Abstract
A negative consequence of biodiversity loss is reduced rates of ecosystem functions. Phylogenetic-based biodiversity indices have been claimed to provide more accurate predictions of ecosystem functioning than species diversity alone. This approach assumes that the most relevant traits for ecosystem functioning present a phylogenetic signal. Yet, traits-mediating niche partitioning and resource uptake efficiency in animals can be labile. To assess the relative power of a key trait (body size) and phylogeny to predict zooplankton top-down control on phytoplankton, we manipulated trait and phylogenetic distances independently in microcosms while holding species richness constant. We found that body size provided strong predictions of top-down control. In contrast, phylogeny was a poor predictor of grazing rates. Size-related grazing efficiency asymmetry was mechanistically more important than niche differences in mediating ecosystem function in our experimental settings. Our study demonstrates a strong link between a single functional trait (i.e. body size) in zooplankton and trophic interactions, and urges for a cautionary use of phylogenetic information and taxonomic diversity as substitutes for trait information to predict and understand ecosystem functions.
Keywords: functional ecology, trait diversity, trait divergence, niche partitioning, competitive asymmetry, selection effect
1. Background
The impact of humans on Earth has strongly increased in recent decades, leading to accelerated rates of species extinctions [1]. One important consequence of species losses is reduced rates of ecosystem processes, which can be detrimental for ecosystem functioning and the provisioning of ecosystem services [2–4]. Several studies manipulating the number of species within communities have, indeed, demonstrated a positive relationship between biodiversity and ecosystem functioning [3–5]. However, ecosystem functioning can vary when richness is held constant [5]. This variance likely results from niche and evolutionary differences among species [6]. Biodiversity indices that integrate species ecological similarities and differences should therefore improve predictions of ecosystem functioning [6,7].
Species often not only differ in their resource use, but may also differ in resource uptake efficiency [8]. Because of such functional differences, the local extinction of certain species from an ecosystem can have a greater impact on ecosystem functioning than the extinction of other species [9,10]. Correctly identifying and accounting for relevant species differences in biodiversity measurements is thus a key step towards more predictive biodiversity–ecosystem functioning (BEF) research. Functional diversity indices incorporate information on one or more key functional traits and thus provide a single metric to quantify ecological similarities and differences among species, which can help elucidate the mechanisms underlying BEF [11]. However, it is often difficult to ascertain that the most relevant traits were included or to measure all relevant traits for multiple species. Phylogenetic distance has been suggested as an alternative way to account for overall species similarities and differences [6]. This approach assumes that relevant traits for ecosystem functioning are conserved along the phylogeny, with more closely related species resembling each other more in trait values than distantly related species [12]. A number of recent studies have, indeed, provided evidence that evolutionary diversification could lead to more diversification in traits, and thus enhance ecosystem functioning [7,13]. Nevertheless, traits mediating resource use in animals can be labile, which might obscure the relationship between phylogenetic diversity and the functioning of ecosystems, leading to inaccurate predictions [6,14,15]. Consequently, the relative predictive power of traits and phylogenies on ecosystem functioning will ultimately depend on which traits are measured and which traits are phylogenetically conserved or labile.
There are several ways by which trait and phylogenetic information might vary in their relative importance for explaining variation in ecosystem functioning. First, if all relevant traits for the ecosystem function are conserved along the phylogeny and were properly accounted for in a trait-based analysis, then phylogenetic- and trait-based indices should provide similar and strong explanatory power. Yet, to the extent that traits are labile, directly accounting for trait information might provide more explanatory power than phylogenetic information. If additional traits that are relevant to the ecosystem function are phylogenetically conserved, then phylogenetic information may still add to the explanatory power provided by the measured (labile) traits, such that a combined analysis of trait and phylogenetic information would be ideal [16]. Finally, if none of the traits relevant to ecosystem functioning are phylogenetically conserved, then phylogenetic information will have no explanatory power for the studied ecosystem function [12]. An experimental system that lacks conservatism in a key trait relevant to ecosystem function presents an ideal setting to test whether phylogenetic distances can represent variation in additional ecological information relevant for ecosystem functioning. In such cases, a priori selected traits and unmeasured, evolutionarily conserved traits could complementarily explain ecosystem functioning [15–17].
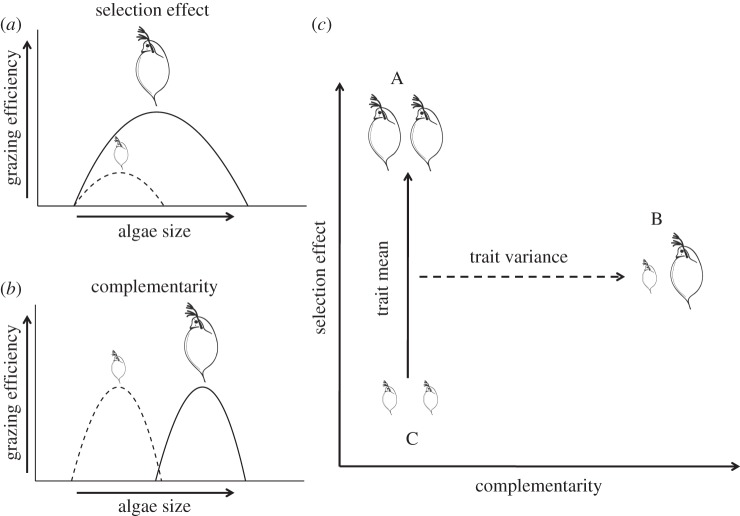
Most studies assessing the role of phylogenetic diversity on ecosystem functioning have focused on a single trophic level, primary producers [7,16], thus hindering generalizations. Given the relevance of trophic interactions for ecosystem functioning [18], it is an important next step to test the capacity for phylogenetic distances to predict ecosystem functions at higher trophic levels. In freshwater ecosystems, herbivorous zooplankton grazing plays an important functional role by controlling algal blooms and transferring energy and matter upwards through the food chain [18]. Body size of zooplankton has been repeatedly suggested to play a key role in top-down control of algae [19,20], and this trait was found to be evolutionarily labile in cladoceran zooplankton (see electronic supplementary material for details). The importance of body size for top-down control is generally attributed to grazing efficiency asymmetry (selection effect), where larger species are superior grazers (figure 1a) [19,21] and therefore increasing community average size (CAS) might enhance grazing rates (GRs; figure 1c) [22]. Alternatively, it has been suggested that feeding niche partitioning (complementarity) between large and small zooplankton species may (co)determine top-down control on phytoplankton (figure 1b). This latter hypothesis is based on the assumption that larger zooplankton species would be more efficient grazers on larger phytoplankton, whereas small grazers would deplete small phytoplankton more efficiently (figure 1b) [22–24]. In such a case of size-related diet partitioning (complementarity), it is expected that body size diversity, not CAS, would better explain variation in GRs among systems (figure 1c).
Figure 1.
Schematic of alternative mechanisms of how community trait mean and variance can impact ecosystem functioning through selection effects and complementarity. (a) Selection effects (grazing efficiency asymmetry in our system) happen when dominant trait values in a community enhance ecosystem functioning. For instance, in freshwater systems, zooplankton species may strongly overlap in resource use, but larger zooplankton species are often superior grazers [19]. (b) Size-mediated complementarity happens when large and small zooplankton species explore alternative resources (e.g. large versus small phytoplankton [22,24]). (c) Selection effects and complementarity result in differences in the expected relationship between community trait mean and variance and ecosystem functioning. Three hypothetical communities are represented as A, B and C. Increasing average body size is expected to affect ecosystem functioning when grazing efficiency asymmetry (selection effect) is the dominant mechanism that explains differences in ecosystem functioning. This may happen independently of size diversity (i.e. from community C to A, marked by solid line). Alternatively, increasing size diversity (trait variance) within communities may enhance ecosystem functioning when complementarity is important (dashed line) [22].
While the relationship between zooplankton body size and GRs has been previously documented [19,22], the role of zooplankton phylogenetic diversity in this context has been overlooked, mainly because most previous studies focused on species of the genus Daphnia. We here broadened the taxonomic scope to test whether phylogenetic diversity provides a more comprehensive and accurate representation of species similarities and differences over multiple niche axes than body size alone, complements the information obtained from body size, or does not add any relevant information to predict grazing pressure. A strong signal of zooplankton phylogeny on top-down control would indicate evolutionary conservatism in additional, less explored functional traits that impact grazing efficiency in this system [25].
In order to assess the relative power of functional (body size) and phylogenetic distances to predict top-down control of cladoceran zooplankton on unicellular algae, we designed a microcosm grazing experiment in which zooplankton species pairs varied independently in body size and evolutionary relationships. We combined species based on their trait and phylogenetic distances in order to reflect body size convergence (i.e. distantly related species overlapping in body size), body size conservatism (i.e. proportional trait and phylogenetic distances) or body size divergence (figure 2). Specifically, we tested three key hypotheses: (i) phylogenetic distances better approximate species similarities and differences and thus provide more accurate predictions of top-down control than accounting solely for body size. (ii) Body size and phylogeny provide complementary information on top-down control, thus higher predictive power is achieved when both body size and phylogenetic information is taken into account. (iii) Zooplankton body size is the most relevant trait and better predicts top-down control than phylogeny, and therefore, phylogenetic information does not contribute to predict grazing pressure. For each species pair, we measured GRs under standardized conditions. By comparing these community GRs with those of all species in monoculture, we could assess to what extent the observed patterns were explained by grazing efficiency asymmetry (selection effect) or by niche differences (complementarity) among species (figure 1a,b). If selection effects (figure 1a) represent the dominant mechanism driving top-down control, then we expect that CAS will better predict top-down control than size diversity (figure 1c). Conversely, if size-related complementarity is the prevalent mechanism (figure 1b), then we expect that differences in size diversity will better predict top-down control (figure 1c).
Figure 2.
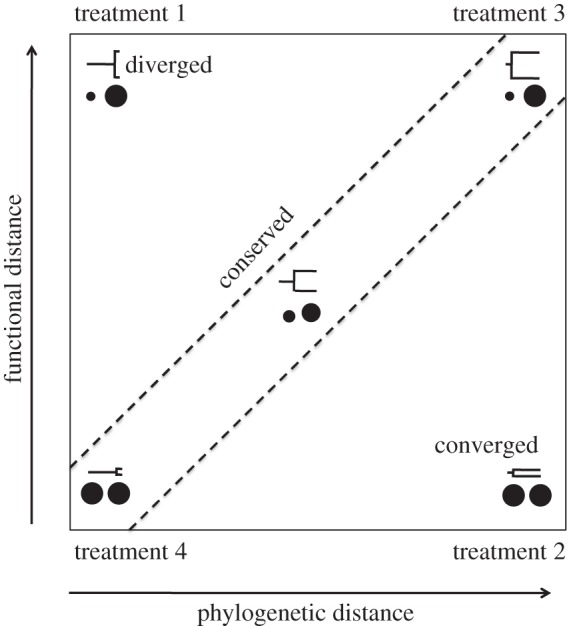
Scheme depicts the different treatments creating combinations of species pairs that differ in body size and/or phylogenetic distances. Treatment (i) represents closely related species that have diverged in size; treatment (ii) refers to distantly related species that have converged in size; treatment (iii) represents a situation of trait conservatism along the phylogeny, in which distantly related species are dissimilar in size; treatment (iv) refers to closely related species that are similar in body size. Adapted from Cadotte et al. [15].
2. Material and methods
(a). Experimental organisms and trait/phylogenetic diversity treatments
We selected species pairs from a pool of eight freshwater cladoceran species from two speciose families (the Daphniidae and the Chydoridae) that vary widely in body size and phylogenetic distances (see electronic supplementary material). Cladocerans are, in general, quite similar in their main food sources, but there are clear differences in feeding habits of Daphniidae and the macrophyte-associated Chydoride [26]. The eight species that we used in our experiments are widespread and common in Belgian ponds and shallow lakes [27] (see also electronic supplementary material). In order to test the influence of trait and phylogenetic diversity on top-down control in zooplankton communities independently of species richness, we held species richness constant at two per microcosm.
We measured the GRs of different zooplankton pairs that represented four crossed levels of body size diversity (functional trait diversity, FD) and phylogenetic diversity (PD; figure 2). The four treatments were: (i) HIGH FD–LOW PD; (ii) LOW FD–HIGH PD; (iii) HIGH FD–HIGH PD; and (iv) LOW FD–LOW PD. For each of these four treatments, there were two pairs of species, and for each species combination, there were three replicates (for details, see electronic supplementary material, figure S2 and table S2). This orthogonal experimental design allowed us to test whether variation in GRs occurred mainly along the trait axis, mainly along the phylogenetic axis, or rather depended on the combined information of both axes (figure 2). It is important to note that different communities may have exactly the same trait diversity pattern (i.e. homogeneous size distribution; LOW FD) but differ considerably in average size (e.g. represented by communities A and C in figure 1c). For the two treatments involving LOW FD (i.e. treatments (ii) and (iv), with low variation in body size), we contrasted two large versus two small species (see electronic supplementary material for details, table S2). This design allows us to separate the effects of CAS (i.e. large versus small; communities A and C in figure 1c) from the effects of size diversity (represented by community B in figure 1c).
All species were collected from communities sampled in farmland ponds and shallow lakes in Belgium, and were isolated and cultured as single clones. The experimental populations of all species were cultured for multiple generations under standardized conditions (in 500-ml glass jars, dechlorinated tap water, 20°C, 14 h light/10 h dark photoperiod) prior to the grazing experiments. All studied cladoceran species are cyclical parthenogens. They produce a number of clonal offspring at intervals of approximately 2–4 days. For the grazing experiments, we used cohorts with similar-sized mature individuals to rule out trait variability within species. Body size values for adult individuals of each species were measured after each grazing experiment (see electronic supplementary material, table S3). Average body sizes of the different species used ranged from 0.52 to 3.60 mm. The species were fed with a mixture (1 : 1 ratio) of two different algae species that vary considerably in cell size, Chlorella vulgaris (approx. 3 µm) and Scenedesmus obliquus (approx. 15 µm). Both algae species were cultured in isolation with the addition of trace elements.
(b). Experimental set-up
To measure feeding rates by zooplankton assemblages on algae, which we use as a proxy for the capacity of top-down control on algae, we calculated the gross change in phytoplankton biomass in experimental units over a 24 h period. Grazing experiments were carried out in clear 250 ml glass Duran bottles. The number of individuals for each species per experimental bottle was standardized based on biomass (dry weight over 24 h/70°C; see electronic supplementary material, table S3). To begin each grazing experiment, we transferred the required number of individuals (see electronic supplementary material, table S3) to experimental bottles then added an algal solution with two species, the small algae C. vulgaris and the large S. obliquus in equal proportions (1 : 1) with an initial concentration of approximately 150 000 cells per ml. We choose those algae species because they are widespread and common in freshwater systems, easy to culture under standardized laboratory conditions and because they vary considerably in cell size. Bottles were placed in a roller machine (four turns per minute along the longitudinal axis of the experimental jars) to avoid algae settlement during the grazing experiment. Control bottles (n = 3 per experiment) were identical to experimental bottles but had no zooplankton. We also performed grazing experiments for all species in monocultures replicated three times.
At the start and at the end of each grazing experiment, we took a 2 ml water sample from the centre of experimental and control bottles and fixated with 110 µl formalin to count algae cell numbers afterwards. Cells were counted with the Attune® Acoustic Focusing Cytometer (Applied Biosystems™ by Life Technologies™, Carlsbad, CA), which allowed us to measure the total number of algae cells per millilitre and to differentiate between small and large algae cells. The latter information was crucial to test whether large zooplankton species are more efficient grazers on large phytoplankton, whereas smaller zooplankton species are more efficient grazers on small algae cells, thus leading to size-related complementarity via diet niche partitioning [22,24].
After each grazing experiment, we removed the zooplankton from the bottles and measured total body length (excluding tail spines) of 10 individuals from each replicate using a dissecting microscope. For larger species with less than 10 individuals per replicate (see electronic supplementary material, table S3), all the individuals in a replicate were measured. To compare GRs among zooplankton assemblages, we calculated phytoplankton consumption over the period of 24 h.
(c). Statistical analysis
We calculated GRs as the total gross change in phytoplankton biomass using the following equation
where ‘Δphytoplankton control’ is the logarithmic difference between the phytoplankton concentration over 24 h in the control jars (i.e. control phytoplanktont0 − control phytoplanktont1) and ‘Δphytoplankton treatment’ is the logarithmic difference between the phytoplankton concentration over 24 h in the jars with zooplankton (i.e. treatment phytoplanktont0 − treatment phytoplanktont1). Using the same equation, we also calculated: (i) GRsmall as the gross change in small phytoplankton (C. vulgaris) over 24 h and (ii) GRlarge as the gross change in large phytoplankton (S. obliquus) cells over 24 h.
We first calculated phylogenetic and functional trait (body size) distances among all pairs of species used in the experiment. Phylogenetic distances among species were estimated from a reconstructed molecular–phylogenetic tree (based on maximum-likelihood) for cladocerans occurring in Belgium (for details on the phylogenetic tree reconstruction, see electronic supplementary material). Based on this phylogenetic tree, we calculated a phylogenetic (cophenetic) distance (PDist) matrix between all species pairs based on branch lengths [28]. Functional trait distances among species, the Euclidean distance between the average measured body size values, were used to construct a functional distance (FDist) matrix.
For each distance matrix, we calculated the mean pairwise distance among species in a given assemblage using the package Picante in R [29] (note that the mean pairwise distance for communities with only two species is equal to FDist and PDist). This results in a measure of trait (FD) or phylogenetic (PD) diversity within experimental bottles. The diversity measures FD and PD may reflect trait or phylogenetic complementarity if species that vary in size or evolutionary relationships exploit alternative resources (figure 1b,c) [7,22,30]. In addition to the variance component of size (i.e. size diversity, FD), we also calculated CAS (figure 1c). CAS may reflect grazing efficiency asymmetry (i.e. a selection effect) if differences in grazing efficiency between large versus small species (figure 1a) among communities results in different top-down control (represented by solid line in figure 1c).
As explained above, the information captured by FD, PD and CAS reflects different mechanisms of ecosystem function, i.e. complementarity (FD and PD) versus grazing efficiency asymmetry (CAS; figure 1). Yet, they can also overlap in their role in explaining ecosystem functioning if they are correlated. For example, starting from a community composed of only small species (community C in figure 1c), both trait diversity and CAS can increase in the same direction (captured in the shift to community B in figure 1c). In order to quantify the unique and shared effects of CAS, community size diversity and community phylogenetic diversity on GRs, we applied variation partitioning based on partial regressions [31–33] using PD, FD and CAS as predictors, and GR as a response variable. This statistical method separates the variation in a response variable(s) into independent contributions of predictors, shared contributions among all combinations of predictors (i.e. variation that is associated with multiple predictors, which results when predictors covary with one another), and unexplained causes of variation [31,34]. The proportion of explained variation for each component is given by the adjusted R2 (adj.R2, an R2 statistic adjusted to avoid type I error and consequent overestimation of the amount of explained variation [35]). Because adj.R2 is an estimate based on the number of variables included, negative values may occur but are interpretable as not significantly different from zero. The proportion of variation explained by each component is additive and sums to one. We considered each replicate separately and used trait–phylogenetic distances to control for non-independence among replicates (i.e. each replicate had the same value for CAS; FD and PD).
Zooplankton species show feeding niche complementarity if small zooplankton species are more efficient grazers on small algae cells and large zooplankton species are more efficient grazers on large algae cells (figure 1b [22,24]). To test whether this was observed in our experiment, we classified each zooplankton species as large (more than 1.5 mm; n = 4) or small (less than or equal to 1.5 mm; n = 4) then tested the effect of body size class on GRs (as observed in monocultures). To do this, we ran three separate analysis of variance (ANOVA), using body size as a factor and three response variables, one at a time: (i) total GRs (i.e. the entire spectrum of phytoplankton cell sizes), (ii) GRs on small algae cells, and (iii) GRs on large algae cells.
Species interactions, such as facilitation and competition, may have non-additive consequences on ecosystem functions. To test the hypothesis that the performances of two-species communities equalled the average performance of the contributing species in monoculture we first calculated the expected GRs (GREXP) for each species pair by averaging the GR values of each species in monoculture. We subtracted this from the observed GRs in combination (Spint = GROBS − GREXP), where Spint refers to species interactions and GROBS refers to observed GRs for a given species pair. Thus, a value of zero indicates that observed GRs are equal to what would be expected for a given species combination without any interaction, whereas positive values indicate synergistic interactions, where the two-species communities perform better than the average of the two monocultures, e.g. because of niche complementarity. Negative values indicate antagonistic species interactions, e.g. owing to interference competition. Values were transformed to percentages to allow comparisons among species pairs. We used two-way permutation univariate ANOVA [36] (package lmPerm in R) to test the effect of FD and PD on species interactions Spint. All analyses were run in R [36].
3. Results
Variation partitioning revealed that the explanatory variables PD, FD and CAS and their intersections accounted for approximately 64% of observed variation in GRs (table 1). CAS accounted for approximately 93% of the total amount of explained variation, either as a pure effect (41% of explained variation) or shared with FD (52% of explained variation). Specifically, communities composed of larger species depleted algae more efficiently than communities composed of smaller species (see also electronic supplementary material, figure S4). The pure effects of PD and FD were not significant, whereas the pure effect of CAS was highly significant (table 1). Additionally, FD and CAS shared a large fraction of overlapping information (table 1). Phylogenetic diversity was uninformative either as a pure effect or shared with other variables (table 1).
Table 1.
Proportion of explained variation obtained through variation partitioning analysis based on partial regressions on top-down control given by each predictor and their shared effects. Predictors are as follows: PD, phylogenetic diversity; FD, functional trait (body size) diversity and CAS, community average body size. Three asterisks represent p < 0.001. Note that it is not possible to calculate p-values for intersections, i.e. shared effects.
| components | adj.R2 |
|---|---|
| pure effect of PD | 0.000 |
| pure effect of FD (body size) | 0.043 |
| pure effect of CAS | 0.262*** |
| shared effects between PD and FD | 0.003 |
| shared effects between FD and CAS | 0.330 |
| shared effects between PD and CAS | 0.000 |
| shared effects between PD, FD and CAS | 0.000 |
| total explained variance | 0.638*** |
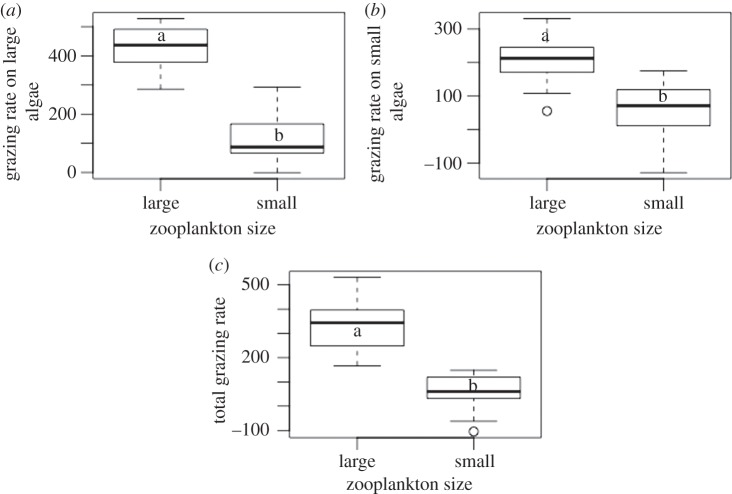
The ANOVA based on monocultures revealed that larger zooplankton species depleted both large and small phytoplankton more efficiently than small zooplankton (figure 3a,b; large algae: F = 103, p < 0.001; small algae: F = 19.42, p < 0.001). The larger species were therefore superior grazers over the entire spectrum of algae sizes (F = 49.73, p < 0.001; figure 3c; see electronic supplementary material).
Figure 3.
Box plots depict algae consumption by small and large zooplankton species on: (a) large algae cells; (b) small algae cells; (c) the entire spectrum of algae cell sizes. Distinct letters (a versus b) indicate significant differences (p < 0.001) based on one-way ANOVA.
Tests of species interactions on observed GRs indicate that species combinations with high functional trait diversity performed better than expected from the measurements on individual species (table 2, see also electronic supplementary material, figure S5). In contrast, low functional trait diversity resulted on average in more negative species interactions (table 2, see also electronic supplementary material, figure S5). There was no clear effect of phylogenetic diversity on species interactions (table 2 and electronic supplementary material, figure S5), but we found a significant interaction between functional and phylogenetic diversity on species interactions.
Table 2.
Two-way permutation ANOVA results, testing for the effect of phylogenetic diversity (PD) and functional diversity (FD) on species interactions (quantified as the deviation from the average performance of the species in monoculture) for grazing rates. d.f. refers to degrees of freedom; SS means sum of squares.
| factors | d.f. | SS | p-values |
|---|---|---|---|
| PD | 1 | 140.3 | 0.673 |
| FD | 1 | 4810.5 | <0.001*** |
| PD : FD | 1 | 2633.7 | 0.028* |
4. Discussion
Biodiversity metrics based on phylogenetic distances are increasingly being used as a surrogate for trait differentiation in ecosystem functioning research. While there is some support for this approach [16,17], most of these examples are based on reanalysis of studies that were originally designed to test the effect of species richness on plant biomass production. As a consequence, previous studies had limited power to unequivocally assign the role of phylogeny independently of species and trait diversity, because all three biodiversity dimensions were somewhat correlated [16,37,38] (but see Cadotte [7]). By varying trait and phylogenetic distances independently and holding species richness constant in our experimental study, we could disentangle the role of phylogeny and a key trait (body size) in determining zooplankton top-down control on phytoplankton, thus achieving strong inference power to detect the role of phylogeny if a key trait is accounted for. We clearly demonstrated that body size diversity, but especially CAS, were much better predictors of cladoceran GRs than phylogenetic diversity. We thus found no support for our first and second hypothesis that phylogenetic diversity could provide more accurate or complementary predictions of top-down control, respectively, than accounting solely for body size information in freshwater herbivorous zooplankton. We also demonstrated that large zooplankton species were superior grazers of both small and large phytoplankton cell sizes, indicating that the difference in grazing efficiency (selection effect) between small and large zooplankton species (figure 1a) was more important than niche differences (complementarity; figure 1b) in mediating trophic interactions in our experimental setting.
Studies detecting a relationship between species richness and ecosystem functions often observed a high variance in ecosystem functioning for the same number of species and could not always mechanistically explain this relationship [3,5]. These variable results could derive from trait and evolutionary differences among species, which are ignored in taxonomic indices of diversity [6,30]. Our study confirmed that top-down control of algae can differ substantially among species pairs and this can be explained by interspecific trait differences. By systematically contrasting functional trait and phylogenetic distances, we demonstrate that variation in a single trait, body size, predicted GRs better than measures of phylogenetic distance in this system. Thus, we found strong support for our third hypothesis that body size is a key trait determining cladoceran top-down control on unicellular algae and that phylogenetic distances do not add important information to predict grazing pressure in this system. A study of herbivorous marine amphipods where traits were not conserved along the phylogeny also found that directly accounting for traits provided better estimates of ecosystem function (resistance to invasion) than phylogeny [14]. However, our findings contrast with the results of Thompson et al. [25], who concluded that zooplankton phylogenetic diversity was a better predictor of top-down control than diversity in body size and other functional traits in freshwater shallow lakes. One potential explanation for this discrepancy could be due to the fact that the study of Thompson et al. [25] included copepods, whereas our study focused on variation within the cladocerans. Copepods are known to differ substantially in food niche from cladocerans [26,39]. In addition, it is often difficult to differentiate causes from consequences using observational data. Thus, two interpretations of the results of Thompson et al. [25] are actually possible: phylogenetically diverse communities, indeed, reduced chlorophyll a more strongly in natural systems, or phylogenetically diverse communities are more likely to occur in ponds with lower chlorophyll a.
Although some studies have demonstrated that phylogenetic distances can predict ecosystem functioning in experimental settings [7,13], it is important to note that phylogenetic distinctiveness is not an ecological mechanism per se. The use of phylogenetic distances as a predictor of ecosystem functioning relies on the assumption that relevant traits are conserved along the phylogeny [12]. Ultimately, trait differentiation is the main currency linking biodiversity to ecosystem functioning [11]. Studies demonstrating that phylogeny is more important than the measured traits [17,40] either reflect a failure to correctly identifying which traits translate into enhanced ecosystem functioning or reflect that relationships are complex and thus cannot be captured by measuring just a few traits. Flynn and collaborators [16] found that trait and phylogenetic distances complementarily explain ecosystem functioning, suggesting that measured labile traits and unmeasured phylogenetically conserved traits were both important in determining biomass production in grassland plants. In our system of herbivorous zooplankton, phylogenetic diversity became an important predictor of ecosystem functioning, along with diversity in body size, only after removing all cases of body size convergences and divergences and focusing exclusively on species combinations presenting trait conservatism (i.e. treatments (iii) and (iv) only; see electronic supplementary material). This illustrates that the potential for phylogenetic distances to predict top-down control depends entirely on body size conservatism and does not add information on relevant unmeasured traits. On the other hand, our results also show that under such circumstances phylogenetic diversity is indeed superior to taxonomic diversity in explaining ecosystem functions.
Niche differentiation (complementarity) in resource use is often invoked to explain the relationship between biodiversity and ecosystem functioning. Another plausible mechanism, however, is selection effects, where more diverse communities are more likely to include at least one dominant species [8,41]. A recent observational study found that zooplankton body size diversity enhanced top-down control and suggested that this occurred because of niche partitioning in resource use [22]. Ye et al. [23] suggest that such a relationship emerged, because smaller zooplankton potentially consumed smaller algae more efficiently while large zooplankton predominantly grazed upon large phytoplankton. Our experiments demonstrate that the same relationship between size diversity and GRs can emerge via grazing efficiency asymmetry (figure 1a) in the absence of niche differences. This occurs because in order to have a higher value for the index of size diversity, at least one large zooplankton species must be present, and these tend to be superior grazers. In this study, we used variation partitioning and demonstrated that accounting for community average body size rather than size diversity within assemblages (FD), enabled us to better predict differences in top-down control. Specifically, communities composed of larger species performed better than communities of smaller species. The majority of studies relating trait and phylogenetic diversity to ecosystem functioning have only used variance metrics, such as FD and PD, as predictors and often suggested niche partitioning as the main mechanism determining ecosystem functioning [7,25,42]. As we have shown, combining variance and mean trait metrics enables better insight into the relative importance of complementarity and selection effect as determinants of ecosystem functioning [30].
Our results also demonstrate that species interactions often have non-additive consequences on ecosystem functioning. Overall, we observed that functionally dissimilar species presented positive interactions (facilitation), while species overlapping in body size tend to present antagonistic interactions, probably owing to interference competition. In Daphnia, reduced GRs among species has been shown to result from chemical and mechanical inhibitory mechanisms [43]. Although we do not have data to further test this, our results support the idea that whatever inhibitory mechanisms of competition are at play, they are stronger among species that overlap in body size. Our study thus suggests that size-related grazing efficiency and size-related complementarity co-determine zooplankton top-down control on algae, but with a more important role for the former mechanism.
We used microcosm experiments to investigate the effect of functional and phylogenetic distances on top-down control. Several recent studies provided evidence that results from microscale BEF experiments are often representative of large-scale natural systems [44,45]. The mechanisms on which our interpretation relies, which are based on trait-based selection effects and non-additive species interactions, can be easily extrapolated to other systems. Our findings may apply directly to herbivorous marine zooplankton [14] and to herbivorous terrestrial insects [30], in which the impact of key traits on the biomass of lower trophic levels has been recently demonstrated. Additionally, our results may also be representative of other ecosystem functions, such as pollination, in which competitive asymmetry plays a larger role than complementarity [10]. The experimental approach we used, separating the role of a key functional trait and phylogenetic distances, provides exciting and promising avenues to understand how evolutionary convergences and divergences affect trophic interactions and shape ecosystem functioning. A potential limitation of our study was that it was not possible to design it so as to completely separate the role of CAS from that of phylogenetic composition. However, we performed ad hoc analysis and found that phylogenetic composition per se is not the driving force of top-down control in our experimental setting (for a full analysis of the effect of phylogenetic composition, see electronic supplementary material, figure S6).
In order to effectively inform management in an era of accelerated extinction rates, it has been suggested that BEF research should shift from approaches based on the number of species to more accurate trait and/or evolutionary-based approaches [6,11,30], which could provide more mechanistic understanding of ecosystem functioning. Our study partially supports this idea, as we have shown that body size diversity, and especially CAS, provided strong predictions of top-down control, despite constant species richness. This happens because large species are superior grazers across a range of algae cell sizes (figure 1a). Yet, we refuted our first two hypotheses, which are based on the idea that additional, evolutionarily conserved traits could provide added power to explain top-down control on phytoplankton. Our results illustrate the importance of pinpointing specific relevant traits to mechanistically understand and predict ecosystem functioning. Zooplankton top-down control on phytoplankton represents an important ecosystem service worldwide by controlling algal blooms, including toxic algae. Recent studies have demonstrated that increasing anthropogenic impacts, such as climate warming and chemical pollution will affect the size structure of zooplankton communities, potentially leading to a predominance of smaller zooplankton [46]. Based on our results, we suggest that to maintain zooplankton top-down control on algae, ecological conditions that favour combinations of larger, superior grazer species should be prioritized. One mechanism to do so is to reduce fish predation pressure (cf. biomanipulation as a management strategy [46]). In summary, our study provides evidence for strong trait-based top-down control and warns against a blind use of phylogenetic and taxonomic diversity as surrogates for trait differentiation to predict and explain ecosystem functions.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Berthe Vastenhoud and Luz Amadei for their help with data collection during the experiment. Patrick Venail, Pieter Lemmens and an anonymous referee provided valuable comments on an earlier version of the manuscript.
Data accessibility
Data are available in the electronic supplementary material.
Authors' contributions
A.G. and L.D.M. developed the ideas and the concept of this study. A.G. designed the experiment with input of L.D.M. A.G. conducted the experiment and performed the statistical analyses. J.H.P. and L.D.M. contributed to discussions on data-analysis and interpretation. A.G. wrote the first version of the manuscript and all authors contributed substantially to the final version.
Competing interests
We have no competing interests.
Funding
A.G. received a doctoral scholarship from Brazil through ‘Science Without Borders - CNPq’, grant number 245629/2012-2, and J.H.P. a postdoctoral fellowship (F+) from the KU Leuven Research Fund. Financial support was obtained from KU Leuven Research Fund Excellence Center financing (PF/2010/07), ERA-NET BIOdivERsA project TIPPINGPOND (nationally funded by Belspo) and Belspo IAP SPEEDY.
References
- 1.Pimm SL, Jenkins CN, Abell R, Brooks TM, Gittleman JL, Joppa LN, Raven PH, Roberts CM, Sexton JO. 2014. The biodiversity of species and their rates of extinction, distribution, and protection. Science 344, 1246752 ( 10.1126/science.1246752) [DOI] [PubMed] [Google Scholar]
- 2.Naeem S, Duffy JE, Zavaleta E. 2012. The functions of biological diversity in an age of extinction. Science 336, 1401–1406. ( 10.1126/science.1215855) [DOI] [PubMed] [Google Scholar]
- 3.Cardinale BJ, et al. 2012. Biodiversity loss and its impact on humanity. Nature 486, 59–67. ( 10.1038/nature11148) [DOI] [PubMed] [Google Scholar]
- 4.Lefcheck JS, et al. 2015. Biodiversity enhances ecosystem multifunctionality across trophic levels and habitats. Nat. Commun. 6, 6936 ( 10.1038/ncomms7936) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardinale BJ, Srivastava DS, Emmett Duffy J, Wright JP, Downing AL, Sankaran M, Jouseau C. 2006. Effects of biodiversity on the functioning of trophic groups and ecosystems. Nature 443, 989–992. ( 10.1038/nature05202) [DOI] [PubMed] [Google Scholar]
- 6.Srivastava DS, Cadotte MW, MacDonald AAM, Marushia RG, Mirotchnick N, Mooers A. 2012. Phylogenetic diversity and the functioning of ecosystems. Ecol. Lett. 15, 637–648. ( 10.1111/j.1461-0248.2012.01795.x) [DOI] [PubMed] [Google Scholar]
- 7.Cadotte MW. 2013. Experimental evidence that evolutionarily diverse assemblages result in higher productivity. Proc. Natl Acad. Sci. USA 110, 8996–9000. ( 10.1073/pnas.1301685110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll IT, Cardinale BJ, Nisbet RM. 2011. Niche and fitness differences relate the maintenance of diversity to ecosystem function. Ecology 92, 1157–1165. ( 10.1890/10-0302.1) [DOI] [PubMed] [Google Scholar]
- 9.Larsen TH, Williams NM, Kremen C. 2005. Extinction order and altered community structure rapidly disrupt ecosystem functioning. Ecol. Lett. 8, 538–547. ( 10.1111/j.1461-0248.2005.00749.x) [DOI] [PubMed] [Google Scholar]
- 10.Winfree R, Fox J, Williams NM, Reilly JR, Cariveau DP, Shea K. 2015. Abundance of common species, not species richness, drives delivery of a real-world ecosystem service. Ecol. Lett. ( 10.1111/ele.12424) [DOI] [PubMed] [Google Scholar]
- 11.Hillebrand H, Matthiessen B. 2009. Biodiversity in a complex world: consolidation and progress in functional biodiversity research. Ecol. Lett. 12, 1405–1419. ( 10.1111/j.1461-0248.2009.01388.x) [DOI] [PubMed] [Google Scholar]
- 12.Wiens JJ, et al. 2010. Niche conservatism as an emerging principle in ecology and conservation biology. Ecol. Lett. 13, 1310–1324. ( 10.1111/j.1461-0248.2010.01515.x) [DOI] [PubMed] [Google Scholar]
- 13.Gravel D, Bell T, Barbera C, Combe M, Pommier T, Mouquet N. 2012. Phylogenetic constraints on ecosystem functioning. Nat. Commun. 3, 1117 ( 10.1038/ncomms2123) [DOI] [PubMed] [Google Scholar]
- 14.Best RJ, Caulk NC, Stachowicz JJ, Emmett Duffy J. 2013. Trait vs. phylogenetic diversity as predictors of competition and community composition in herbivorous marine amphipods. Ecol. Lett. 16, 72–80. ( 10.1111/ele.12016) [DOI] [PubMed] [Google Scholar]
- 15.Cadotte M, Albert CH, Walker SC. 2013. The ecology of differences: assessing community assembly with trait and evolutionary distances. Ecol. Lett. 16, 1234–1244. ( 10.1111/ele.12161) [DOI] [PubMed] [Google Scholar]
- 16.Flynn DFB, Mirotchnick N, Jain M, Palmer MI, Naeem S. 2011. Functional and phylogenetic diversity as predictors of biodiversity–ecosystem-function relationships. Ecology 92, 1573–1581. ( 10.1890/10-1245.1) [DOI] [PubMed] [Google Scholar]
- 17.Cadotte MW, Cardinale BJ, Oakley TH. 2008. Evolutionary history and the effect of biodiversity on plant productivity. Proc. Natl Acad. Sci. USA 105, 17 012–17 017. ( 10.1073/pnas.0805962105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Persson J, Brett MT, Vrede T, Ravet JL. 2007. Food quantity and quality regulation of trophic transfer between primary producers and a keystone grazer (Daphnia) in pelagic freshwater food webs. Oikos 116, 1152–1163. ( 10.1111/j.0030-1299.2007.15639.x) [DOI] [Google Scholar]
- 19.Mourelatos S, Lacroix G. 1990. In situ filtering rates of Cladocera: effect of body length, temperature, and food concentration. Limnol. Oceanogr. 35, 1101–1111. ( 10.4319/lo.1990.35.5.1101) [DOI] [Google Scholar]
- 20.Lampert W, Sommer U. 2007. Limnoecology, 2nd edn New York, NY: Oxford University Press. [Google Scholar]
- 21.Brooks JL, Dodson SI. 1965. Predation, body size, and composition of plankton. Science 150, 28–35. ( 10.1126/science.150.3692.28) [DOI] [PubMed] [Google Scholar]
- 22.Ye L, Chang C-Y, García-Comas C, Gong G-C, Hsieh C-H, Beckerman A. 2013. Increasing zooplankton size diversity enhances the strength of top-down control on phytoplankton through diet niche partitioning. J. Anim. Ecol. 82, 1052–1061. ( 10.1111/1365-2656.12067) [DOI] [PubMed] [Google Scholar]
- 23.Hansen B, BjØRnsen PK, Hansen PJ. 1994. The size ratio between planktonic predators and their prey. Limnol. Oceanogr. 39, 395–403. ( 10.4319/lo.1994.39.2.0395) [DOI] [Google Scholar]
- 24.Cyr H, Curtis JM. 1999. Zooplankton community size structure and taxonomic composition affects size-selective grazing in natural communities. Oecologia 118, 306–315. ( 10.1007/s004420050731) [DOI] [PubMed] [Google Scholar]
- 25.Thompson PL, Davies TJ, Gonzalez A. 2015. Ecosystem functions across trophic levels are linked to functional and phylogenetic diversity. PLoS ONE 10, e0117595 ( 10.1371/journal.pone.0117595) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnett AJ, Finlay K, Beisner BE. 2007. Functional diversity of crustacean zooplankton communities: towards a trait-based classification. Freshw. Biol. 52, 796–813. ( 10.1111/j.1365-2427.2007.01733.x) [DOI] [Google Scholar]
- 27.Louette G, De Bie T, Vandekerkhove J, Declerck S, De Meester L. 2007. Analysis of the inland cladocerans of Flanders (Belgium). Inferring changes over the past 70 years. Belgian J. Zool. 137, 117–123. [Google Scholar]
- 28.Swenson NG. 2014. Functional phylogenetic ecology in R, p. 212 New York, NY: Springer. [Google Scholar]
- 29.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. ( 10.1093/bioinformatics/btq166) [DOI] [PubMed] [Google Scholar]
- 30.Deraison H, Badenhausser I, Loeuille N, Scherber C, Gross N, Haddad N. 2015. Functional trait diversity across trophic levels determines herbivore impact on plant community biomass. Ecol. Lett. 18, 1346–1355. ( 10.1111/ele.12529) [DOI] [PubMed] [Google Scholar]
- 31.Borcard D, Legendre P, Drapeau P. 1992. Partialling out the spatial component of ecological variation. Ecology 73, 1045 ( 10.2307/1940179) [DOI] [Google Scholar]
- 32.Legendre P, Legendre L. 2012. Numerical ecology. Dev. Environ. Model. 24, 990 ( 10.1016/b978-0-444-53868-0.50016-2) [DOI] [Google Scholar]
- 33.Whittaker J. 1984. Model interpretation from the additive elements of the likelihood function. Appl. Stat. 33, 52 ( 10.2307/2347663) [DOI] [Google Scholar]
- 34.Borcard D, Gillet F, Legendre P. 2011. Numerical ecology with R. New York, NY: Springer. [Google Scholar]
- 35.Peres-Neto PR, Legendre P, Dray S, Borcard D. 2006. Variation partitioning of species data matrices: estimation and comparison of fractions. Ecology 87, 2614–2625. ( 10.1890/0012-9658(2006)872614:Vposdm%5D2.0.Co;2) [DOI] [PubMed] [Google Scholar]
- 36.Anderson MJ. 2001. Permutation tests for univariate or multivariate analysis of variance and regression. Can. J. Fish Aquat. Sci. 58, 626–639. ( 10.1139/f01-004) [DOI] [Google Scholar]
- 37.Venail P, et al. 2015. Species richness, but not phylogenetic diversity, influences community biomass production and temporal stability in a re-examination of 16 grassland biodiversity studies. Funct. Ecol. 29, 615–626. ( 10.1111/1365-2435.12432) [DOI] [Google Scholar]
- 38.Cadotte MW. 2015. Phylogenetic diversity and productivity: gauging interpretations from experiments that do not manipulate phylogenetic diversity. Funct. Ecol. ( 10.1111/1365-2435.12543) [DOI] [Google Scholar]
- 39.DeMott WR. 1986. The role of taste in food selection by freshwater zooplankton. Oecologia 69, 334–340. ( 10.1007/bf00377053) [DOI] [PubMed] [Google Scholar]
- 40.Cadotte MW, Cavender-Bares J, Tilman D, Oakley TH. 2009. Using phylogenetic, functional and trait diversity to understand patterns of plant community productivity. PLoS ONE 4, e5695 ( 10.1371/journal.pone.0005695) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hector A, Bazeley-White E, Loreau M, Otway S, Schmid B. 2002. Overyielding in grassland communities: testing the sampling effect hypothesis with replicated biodiversity experiments. Ecol. Lett. 5, 502–511. ( 10.1046/j.1461-0248.2002.00337.x) [DOI] [Google Scholar]
- 42.Venail PA, Vives MJ. 2013. Phylogenetic distance and species richness interactively affect the productivity of bacterial communities. Ecology 94, 2529–2536. ( 10.1890/12-2002.1) [DOI] [PubMed] [Google Scholar]
- 43.Hargrave CW, Hambright KD, Weider LJ. 2011. Variation in resource consumption across a gradient of increasing intra- and interspecific richness. Ecology 92, 1226–1235. ( 10.1890/09-1948.1) [DOI] [PubMed] [Google Scholar]
- 44.Duffy JE, et al. 2015. Biodiversity mediates top-down control in eelgrass ecosystems: a global comparative-experimental approach. Ecol. Lett. 18, 696–705. ( 10.1111/ele.12448) [DOI] [PubMed] [Google Scholar]
- 45.Maestre FT, et al. 2012. Plant species richness and ecosystem multifunctionality in global drylands. Science 335, 214–218. ( 10.1126/science.1215442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moss B, et al. 2011. Allied attack: climate change and eutrophication. Inland Waters 1, 101–105. ( 10.5268/iw-1.2.359) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in the electronic supplementary material.