned that SLN might have to undergo a displacement to be able to remain bound to SERCA during the E1 to Ca2E1-P transition. On the other hand, the Jones group studied the crystal structure of PLB with SERCA and suggested that PLB binding to SERCA collapses the Ca2+ binding sites and inhibits the pump from Ca2+ binding and transport (35). Overall, these structural studies suggest that PLB and SLN may employ a different set of residues for their respective interaction with SERCA and may operate via different mechanisms. The crystal structures were however unable to resolve the interactions at the N and C-termini of SLN owing to the dynamic nature of these regions.
5. Structural differences in SLN and PLB are the basis for their unique functions
5.1. SLN and PLB differ significantly at their N and C-termini
Both SLN and PLB are α-helical TM proteins with unique cytosolic and luminal domains (Fig. 5) (91). In SLN, residues 1–7 form an unstructured N-terminus andresidues 27–31 form an unstructured C-terminus, whereas the TM region (that constitutes an α-helix) comprises residues 8–26. SLN has a unique C-terminal RSYQY sequence that protrudes in to the lumen of the SR. PLB, however, has no such tail sequence; its last three residues (MLL) continue as a part of the TM α-helix (65, 92, 93). Alanine mutagenesis studies have shown that the C-terminal tail of SLN, especially residues Tyr29 and Tyr31, is critical for regulation of SERCA (65). It has also been suggested that the C-terminus plays a role in SLN localization and interaction with SERCA. Studies by Gramolini et. al. showed that attaching a C-terminal FLAG sequence to SLN C-terminus generated a ‘super-inhibitor’ of SERCA, while an N-terminal FLAG sequence did not affect the function of SLN (85). They also showed that the C-terminal tail region of SLN, from Trp22 to Tyr31, is essential to maintaining the stability and retention of SLN in the ER membrane, in the absence of SERCA (85). Other studies have suggested that the C-terminal sequence is important for retrieval of SLN from the ER-Golgi intermediate compartment and its maintenance in the SR membrane (85, 94).
Figure 5. Comparison of SLN and PLB protein sequences in mammals.
SLN is 31 aa long with 7 unique cytosolic residues and a highly conserved C-terminus (RSYQY). PLB is 52 aa long, and has an extended cytosolic domain of 30 aa that includes the phosphorylation sites Ser16 and Thr17 (**). PLB and SLN sequences are highly conserved within the same family but not between the two proteins except in the TM region. Identical residues between SLN and PLB in the TM region are highlighted in red. The conserved residues in PLB and SLN are highlighted in cyan. Note that the major differences between PLB and SLN occur at the N and C-termini.
Further, NMR and mutagenesis approaches revealed that Tyr29 and Tyr31 in the SLN C-terminal region provide additional anchorage by hydrophobically interacting with the aromatic residues in the M1–M2 luminal loop of SERCA (84, 95). A recent study by Gorski et. al. showed that the C-terminal domain of SLN could be functionally transferred to PLB to further enhance SERCA inhibition (96). However, our studies using chimeras of SLN and PLB (involving C-terminal exchange) suggested that the SLN C-terminus, when transferred to PLB, can increase binding affinity to SERCA but fails to uncouple the SERCA pump (80).
5.2. SLN and PLB share sequence homology in their TM domain
Structural comparison of SLN and PLB shows significant homology only in their TM region (Fig. 5); the SLN TM domain is 19 residues long whereas the PLB TM has 21 residues. Interestingly, they share 7 identical residues and 7 hydrophobically conserved residues (97). High homology in this region and conserved gene structure suggests that both molecules evolved from a common ancestral protein (66, 98). The TM of PLB has a unique arrangement of residues that allow pentamer formation with other PLB molecules. One side of PLB TM residues is involved in forming a leucine zipper with other PLB monomers while residues on the other side of the helix directly interact with SERCA to regulate its function (5, 17). Owing to the TM homology of SLN and PLB, it has been proposed that, similar to the pentamer of PLB, SLN monomers may be able to form oligomers. Hellstern et. al. showed that synthetic SLN did not form oligomers in liposomes as readily as PLB. It could be coerced into oligomerization only on drastically increasing the concentration of SLN, however. Using FRET analysis of SLN and SERCA fluorescent fusion proteins, Autry et. al. also demonstrated that SLN forms dimers and oligomers; however, only the monomer interacts with SERCA (99).
Recent studies suggest that TM regions in PLB and SLN play important roles in their interaction with SERCA and the regulation of SERCA activity. The nature of PLB TM interactions with SERCA has been thoroughly mapped using mutagenesis, co-immunoprecipitation and cross-linking. These studies showed that the PLB TM helps to bind and localize onto the TM groove on the surface of SERCA, within the SR membrane. Although it is well known that the cytosolic domain of PLB contains the phosphorylation sites Ser16 and Thr17, both TM (domain II) and cytosolic domain of PLB play important roles in SERCA inhibition. At high Ca2+ concentrations, PLB may be dislodged from the SERCA groove due to conformational change and closure of the groove upon Ca2+ binding (89, 90). Mutation within the TM region greatly influenced PLB regulation of SERCA, which suggested that the TM is equally important for PLB function (80). In contrast, it has been shown that the TM region in SLN is critical for its localization and interaction with SERCA, but that SLN having only the TM and C-terminal residues did not affect SERCA function despite the protein remaining bound to SERCA (13, 90). These studies therefore highlight that the TM region is critical for protein binding and interaction with SERCA.
5.3. The N-terminal interactions of PLB and SLN with SERCA
The N-terminal domain (Ia and Ib) of PLB consists of 30 residues, assumes an α-helical structure, and is fairly well-conserved across species (Fig. 5). However, its nature of interaction with SERCA still remains a mystery. The PLB N-terminal domain can be phosphorylated at Ser16 and Thr17 by PKA and by Ca/CaM kinaseII (CaMKII), respectively, and the phosphorylation status modulates PLB interaction with SERCA (5, 17). The phosphorylated PLB is unable to inhibit SERCA but studies have shown that it remains physically associated with the pump (35, 100–103). This altered interaction remains to be investigated in detail (36). Phosphorylation causes an allosteric change in PLB that relieves its inhibition of SERCA and promotes the pentamer formation (101,104). NMR studies have suggested that PLB phosphorylation causes unwinding of the helix in domain Ib that converts the L-shaped, ground ‘T’ state or an excited R state of PLB to a more extended and SERCA bound ‘B’ state (Fig. 6). Only the ‘T and R’ state of PLB are able to inhibit SERCA, while the ‘B’ state is noninhibitory and increases SERCA activity (101, 103). The detailed mechanism of the PLB N-terminal interaction with SERCA is still unknown. Cross-linking and mutagenesis studies of PLB N-terminal residues have shown physical interaction between Lys3 in domain 1a of PLB with residues 397–400 in the N domain of SERCA (105). Further, domain Ib of PLB and the cytosolic loop between M6 and M7 of SERCA have been shown to interact with each other (47). However, these interactions could not be verified and studied in detail (41). Moreover, the location of the PLB N-terminus on SERCA could not be resolved in the recent crystal structure (35). Thus we still do not know which residues in SERCA physically interact with the PLB N-terminus and how these interactions are regulated by Ca2+.
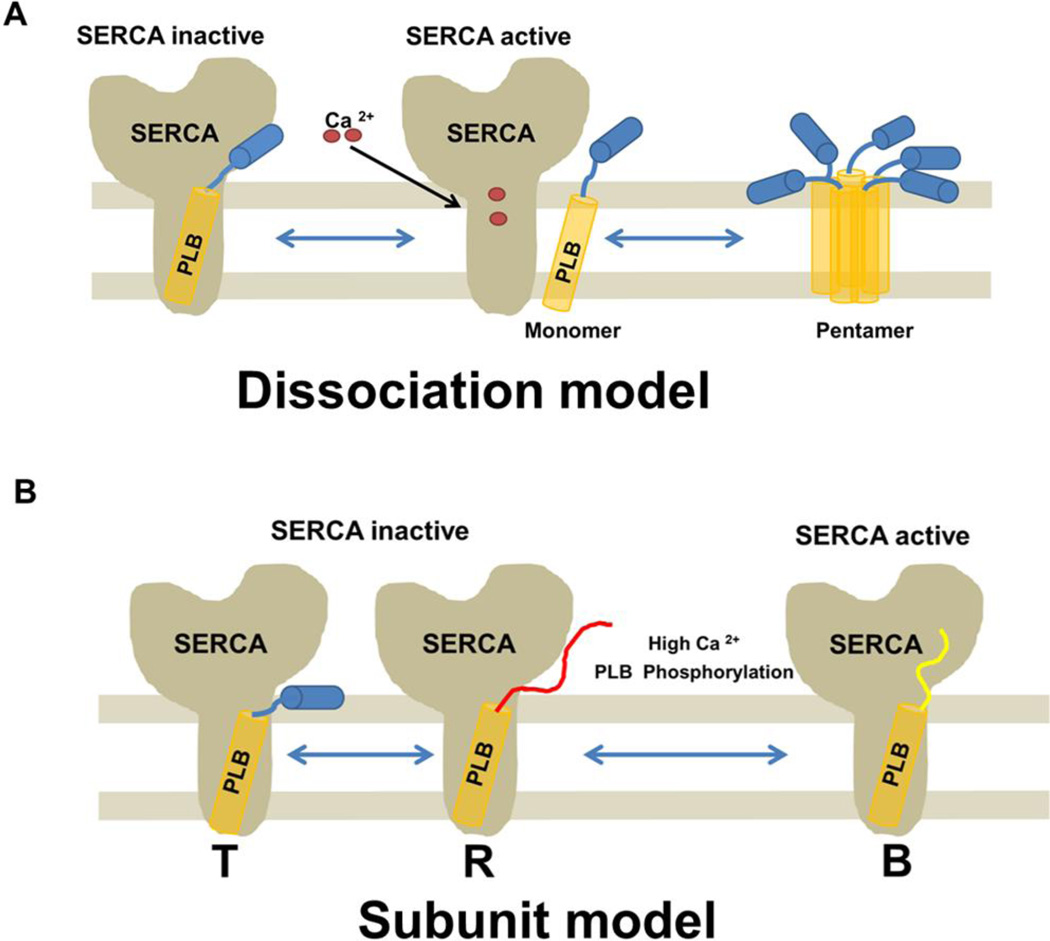
Figure 6. Two different models showing PLB interaction with SERCA.
A, Dissociation Model: cross-linking and co-immunoprecipitation studies suggest that PLB dissociates from the SERCA groove upon Ca2+ binding. In this model the PLB monomer binds to Ca2+-free SERCA, thereby inhibiting SERCA, but dissociates upon Ca2+ binding causing pump activation. The free PLB monomers then re-associate to form pentamers. B, Subunit model: This model is based on NMR studies in reconstituted vesicles. This model shows that PLB remains bound to SERCA but that the cytoplasmic regulatory domain in PLB interconverts between three different states: a ground ‘T’ state (helical and membrane associated), an excited ‘R’ state (unfolded and membrane detached), and a ‘B’ state (extended, enzyme bound and noninhibitory). Phosphorylation at Ser16 of PLB shifts the populations toward the ‘B’ state, increasing SERCA activity.
The relatively short, 7 residue SLN N-terminus varies across species. The 2ERSTQE sequence in rodents changes to 2ERSTRE in rabbit and to 2GINTRE in primates. Unlike the PLB cytosolic domain, the functional relevance of the SLN N-terminus is not well characterized. SLN contains a Thr5 residue that has been proposed to be the phosphorylation site; this could affect its function in a reversible manner as found for PLB. Initial studies by Gramolini et. al. showed that Serine/Threonine Kinase-16 (STK-16) could phosphorylate SLN and reverse the SLN inhibition of SERCA (71). These studies suggested that SLN could be phosphorylated either at Ser4 and Thr5 but did not successfully distinguish which specific residue was phosphorylated. Using adenoviral gene transfers of mutant SLN (T5A and T5E) in rat ventricular myocytes, Bhupathy et. al. showed that SLN inhibition of SERCA could be relieved on phosphorylation at Thr5 of SLN by CaMKII; however, evidence for such phosphorylation is lacking in vivo and the relevance of SLN phosphorylation to function is questionable (106). Thus far, the mechanism of SLN interaction through its N-terminal residues with SERCA is unclear. The recent SLN/SERCA co-crystal studies have also failed to localize the N-terminal interactions on SERCA.
5.4. The N-terminus of SLN is an important domain for the uncoupling of SERCA
A number of groups have tried to understand how SLN regulates SERCA activity using mutagenesis and chimeric proteins made between SLN and PLB. Domain swapping between SLN and PLB can be exploited to better understanding if functions can be assigned to individual domains/regions and transferred between them. Using this and the mutagenesis approach, we have recently shown that SLN requires its N-terminus for its uncoupling function (80). Deletion of the N-terminus caused SLN to constitutively remain in the SERCA groove. This truncated SLN was insensitive to an increase in Ca2+ concentration and failed to affect SERCA function. This suggests that the TM and C-terminus are sufficient for SLN to bind to SERCA; however, mere occupation of the groove is insufficient for its function, which may require dynamic interaction between the N-terminus, the TM, and the C-terminus of SLN for its regulation of SERCA.
Swapping of the SLN N-terminus with that of PLB caused the chimeric protein to behave much like PLB, suggesting that the N-terminal domain of PLB plays a dominant role. On the other hand, replacing the PLB N-terminus with the SLN N-terminus promoted SLN-like activity, pointing towards an important role for the N-terminus. Molecular dynamics simulations of SLN bound to SERCA further support our finding that the SLN N-terminus is crucial for SLN function. Significant changes in interactions between SERCA residues due to altered dynamics of Glu45 and Arg324 in SERCA were observed on deletion of the SLN N-terminus. We therefore propose that interaction of SERCA with N-terminal residues of SLN could interfere with Ca2+ occlusion and promote slippage. The next crucial step would be to determine the residues in SERCA that interact with SLN and lead to uncoupling.
6. Perspectives
Emerging data has revealed new insights on the mechanism of SLN interaction with SERCA and suggest that it is quite different from the PLB-SERCA interaction. PLB acts as an inhibitor and affinity modulator of SERCA whereas SLN is an uncoupler of SERCA resulting in increased ATP hydrolysis and heat production (Fig. 7). The recently published data using protein cross-linking provide new details on SLN interaction with SERCA; the functional differences seem mainly to stem from the presence of a short unstructured 7-residue N-terminal stretch in SLN compared to an extended cytosolic α-helical domain in PLB. Although we do not have a complete understanding of how SLN binding to SERCA promotes uncoupling, the existing data suggest that SLN binds to Ca2+ bound SERCA and remains in the SERCA TM groove during the Ca2+ transport cycle, and that the ability of SLN to remain in the TM groove may be responsible for its uncoupling of Ca2+ transport.
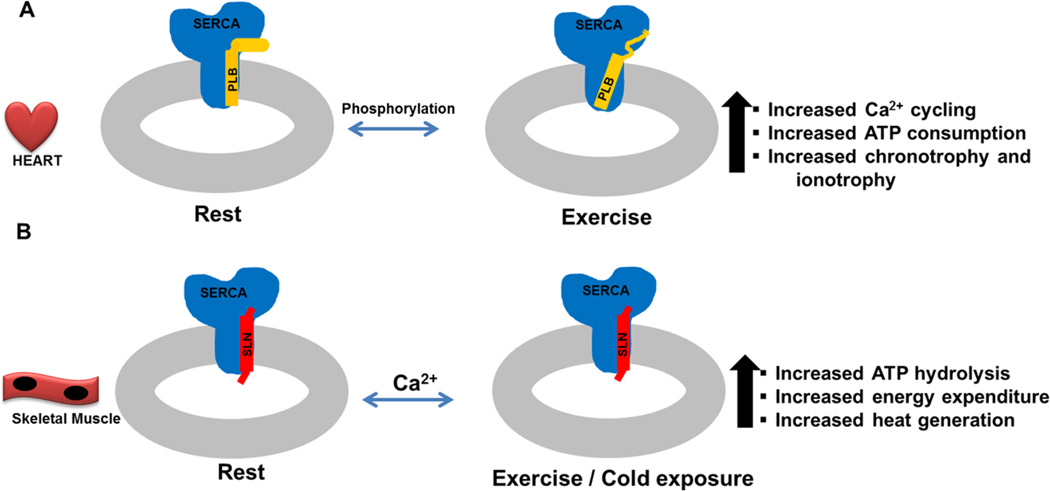
Figure 7. Regulation of the SERCA pump by PLB and SLN in the heart and skeletal muscle.
The heart and skeletal muscle express different isoforms of SERCA, ryanodine receptor, L-type Ca2+ channel and calsequestrin. The mechanisms regulating EC-coupling in the heart are fundamentally different in the Ca2+ entry through L-type channels that are essential for SR Ca2+ release and overall Ca2+ homeostasis. A, In the heart muscle β-adrenergic signaling plays a dominant role in regulating Ca2+ cycling and beat-to-beat function of the heart. PLB phosphorylation is central to the increased ionotrophy and chronotrophy of the heart. B, Skeletal muscle largely relies on SR Ca2+ store for EC coupling, and Ca2+ cycling in skeletal muscle is not dependent on β-adrenergic signaling. SLN expression is tissue specific (high in oxidative, while low in glycolytic muscle), therefore its effect on SERCA is modulated by its expression level rather than phosphorylation status as is known for PLB. SLN uncoupling of SERCA increases during exercise and cold adaptation resulting in increased heat production and energy expenditure.
The mechanism of PLB inhibition of SERCA is far from completely understood. Although phosphorylation of PLB at Ser16 and Thr17 play important roles in relieving inhibition of SERCA, it is unclear where exactly the PLB N-terminus interacts with SERCA and what happens to this interaction upon phosphorylation. While cross-linking and co-immunoprecipitation studies suggest that the PLB dissociates from the SERCA TM groove upon Ca2+ binding/phosphorylation, the FRET analyses indicate it may assume a different conformation and bind to a different site (100, 104, 107). Thus there is still disagreement about whether PLB dissociates from SERCA or moves to a different site upon Ca2+ binding and/or phosphorylation (13, 36, 37, 40–42, 100, 103, 105, 107, 108).
Our studies in whole animal models have shown that SLN is an important player in muscle thermogenesis whereas PLB is not required for muscle-based thermogenesis. In a recently published study we have also shown that SLN plays a role in muscle energy expenditure and whole body metabolism; overexpression of SLN protects against diet-induced obesity in mice by enhancing oxidative metabolism in muscle (15, 16). These data suggest that the uncoupling of SERCA by SLN leads to increased energy demand. However, the detailed signaling mechanisms by which the increased cytosolic Ca2+ levels and an increased ATP demand caused by the SLN lead to these metabolic changes remains to be investigated.
The interaction of SLN with SERCA has given us a new perspective toward the role of the SERCA pump, not only as a pump that regulates cytosolic Ca2+ levels in all tissues, but as a means to modulate energy metabolism in muscles. These studies led us to propose that SLN is an attractive target for increasing energy expenditure in muscle to combat weight gain and obesity.
Highlights.
SERCA regulation by PLB and SLN is important for muscle function.
PLB inhibits the pump by reducing SERCA affinity for Ca2+.
SLN uncouples the SERCA pump ATP hydrolysis from Ca2+ transport.
SERCA uncoupling by SLN promotes futile cycling and increases thermogenesis.
Acknowledgments
This work was supported by grants from the NIH (R01 DK098240) to M.P. S.A.S was supported by a Predoctoral fellowship 13PRE17000059 from the American Heart Association and S.K.S is supported by a Postdoctoral fellowship 14POST200380847 from the American Heart Association. We thank Naresh C. Bal and Subash C. Gupta for help in preparation of Figs.1 and 5. We thank Kathy Huson for editing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures:
None
References
- 1.Bers DM. Cardiac excitation–contraction coupling. Nature. 2002;415(6868):198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- 2.Berchtold MW, Brinkmeier H, Müntener M. Calcium ion in skeletal muscle: its crucial role for muscle function, plasticity, and disease. Physiological reviews. 2000;80(3):1215–1265. doi: 10.1152/physrev.2000.80.3.1215. [DOI] [PubMed] [Google Scholar]
- 3.Aubier M, Viires N. Calcium ATPase and respiratory muscle function. European Respiratory Journal. 1998;11(3):758–766. [PubMed] [Google Scholar]
- 4.Periasamy M, Kalyanasundaram A. SERCA pump isoforms: their role in calcium transport and disease. Muscle & nerve. 2007;35(4):430–442. doi: 10.1002/mus.20745. [DOI] [PubMed] [Google Scholar]
- 5.MacLennan DH, Kranias EG. Phospholamban: a crucial regulator of cardiac contractility. Nature reviews Molecular cell biology. 2003;4(7):566–577. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- 6.Traaseth NJ, et al. Structural and dynamic basis of phospholamban and sarcolipin inhibition of Ca2+-ATPase. Biochemistry. 2008;47(1):3–13. doi: 10.1021/bi701668v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haghighi K, Gregory KN, Kranias EG. Sarcoplasmic reticulum Ca-ATPase–phospholamban interactions and dilated cardiomyopathy. Biochemical and biophysical research communications. 2004;322(4):1214–1222. doi: 10.1016/j.bbrc.2004.07.164. [DOI] [PubMed] [Google Scholar]
- 8.Kranias E, Bers D. Calcium Signalling and Disease. Springer; 2007. Calcium and cardiomyopathies; pp. 523–537. [DOI] [PubMed] [Google Scholar]
- 9.Kranias EG, Hajjar RJ. Modulation of cardiac contractility by the phopholamban/SERCA2a regulatome. Circulation research. 2012;110(12):1646–1660. doi: 10.1161/CIRCRESAHA.111.259754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo W, et al. Targeted ablation of the phospholamban gene is associated with markedly enhanced myocardial contractility and loss of beta-agonist stimulation. Circulation research. 1994;75(3):401–409. doi: 10.1161/01.res.75.3.401. [DOI] [PubMed] [Google Scholar]
- 11.Mall S, et al. The presence of sarcolipin results in increased heat production by Ca2+-ATPase. Journal of Biological Chemistry. 2006;281(48):36597–36602. doi: 10.1074/jbc.M606869200. [DOI] [PubMed] [Google Scholar]
- 12.Smith W, Broadbridge R, EAST J, LEE A. Sarcolipin uncouples hydrolysis of ATP from accumulation of Ca2+ by the Ca2+-ATPase of skeletal-muscle sarcoplasmic reticulum. Biochem. J. 2002;361:277–286. doi: 10.1042/0264-6021:3610277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sahoo SK, Shaikh SA, Sopariwala DH, Bal NC, Periasamy M. Sarcolipin protein interaction with sarco (endo) plasmic reticulum Ca2+ ATPase (SERCA) is distinct from phospholamban protein, and only sarcolipin can promote uncoupling of the SERCA pump. Journal of Biological Chemistry. 2013;288(10):6881–6889. doi: 10.1074/jbc.M112.436915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bal NC, et al. Sarcolipin is a newly identified regulator of muscle-based thermogenesis in mammals. Nature medicine. 2012;18(10):1575–1579. doi: 10.1038/nm.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maurya SK, et al. Sarcolipin Is a Key Determinant of the Basal Metabolic Rate, and Its Overexpression Enhances Energy Expenditure and Resistance against Diet-induced Obesity. Journal of Biological Chemistry. 2015;290(17):10840–10849. doi: 10.1074/jbc.M115.636878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sopariwala DH, et al. Sarcolipin overexpression improves muscle energetics and reduces fatigue. Journal of Applied Physiology. 2015;118(8):1050–1058. doi: 10.1152/japplphysiol.01066.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmerman HK, Jones LR. Phospholamban: protein structure, mechanism of action, and role in cardiac function. Physiological reviews. 1998;78(4):921–947. doi: 10.1152/physrev.1998.78.4.921. [DOI] [PubMed] [Google Scholar]
- 18.Olesen C, et al. The structural basis of calcium transport by the calcium pump. Nature. 2007;450(7172):1036–1042. doi: 10.1038/nature06418. [DOI] [PubMed] [Google Scholar]
- 19.Olesen C, Sørensen TL-M, Nielsen RC, Møller JV, Nissen P. Dephosphorylation of the calcium pump coupled to counterion occlusion. Science. 2004;306(5705):2251–2255. doi: 10.1126/science.1106289. [DOI] [PubMed] [Google Scholar]
- 20.Toyoshima C, Mizutani T. Crystal structure of the calcium pump with a bound ATP analogue. Nature. 2004;430(6999):529–535. doi: 10.1038/nature02680. [DOI] [PubMed] [Google Scholar]
- 21.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature. 2000;405(6787):647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 22.Toyoshima C, Nomura H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature. 2002;418(6898):605–611. doi: 10.1038/nature00944. [DOI] [PubMed] [Google Scholar]
- 23.Toyoshima C, Nomura H, Sugita Y. Structural basis of ion pumping by Ca 2+-ATPase of sarcoplasmic reticulum. FEBS letters. 2003;555(1):106–110. doi: 10.1016/s0014-5793(03)01086-x. [DOI] [PubMed] [Google Scholar]
- 24.Toyoshima C, Nomura H, Tsuda T. Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature. 2004;432(7015):361–368. doi: 10.1038/nature02981. [DOI] [PubMed] [Google Scholar]
- 25.Møller JV, Olesen C, Winther A-ML, Nissen P. The sarcoplasmic Ca 2+-ATPase: design of a perfect chemi-osmotic pump. Quarterly reviews of biophysics. 2010;43(04):501–566. doi: 10.1017/S003358351000017X. [DOI] [PubMed] [Google Scholar]
- 26.Toyoshima C, Inesi G. Structural basis of ion pumping by Ca2+-ATPase of the sarcoplasmic reticulum. Biochemistry. 2004;73(1):269. doi: 10.1146/annurev.biochem.73.011303.073700. [DOI] [PubMed] [Google Scholar]
- 27.Inesi G. Mechanism of calcium transport. Annual review of physiology. 1985;47(1):573–601. doi: 10.1146/annurev.ph.47.030185.003041. [DOI] [PubMed] [Google Scholar]
- 28.Hasselbach W. The reversibility of the sarcoplasmic calcium pump. Biochimica et Biophysica Acta (BBA)-Reviews on Biomembranes. 1978;515(1):23–53. doi: 10.1016/0304-4157(78)90007-2. [DOI] [PubMed] [Google Scholar]
- 29.de Meis L. Role of the sarcoplasmic reticulum Ca2+-ATPase on heat production and thermogenesis. Bioscience reports. 2001;21:113–137. doi: 10.1023/a:1013640006611. [DOI] [PubMed] [Google Scholar]
- 30.Meis L. Ca2+-ATPases (SERCA): energy transduction and heat production in transport ATPases. The Journal of membrane biology. 2002;188(1):1–9. doi: 10.1007/s00232-001-0171-5. [DOI] [PubMed] [Google Scholar]
- 31.Meis Ld, Vianna AL. Energy interconversion by the Ca2+-dependent ATPase of the sarcoplasmic reticulum. Annual review of biochemistry. 1979;48(1):275–292. doi: 10.1146/annurev.bi.48.070179.001423. [DOI] [PubMed] [Google Scholar]
- 32.Tada M, Kirchberger MA, Katz AM. Phosphorylation of a 22,000-dalton component of the cardiac sarcoplasmic reticulum by adenosine 3': 5'-monophosphate-dependent protein kinase. Journal of Biological Chemistry. 1975;250(7):2640–2647. [PubMed] [Google Scholar]
- 33.Tada M, Toyofuku T. Molecular regulation of phospholamban function and expression. Trends in cardiovascular medicine. 1998;8(8):330–340. doi: 10.1016/s1050-1738(98)00032-2. [DOI] [PubMed] [Google Scholar]
- 34.Akin BL, Chen Z, Jones LR. Superinhibitory phospholamban mutants compete with Ca2+ for binding to SERCA2a by stabilizing a unique nucleotide-dependent conformational state. Journal of Biological Chemistry. 2010;285(37):28540–28552. doi: 10.1074/jbc.M110.151779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akin BL, Hurley TD, Chen Z, Jones LR. The structural basis for phospholamban inhibition of the calcium pump in sarcoplasmic reticulum. Journal of Biological Chemistry. 2013;288(42):30181–30191. doi: 10.1074/jbc.M113.501585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Z, Akin BL, Jones LR. Mechanism of reversal of phospholamban inhibition of the cardiac Ca2+-ATPase by protein kinase A and by anti-phospholamban monoclonal antibody 2D12. Journal of Biological Chemistry. 2007;282(29):20968–20976. doi: 10.1074/jbc.M703516200. [DOI] [PubMed] [Google Scholar]
- 37.Chen Z, Akin BL, Jones LR. Ca2+ binding to site I of the cardiac Ca2+ pump is sufficient to dissociate phospholamban. Journal of Biological Chemistry. 2010;285(5):3253–3260. doi: 10.1074/jbc.M109.080820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Z, Akin BL, Stokes DL, Jones LR. Cross-linking of C-terminal residues of phospholamban to the Ca2+ pump of cardiac sarcoplasmic reticulum to probe spatial and functional interactions within the transmembrane domain. Journal of Biological Chemistry. 2006;281(20):14163–14172. doi: 10.1074/jbc.M601338200. [DOI] [PubMed] [Google Scholar]
- 39.Chen Z, Stokes DL, Jones LR. Role of leucine 31 of phospholamban in structural and functional interactions with the Ca2+ pump of cardiac sarcoplasmic reticulum. Journal of Biological Chemistry. 2005;280(11):10530–10539. doi: 10.1074/jbc.M414007200. [DOI] [PubMed] [Google Scholar]
- 40.Chen Z, Stokes DL, Rice WJ, Jones LR. Spatial and dynamic interactions between phospholamban and the canine cardiac Ca2+ pump revealed with use of heterobifunctional cross-linking agents. Journal of Biological Chemistry. 2003;278(48):48348–48356. doi: 10.1074/jbc.M309545200. [DOI] [PubMed] [Google Scholar]
- 41.Jones LR, Cornea RL, Chen Z. Close proximity between residue 30 of phospholamban and cysteine 318 of the cardiac Ca2+ pump revealed by intermolecular thiol cross-linking. Journal of Biological Chemistry. 2002;277(31):28319–28329. doi: 10.1074/jbc.M204085200. [DOI] [PubMed] [Google Scholar]
- 42.Wegener A, Jones L. Phosphorylation-induced mobility shift in phospholamban in sodium dodecyl sulfate-polyacrylamide gels. Evidence for a protein structure consisting of multiple identical phosphorylatable subunits. Journal of Biological Chemistry. 1984;259(3):1834–1841. [PubMed] [Google Scholar]
- 43.Chu G, et al. Monomeric phospholamban overexpression in transgenic mouse hearts. Circulation research. 1997;81(4):485–492. doi: 10.1161/01.res.81.4.485. [DOI] [PubMed] [Google Scholar]
- 44.Koss KL, Kranias EG. Phospholamban: a prominent regulator of myocardial contractility. Circulation research. 1996;79(6):1059–1063. doi: 10.1161/01.res.79.6.1059. [DOI] [PubMed] [Google Scholar]
- 45.Schmitt JP, et al. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299(5611):1410–1413. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- 46.Zhai J, et al. Cardiac-specific Overexpression of a Superinhibitory Pentameric Phospholamban Mutant Enhances Inhibition of Cardiac Functionin Vivo. Journal of Biological Chemistry. 2000;275(14):10538–10544. doi: 10.1074/jbc.275.14.10538. [DOI] [PubMed] [Google Scholar]
- 47.Asahi M, Green NM, Kurzydlowski K, Tada M, MacLennan DH. Phospholamban domain IB forms an interaction site with the loop between transmembrane helices M6 and M7 of sarco (endo) plasmic reticulum Ca2+ ATPases. Proceedings of the National Academy of Sciences. 2001;98(18):10061–10066. doi: 10.1073/pnas.181348298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Odermatt A, Kurzydlowski K, MacLennan DH. The vmax of the Ca2+-ATPase of cardiac sarcoplasmic reticulum (SERCA2a) is not altered by Ca2+/calmodulin-dependent phosphorylation or by interaction with phospholamban. Journal of Biological Chemistry. 1996;271(24):14206–14213. doi: 10.1074/jbc.271.24.14206. [DOI] [PubMed] [Google Scholar]
- 49.Toyofuku T, Kurzydlowski K, Tada M, MacLennan DH. Amino acids Lys-Asp-Asp-Lys-Pro-Val402 in the Ca (2+)-ATPase of cardiac sarcoplasmic reticulum are critical for functional association with phospholamban. Journal of Biological Chemistry. 1994;269(37):22929–22932. [PubMed] [Google Scholar]
- 50.Toyoshima C, et al. Modeling of the inhibitory interaction of phospholamban with the Ca2+ ATPase. Proceedings of the National Academy of Sciences. 2003;100(2):467–472. doi: 10.1073/pnas.0237326100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones LR, Besch H, Fleming JW, McConnaughey MM, Watanabe AM. Separation of vesicles of cardiac sarcolemma from vesicles of cardiac sarcoplasmic reticulum. Comparative biochemical analysis of component activities. Journal of Biological Chemistry. 1979;254(2):530–539. [PubMed] [Google Scholar]
- 52.Lamers J, Stinis J. Phosphorylation of low molecular weight proteins in purified preparations of rat heart sarcolemma and sarcoplasmic reticulum. Biochimica et Biophysica Acta (BBA)-Protein Structure. 1980;624(2):443–459. doi: 10.1016/0005-2795(80)90086-0. [DOI] [PubMed] [Google Scholar]
- 53.Bidlack JM, Shamoo AE. Adenosine 3′, 5′-monophosphate-dependent phosphorylation of a 6000 and A 22000 dalton protein from cardiac sarcoplasmic reticulum. Biochimica et Biophysica Acta (BBA)-General Subjects. 1980;632(2):310–325. doi: 10.1016/0304-4165(80)90089-6. [DOI] [PubMed] [Google Scholar]
- 54.Will H, Levchenko TS, Levitsky DO, Smirnov VN, Wollenberger A. Partial characterization of protein kinase-catalyzed phosphorylation of low molecular weight proteins in purified preparations of pigeon heart sarcolemma and sarcoplasmic reticulum. Biochimica et Biophysica Acta (BBA)-General Subjects. 1978;543(2):175–193. doi: 10.1016/0304-4165(78)90063-6. [DOI] [PubMed] [Google Scholar]
- 55.Nicolaou P, Hajjar RJ, Kranias EG. Role of protein phosphatase-1 inhibitor-1 in cardiac physiology and pathophysiology. Journal of molecular and cellular cardiology. 2009;47(3):365–371. doi: 10.1016/j.yjmcc.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Macdougall LK, Jones LR, Cohen P. Identification of the major protein phosphatases in mammalian cardiac muscle which dephosphorylate phospholamban. European journal of biochemistry. 1991;196(3):725–734. doi: 10.1111/j.1432-1033.1991.tb15871.x. [DOI] [PubMed] [Google Scholar]
- 57.Haghighi K, et al. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(5):1388–1393. doi: 10.1073/pnas.0510519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ha KN, et al. Lethal Arg9Cys phospholamban mutation hinders Ca2+-ATPase regulation and phosphorylation by protein kinase A. Proceedings of the National Academy of Sciences. 2011;108(7):2735–2740. doi: 10.1073/pnas.1013987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Young HS, Ceholski DK, Trieber CA. Deception in simplicity: Hereditary phospholamban mutations in dilated cardiomyopathy 1. Biochemistry and Cell Biology. 2014;93(1):1–7. doi: 10.1139/bcb-2014-0080. [DOI] [PubMed] [Google Scholar]
- 60.Haghighi K, et al. Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. Journal of Clinical Investigation. 2003;111(6):869. doi: 10.1172/JCI17892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.MacLennan DH. Isolation of proteins of the sarcoplasmic reticulum. Methods in enzymology. 1974;32:291. doi: 10.1016/0076-6879(74)32030-7. [DOI] [PubMed] [Google Scholar]
- 62.Racker E, Eytan E. A coupling factor from sarcoplasmic reticulum required for the translocation of Ca2+ ions in a reconstituted Ca2+ ATPase pump. Journal of Biological Chemistry. 1975;250(18):7533–7534. [PubMed] [Google Scholar]
- 63.MacLennan DH, et al. Ion pathways in proteins of the sarcoplasmic reticulum. Annals of the New York Academy of Sciences. 1980;358(1):138–148. doi: 10.1111/j.1749-6632.1980.tb15392.x. [DOI] [PubMed] [Google Scholar]
- 64.Wawrzynow A, et al. Sarcolipin, the “proteolipid” of skeletal muscle sarcoplasmic reticulum, is a unique, amphipathic, 31-residue peptide. Archives of biochemistry and biophysics. 1992;298(2):620–623. doi: 10.1016/0003-9861(92)90457-8. [DOI] [PubMed] [Google Scholar]
- 65.Odermatt A, et al. Sarcolipin regulates the activity of SERCA1, the fast-twitch skeletal muscle sarcoplasmic reticulum Ca2+-ATPase. Journal of Biological Chemistry. 1998;273(20):12360–12369. doi: 10.1074/jbc.273.20.12360. [DOI] [PubMed] [Google Scholar]
- 66.Odermatt A, et al. Characterization of the gene encoding human sarcolipin (SLN), a proteolipid associated with SERCA1: absence of structural mutations in five patients with Brody disease. Genomics. 1997;45(3):541–553. doi: 10.1006/geno.1997.4967. [DOI] [PubMed] [Google Scholar]
- 67.Babu GJ, et al. Targeted overexpression of sarcolipin in the mouse heart decreases sarcoplasmic reticulum calcium transport and cardiac contractility. Journal of Biological Chemistry. 2006;281(7):3972–3979. doi: 10.1074/jbc.M508998200. [DOI] [PubMed] [Google Scholar]
- 68.Babu GJ, et al. Ablation of sarcolipin enhances sarcoplasmic reticulum calcium transport and atrial contractility. Proceedings of the National Academy of Sciences. 2007;104(45):17867–17872. doi: 10.1073/pnas.0707722104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Babu GJ, et al. Overexpression of sarcolipin decreases myocyte contractility and calcium transient. Cardiovascular research. 2005;65(1):177–186. doi: 10.1016/j.cardiores.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 70.Asahi M, et al. Cardiac-specific overexpression of sarcolipin inhibits sarco (endo) plasmic reticulum Ca2+ ATPase (SERCA2a) activity and impairs cardiac function in mice. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(25):9199–9204. doi: 10.1073/pnas.0402596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gramolini AO, et al. Cardiac-specific overexpression of sarcolipin in phospholamban null mice impairs myocyte function that is restored by phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(7):2446–2451. doi: 10.1073/pnas.0510883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Houser SR, Piacentino V, III, Weisser J. Abnormalities of calcium cycling in the hypertrophied and failing heart. Journal of molecular and cellular cardiology. 2000;32(9):1595–1607. doi: 10.1006/jmcc.2000.1206. [DOI] [PubMed] [Google Scholar]
- 73.Pant M, Bal NC, Periasamy M. Cold adaptation overrides developmental regulation of sarcolipin expression in mice skeletal muscle: SOS for muscle-based thermogenesis? The Journal of Experimental Biology. 2015:119164. doi: 10.1242/jeb.119164. :jeb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Babu GJ, Bhupathy P, Carnes CA, Billman GE, Periasamy M. Differential expression of sarcolipin protein during muscle development and cardiac pathophysiology. Journal of molecular and cellular cardiology. 2007;43(2):215–222. doi: 10.1016/j.yjmcc.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schneider JS, et al. Increased sarcolipin expression and decreased sarco (endo) plasmic reticulum Ca2+ uptake in skeletal muscles of mouse models of Duchenne muscular dystrophy. Journal of muscle research and cell motility. 2013;34(5–6):349–356. doi: 10.1007/s10974-013-9350-0. [DOI] [PubMed] [Google Scholar]
- 76.Zheng J, et al. Increased sarcolipin expression and adrenergic drive in humans with preserved left ventricular ejection fraction and chronic isolated mitral regurgitation. Circulation: Heart Failure. 2014;7(1):194–202. doi: 10.1161/CIRCHEARTFAILURE.113.000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shanmugam M, et al. Ablation of phospholamban and sarcolipin results in cardiac hypertrophy and decreased cardiac contractility. Cardiovascular research. 2010 doi: 10.1093/cvr/cvq294. :cvq294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shanmugam M, et al. Decreased sarcolipin protein expression and enhanced sarco (endo) plasmic reticulum Ca 2+ uptake in human atrial fibrillation. Biochemical and biophysical research communications. 2011;410(1):97–101. doi: 10.1016/j.bbrc.2011.05.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xie L-H, et al. Ablation of sarcolipin results in atrial remodeling. American Journal of Physiology-Cell Physiology. 2012;302(12):C1762–C1771. doi: 10.1152/ajpcell.00425.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sahoo SK, et al. The N Terminus of Sarcolipin Plays an Important Role in Uncoupling Sarco-endoplasmic Reticulum Ca2+-ATPase (SERCA) ATP Hydrolysis from Ca2+ Transport. Journal of Biological Chemistry. 2015;290(22):14057–14067. doi: 10.1074/jbc.M115.636738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee AG. A calcium pump made visible. Current opinion in structural biology. 2002;12(4):547–554. doi: 10.1016/s0959-440x(02)00360-3. [DOI] [PubMed] [Google Scholar]
- 82.Asahi M, Kurzydlowski K, Tada M, MacLennan DH. Sarcolipin inhibits polymerization of phospholamban to induce superinhibition of sarco (endo) plasmic reticulum Ca2+-ATPases (SERCAs) Journal of Biological Chemistry. 2002;277(30):26725–26728. doi: 10.1074/jbc.C200269200. [DOI] [PubMed] [Google Scholar]
- 83.Asahi M, Nakayama H, Tada M, Otsu K. Regulation of sarco (endo) plasmic reticulum Ca 2+ adenosine triphosphatase by phospholamban and sarcolipin: implication for cardiac hypertrophy and failure. Trends in cardiovascular medicine. 2003;13(4):152–157. doi: 10.1016/s1050-1738(03)00037-9. [DOI] [PubMed] [Google Scholar]
- 84.Asahi M, et al. Sarcolipin regulates sarco (endo) plasmic reticulum Ca2+-ATPase (SERCA) by binding to transmembrane helices alone or in association with phospholamban. Proceedings of the National Academy of Sciences. 2003;100(9):5040–5045. doi: 10.1073/pnas.0330962100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gramolini AO, et al. Sarcolipin retention in the endoplasmic reticulum depends on its C-terminal RSYQY sequence and its interaction with sarco (endo) plasmic Ca2+-ATPases. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(48):16807–16812. doi: 10.1073/pnas.0407815101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morita T, et al. Interaction sites among phospholamban, sarcolipin, and the sarco (endo) plasmic reticulum Ca 2+-ATPase. Biochemical and biophysical research communications. 2008;369(1):188–194. doi: 10.1016/j.bbrc.2007.11.098. [DOI] [PubMed] [Google Scholar]
- 87.Inesi G, Lewis D, Ma H, Prasad A, Toyoshima C. Concerted conformational effects of Ca2+ and ATP are required for activation of sequential reactions in the Ca2+ ATPase (SERCA) catalytic cycle. Biochemistry. 2006;45(46):13769–13778. doi: 10.1021/bi061255d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Clarke DM, Loo TW, Inesi G, MacLennan DH. Location of high affinity Ca2+-binding sites within the predicted transmembrahe domain of the sarco-plasmic reticulum Ca2+-ATPase. 1989 doi: 10.1038/339476a0. [DOI] [PubMed] [Google Scholar]
- 89.Toyoshima C, et al. Crystal structures of the calcium pump and sarcolipin in the Mg2+-bound E1 state. Nature. 2013;495(7440):260–264. doi: 10.1038/nature11899. [DOI] [PubMed] [Google Scholar]
- 90.Winther A-ML, et al. The sarcolipin-bound calcium pump stabilizes calcium sites exposed to the cytoplasm. Nature. 2013;495(7440):265–269. doi: 10.1038/nature11900. [DOI] [PubMed] [Google Scholar]
- 91.Hellstern S, et al. Sarcolipin, the shorter homologue of phospholamban, forms oligomeric structures in detergent micelles and in liposomes. Journal of Biological Chemistry. 2001;276(33):30845–30852. doi: 10.1074/jbc.M102495200. [DOI] [PubMed] [Google Scholar]
- 92.Buffy JJ, et al. Two-dimensional solid-state NMR reveals two topologies of sarcolipin in oriented lipid bilayers. Biochemistry. 2006;45(36):10939–10946. doi: 10.1021/bi060728d. [DOI] [PubMed] [Google Scholar]
- 93.Mascioni A, Karim C, Barany G, Thomas DD, Veglia G. Structure and orientation of sarcolipin in lipid environments. Biochemistry. 2002;41(2):475–482. doi: 10.1021/bi011243m. [DOI] [PubMed] [Google Scholar]
- 94.Butler J, et al. Phospholamban and sarcolipin are maintained in the endoplasmic reticulum by retrieval from the ER-Golgi intermediate compartment. Cardiovascular research. 2007;74(1):114–123. doi: 10.1016/j.cardiores.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 95.Hughes E, Clayton JC, Kitmitto A, Esmann M, Middleton DA. Solid-state NMR and functional measurements indicate that the conserved tyrosine residues of sarcolipin are involved directly in the inhibition of SERCA1. Journal of Biological Chemistry. 2007;282(36):26603–26613. doi: 10.1074/jbc.M611668200. [DOI] [PubMed] [Google Scholar]
- 96.Gorski PA, Glaves JP, Vangheluwe P, Young HS. Sarco (endo) plasmic reticulum calcium ATPase (SERCA) inhibition by sarcolipin is encoded in its luminal tail. Journal of Biological Chemistry. 2013;288(12):8456–8467. doi: 10.1074/jbc.M112.446161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bhupathy P, Babu GJ, Periasamy M. Sarcolipin and phospholamban as regulators of cardiac sarcoplasmic reticulum Ca 2+ ATPase. Journal of molecular and cellular cardiology. 2007;42(5):903–911. doi: 10.1016/j.yjmcc.2007.03.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Magny EG, et al. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science. 2013;341(6150):1116–1120. doi: 10.1126/science.1238802. [DOI] [PubMed] [Google Scholar]
- 99.Autry JM, et al. Oligomeric interactions of sarcolipin and the Ca-ATPase. Journal of Biological Chemistry. 2011;286(36):31697–31706. doi: 10.1074/jbc.M111.246843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Karim CB, Zhang Z, Howard EC, Torgersen KD, Thomas DD. Phosphorylation-dependent conformational switch in spin-labeled phospholamban bound to SERCA. Journal of molecular biology. 2006;358(4):1032–1040. doi: 10.1016/j.jmb.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 101.Gustavsson M, et al. Allosteric regulation of SERCA by phosphorylation-mediated conformational shift of phospholamban. Proceedings of the National Academy of Sciences. 2013;110(43):17338–17343. doi: 10.1073/pnas.1303006110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Metcalfe EE, Traaseth NJ, Veglia G. Serine 16 phosphorylation induces an order-to-disorder transition in monomeric phospholamban. Biochemistry. 2005;44(11):4386–4396. doi: 10.1021/bi047571e. [DOI] [PubMed] [Google Scholar]
- 103.Traaseth N, Thomas D, Veglia G. Effects of Ser16 phosphorylation on the allosteric transitions of phospholamban/Ca 2+-ATPase complex. Journal of molecular biology. 2006;358(4):1041–1050. doi: 10.1016/j.jmb.2006.02.047. [DOI] [PubMed] [Google Scholar]
- 104.Bidwell P, Blackwell DJ, Hou Z, Zima AV, Robia SL. Phospholamban binds with differential affinity to calcium pump conformers. Journal of Biological Chemistry. 2011;286(40):35044–35050. doi: 10.1074/jbc.M111.266759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.James P, Inui M, Tada M, Chiesi M, Carafoli E. Nature and site of phospholamban regulation of the Ca2+ pump of sarcoplasmic reticulum. Nature. 1989;342:90–92. doi: 10.1038/342090a0. [DOI] [PubMed] [Google Scholar]
- 106.Bhupathy P, Babu GJ, Ito M, Periasamy M. Threonine-5 at the N-terminus can modulate sarcolipin function in cardiac myocytes. Journal of molecular and cellular cardiology. 2009;47(5):723–729. doi: 10.1016/j.yjmcc.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li J, Bigelow DJ, Squier TC. Conformational changes within the cytosolic portion of phospholamban upon release of Ca-ATPase inhibition. Biochemistry. 2004;43(13):3870–3879. doi: 10.1021/bi036183u. [DOI] [PubMed] [Google Scholar]
- 108.Mahaney JE, Albers RW, Waggoner JR, Kutchai HC, Froehlich JP. Intermolecular conformational coupling and free energy exchange enhance the catalytic efficiency of cardiac muscle SERCA2a following the relief of phospholamban inhibition. Biochemistry. 2005;44(21):7713–7724. doi: 10.1021/bi048011i. [DOI] [PubMed] [Google Scholar]