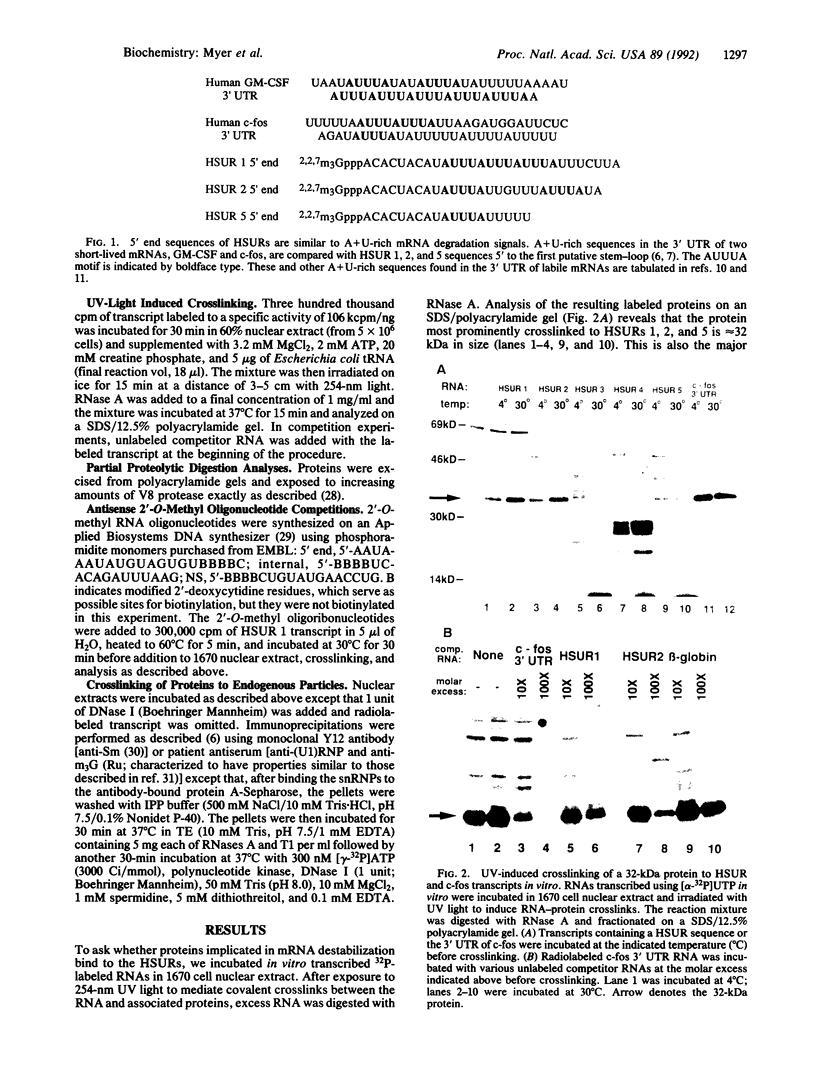
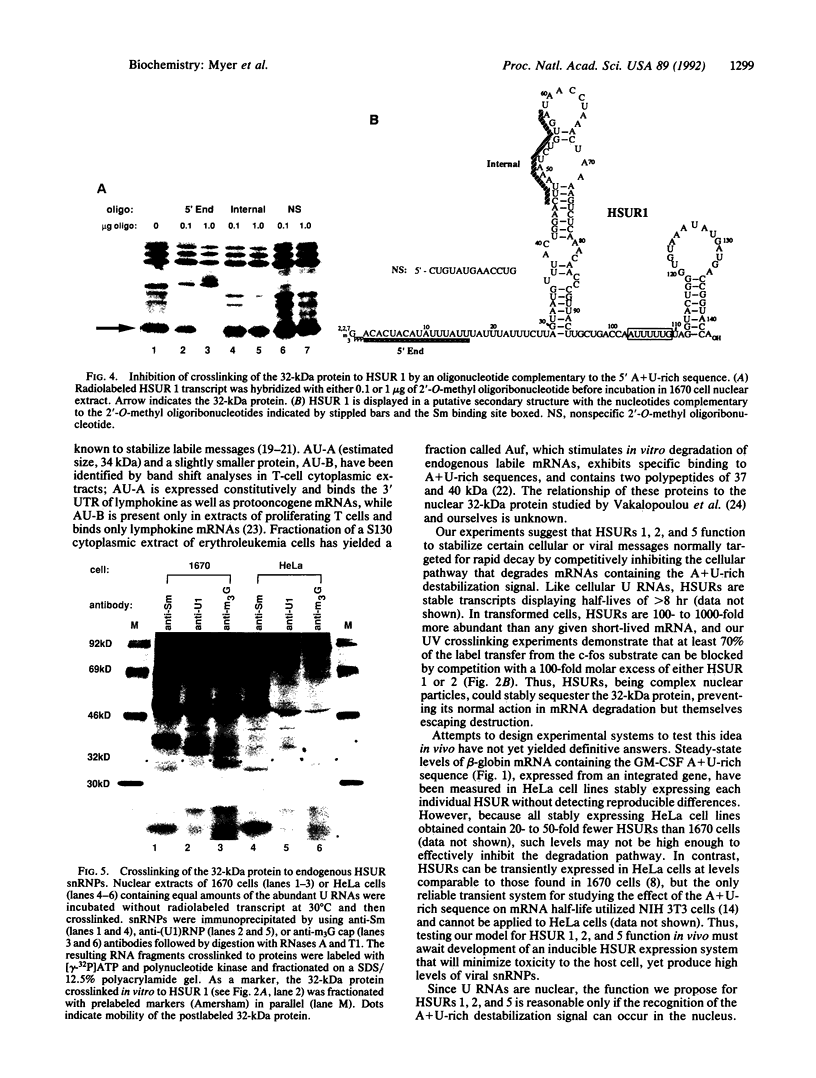
Abstract
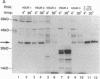
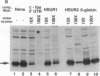
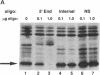
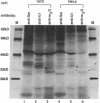
Herpesvirus saimiri (HVS) is one of several primate viruses that carry genes for small RNAs. The five H. saimiri-encoded U RNAs (HSURs) are the most abundant viral transcripts expressed in transformed marmoset T lymphocytes. They assemble with host proteins common to spliceosomal small nuclear ribonucleoproteins (snRNPs). HSURs 1, 2, and 5 exhibit sequences at their 5' ends identical to the AUUUA motif, which targets a number of protooncogene, cytokine, and lymphokine mRNAs for rapid degradation. We show that a 32-kDa protein previously demonstrated to bind to the 3' untranslated region of several unstable messages can be UV crosslinked specifically to HSUR 1, 2, and 5 transcripts in vitro, as well as to endogenous HSUR snRNPs. Our results suggest an unusual role for these viral snRNPs: HSURs may function to attenuate the rapid degradation of certain cellular mRNAs, thereby facilitating viral transformation of host T lymphocytes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biesinger B., Trimble J. J., Desrosiers R. C., Fleckenstein B. The divergence between two oncogenic Herpesvirus saimiri strains in a genomic region related to the transforming phenotype. Virology. 1990 Jun;176(2):505–514. doi: 10.1016/0042-6822(90)90020-r. [DOI] [PubMed] [Google Scholar]
- Bohjanen P. R., Petryniak B., June C. H., Thompson C. B., Lindsten T. An inducible cytoplasmic factor (AU-B) binds selectively to AUUUA multimers in the 3' untranslated region of lymphokine mRNA. Mol Cell Biol. 1991 Jun;11(6):3288–3295. doi: 10.1128/mcb.11.6.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991 May;11(5):2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G., Ross J. Poly(A) shortening and degradation of the 3' A+U-rich sequences of human c-myc mRNA in a cell-free system. Mol Cell Biol. 1988 Apr;8(4):1697–1708. doi: 10.1128/mcb.8.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caput D., Beutler B., Hartog K., Thayer R., Brown-Shimer S., Cerami A. Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Desrosiers R. C., Bakker A., Kamine J., Falk L. A., Hunt R. D., King N. W. A region of the Herpesvirus saimiri genome required for oncogenicity. Science. 1985 Apr 12;228(4696):184–187. doi: 10.1126/science.2983431. [DOI] [PubMed] [Google Scholar]
- Desrosiers R. C., Silva D. P., Waldron L. M., Letvin N. L. Nononcogenic deletion mutants of herpesvirus saimiri are defective for in vitro immortalization. J Virol. 1986 Feb;57(2):701–705. doi: 10.1128/jvi.57.2.701-705.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis P., Malter J. S. The adenosine-uridine binding factor recognizes the AU-rich elements of cytokine, lymphokine, and oncogene mRNAs. J Biol Chem. 1991 Feb 15;266(5):3172–3177. [PubMed] [Google Scholar]
- Kabnick K. S., Housman D. E. Determinants that contribute to cytoplasmic stability of human c-fos and beta-globin mRNAs are located at several sites in each mRNA. Mol Cell Biol. 1988 Aug;8(8):3244–3250. doi: 10.1128/mcb.8.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird-Offringa I. A., Elfferich P., van der Eb A. J. Rapid c-myc mRNA degradation does not require (A + U)-rich sequences or complete translation of the mRNA. Nucleic Acids Res. 1991 May 11;19(9):2387–2394. doi: 10.1093/nar/19.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. I., Murthy S. C., Trimble J. J., Desrosiers R. C., Steitz J. A. Four novel U RNAs are encoded by a herpesvirus. Cell. 1988 Aug 26;54(5):599–607. doi: 10.1016/s0092-8674(88)80004-7. [DOI] [PubMed] [Google Scholar]
- Lee S. I., Steitz J. A. Herpesvirus saimiri U RNAs are expressed and assembled into ribonucleoprotein particles in the absence of other viral genes. J Virol. 1990 Aug;64(8):3905–3915. doi: 10.1128/jvi.64.8.3905-3915.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner E. A., Lerner M. R., Janeway C. A., Jr, Steitz J. A. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci U S A. 1981 May;78(5):2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malter J. S., Hong Y. A redox switch and phosphorylation are involved in the post-translational up-regulation of the adenosine-uridine binding factor by phorbol ester and ionophore. J Biol Chem. 1991 Feb 15;266(5):3167–3171. [PubMed] [Google Scholar]
- Malter J. S. Identification of an AUUUA-specific messenger RNA binding protein. Science. 1989 Nov 3;246(4930):664–666. doi: 10.1126/science.2814487. [DOI] [PubMed] [Google Scholar]
- Meijlink F., Curran T., Miller A. D., Verma I. M. Removal of a 67-base-pair sequence in the noncoding region of protooncogene fos converts it to a transforming gene. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4987–4991. doi: 10.1073/pnas.82.15.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy S. C., Trimble J. J., Desrosiers R. C. Deletion mutants of herpesvirus saimiri define an open reading frame necessary for transformation. J Virol. 1989 Aug;63(8):3307–3314. doi: 10.1128/jvi.63.8.3307-3314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy S., Kamine J., Desrosiers R. C. Viral-encoded small RNAs in herpes virus saimiri induced tumors. EMBO J. 1986 Jul;5(7):1625–1632. doi: 10.1002/j.1460-2075.1986.tb04405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei R., Calame K. Differential stability of c-myc mRNAS in a cell-free system. Mol Cell Biol. 1988 Jul;8(7):2860–2868. doi: 10.1128/mcb.8.7.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Shyu A. B., Belasco J. G., Greenberg M. E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991 Feb;5(2):221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- Shyu A. B., Greenberg M. E., Belasco J. G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989 Jan;3(1):60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- Sproat B. S., Lamond A. I., Beijer B., Neuner P., Ryder U. Highly efficient chemical synthesis of 2'-O-methyloligoribonucleotides and tetrabiotinylated derivatives; novel probes that are resistant to degradation by RNA or DNA specific nucleases. Nucleic Acids Res. 1989 May 11;17(9):3373–3386. doi: 10.1093/nar/17.9.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolle C. A., Payne M. S., Benz E. J., Jr Equal stabilities of normal beta globin and nontranslatable beta0 -39 thalassemic transcripts in cell-free extracts. Blood. 1987 Jul;70(1):293–300. [PubMed] [Google Scholar]
- Trimble J. J., Desrosiers R. C. Transformation by herpesvirus saimiri. Adv Cancer Res. 1991;56:335–355. doi: 10.1016/s0065-230x(08)60485-6. [DOI] [PubMed] [Google Scholar]
- Vakalopoulou E., Schaack J., Shenk T. A 32-kilodalton protein binds to AU-rich domains in the 3' untranslated regions of rapidly degraded mRNAs. Mol Cell Biol. 1991 Jun;11(6):3355–3364. doi: 10.1128/mcb.11.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman D. A., Lee S. I., Steitz J. A. Nucleotide sequence of HSUR 5 RNA from herpesvirus saimiri. Nucleic Acids Res. 1989 Feb 11;17(3):1258–1258. doi: 10.1093/nar/17.3.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittemore L. A., Maniatis T. Postinduction repression of the beta-interferon gene is mediated through two positive regulatory domains. Proc Natl Acad Sci U S A. 1990 Oct;87(20):7799–7803. doi: 10.1073/pnas.87.20.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittemore L. A., Maniatis T. Postinduction turnoff of beta-interferon gene expression. Mol Cell Biol. 1990 Apr;10(4):1329–1337. doi: 10.1128/mcb.10.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisdom R., Lee W. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 1991 Feb;5(2):232–243. doi: 10.1101/gad.5.2.232. [DOI] [PubMed] [Google Scholar]
- Wodnar-Filipowicz A., Moroni C. Regulation of interleukin 3 mRNA expression in mast cells occurs at the posttranscriptional level and is mediated by calcium ions. Proc Natl Acad Sci U S A. 1990 Jan;87(2):777–781. doi: 10.1073/pnas.87.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]