Abstract
Using the single-chain due ferri (DFsc) peptide scaffold, the differential oxidase and oxygenase reactivities of two 4A→4G variants, one with two histidines at the diiron center (G4DFsc) and the other with three histidines (3His-G4DFsc(Mut3)), are explored. By controlling the reaction conditions, the active form responsible for 4-aminophenol (4-AP) oxidase activity in both G4DFsc and 3His-G4DFsc(Mut3) is determined to be the substrate-bound biferrous site. Using circular dichroism (CD), magnetic CD (MCD), and variable-temperature, variable-field (VTVH) MCD spectroscopies, 4-AP is found to bind directly to the biferrous sites of the DF proteins. In G4DFsc, 4-AP increases the coordination of the biferrous site, while in 3His-G4DFsc(Mut3), the coordination number remains the same and the substrate likely replaces the additional bound histidine. This substrate binding enables a two-electron process where 4-AP is oxidized to benzoquinone imine and O2 is reduced to H2O2. In contrast, only the biferrous 3His variant is found to be active in the oxygenation of p-anisidine to 4-nitroso-methoxybenzene. From CD, MCD, and VTVH MCD, p-anisidine addition is found to minimally perturb the biferrous centers of both G4DFsc and 3His-G4DFsc(Mut3), indicating that this substrate binds near the biferrous site. In 3His-G4DFsc(Mut3), the coordinative saturation of one iron leads to the two-electron reduction of O2 at the second iron to generate an end-on hydroperoxo-Fe(III) active oxygenating species.
INTRODUCTION
Binuclear non-heme iron enzymes catalyze a variety of biologically significant reactions with relevance to biotechnology,1,2 environmental remediation,3 and drug design.4,5 The catalyzed reactions are usually O2-dependent and generally include oxidation and oxygenation of substrates.5-8 Representative binuclear non-heme iron enzymes that function as oxidases include alternative oxidase (AOX) and flavin diiron protein (FDP).9-14 AOX has a diiron center that participates in the mitochondrial electron transport chain in plants and bacteria, catalyzing ubiquinol oxidation with O2 (which is reduced to H2O2) and allowing for cyanide-resistant respiration.14 Flavin diiron proteins are oxidases that protect anaerobes from oxidative stress.9-13 Among the many binuclear non-heme iron enzymes that function as oxygenases are methane monooxygenase (MMO, methane to methanol),15-18 toluene-4-monooxygenase (T4MO, toluene to methylphenol),19 and p-aminobenzoate N-oxygenase (AurF, p-aminobenzoate to p-nitrobenzoate).20,21
The diiron active sites of binuclear non-heme iron enzymes are coordinated to the protein by histidine and carboxylate residues and are often contained within common four-helix bundle motifs (Figure 1).5-8,15,16,22-25 While a 2-histidine and 4-carboxylate coordination of the diiron center (two of the carboxylates bridge the ferrous centers) is observed in many cases, such as in MMO,15-17 ribonucleotide reductase,5,25 and Δ-9 desaturase,23,24 the number of coordinating histidine and carboxylate residues can vary.6,20,26 An additional histidine binds to one of the iron sites (3His/4-carboxylate ligand set) in AurF and myo-inositol oxygenase (MIOX) has a diiron center with a 2-histidine and 4-carboxylate coordination.20,27-31 Many binuclear non-heme iron enzymes react with O2 in their biferrous states to form activated intermediates.5-8,32-39 MMO reacts with O2 to form a high-valent intermediate (Q) that acts as the oxygenating species.18,40-42 Alternatively in AurF, a peroxy intermediate species has been proposed as the N-oxygenating species.29,38
Figure 1.
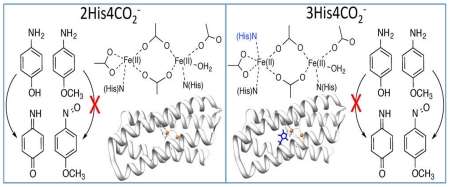
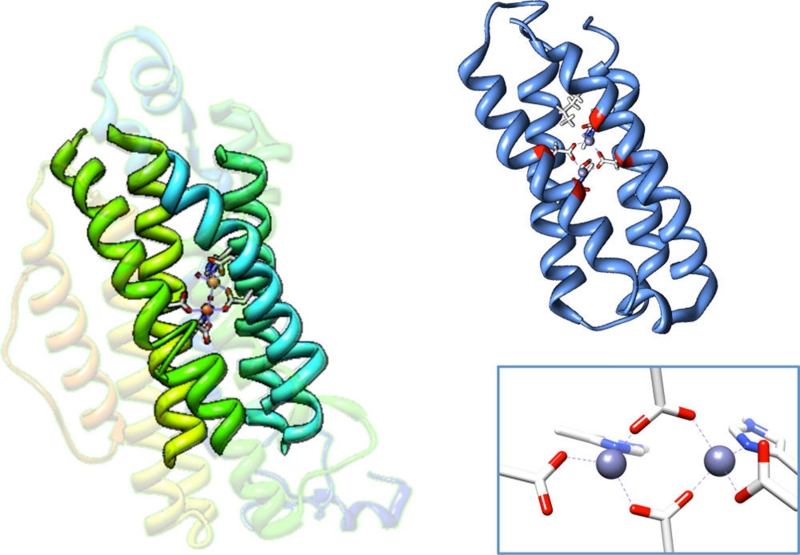
Crystal structure of ribonucleotide reductase (left) (PDB 1RIB)43 with the four-helix bundle motif bolded and the NMR structure of Zn-substituted DFsc (right) (PDB 2HZ8).44 A closeup of the active site with 2 histidines + 4 carboxylate coordination is shown in the bottom right. The 4A’s shown in red of the NMR structure of DFsc are mutated to 4G’s (A10G, A14G, A43G, A47G) in G4DFsc.
Due ferri (DF) de novo proteins, peptides that self-assemble as four-helix bundle units (Figure 1, right) with binuclear non-heme iron centers, are used to model native binuclear non-heme iron enzymes. They offer the simplicity of small molecules while still maintaining the native protein environment.45-47 The single chain DF peptides (DFsc), a subset of DF, form scaffolds that mimic the folding properties and asymmetry of native diiron proteins.48,49 The original DFsc variant, which was constructed to contain the common 2-His/4-carboxylate ligand set (found in ribonucleotide reductase, MMO, Δ-9 desaturase, etc.), displayed O2 reaction rates similar to natural diiron proteins.50 In order to increase solvent and substrate accessibility, a 4A→4G variant (G4DFsc, DFsc with mutations = A10G, A14G, A43G, A47G) was made.51 While still retaining the 2-His/4-carboxylate ligand set, the new variant displayed a decrease in the O2 reaction rate.52 Additionally, a version of G4DFsc where three (rather than two) histidine residues are near the diiron center (3His-G4DFsc-(Mut3)) was constructed to model the ligation of AurF.51 In order to incorporate the third His residue at the diiron site (I100H), three additional mutations (Mut3 = Y18L, I37N, and L81H) were required for protein stability.
The perturbation of G4DFsc to incorporate an additional histidine at the diiron active site was found to significantly alter its reactivity.51 Our previous studies evaluated arylamine oxidase and oxygenase activities of G4DFsc and 3His-G4DFsc(Mut3) using 4-aminophenol (4-AP) and p-anisidine as substrates. While G4DFsc and 3His-G4DFsc(Mut3) were shown to have comparable O2 reaction rates,52 the activity studies revealed that G4DFsc catalyzed the oxidation of 4-AP, while the 3His form only had minimal activity.51 The 3His form did, however, show oxygenase reactivity toward the amine group of p-anisidine, while G4DFsc appeared inactive in this reaction. Thus, 3His-G4DFsc(Mut3) was demonstrated to be a functional model of AurF.
The present study elucidates how incorporation of an additional histidine at the diiron center modifies the reactivities of G4DFsc. These investigations define the active species involved and give insight into the differential oxidase and oxygenase activities of 2His (G4DFsc) and 3His (3His-G4DFsc(Mut3)) forms of diiron proteins. The structures and reactivities of the substrate bound biferrous species are probed using near-infrared (NIR) circular dichroism (CD) and magnetic circular dichroism (MCD) spectroscopies, and variable-temperature, variable-field (VTVH) MCD. NIR CD and MCD are used to probe the ferrous d→d transitions, which correlate to the geometry of each Fe(II) center.6 VTVH MCD uses these excited-state transitions to study the ground state and extract information on the magnetic coupling between the two iron sites, which correlates with the bridging ligands. This also provides the zero-field splitting (ZFS) of the individual iron centers, which complements the geometric information obtained from CD and MCD spectroscopies. On the basis of these studies, we elucidate how the properties of substrate binding to the biferrous sites define the differences in reactivities observed between the 2His (G4DFsc) and the 3His (3His-G4DFsc(Mut3)) forms of de novo designed binuclear non-heme iron proteins.
RESULTS AND ANALYSIS
Oxidase Activity
Reactivity
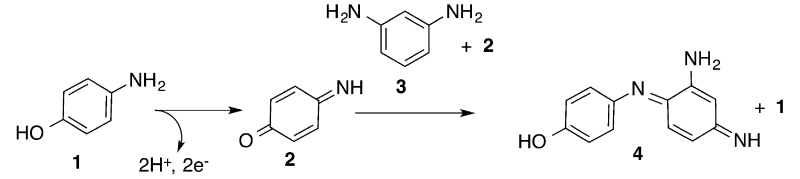
Based on our prior studies, the biferrous G4DFsc reacts with O2 to first form a paramagnetic species (attributed to a biferric site with only μ-1,3 carboxylate bridges, Species B) that decays into a diamagnetic species (attributed to a hydroxo-bridged antiferromagnetically coupled biferric site, Species C).52 However, 3His-G4DFsc(Mut3) only formed the biferric paramagnetic species B due to the additional His coordinatively saturating one iron center. In our prior studies that showed oxidase reactivity for G4DFsc, but not for the 3His form, Fe(II) was added under aerobic conditions to a solution of apo-protein and 4-AP (1) in the presence of m-phenylenediamine (3), which couples to the oxidized product (quinone imine) (2) to yield an aminoindoaniline dye (4) with λmax = 486 nm (ε = 13 200 cm−1 M−1) at pH 7 (Figure 2).51 The conditions of the prior studies, which allowed for the simultaneous presence of the biferric (Species B and C)52 and biferrous forms and may have included contributions from Fe(II) binding to the protein, complicated our ability to identify the catalytically active species. Using more controlled reaction conditions, the prior studies51 were thus extended to identify the reactive species of G4DFsc responsible for catalyzing the oxidation of 4-AP, as well as address the apparent lack of activity in the 3His form.
Figure 2.
Two-electron oxidation of 4-aminophenol followed by coupling with m-phenylenediamine to form an aminoindoaniline dye.
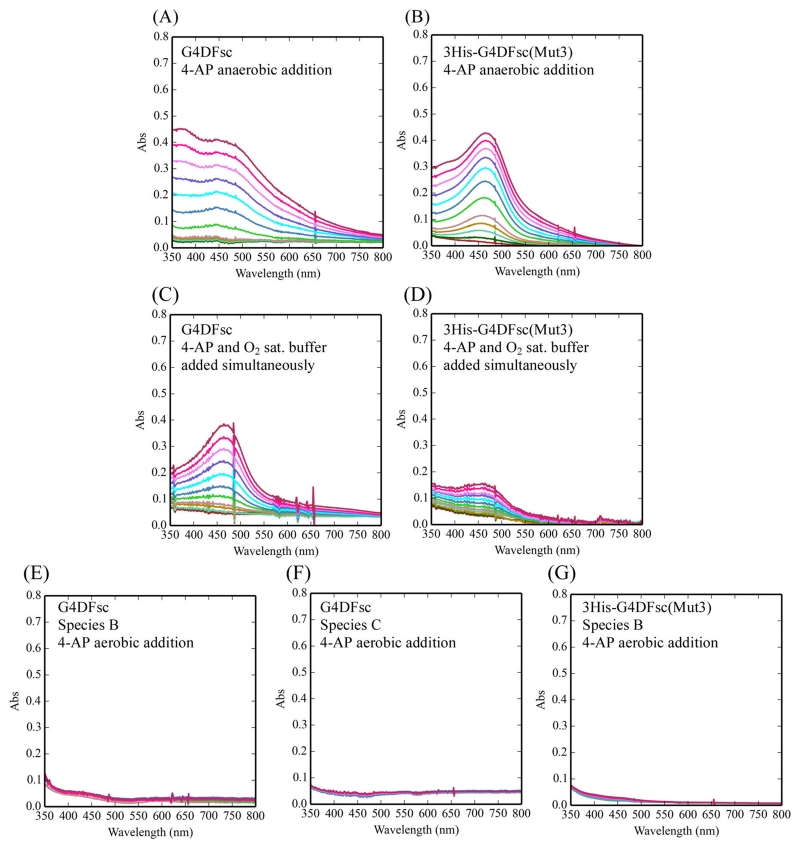
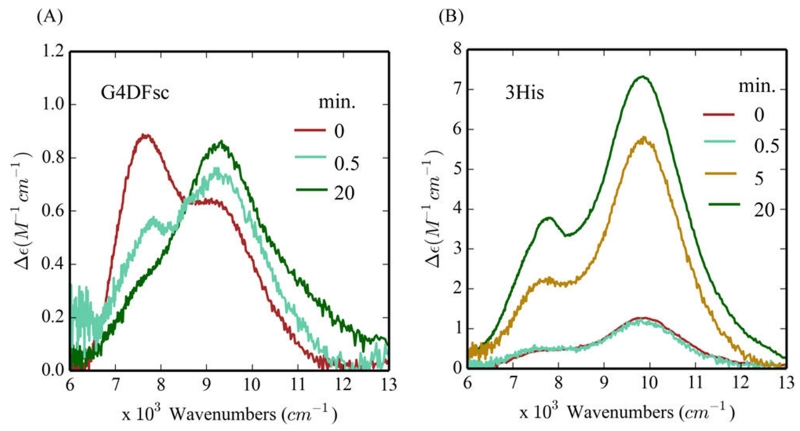
To test the biferrous forms of G4DFsc and 3His-G4DFsc-(Mut3), Fe(II) was added to the apo-protein in an anaerobic environment (glovebox) to exclude possible iron binding effects that may complicate the reactivity. 4-AP was also added to the Fe(II) protein under anaerobic conditions and incubated for 15 min. This was followed by the anaerobic addition of m-phenylenediamine and then O2-saturated buffer at 4 °C. Under these conditions, 4-AP oxidation was observed in both G4DFsc and 3His-G4DFsc(Mut3) (Figure 3A,B). Since these results were not consistent with our prior study, 4-AP (with m-phenylenediamine) was also added simultaneously with O2-saturated buffer to biferrous protein. These conditions reproduced the observations of prior studies, where the 3His form showed much reduced activity compared to G4DFsc (Figure 3C,D). Biferric species B and C in G4DFsc, as well as the biferric form of 3His-G4DFsc(Mut3), were also tested and showed no activity (Figure 3E–G). The 4-AP oxidation with both substrate loaded G4DFsc and 3His-G4DFsc(Mut3) biferrous forms produced multiple equivalents of the indoaniline dye per diiron site over time (5 equiv after 20 min, Figure S9). These data indicate that the biferrous forms of G4DFsc and 3His-G4DFsc(Mut3) are the catalytically active species and that in the case of the 3His form, substrate addition prior to oxidation is required for reactivity.
Figure 3.
Oxidation of 4-aminophenol (4-AP) for various reaction conditions over 10 min. The reaction conditions include the following: (1) the addition of O2-saturated buffer to biferrous G4DFsc (A) and 3His-G4DFsc(Mut3) (B) with 4-AP (in the presence of m-phenylenediamine) already loaded; (2) the simultaneous addition of O2-saturated buffer and 4-AP (in the presence of m-phenylenediamine) to biferrous G4DFsc (C) and 3His-G4DFsc(Mut3) (D); and (3) the addition of 4-AP (in the presence of m-phenylenediamine) to the biferric paramagnetic Species B (E) and diamagnetic Species C (F) for G4DFsc and the biferric diamagnetic Species B for 3His-G4DFsc(Mut3) (G). Aminoindoaniline dye formation was used to identify the oxidation of 4-AP (see Figure 2). Final concentrations (after O2-saturated buffer addition) were 10 μM protein, 3.9 mM 4-AP, and 10 mM m-phenylenediamine.
Spectroscopic Definition of Substrate Binding
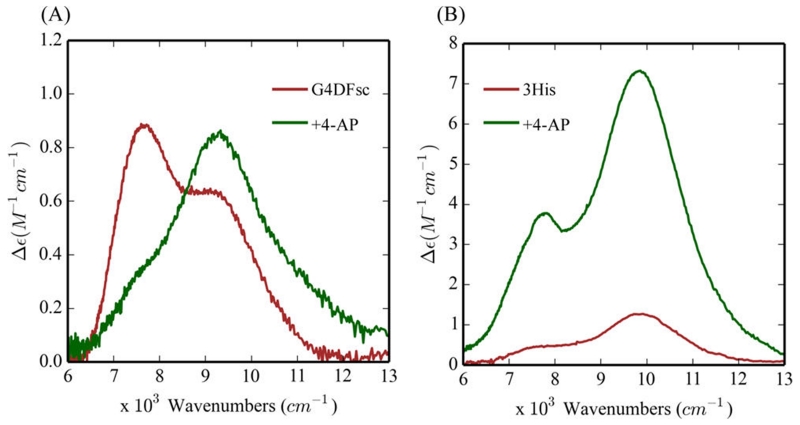
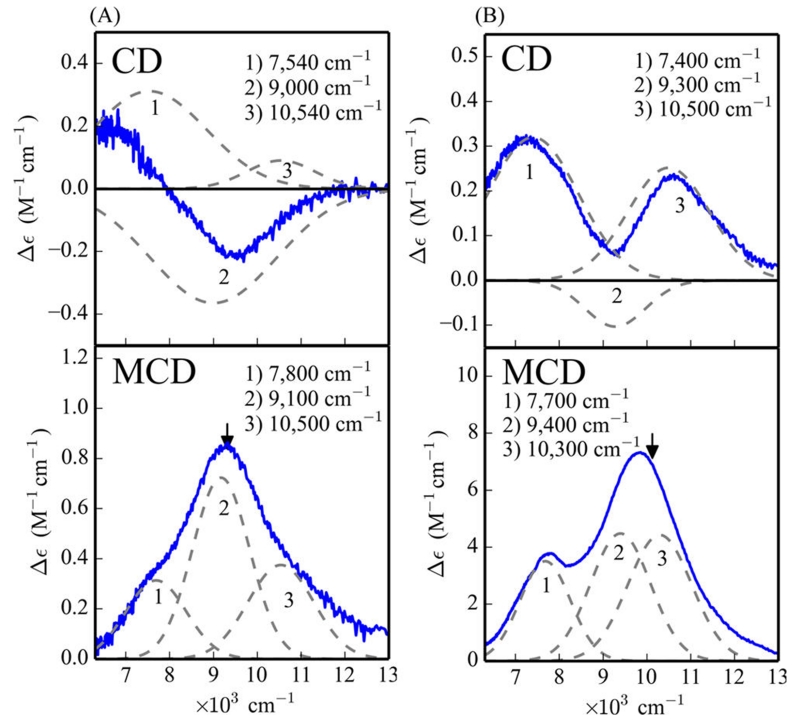
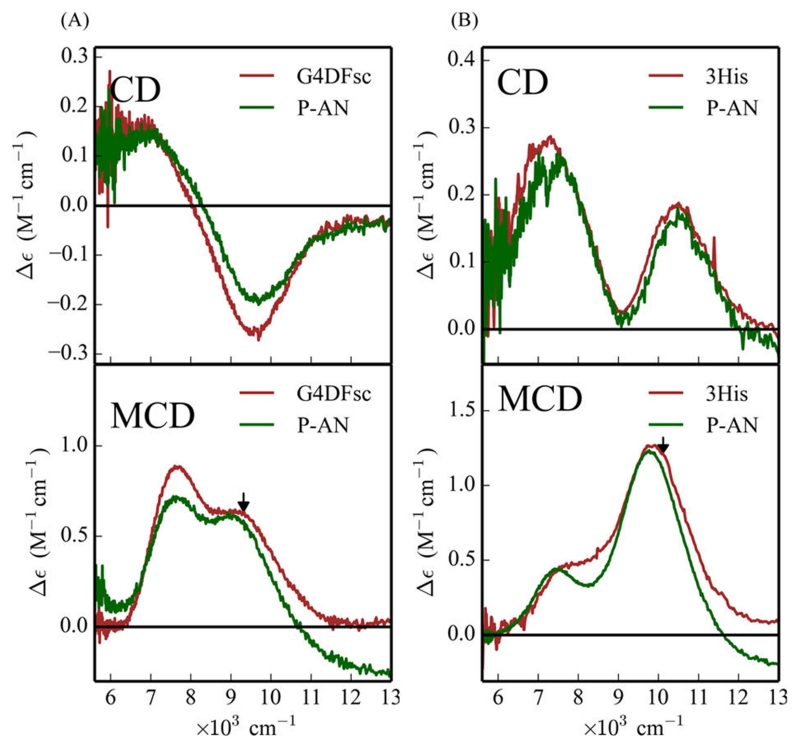
To understand the effects of substrate loading on the biferrous sites, MCD and VTVH MCD spectroscopies were employed to determine the structures of the Fe(II)Fe(II) forms of these DF proteins in the presence of 4-AP. While addition of 4-AP to the Fe(II)Fe(II) states of G4DFsc and 3His-G4DFsc(Mut3) only slightly perturbs their CD spectra (Figure S3), the substrate addition significantly alters the MCD spectra (Figure 4) for both DF forms. In the presence of excess 4-AP (15-fold), an additional higher energy feature appears in the low temperature MCD spectrum of biferrous G4DFsc (Figure 4A) and the MCD intensity significantly increases for biferrous 3His-G4DFsc-(Mut3) (Figure 4B). These changes indicate that 4-AP is appreciably perturbing the biferrous site in each protein and likely binding to an Fe(II) center (Figure S18). The CD and MCD features for G4DFsc in the presence of 4-AP were Gaussian resolved into three bands at 7540(+), 9000(−), and 10 540(+) cm−1 in the CD spectrum, and at 7800, 9100, and 10 500 cm−1 in the MCD spectrum (Figure 5A). The CD and MCD spectra for 3His-G4DFsc(Mut3) with excess 4-AP were also resolved into three bands (Figure 5B) that have the same signs and energies as those of the 3His form without 4-AP but with ~7 times greater intensity with substrate bound.52 Note that the small shifts in energy and narrowing of the band shapes in the MCD spectra are due to the lower temperature of data collection. The three bands for the 3His form (7400(+), 9300(−), and 10 500(+) cm−1 for CD and 7700, 9400, and 10 300 cm−1 for MCD) also have the same sign and are at energies very similar to those for G4DFsc with 4-AP bound (G4DFsc+4-AP). Biferrous 3His-G4DFsc(Mut3) was previously described as having a 5-coordinate + 6-coordinate site based on its CD and MCD spectra, while G4DFsc was described as having a 5-coordinate + 5-coordinate site.52 The similarities in energy and sign of these CD and MCD features for G4DFsc+4-AP, 3His-G4DFsc-(Mut3)+4-AP, and 3His-G4DFsc(Mut3) indicate that all three forms have equivalent coordination environments with one 6-coordinate and one 5-coordinate trigonal bipyramidal Fe(II) center. This would suggest that 4-AP addition increases the coordination of G4DFsc (5-coordinate + 5-coordinate to 5-coordinate + 6-coordinate), while maintaining 5-coordinate + 6-coordinate for 4-AP bound 3His-G4DFsc(Mut3) but with a significant perturbation of the diiron site (i.e., the increased MCD intensity).
Figure 4.
4-Aminophenol (4-AP) perturbation of the MCD spectra of G4DFsc (A) and 3His-G4DFsc(Mut3) (B) in the absence (red) and presence (green) of 4-AP. All spectra taken at 2 K and 7 T (baseline subtracted). Protein concentrations were ~1 mM with 15-fold excess substrate.
Figure 5.
Gaussian resolution of the CD and MCD spectra of G4DFsc (A) and 3His-G4DFsc(Mut3) (B) with the addition of 4-aminophenol. Black arrows denote where VTVH MCD isotherms were taken. CD was taken at 4 °C. MCD was taken at 2 K and 7 T (baseline subtracted). Protein concentrations are ~1 mM with 15-fold excess substrate.
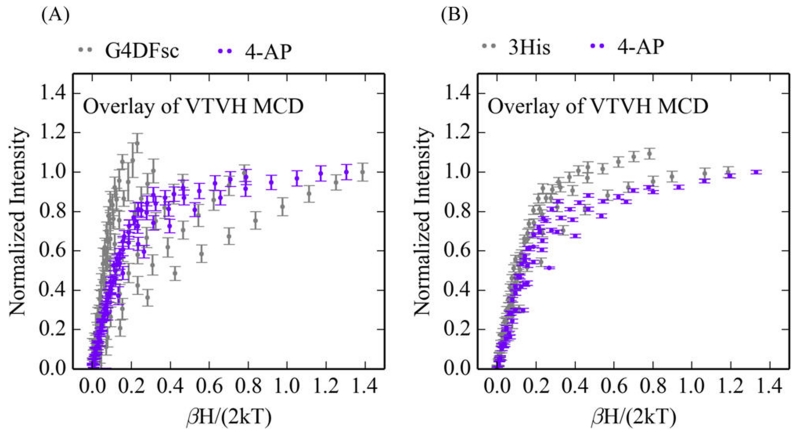
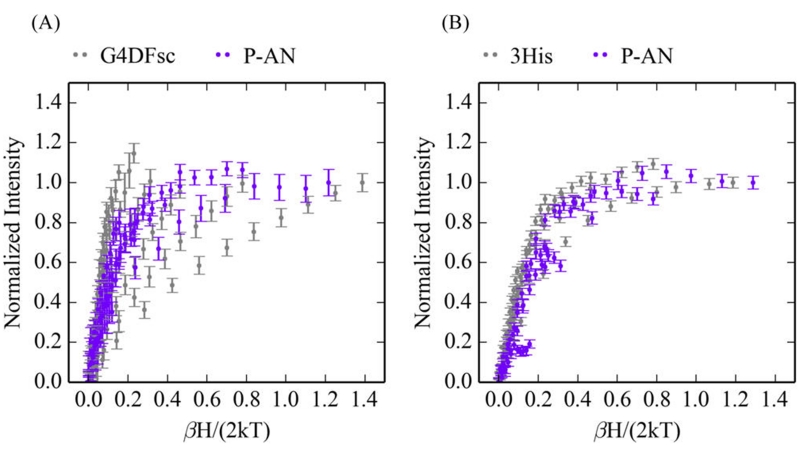
The VTVH MCD data for both G4DFsc and 3His-G4DFsc(Mut3) are perturbed with the addition of 4-AP (Figure 6), which further substantiates the above results that suggest 4-AP binds to these biferrous sites, in this case perturbing their ground states. Interestingly, the VTVH MCD data for G4DFsc +4-AP and 3His-G4DFsc(Mut3)+4-AP are similar to each other (Figure 7A).
Figure 6.
Effect of 4-aminophenol (4-AP) addition on VTVH MCD data. Overlay with (purple) and without (gray) 4-AP for G4DFsc at 9100 cm−1 (A) and 3His-G4DFsc(Mut3) at 10 400 cm−1 (B). Isotherms at 2, 3, 5, 7.5, 10, 15, 20, and 25 K are shown for all data sets.
Figure 7.
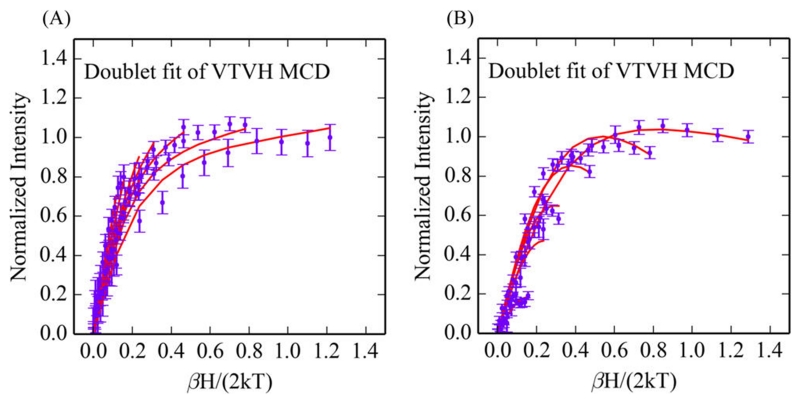
VTVH MCD data for biferrous G4DFsc and 3His-G4DFsc(Mut3) following 4-aminophenol (4-AP) addition. (A) Overlay of the VTVH MCD for G4DFsc (purple) (at 9100 cm−1) and 3His-G4DFsc(Mut3) (orange) (at 10 400 cm−1) with addition of 4-AP. (B) Doublet fit to the VTVH MCD of 3His-G4DFsc(Mut3)+4-AP. Isotherms at 2, 3, 5, 7.5, 10, 15, and 20 K are shown.
The spin-manifold for a biferrous system is described by the spin-Hamiltonian (eq 1):6
| (1) |
where J, D1, D2, E1, and E2 quantify the magnetic coupling between Fe1 and Fe2, and the axial and rhombic ZFS of Fe1 and Fe2, respectively. As previously described,6 this equation is applied to the uncoupled basis set |S1,S2,Ms1,Ms2⟩ of Fe1 and Fe2 to obtain eigenvectors and eigenvalues from a 25 × 25 matrix. VTVH MCD data can be fit using a model, also previously described,6 that expresses the MCD intensity for a non-Kramers system (i.e., the biferrous site) as a series of doublets where each doublet has an effective g-value (g∥), rhombic ZFS (δ), and C-term (Atot) and B-term MCD intensities. These doublets correlate to the sublevels given by the eigenvectors and eigenvalues obtained from eq 1, and thus allow determination of the exchange-coupling associated with bridging ligation and the ZFS associated with the ligand field of each iron center. The VTVH MCD data for both 3His-G4DFsc(Mut3)+4-AP (Figure 7B) and G4DFsc+4-AP (Figure S5) were fit to the doublet model with similar parameters, as described in the Supporting Information. This gives a system that is antiferromagnetically coupled with oppositely signed ZFS values for each Fe(II) center (−J < 1 cm−1, D1 = 5–15 cm−1 with (E/D)1 = 0.33, and −D2 = 5–15 cm−1 with (E/D)2 = 0.33, see Table 1). These parameter ranges are similar to those for 3His-G4DFsc(Mut3) found previously52 (Table 1) that describe a weakly antiferromagnetic exchange-coupled site, indicating μ-1,3 carboxylate bridges.
Table 1. Spin-Hamiltonian Parameters for G4DFsc and 3His-G4DFsc(Mut3).
| G4DFsc | 3His | G4DFsc+4-AP | 3His+4-AP | |
|---|---|---|---|---|
| −J (cm−1) | 3–4 | 1–3 | <1 | <1 |
| D1 (cm−1) | 5–10 | 10–15 | 5–15 | 5–15 |
| (E/D)1 | 0.33 | 0.33 | 0.33 | 0.33 |
| D2 (cm−1) | −7 to −14 | −10 to −15 | −5 to −15 | −5 to −15 |
| (E/D)2 | 0.15 | 0.33 | 0.33 | 0.33 |
The 4-AP likely binds to the Fe(II)Fe(II) site as a phenolate with the oxygen as the donor atom (pKa of hydroxo group ~10.8). While this phenolate is likely binding to an open coordinate position of the 2His DF form (G4DFsc), it appears to be replacing a ligand in the 3His form (based on the increase in MCD intensity). Given the spectroscopic similarities between G4DFsc+4-AP and 3His-G4DFsc(Mut3)+4-AP, as described above, we propose that the 4-aminophenolate has replaced the additional active site His at position 100 in 3His-G4DFsc(Mut3).
Kinetics
The change in the MCD spectrum for both G4DFsc and 3His-G4DFsc(Mut3) upon the addition of 4-AP provides a spectroscopic approach to determine the rate of substrate binding. A solution of 4-AP (in 54% d-glycerol) was anaerobically mixed into a sample of biferrous protein (also in 54% d-glycerol) in an MCD cell and frozen at different time intervals. The intermediate spectrum of G4DFsc taken from a sample frozen (in liquid nitrogen) at 0.5 min after addition of 4-AP (Figure 8A, teal) is a combination of the final spectrum (after 20 min, dark green) and the initial spectrum (red), and overlays with a composite spectrum generated using 55–70% of the G4DFsc +4-AP final MCD spectrum and 30–45% of the G4DFsc MCD spectrum (Figure S6). Assuming a simple A→B binding scheme and using this 0.5 min interval spectrum, the rate of 4-AP binding is estimated to be kB = 0.027–0.042 s−1. While the MCD spectrum for 3His-G4DFsc(Mut3) after 4-AP addition at the same intermediate time interval (0.5 min) (Figure 8B, teal) shows little perturbation from its initial spectrum (red), the spectrum at 5 min after 4-AP addition shows an increase in intensity (dark yellow) that results in a final spectrum after 20 min (dark green). The intermediate spectrum (5 min) overlays with a composite spectrum generated using 60–75% of the final and 25–40% of the initial spectrum (Figure S8). As calculated above for G4DFsc, the rate for 4-AP binding to 3His-G4DFsc(Mut3) is kB = 0.003–0.0046 s−1. These data demonstrate that 4-AP binds an order of magnitude more rapidly to the biferrous site of the 2His form than to the biferrous site of the 3His DF variant. This result would be consistent with a binding mechanism where the 4-AP displaces the third His at the diiron site in 3His-G4DFsc(Mut3), while simply increasing the coordination number of one Fe(II) center of the 2His form.
Figure 8.
MCD spectra monitoring 4-aminophenol binding at different time intervals for G4DFsc (A) and 3His-G4DFsc(Mut3) (B). All time points taken at 2 K and 7 T. The 0.5 min time interval in (A) is a composite of the initial (red) and end (dark green) spectra (30/70). The 5 min time interval in (B) has intensity consistent with a composition of the initial (red) and end (dark green) spectra (25/75).
Based on previous studies, the rates for O2 reactivity of the biferrous G4DFsc and 3His-G4DFsc(Mut3) forms without substrate are ~0.02 and ~0.04 s−1, respectively.52 Thus, the rate of O2 oxidation is greater than the rate of substrate binding for the biferrous 3His form, but comparable for biferrous G4DFsc. These substrate binding and O2 reaction rates thus provide an explanation for the difference in observed reactivity where the 3His form is active only when the reduced protein has been incubated with substrate leading to replacement/substitution of the third active site His ligand, while incubation is not required for activity in the 2His form.
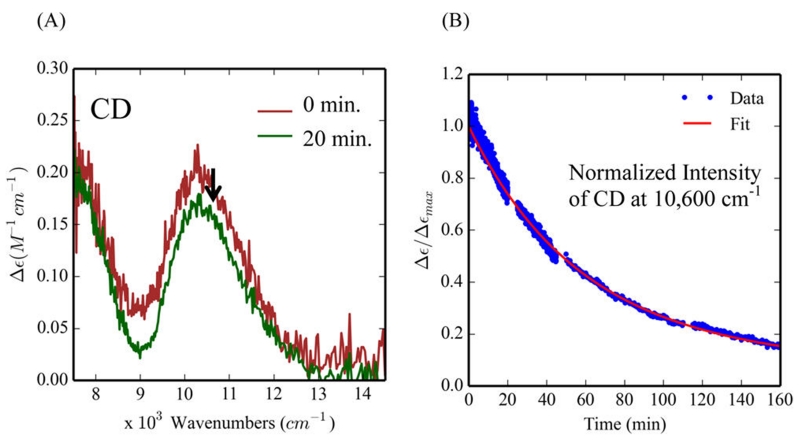
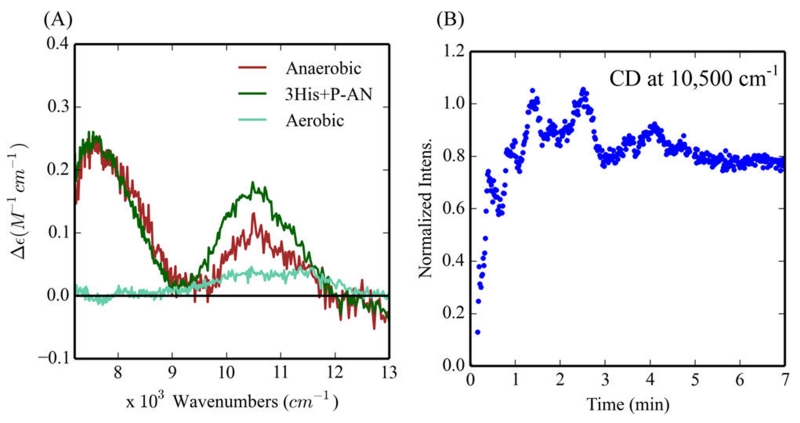
CD spectroscopy was used to detect the amount of the biferrous form of the 3His variant present during 4-AP oxidation under turnover conditions. After maintaining 90–100% intensity for ~10 min (i.e., close to steady state) (Figures 9A and Figure S10), there is a very slow decay of these NIR CD (the decay can be fit with two rates, kd1 = 3 × 10−4 s−1 and kd2 =1.5 × 10−5 s−1, Figure 9B). The decay may be correlated with the consumption of substrate and the oxidation of 3His-G4DFsc(Mut3) under substrate limited conditions. Given that the resting biferric form of the 3His does not show oxidase activity over 10 min (Figure 3G) and the sustained presence of the biferrous form during turnover of 4-AP oxidation, a two-electron process may be invoked where O2 is reduced by two electrons to H2O2 and 4-AP is oxidized to quinone imine at the biferrous site. Any 2Fe(III) peroxy intermediate generated during turnover should be rapidly reduced to the biferrous form. These data also suggest that 4-AP inhibits the rebinding of the third His residue to the biferrous site during steady state conditions. As substrate is depleted, the slow decay of the biferrous site (over ~2.7 h, Figure 9B) thus reflects rebinding of the third His and oxidation of the 3His site to the minimally reactive 3His biferric species. Thus, after substrate binding, the 3His and 2His forms have similar active sites that are competent for 4-AP oxidase reactivity where the bound substrate is oxidized and O2 is reduced in a two-electron process.
Figure 9.
Biferrous CD features during the 4-aminophenol (4-AP) oxidation reaction for 3His-G4DFsc(Mut3). (A) CD of 3His-G4DFsc-(Mut3)+4-AP before (red) and after (green) addition of O2-saturated buffer. (B) The time course over 160 min of the normalized intensity of the CD feature at 10 600 cm−1 (red line is the kinetic fit). CD data collected at 4 °C in the presence of ~15-fold excess substrate (~0.25 mM protein).
Oxygenase Activity
Reactivity
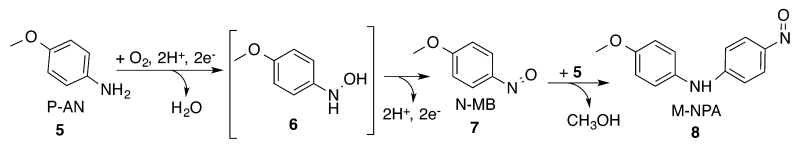
Our prior studies found that 3His-G4DFsc(Mut3), but not G4DFsc, was active in the oxygenation of p-anisidine.51 This involved the oxidation of p-anisidine (P-AN, 5) to the corresponding hydroxylamine (6). The hydroxylamine was directly observed when p-amino-benzonitrile was used as the substrate. However, in the case of the p-anisidine, the hydroxylamine product was instead oxidized to 4-nitroso-methoxybenzene (N-MB, 7), which coupled with a second molecule of P-AN to form 4-methoxy-N-(4-nitrosophenyl)aniline (M-NPA, 8) (Figure 10). The experimental conditions for these studies involved adding Fe(II) to a solution of apo-protein and substrate under aerobic conditions. While Fe(II) should be bound, these conditions allowed for either the biferric or biferrous forms of 3His-G4DFsc(Mut3) as the active species. This work extends on prior results to identify which form of 3His-G4DFsc(Mut3) is active.
Figure 10.
Reaction scheme of the oxidation of p-anisidine (P-AN) (5) to 4-hydroxylamine-methoxybenzene (6), with addition oxidation to 4-nitroso-methoxybenzene (N-MB) (7), and the coupling of N-MB and P-AN to form 4-methoxy-N-(4-nitrosophenyl)aniline (M-NPA) (8).
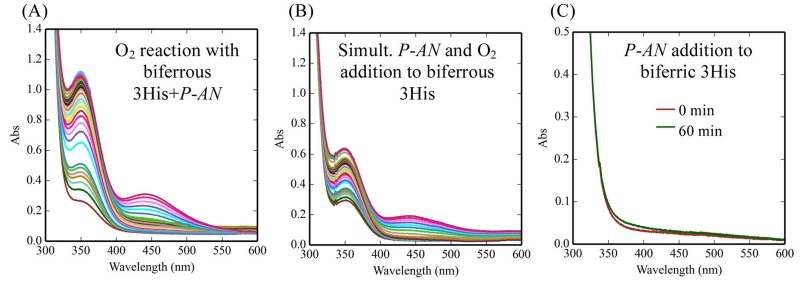
When p-anisidine substrate was incubated with biferrous 3His-G4DFsc(Mut3) anaerobically and then reacted with O2, the UV–vis absorption spectra showed the production of N-MB at 342 nm and the coupled product M-NPA at 431 nm (Figure 11A). The UV–vis absorption spectra for a sample in which O2 and p-anisidine were added simultaneously to biferrous 3His-G4DFsc(Mut3) also showed reactivity but only about half that of the biferrous protein with substrate preloaded (Figure 11B). The biferric form of 3His-G4DFsc(Mut3) showed no activity (Figure 11C) provided the buffer was exchanged after the biferric state was formed and prior to p-anisidine addition. [Note that without exchanging the buffer of the biferric protein, p-anisidine oxidation was observed (Figure S11A). However, this oxidation can be attributed to the presence of hydrogen peroxide (where the protein has generated a stoichiometric amount), which independently did test positively for p-anisidine oxidation (Figure S11B) and showed similar absorption changes (Figure S11C), as exhibited by the biferric form if the buffer was not exchanged.] These UV–vis absorption results were confirmed by LCMS, where anaerobic addition of p-anisidine to biferrous 3His-G4DFsc(Mut3) followed by addition of O2-saturated buffer (Figure S12A) led to greater amounts of N-MB (m/z = 138, retention time 12.6 min) and product M-NPA (m/z = 229, retention time 24.21 min) than the H2O2 present in the biferric reaction mixture (Figure S12B) (2 times the area of the peak associated with nitroso product and ~18 times the area of the peak associated with the coupled product). These results show that substrate binding to the biferrous 3His protein initiates the amino-oxygenation reaction.
Figure 11.
Oxidation of p-anisidine as followed by UV–vis absorption spectroscopy. (A) p-Anisidine was added anaerobically to biferrous 3His-G4DFsc(Mut3) followed by addition of O2-saturated buffer. (B) p-Anisidine and O2-saturated buffer were added simultaneously to biferrous 3His-G4DFsc(Mut3). (C) p-Anisidine was added to biferric 3His-G4DFsc(Mut3) (buffer exchanged prior to p-anisidine addition to eliminate any presence of H2O2 in solution). (A) and (B) are data collected over 180 min. Data taken at 20 °C. Final samples contained 65 μM protein and 1 mM substrate.
Alternatively, for biferrous G4DFsc, the anaerobic addition of the p-anisidine substrate followed by O2-saturated buffer led to no discernible peaks in the LCMS associated with product formation (Figure S12C). The competition between substrate binding and O2 reactivity can thus be excluded as a possible explanation for the lack of activity of the 2His form that was previously reported.51
Spectroscopic Definition of Substrate Binding
Since the reactivity studies indicate that substrate binding to the reduced 3His protein is necessary for amino-oxygenation reactivity, the effect of p-anisidine on the biferrous site was investigated with CD, MCD and VTVH MCD spectroscopy. Substrate addition (+P-AN) to G4DFsc and 3His-G4DFsc(Mut3) only minimally perturbs the CD (Figure 12A,B, top) and MCD (Figure 12A,B, bottom) spectra with slight shifts in the energies and intensities of the bands. The most notable change is that the lower energy feature of the MCD spectrum of the 3His form shifts from ~7700 cm−1 (shoulder in red spectrum, Figure 12B, bottom) to ~7400 cm−1 (green spectrum in Figure 12B, bottom). These small changes indicate that the substrate binding affects the biferrous site, but does not change the coordination, and thus, is not likely bound directly to the diiron center.
Figure 12.
Effect of p-anisidine addition on the CD (top) and MCD (bottom) of G4DFsc (A) and 3His-G4DFsc(Mut3) (B) with unbound (red) and bound (green) forms. Protein concentration is ~1 mM with 20–30-fold excess substrate. MCD data taken at 2 K and 7 T (baseline subtracted). CD data taken at 4 °C. Arrows in bottom spectra indicate where VTVH MCD data were taken.
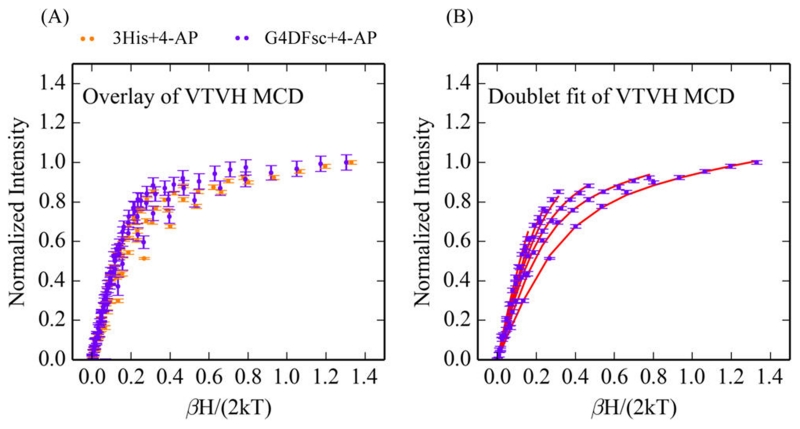
While only slightly perturbing the CD and MCD features of both biferrous G4DFsc and 3His-G4DFsc(Mut3), p-anisidine addition does markedly change the VTVH MCD behavior for both forms (VTVH MCD curves change from gray to purple upon substrate binding in Figure 13). For G4DFsc+P-AN, the VTVH MCD data (Figure 14A) were fit using the doublet model (vide supra) as described in Supporting Information. The VTVH MCD data of G4DFsc+P-AN indicate that −J < 2 cm−1 (Table 2) for this form, which is lower in magnitude than that found for G4DFsc without substrate (−J ≈ 3–4 cm−1). For 3His-G4DFsc(Mut3)+P-AN, doublet model fitting (see Supporting Information) of the VTVH MCD data (Figure 14B) indicates that there is a slight increase in the magnitude of the antiferromagnetic coupling (−J ≈ 2–3 cm−1 3His-G4DFsc-(Mut3)+P-AN, and −J ≈ 1–2 cm−1 3His-G4DFsc(Mut3)-P-AN) (Table 2).
Figure 13.
Effect of p-anisidine addition on the VTVH MCD isotherms. (A) VTVH MCD of G4DFsc (gray) and G4DFsc+P-AN (purple) at 9,100 cm−1. (B) VTVH MCD of 3His-G4FDsc(Mut3) (gray) and 3His-G4FDsc(Mut3)+P-AN (purple) at 10 400 cm−1. Isotherms at 2, 3, 5, 7.5, 10, 15, and 20 K are shown.
Figure 14.
Doublet fits for the VTVH MCD data for G4DFsc+P-AN (A) and 3His-G4DFsc(Mut3)+P-AN (B). Data for G4DFsc were collected at 9100 cm−1, and those for the 3His form were collected at 10 500 cm−1.
Table 2. Spin-Hamiltonian Parameters for G4DFsc and 3His-G4DFsc(Mut3) in the Presence of p-Anisidine.
| G4DFsc | 3His-G4DFsc(Mut3) | |
|---|---|---|
| −J(cm−1) | <2 | 2–3 |
| D1 (cm−1) | 5–15 | 5–10 |
| (E/D)1 | 0.33 | 0.33 |
| D2 (cm−1) | −5 to −15 | −5 to −10 |
| (E/D)2 | 0.33 | 0.33 |
The antiferromagnetic exchange-coupling values for both G4DFsc+P-AN and 3His-G4DFsc+P-AN are consistent with μ-1,3 carboxylate bridges between the two Fe(II) sites. However, the change in their magnitude suggests that p-anisidine is binding near the biferrous site and perturbing the carboxylate bridges between the ferrous centers.
Kinetics
Our previous studies of the O2 reactivity of biferrous G4DFsc and 3His-G4DFsc(Mut3) suggest that the exchange-coupling and O2 reaction rates are correlated, resulting from the O2 reaction occurring at only one Fe(II) site but being reduced by two electrons.52 The transfer of the second electron from the remote Fe(II) site would then occur through the carboxylate bridges. In this model, the decrease in the carboxylate-bridge-mediated exchange-coupling of G4DFsc upon addition of p-anisidine would result in decreased O2 reactivity. This is confirmed where the rate of the O2 reaction for G4DFsc+P-AN is 0.002 s−1 (Figure S14) (relative to ~0.02 s−1 without p-anisidine).52 In contrast, the binding of p-anisidine to the 3His form results in only a small change in exchange-coupling (−J from 1–2 cm−1 to 2–3 cm−1), and the rate of obtaining steady state for nitroso production (k ≈ 0.045 s−1, Figure S15) is close to the O2 reaction rate of the 3His form without substrate (~0.04 s−1).52
NIR CD and MCD spectroscopy were used to observe the biferrous bands of 3His-G4DFsc(Mut3) during the arylamine oxygenation reaction. Under aerobic conditions (i.e., turnover conditions), there are no NIR CD features present during production of the nitroso product (Figure 15A, teal), which indicates that 3His-G4DFsc(Mut3) is mostly present in the biferric form at steady state, and thus, any reduction of the diiron center during turnover (Figure S16) must be rate limiting. In oxygen-depleted environments during nitroso production, the NIR CD and MCD features associated with biferrous 3His-G4DFsc(Mut3) are regenerated (Figure 15A, red, and B, and Figure S17(red)). Since p-anisidine does not reduce the biferric form of 3His-G4DFsc(Mut3) (Figure S17(green)), the regeneration of Fe(II)Fe(II) state suggests that the biferric 3His form is reduced by a reaction intermediate during catalysis.
Figure 15.
NIR CD spectra of the p-anisidine reaction with 3His-G4DFsc(Mut3). (A) CD spectra of the 3His protein in the presence of p-anisidine before exposure to O2 (green), under aerobic conditions (teal), and 30 min after first allowing O2 containing buffer (~250 μM O2) to react for 60 s and then sealing the CD cell to create a closed system (red). Protein concentrations for red and teal are 250 μM. (B) Time dependence of the 10 500 cm−1 feature after first allowing 3His-G4DFsc(Mut3)+P-AN to react with O2 followed by sealing the CD cell to create a closed system (see red in (A)). Data collected at 20 °C.
The oxygenation of p-anisidine to the nitroso product is a four-electron process where the first two-electron step should be the hydroxylation of the amine group followed by oxidation to the nitroso product. The above results suggest that the biferrous 3His-G4DFsc(Mut3) variant catalyzes the oxygenation of the amine group to a hydroxylamine and results in a biferric species. This hydroxylamine may then be oxidized to the nitroso product by the Fe(III)Fe(III) state of the 3His protein, regenerating the Fe(II)Fe(II) state in the rate-limiting step.
DISCUSSION
Our previous studies identified markedly different reactivities in the 2His and 3His 4A→4G variants of DFsc, where 4-aminophenol oxidase activity was observed in the 2His variant (G4DFsc), but not in the 3His variant (3His-G4DFsc(Mut3)), and arylamine oxygenase activity was detected for the 3His variant, but not the 2His form.51 This study has extended these results to understand the molecular basis for these reactivity differences.
Oxidase
By systematically controlling the reaction conditions, we have identified the active species for the 4-AP oxidation. Both the 2His and 3His variants display oxidase activity when substrate binding to the biferrous site preceded the reaction with O2. The CD, MCD and VTVH MCD studies show substantial perturbation of the biferrous sites of both G4DFsc and 3His-G4DFsc(Mut3) by 4-AP. While 4-AP addition to G4DFsc resulted in a new higher energy feature (~10 500 cm−1) in the MCD spectrum, its addition to 3His-G4DFsc(Mut3) causes a marked increase in MCD intensity (Δε from ~1 to ~7 cm−1 M−1). These perturbations suggest that this substrate binds directly to the biferrous sites of both forms.
Gaussian resolution of the CD and MCD features for both G4DFsc and 3His-G4DFsc(Mut3) in the presence of 4-AP reveal three bands that are similar in sign and energy to 3His-G4DFsc(Mut3) without 4-AP and suggest that all three forms have a 5-coordinate + 6-coordinate trigonal bipyramidal biferrous site. Given the lack of increased coordination in the 3His form upon substrate addition, it may be inferred that structural limitations prevent the substrate from simply binding to the open site on the 5-coordinate iron center of the 3His variant.
The VTVH MCD data of biferrous G4DFsc and 3His-G4DFsc(Mut3) were also perturbed by the binding of 4-AP. The associated spin-Hamiltonian parameters show that both forms are weakly antiferromagnetic coupled, indicating they still have μ-1,3 carboxylates bridging the biferrous site. These spectral data show that G4DFsc+4-AP and 3His-G4DFsc(Mut3)+4-AP have similar site structures where 4-AP binds to and increases the coordination of one Fe(II) in G4DFsc while replacing a ligand (likely the additional His) in 3His-G4DFsc(Mut3).
Substrate binding to the biferrous site initiates the oxidase activity. Therefore, the kinetics of 4-AP binding to the biferrous sites of G4DFsc and 3His-G4DFsc(Mut3) were explored with “freeze–quench” MCD spectroscopy. The binding rate of 4-AP for G4DFsc (kB = 0.027–0.042 s−1) was found to be an order of magnitude greater than that for the 3His variant (kB = 0.003–0.0046 s−1). However, the O2 reaction rates of both proteins are comparable at ~10−2 s−1.52 Based on the kinetics, 4-AP binding to the 3His variant is an order of magnitude lower than the O2 reaction rate. Thus, when substrate and O2 are simultaneously added, the biferrous 3His form first reacts with O2 to generate an inactive biferric species prior to substrate binding, which renders the protein ineffective for substrate oxidation. Since the 2His form exhibits faster 4-AP binding (likely due to the decreased coordination of the biferrous site), the slow substrate binding in the 3His variant may be attributed to its competition with the additional His ligand. The 3His-G4DFsc(Mut3) variant therefore becomes active only upon anaerobic incubation of substrate with the biferrous form, presumably following replacement of the additional His ligand at the active site by 4-AP.
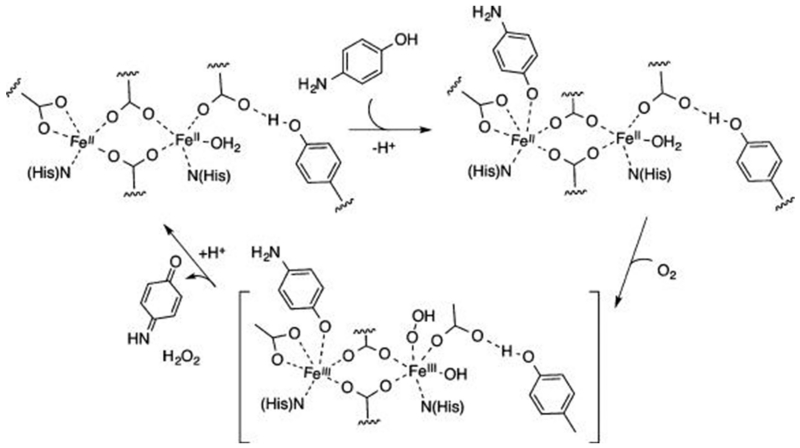
NIR CD spectroscopy was used to monitor the ferrous d→d transitions during 4-AP oxidation and showed the dominant presence of the biferrous 3His-G4DFsc(Mut3) species during steady state. Given that the resting biferric forms were inactive for substrate oxidation, a two-electron process may be invoked where O2 is reduced to H2O2 (which is lost) and 4-AP is oxidized to the benzoquinone imine at the biferrous site (Figure 16). The presence of 4-AP likely inhibits rebinding of the third His ligand to the biferrous site during turnover. There are two possible mechanisms for inhibition of the rebinding of the third His: (1) one 4-AP molecule stays bound during turnover, or (2) the 4-AP out-competes the third His protein residue in binding to the diiron center. The former possibility would involve a ping-pong mechanism since only one open coordination site is available for O2 reduction and 4-AP oxidation, while the latter would allow for direct O2 reduction at one iron site and 4-AP oxidation at the other.
Figure 16.
Mechanistic scheme of 4-aminophenol oxidation with biferrous protein.
The observed lack of oxidase activity of the biferric species in G4DFsc and 3His-G4DFsc(Mut3) is in contrast to other DF constructs.53 Since the NIR CD and MCD data demonstrate that substrate is accessible to the diiron sites, the lack of activity may be related to either unfavorable thermodynamics (redox potentials of G4DFsc and 3His-G4DFsc(Mut3) are unknown, but the two-electron potential for 4-AP is ~300 mV (converted to SHE, pH 7–9))54,55 or the coordinative saturation that is proposed for the ferric centers in G4DFsc and 3His-G4DFsc-(Mut3).52
Oxygenase
The oxygenation of p-anisidine to 4-nitroso-methoxybenzene (N-MB) by the 4A→4G DF variants observed in prior studies51 was further explored to understand the differential reactivity between G4DFsc and 3His-G4DFsc-(Mut3) as well as to identify the catalytically relevant species. In particular, the effects of anaerobic p-anisidine addition to the biferrous site of both G4DFsc and 3His-G4DFsc(Mut3) were spectroscopically studied to correlate geometric perturbations with reactivity.
Similar to prior results, these studies confirm that G4DFsc lacks oxygenase activity even with anaerobic addition of p-anisidine to the biferrous form. The NIR CD and MCD spectra show slight shifts in energies and intensities for the features associated with Fe(II) d→d transitions upon substrate binding to biferrous G4DFsc. These minimal spectral changes suggest that while the substrate does not bind directly to the biferrous site, it does bind near enough to perturb the biferrous site geometry. The VTVH MCD data of G4DFsc+P-AN indicate a decrease in antiferromagnetic coupling. Our prior results on the O2 reactivity of these DF variants indicate that O2 reduction occurs at a single iron center.52 Thus, the rate of the two-electron reduction of O2 may be influenced by the exchange-coupling of the two Fe(II) sites due to μ-1,3 carboxylate bridges. The decrease in antiferromagnetic coupling in G4DFsc upon P-AN addition may thus slow the rate of O2 reduction. However, the lack of oxygenase reactivity likely reflects an improper orientation of the substrate in relation to the reactive hydroperoxy species that would be produced upon O2 reduction (vide infra).
For 3His-G4DFsc(Mut3), substrate binding to the reduced protein was determined to be important for oxygenase activity. Similar to G4DFsc, the NIR CD and MCD spectra of the 3His form show only slight energy shifts and intensity changes for the Fe(II) d→d transitions upon the addition of p-anisidine, which thus indicates that substrate binds near (but not directly to) the biferrous site. In contrast to G4DFsc, substrate addition to 3His-G4DFsc(Mut3) slightly increases the antiferromagnetic coupling, which would not slow the rate of O2 reduction at a single iron center. These results highlight a differential perturbation of the p-anisidine substrate on the biferrous centers of G4DFsc and 3His-G4DFsc(Mut3) that may influence oxygen-dependent reactivities.
The oxygenation of p-anisidine to the nitroso product N-MB is a four-electron process, requiring a two-electron exogenous reductant for single turnover in the proposed mechanisms of AurF.20,29 Interestingly, the catalytic turnover of N-MB is detected in the 3His-G4DFsc(Mut3) system without the addition of exogenous reducing equivalents. The appearance of Fe(II) d→d transitions of 3His-G4DFsc(Mut3) during p-anisidine oxygenation upon O2 depletion indicates that the Fe(II)Fe(II) state is regenerated during turnover. These observations are consistent with the formation of an initial two-electron oxidized hydroxylamine product that subsequently reduces the biferric state to regenerate the biferrous site and form the 4-nitroso-methoxybenzene product. A lack of Fe(II) d→d transitions during steady state suggests that this reduction of the biferric site by the hydroxylamine intermediate would be the slow step in turnover.
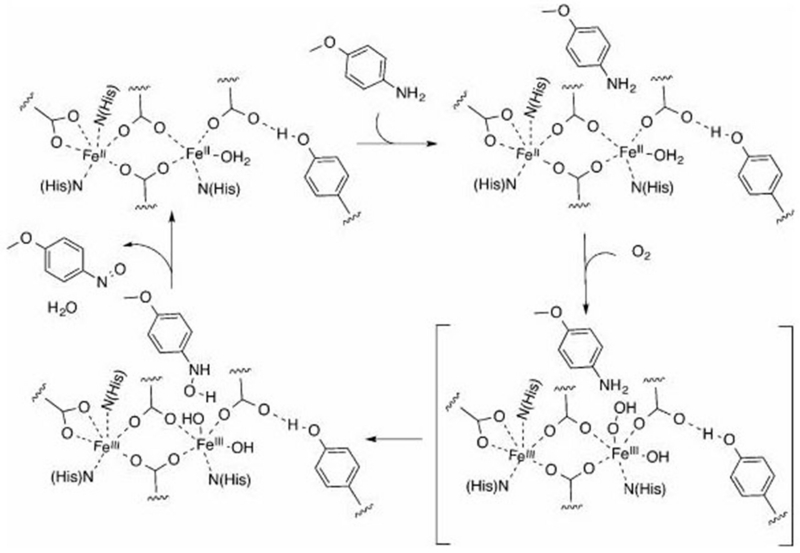
Based on the above results, a mechanism for the oxygenation of p-anisidine by 3His-G4DFsc(Mut3) is proposed in Figure 17. First, p-anisidine binds near the biferrous site (perhaps through hydrogen-bonding interactions between the amine group of p-anisidine and a residue near the active site). This is followed by O2 end-on binding to the unsaturated Fe(II) site, leading to its reduction by two electrons to generate a hydroperoxy biferric species. Then, hydroxylation of the amine group of p-anisidine is likely to proceed via an electrophilic attack on the nitrogen lone pair by the hydroperoxy species. This hypothesis is consistent with studies of amine hydroxylation catalyzed by AurF, where protonation of the bridged peroxy species was found to drive this electrophilic attack.56 However, in the case of AurF, the hydroperoxy species is bridged rather than end-on. In our case, the third His saturates one iron center, preventing bridging of the peroxide, but still allowing for protonation and electrophilic attack. The biferric species formed upon substrate oxygenation would then be reduced by the resultant hydroxylamine, forming the nitroso product and regenerating the biferrous site for turnover.
Figure 17.
Mechanistic scheme for oxygenation of p-anisidine by 3His-G4DFsc(Mut3).
CONCLUSIONS
These studies on de novo designed proteins offer molecular level insight into the O2-dependent chemistry of binuclear non-heme iron enzymes. Contrary to our prior results, 3His-G4DFsc-(Mut3) does equivalent oxidase reactivity to that of the 2His form, provided enough time is given for substrate binding. This activates the 3His-G4DFsc(Mut3) through substitution of a ligand. The results illustrate that protein design may be used to inhibit oxidase reactivity by altering the substrate-binding rate through saturating the coordination of an iron site. While G4DFsc lacks arylamine oxygenase activity, p-anisidine still binds near the diiron sites in both 3His-G4DFsc(Mut3) and G4DFsc. However, there is a differential perturbation that may reflect reactivity differences. Saturating one Fe(II) of the biferrous site but enabling two-electron transfer through μ-1,3 carboxylate bridges appears to facilitate the formation of an end-on hydroperoxy species at one Fe center that is competent in electrophilic attack on the lone pair of the amine group. In contrast to some mechanisms proposed for AurF,20,29 3His-G4DFsc(Mut3) appears to be a mixed function oxidase where it performs both oxygenation and oxidation of the p-anisidine substrate. The apparent necessity of direct substrate binding to the diiron site suggests the importance of a sufficient electron transfer pathway in oxidase reactivity, while the presence of substrate in a protein-binding pocket may facilitate oxygenase reactivity. These studies demonstrate how modification of substrate binding through additional protein ligation may tune the function of the active site in binuclear non-heme iron enzymes.
MATERIALS AND METHODS
Preparation of Apo-Protein
Apo-protein of G4DFsc and 3His-G4DFsc(Mut3) were prepared as previously described.51
Preparation of Biferrous Protein
Biferrous protein of G4DFsc and 3His-G4DFsc(Mut3) were prepared anaerobically as previously described.52 This preparation included degassing apo-protein on a Schlenk line by gentle pump/purge cycles followed by purging the headspace for continuously for several hours. Protein was then transferred to a glovebox and loaded with Fe(II) (FeSO4·6H2O from J.T. Baker) by adding ~1.7–1.8 equiv of ferrous ammonium sulfate (solution prepared anaerobically) and allowing the solution to incubate for ~15–20 min. For the buffer, MOPS and NaCl were obtained from Sigma-Aldrich.
Oxidase Activity Assays
For substrate preloaded samples, 4-AP (in 30% DMF prepared anaerobically) was anaerobically added to biferrous protein (G4DFsc or 3His-G4DFsc(Mut3) in 150 mM MOPS/150 mM NaCl at pH 7) and incubated for 20 min. m-phenylenediamine was then added to the protein sample before being loaded into an anaerobic cell for absorption experiments. An equivalent volume of O2-saturated buffer (spurging buffer with O2 gas in a closed vessel at 4 °C for 15 min) was added to the sample (resulting in dilution of protein by 1/2) and rapidly mixed immediately before collecting data. For simultaneous addition of O2-saturated buffer and 4-AP to the protein, 4-AP and m-phenylenediamine solutions were prepared in the glovebox prior to mixing experiments. Biferrous protein was loaded into an anaerobic cell (in a glovebox). Immediately before addition to the protein sample, 4-AP and m-phenylenediamine were added to the O2-saturated buffer, which was then added to the biferrous protein and the UV–vis absorption spectra were collected after a few seconds of rapid mixing. Resting G4DFsc (Species C) and 3His-G4DFsc(Mut3) biferric forms were prepared by adding O2-saturated buffer (equivalent volume, dilution of protein by one-half), allowing the reaction to proceed for at least 2 h at 4 °C. This sample was then added to a UV–vis absorption cell. 4-AP and m-phenylenediamine were added and mixed immediately before initiating data collection. For biferric Species B of G4DFsc, biferrous protein was prepared followed by O2-saturated buffer addition, which was allowed to react for ~100 s (~50–70% Species B). 4-AP and m-phenylenediamine were then added to the sample and data were collected. Final concentrations (after O2-saturated buffer addition) of protein, 4-AP and m-phenylenediamine were 10 μM for protein, 3.9 mM 4-AP, and 10 mM m-phenylenediamine. The concentrations of DMF in the protein solutions were kept below 3%. In all experiments, after addition of O2-saturated buffer, the samples were held under aerobic conditions. A circulator connected to the UV–vis cell holder was used to maintain the absorption cell at 4–12 °C.
NIR CD and MCD of 4-Aminophenol Addition to Biferrous Protein
For the CD experiments, biferrous protein was prepared in the glovebox. 4-AP was added (in ~15-fold excess) to biferrous protein (~1–2 mM) and the protein sample was incubated for ~20 min. The sample was then loaded into an anaerobic CD cell, and data were measurements were collected. A circulator connected to the CD cell holder was used to maintain the anaerobic quartz CD cell at 4–12 °C. Multiple scans (5–20) were averaged. The CD spectrum of apo-protein was subtracted from the spectra. For MCD experiments, protein samples were prepared with 4-AP (~1 mM in protein incubated for ~20 min with ~15-fold excess 4-AP and in 54% d-glycerol (glycerol-d8 ; 98% 2H from Cambridge Isotope Laboratories) and 46% buffer (150 mM MOPS/150 mM NaCl in D2O (D2O; 99.9% 2H) at pD 7). These samples were loaded into MCD cells under anaerobic conditions and immersed into liquid nitrogen prior to data collection. CD and MCD spectra were fit with the minimum number of Gaussian peaks required for the data using a nonlinear least-squares procedure that allows for variation in peak width and energy positions between the CD and MCD fits. For the time-dependent MCD experiments, a solution of 4-AP in 54% deuterated glycerol was anaerobically prepared. 4-AP solution (in d-glycerol) was injected into an MCD cell with biferrous protein (in 54% d-glycerol and 46% deuterated buffer) in an anaerobic environment, quickly mixed, and incubated for different time intervals (e.g., 0.5 min, 5 min, etc.) prior to being immersed into liquid nitrogen. MCD spectra shown are the averages of 5–20 scans. The averaged spectra taken at the same magnitude but oppositely signed fields were averaged (e.g., ((+7T) – (−7T))/2) as well to subtract out the baseline. Both the near IR (600–1900 nm region) CD and MCD data were collected on a JASCO J730D spectropolarimeter with a cooled InSb detector. The concentrations of DMF in the protein samples were kept below 3%. Deuterium oxide and d-glycerol were obtained from Cambridge Isotope Laboratories (Andover, MA).
CD kinetics of 4-Aminophenol Oxidation
A solution of 4-AP and biferrous protein was prepared (0.5 mM protein with ~15-fold excess of 4-AP in regular H2O buffer) anaerobically where the sample was incubated for ~20 min before being placed into an anaerobic quartz CD cell. O2-saturated buffer was prepared by spurging the buffer with O2 gas in a sealed container for ~15 min at 4 °C. A circulator connected to the CD cell holder was used to maintain temperature at 4–12 °C. Prior to O2-saturated buffer addition, the CD spectrum of the biferrous protein with 4-AP was measured. The JASCO J730D spectropolarimeter was fixed at 10 600 cm−1. The CD intensity at the associated time points were collected upon O2-saturated buffer addition (where the protein sample was diluted by 1/2). A complete CD spectrum was again taken after ~20 min, 1 h, and ~6 h. A cooled InSb detector was used for data collection. The concentrations of DMF in the protein samples were kept below 3%. After O2-saturated buffer addition, samples were kept under aerobic conditions.
Oxygenase Activity Assays
For substrate bound protein experiments, p-anisidine was anaerobically added to biferrous protein and incubated for ~20 min. The sample was then placed in an anaerobic UV–vis absorption cell. O2-saturated buffer was added to the protein solution (dilution of protein by 1/2) at 20 °C, rapidly mixed with a pipet and spectral collection as initiated. Scans were measured every 10 s for the first minute, every minute for the first 10 min, and then every 10 min afterward. After addition of O2-saturated buffer (prepared by spurging O2 gas into buffer in a closed vessel for ~15 min at 20 °C), the sample was maintained under aerobic conditions. For simultaneous addition of substrate and O2 to the protein, biferrous protein was prepared anaerobically and placed into an anaerobic UV–vis absorption cell. Substrate was added to O2-saturated buffer, which was then added to the protein solution (dilution of protein by 1/2) in the UV–vis absorption cell immediately before data collection. Absorption spectra were collected every 10 s for the first minute, every minute for the first 10 min, and then every 10 min. For p-anisidine addition to the biferric form, the biferric species was formed by addition of O2-saturated buffer to biferrous protein and allowed to react for 1–2 h. The sample was then buffer exchanged to remove any H2O2 produced during the reaction. This sample was then loaded into a UV–vis absorption cell, and a single UV–vis absorption spectrum was collected. p-Anisidine was then added to the protein sample, and UV–vis absorption spectra were collected every 10 s for the first minute, every minute for the first 10 min, and then every 10 min. Final samples contained 65 μM protein and 1 mM substrate in 150 mM MOPS/150 mM NaCl buffer at pH 7.
For the experimental details concerning the LCMS of product formation, the H2O2 and non-buffer-exchanged biferric species controls of p-anisidine oxidase assays, and UV–vis absorption monitoring of the O2 reactivity of biferrous G4DFsc in the presence of p-anisidine, see the methods section in the Supporting Information.
NIR CD and MCD of p-Anisidine Addition to Biferrous Protein
For CD experiments, p-anisidine was anaerobically added to biferrous protein (in D2O buffer) and allowed to incubate for ~20 min. The sample (~1–2 mM protein and 20–30-fold excess substrate) was then placed into an anaerobic CD cell. Multiple scans (5–20) of the spectrum were collected on a JASCO J730D spectropolarimeter with a cooled InSb detector and averaged. The CD of apoprotein was subtracted from these data. MCD cells were prepared by mixing d-glycerol with substrate-loaded biferrous protein CD samples (54% glycerol, 46% buffer/protein solution) and injecting the sample into the MCD cells under anaerobic conditions. The samples were then immersed into liquid nitrogen. Multiple scans (5–20) were taken and averaged at varying fields (±7, ±5, ±3, ±1, and 0 T) and temperatures (2, 5, 10, and 20 K). The baseline of the MCD data was subtracted by averaging positive and negative fields (e.g., ((+7T) − (−7T))/2).
CD Kinetics of p-Anisidine Oxygenation
Substrate (p-anisidine) was added to biferrous protein prepared anaerobically (in ~30-fold excess) and allowed to incubate for ~20 min. The protein sample (~0.5 mM protein in H2O buffer) was then loaded into the anaerobic CD cell. For steady state conditions, O2-saturated buffer was added to the sample (dilution of protein by 1/2), the CD features were monitored at 10 600 cm−1 for 60 min and a CD spectrum was collected under aerobic conditions after 1 h (taking ~5 scans and averaging them). This sample was then degassed, concentrated to ~1/2 volume, and mixed with d-glycerol (54% v/v). The sample was then injected into an MCD sample cell under anaerobic conditions and immersed into liquid nitrogen. Multiple scans were taken of the MCD spectrum at 2 K and ±7 T and averaged. The MCD spectra at positive and negative fields were averaged to subtract baseline. For monitoring of the reaction under non-steady-state conditions by CD spectroscopy, a substrate-bound biferrous protein sample was prepared (~30-fold excess substrate, 0.5 mM protein) as above. The protein sample was added to an anaerobic CD cell and the CD spectrum was collected. The CD intensity at 10 600 cm−1 was monitored for ~60 s as an equal volume of O2 containing buffer (~250 μM) was mixed with the protein solution under aerobic conditions. The CD cell was then sealed, and the CD intensity at 10 600 cm−1 was monitored for an additional 30 min. Upon completion, multiple scans of the CD spectrum were taken and averaged. The CD spectrum of apo-protein was subtracted.
Near-IR VTVH MCD Experiments
A total of 100–200 scans of the MCD intensity at specific energies were collected for temperatures of 2, 3, 5, 7.5, 10, 15, 20, and 25 K at fields of 0, ±0.35, ±0.7, ±1.4, ±2.1, ±2.8, ±3.5, ±4.2, ±4.7, ±5.6, ±6.3, and ±7.0 T. These measurements were averaged for each field at each temperature. The reported data are averages of baseline-subtracted measurements for positive and negative field taken at the same temperature (error bars represent the propagation of errors from the standard deviations). The chi-squared value was used to judge the goodness of fit for the VTVH MCD data. The VTVH MCD data were fit using doublet models as described previously.6
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the NSF (MCB-1404866 to E.I.S. and CHE-1413295 to W.F.D.) and the NIH (F32-GM808852 and R15-GM110657 to A.J.R. and GM54616 and GM71628 to W.F.D.). We thank the Vincent Coates Foundation Mass Spectrometry Laboratory (Stanford University Mass Spectrometry), particularly Ludmila Alexandrova, for acquisition of the LCMS data included in the Supporting Information.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
UV–vis absorption spectra of 4-AP oxidation with 3His-G4DFsc(Mut3) without m-phenylenediamine; UV–vis absorption of 4-AP oxidation in the presence of m-phenylenediamine without protein; CD spectra of G4DFsc and 3His-G4DFsc-(Mut3) with and without 4-AP addition; MCD spectra of G4DFsc+4-AP at different temperatures and magnetic fields showing C-term behavior; doublet fitting of the VTVH MCD data for G4DFsc+4-AP and 3His-G4DFsc(Mut3)+4-AP; composite MCD spectrum for 0.6 × G4DFsc+4-AP and 0.4 × G4DFsc; MCD spectra of 4-AP addition to 3His-G4DFsc-(Mut3) at different times; composite MCD spectrum of 0.68 × 3His-G4DFsc(Mut3)+4-AP and 0.32 × 3His-G4DFsc(Mut3); kinetic trace for the 470 nm feature of 3His-G4DFsc(Mut3)+4-AP reacted with O2-saturated buffer; UV–vis absorption spectrum of p-anisidine oxidation by H2O2 and non-buffer-exchanged biferric 3His-G4DFsc(Mut3); LCMS data for O2 reaction with biferric and biferrous 3His-G4DFsc(Mut3)+P-AN and biferrous G4DFsc+PAN; MCD spectra at different temperatures for 3His-G4DFsc(Mut3)+P-AN; doublet fitting of the VTVH MCD data for G4DFsc+P-AN and 3His-G4DFsc-(Mut3)+P-AN; UV–vis absorption spectra of O2 reaction with G4DFsc+P-AN; pre-steady-state kinetic traces (near-IR, CD, and UV–vis absorption) for O2 reaction with 3His-G4DFsc-(Mut3)+P-AN; UV–vis absorption after 48 h of O2 reaction with 3His-G4DFsc(Mut3)+P-AN; MCD of O2-depleted 3His-G4DFsc(Mut3)+P-AN samples; correlation energy diagram for G4DFsc+P-AN and 3His-G4DFsc(Mut3)+P-AN; details of materials and methods for LCMS of product formation, H2O2+P-AN and non-buffer-exchanged biferric 3His-G4DFsc-(Mut3)+P-AN controls; and UV–vis absorption spectra of the O2 reaction with G4DFsc+P-AN. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b03524.
REFERENCES
- (1).Fishman A, Tao Y, Bentley WE, Wood TK. Biotechnol. Bioeng. 2004;87:779–790. doi: 10.1002/bit.20185. [DOI] [PubMed] [Google Scholar]
- (2).Schirmer A, Rude MA, Li X, Popova E, Del Cardayre SB. Science. 2010;329:559–562. doi: 10.1126/science.1187936. [DOI] [PubMed] [Google Scholar]
- (3).Lee SW, Keeney DR, Lim DH, Dispirito AA, Semrau JD. Appl. Environ. Microbiol. 2006;72:7503–7509. doi: 10.1128/AEM.01604-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Yang B, Hodgkinson A, Millward BA, Demaine AG. Int. J. Diabetes Mellitus. 2010;2:169–174. [Google Scholar]
- (5).Shao J, Zhou B, Chu B, Yen Y. Curr. Cancer Drug Targets. 2006;6:409–431. doi: 10.2174/156800906777723949. [DOI] [PubMed] [Google Scholar]
- (6).Solomon EI, Brunold TC, Davis MI, Kemsley JN, Lee SK, Lehnert N, Neese F, Skulan AJ, Yang YS, Zhou J. Chem. Rev. 2000;100:235–349. doi: 10.1021/cr9900275. [DOI] [PubMed] [Google Scholar]
- (7).Andrews SC. Biochim. Biophys. Acta. 2010;1800:691–705. doi: 10.1016/j.bbagen.2010.05.010. [DOI] [PubMed] [Google Scholar]
- (8).Theil ECJ. Nutr. 2003;133:1549S–1553S. doi: 10.1093/jn/133.5.1549S. [DOI] [PubMed] [Google Scholar]
- (9).Fang H, Caranto JD, Mendoza R, Taylor AB, Hart PJ, Kurtz DM., Jr. J. Biol. Inorg. Chem. 2012;17:1231–1239. doi: 10.1007/s00775-012-0938-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kurtz DM., Jr. Dalton Trans. 2007;0:4115–4121. [Google Scholar]
- (11).Hayashi T, Caranto JD, Wampler DA, Kurtz DM, Jr., Moënne-Loccoz P. Biochemistry. 2010;49:7040–7049. doi: 10.1021/bi100788y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Silaghi-Dumitrescu R, Kurtz DM, Jr., Ljungdahl LG, Lanzilotta WN. Biochemistry. 2005;44:6492–6501. doi: 10.1021/bi0473049. [DOI] [PubMed] [Google Scholar]
- (13).Jin S, Kurtz DM, Jr., Liu ZJ, Rose J, Wang BC. Biochemistry. 2004;43:3204–3213. doi: 10.1021/bi0356193. [DOI] [PubMed] [Google Scholar]
- (14).Shiba T, Kido Y, Sakamoto K, Inaoka DK, Tsuge C, Tatsumi R, Takahashi G, Balogun EO, Nara T, Aoki T, Honma T, Tanaka A, Inoue M, Matsuoka S, Saimoto H, Moore AL, Harada S, Kita K. Proc. Natl. Acad. Sci. U.S.A. 2013;110:4580–4585. doi: 10.1073/pnas.1218386110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Whittington DA, Lippard SJ. J. Am. Chem. Soc. 2001;123:827–838. doi: 10.1021/ja003240n. [DOI] [PubMed] [Google Scholar]
- (16).Mitić N, Schwartz JK, Brazeau BJ, Lipscomb JD, Solomon EI. Biochemistry. 2008;47:8386–8397. doi: 10.1021/bi800818w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Lipscomb JD. Annu. Rev. Microbiol. 1994;48:371–399. doi: 10.1146/annurev.mi.48.100194.002103. [DOI] [PubMed] [Google Scholar]
- (18).Baik MH, Newcomb M, Friesner RA, Lippard SJ. Chem. Rev. 2003;103:2385–2420. doi: 10.1021/cr950244f. [DOI] [PubMed] [Google Scholar]
- (19).Bertrand E, Sakai R, Rozhkova-Novosad E, Moe L, Fox BG, Groves JT, Austin RN. J. Inorg. Biochem. 2005;99:1998–2006. doi: 10.1016/j.jinorgbio.2005.06.020. [DOI] [PubMed] [Google Scholar]
- (20).Choi YS, Zhang H, Brunzelle JS, Nair SK, Zhao H. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6858–6863. doi: 10.1073/pnas.0712073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Winkler R, Hertweck C. Angew. Chem., Int. Ed. 2005;44:4083–4087. doi: 10.1002/anie.200500365. [DOI] [PubMed] [Google Scholar]
- (22).Wei PP, Skulan AJ, Mitić N, Yang YS, Saleh L, Bollinger JM, Jr., Solomon EI. J. Am. Chem. Soc. 2004;126:3777–3788. doi: 10.1021/ja0374731. [DOI] [PubMed] [Google Scholar]
- (23).Lindqvist Y, Huang W, Schneider G, Shanklin J, EMBO J. 1996;15:4081–4092. [PMC free article] [PubMed] [Google Scholar]
- (24).Yang YS, Broadwater JA, Pulver SC, Fox BG, Solomon EI. J. Am. Chem. Soc. 1999;121:2770–2783. [Google Scholar]
- (25).Stubbe J. Curr. Opin. Chem. Biol. 2003;7:183–188. doi: 10.1016/s1367-5931(03)00025-5. [DOI] [PubMed] [Google Scholar]
- (26).Xing G, Hoffart LM, Diao Y, Prabhu KS, Arner RJ, Reddy CC, Krebs C, Bollinger JM., Jr. Biochemistry. 2006;45:5393–5401. doi: 10.1021/bi0519607. [DOI] [PubMed] [Google Scholar]
- (27).Simurdiak M, Lee J, Zhao H. Chem BioChem. 2006;7:1169–1172. doi: 10.1002/cbic.200600136. [DOI] [PubMed] [Google Scholar]
- (28).Krebs C, Matthews ML, Jiang W, Bollinger JM., Jr. Biochemistry. 2007;46:10413–10418. doi: 10.1021/bi701060g. [DOI] [PubMed] [Google Scholar]
- (29).Li N, Korboukh VK, Krebs C, Bollinger JM., Jr. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15722–15727. doi: 10.1073/pnas.1002785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Brown PM, Caradoc-Davies TT, Dickson JM, Cooper GJ, Loomes KM, Baker EN. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15032–15037. doi: 10.1073/pnas.0605143103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Thorsell AG, Persson C, Voevodskaya N, Busam RD, Hammarstrom M, Graslund S, Graslund A, Hallberg BM. J. Biol. Chem. 2008;283:15209–15216. doi: 10.1074/jbc.M800348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Liu KE, Wang D, Huynh BH, Edmondson DE, Salifoglou A, Lippard SJ. J. Am. Chem. Soc. 1994;116:7465–7466. [Google Scholar]
- (33).Tong WH, Chen S, Lloyd SG, Edmondson DE, Huynh BH, Stubbe JJ. Am. Chem. Soc. 1996;118:2107–2108. [Google Scholar]
- (34).Bollinger JM, Jr., Krebs C, Vicol A, Chen S, Ley BA, Edmondson DE, Huynh BH. J. Am. Chem. Soc. 1998;120:1094–1095. [Google Scholar]
- (35).Yun D, Garcia-Serres R, Chicalese BM, An YH, Huynh BH, Bollinger JM., Jr. Biochemistry. 2007;46:1925–1932. doi: 10.1021/bi061717n. [DOI] [PubMed] [Google Scholar]
- (36).Broadwater JA, Ai J, Loehr TM, Sanders-Loehr J, Fox BG. Biochemistry. 1998;37:14664–14671. doi: 10.1021/bi981839i. [DOI] [PubMed] [Google Scholar]
- (37).Pereira AS, Small W, Krebs C, Tavares P, Edmondson DE, Theil EC, Huynh BH. Biochemistry. 1998;37:9871–9876. doi: 10.1021/bi980847w. [DOI] [PubMed] [Google Scholar]
- (38).Korboukh VK, Li N, Barr EW, Bollinger JM, Jr., Krebs CJ. Am. Chem. Soc. 2009;131:13608–13609. doi: 10.1021/ja9064969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Murray LJ, Garcia-Serres R, Naik S, Huynh BH, Lippard SJ. J. Am. Chem. Soc. 2006;128:7458–7459. doi: 10.1021/ja062762l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Tinberg CE, Lippard SJ. Acc. Chem. Res. 2011;44:280–288. doi: 10.1021/ar1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Ambundo EA, Friesner RA, Lippard SJ. J. Am. Chem. Soc. 2002;124:8770–8771. doi: 10.1021/ja0265759. [DOI] [PubMed] [Google Scholar]
- (42).Beauvais LG, Lippard SJ. Biochem. Biophys. Res. Commun. 2005;338:262–266. doi: 10.1016/j.bbrc.2005.08.220. [DOI] [PubMed] [Google Scholar]
- (43).Nordlund P, Eklund H. J. Mol. Biol. 1993;232:123–164. doi: 10.1006/jmbi.1993.1374. [DOI] [PubMed] [Google Scholar]
- (44).Calhoun JR, Liu W, Spiegel K, Dal Peraro M, Klein ML, Valentine KG, Wand AJ, DeGrado WF. Structure. 2008;16:210–215. doi: 10.1016/j.str.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).DeGrado WF, Summa CM, Pavone V, Nastri F, Lombardi A. Annu. Rev. Biochem. 1999;68:779–819. doi: 10.1146/annurev.biochem.68.1.779. [DOI] [PubMed] [Google Scholar]
- (46).Bryson JW, Betz SF, Lu HS, Suich DJ, Zhou HX, O’Neil KT, DeGrado WF. Science. 1995;270:935–941. doi: 10.1126/science.270.5238.935. [DOI] [PubMed] [Google Scholar]
- (47).DeGrado WF, Wasserman ZR, Lear JD. Science. 1989;243:622–628. doi: 10.1126/science.2464850. [DOI] [PubMed] [Google Scholar]
- (48).Bell CB, III, Calhoun JR, Bobyr E, Wei P, Hedman B, Hodgson KO, DeGrado WF, Solomon EI. Biochemistry. 2009;48:59–73. doi: 10.1021/bi8016087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Calhoun JR, Kono H, Lahr S, Wang W, DeGrado WF, Saven JG. J. Mol. Biol. 2003;334:1101–1115. doi: 10.1016/j.jmb.2003.10.004. [DOI] [PubMed] [Google Scholar]
- (50).Calhoun JR, Bell CB, III, Smith TJ, Thamann TJ, DeGrado WF, Solomon EI. J. Am. Chem. Soc. 2008;130:9188–9189. doi: 10.1021/ja801657y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Reig AJ, Pires MM, Snyder RA, Wu Y, Jo H, Kulp DW, Butch SE, Calhoun JR, Szyperski T, Solomon EI, DeGrado WF. Nat. Chem. 2012;4:900–906. doi: 10.1038/nchem.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Snyder RA, Betzu J, Butch SE, Reig AJ, DeGrado WF, Solomon EI. Biochemistry. 2015 doi: 10.1021/acs.biochem.5b00324. DOI: 10.1021/acs.biochem.5b00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Kaplan J, DeGrado WF. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11566–11570. doi: 10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Tang HT, Lunte CE, Halsall HB, Heineman WR. Anal. Chim. Acta. 1988;214:187–195. [Google Scholar]
- (55).Safavi A, Maleki N, Moradlou O. Electroanalysis. 2008;20:2158–2162. [Google Scholar]
- (56).Park K, Li N, Kwak Y, Srnec M, Bell CB, III, Liu LV, Wong SD, Yoda Y, Kitao S, Seto M, Hu M, Zhao J, Krebs C, Bollinger JM, Jr., Solomon EI. Manuscript in preparation. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.