Abstract
Intensified surveillance during the 2009 A/H1N1 influenza pandemic in Israel resulted in large virological and serological datasets, presenting a unique opportunity for investigating the pandemic dynamics. We employ a conditional likelihood approach for fitting a disease transmission model to virological and serological data, conditional on clinical data. The model is used to reconstruct the temporal pattern of the pandemic in Israel in five age-groups and evaluate the factors that shaped it. We estimate the reproductive number at the beginning of the pandemic to be R = 1.4. We find that the combined effect of varying absolute humidity conditions and school vacations (SVs) is responsible for the infection pattern, characterized by three epidemic waves. Overall attack rate is estimated at 32% (28–35%) with a large variation among the age-groups: the highest attack rates within school children and the lowest within the elderly. This pattern of infection is explained by a combination of the age-group contact structure and increasing immunity with age. We assess that SVs increased the overall attack rates by prolonging the pandemic into the winter. Vaccinating school children would have been the optimal strategy for minimizing infection rates in all age-groups.
Keywords: disease transmission model, age structure, serology, school vacations, absolute humidity, vaccine allocation
1. Introduction
In recent years, there has been significant progress in employing influenza surveillance data, together with epidemiological modelling, in order to address some of the important questions regarding the human-to-human transmission of influenza and the factors that affect it [1]. An especially promising approach, which also raises methodological challenges, is incorporating different types of datasets, each with its own strengths and weaknesses, in order to piece together a picture of the unfolding of an epidemic and the underlying mechanisms [1]. In this work, we employ several types of data available from the 2009 A/H1N1 pandemic in Israel, together with model-fitting techniques, in order to obtain insights that could not have been obtained from each of these datasets in isolation.
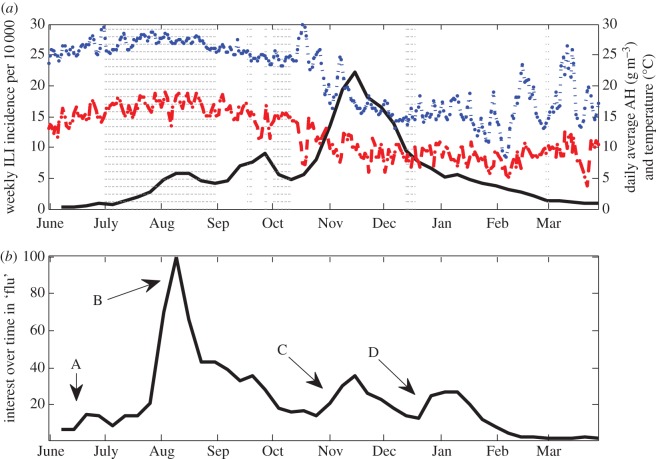
The novel A/H1N1 influenza strain (H1N1pdm) was first identified in Israel at the end of April 2009 [2,3]. By June, an epidemic was already underway across the country. Clinical diagnoses of influenza-like illness (ILI) collected during the pandemic showed a pattern characterized by three apparent waves of infection (figure 1a). This pattern presents an opportunity for investigating the changing factors affecting the transmission rates of influenza within a population. Weather conditions, in particular temperature and absolute humidity (AH), are the main factors currently used to explain the seasonal pattern of interpandemic influenza outbreaks in temperate regions [4–8]. With regards to the 2009 pandemic, several studies have found that varying AH conditions throughout the pandemic period help to explain the spatio-temporal dynamics of the pandemic [9,10], whereas other studies indicated that regional differences in AH conditions can help to explain the observed variation in the pandemic dynamics across different geographical regions [11,12]. Maximum levels of temperature and AH in Israel were reached during the month of August (figure 1a), which could serve to explain the decline in ILI incidence during August and the subsequent rise at the start of September. Reduced contact rates during school vacations (SVs) is another factor thought to have a role in the seasonality of influenza [13,14]. Evaluating the effect of SVs on the spread of influenza is difficult, as typically the influenza season does not overlap with the school summer vacation. The early initiation of the pandemic has enabled one to examine and confirm the curtailing impact of SVs on influenza transmission [10,12,15–20]. In Israel, the early initiation resulted in the pandemic overlapping not only with the summer vacation during July and August, but also with the Jewish New Year holidays in October (figure 1a).
Figure 1.
(a) Weekly ILI incidence rates per 10 000 (solid line), average daily absolute humidity (dashed line), average daily temperature (dotted line) and vacation periods (vertical dotted lines) during the pandemic in Israel. (b) Results of the search term ‘flu’ (in Hebrew) using Google Trends between June 2009 and March 2010. Alphabet letters mark key events during the pandemic in Israel coinciding with peaks in the public interest in the flu. (A) WHO declares ‘swine flu’ a global pandemic (12 June 2009), (B) first cases of mortality from ‘swine flu’ in Israel (27 July 2009, 3 August 2009), Israeli Ministry of Health orders the acquisition of vaccinations for the entire Israeli population (30 July 2009), (C) beginning of national vaccination campaign for individuals belonging to risk groups (2 November 2009), (D) opening of vaccination campaign to the entire population (18 December 2009). (Online version in colour.)
From a research perspective, another opportunity presented by the pandemic relates to the unique datasets collected during this period. In the wake of the pandemic, the Israeli Ministry of Health enhanced its typical influenza surveillance programme, by carrying out more virological tests of suspected influenza cases and by conducting a large seroepidemiological study [21]. Virological tests provide an indication of the fraction of actual influenza cases among patients with influenza-like symptoms. Serological surveys make it possible to assess the actual proportions of the disease spread in the population, and in so doing account for the undiagnosed cases. The availability of these types of data to complement the clinical diagnosis data is particularly important under the circumstances of a worldwide pandemic. As public awareness and attitude towards H1N1pdm changed in time over the pandemic, so did the tendency to consult with physicians upon the appearance of clinical symptoms, which in turn affected the time series of clinical diagnoses [19]. Examining the time series obtained from a Google Trends search for ‘flu’ in Israel during the pandemic period (figure 1b) reveals a different wave pattern from the one observed in the ILI data (figure 1a), including a disproportionate temporary leap in the public interest in flu at the beginning of August, which could serve as an alternative explanation to the observed peak in ILI incidence during this period. Combining all available types of epidemiological and social data should help facilitate a more accurate evaluation of the pandemic's dynamics in Israel and the factors that shaped it.
Several recent studies introduced new frameworks for integrating multiple types of surveillance data, including virological and/or serological data, with a disease transmission model in order to examine different aspects of influenza epidemics [19,20,22–24]. Here, we present an approach based on conditional likelihood, for fitting and parameter estimations of dynamical models using a variety of datasets. Our transmission model incorporates weather data, as well as data on social contacts among age-groups, to project the number of infected individuals in different age-groups. Our observation model uses clinical diagnosis data while attempting to give a realistic description of the processes by which the virological and serological datasets were collected. The combination of the two models is used to fit the uniquely large and detailed virological and serological datasets collected during the pandemic in Israel. By fitting the virological and serological datasets, we infer maximum-likelihood estimates for the transmission model parameters, assessing the impact of SV and weather conditions on the reproductive number as well as the relative susceptibility of individuals in the different age-groups. Using the model, we reconstruct the temporal pattern of the pandemic in Israel and the overall infection rates in different age-groups. We show that the timing of SVs, in relation to the seasonality induced by weather, can have dramatic effect on the outcome of an epidemic. Finally, we investigate the potential effectiveness of vaccination in mitigating the pandemic and assess optimal vaccine allocation among different age-groups.
2. Methods
2.1. The data
The following is a short description of the different datasets used in the modelling (table 1). Detailed information regarding the data can be found in the electronic supplementary material, S1A. The virological data used here are the results of RT-PCR tests conducted on throat and nasal swabs collected from suspected influenza cases visiting clinics belonging to the Israeli Ministry of Health's sentinel network. The serology data contain the results of haemagglutination inhibition (HI) assays performed on serums collected from individuals prior to and during the 2009 pandemic [21]. The clinical surveillance data consist of diagnoses made by physicians belonging to the Macabbi health maintenance organization (HMO), which has a nationwide coverage of approximately 25% of the Israeli population. The dataset is comprised of four different diagnoses that together cover more than 90% of the diagnoses given to patients sampled in the sentinel clinics for virological testing [25]. We use here the term extended influenza-like illness (eILI) to refer to the combined sum of the four diagnoses. The virological, serological and clinical data were aggregated into weekly data in five age-groups: 0–4, 5–19, 20–44, 45–64 and 65+ (electronic supplementary material, tables S1–S3).
Table 1.
List of the data sources used by the model.
| data | description | values | source/reference |
|---|---|---|---|
 , Tj(t) , Tj(t) |
number of positive/total virology tests of eILI diagnosed patients of age-group j on week t of the pandemic | see electronic supplementary material, table S1 | Israeli Center for Disease Control (ICDC) and the central virology laboratory at Sheba medical center |
 , Yj(t) , Yj(t) |
number of positive/total serology tests of individuals in age-group j on week t of the pandemic | see electronic supplementary material, table S2 | Central Virology Laboratory at Sheba Medical Center [18] |
| eILIj(t) | number of ILI, URI, viral infection and fever diagnoses for Maccabi HMO members in age-group j on week t of the pandemic | see electronic supplementary material, table S3 | Maccabi HMO and the ICDC |
 (1 ≤ i, j ≤ 5) |
contact matrices of 5-year band age-groups for eight European countries | see source ref. | POLYMOD study [22] |
| school vacations periods | dates of school vacations | see electronic supplementary material, table S5 | Israeli Ministry of Education |
| Temp(t), RH(t) |
mean temperature and relative humidity on day t for Israel. Used to calculate the mean absolute humidity for Israel AH(t) | see electronic supplementary material, figure S3. Data are available in source ref. | Israel Meteorological Service [23] |
We calculated a 5 × 5 age-group contact frequency matrix for the population of Israel (electronic supplementary material, table S4) based on results of a survey of the number of contacts among age-groups in eight European countries [26]. We found that after accounting for differences in demographics and scaling, the variance obtained from the European matrices is very small (electronic supplementary material, figure S2). By taking the mean of these matrices and fixing it according the demographics of the Israeli population, we obtain, up to a scaling factor, an age-group contact matrix for Israel. The SV dates used in the model are based on the official vacations schedule for the 2009/2010 school year in the Jewish sector (electronic supplementary material, table S5). Daily mean temperature and AH data for Israel used in the model were obtained by averaging measurements made by meteorological stations of the Israeli Meteorological Society [27] in four Israeli cities representing four geographical regions (electronic supplementary material, figure S3). We used Google Trends data in some model variants for modelling trends in the tendency to visit a physician over time upon experiencing influenza-like symptoms. These data (available at [28]) were extracted by searching Google Trends for the term ‘flu’ (in Hebrew) over the pandemic period (June 2009–March 2010) in Israel (figure 1b).
2.2. Model formulation
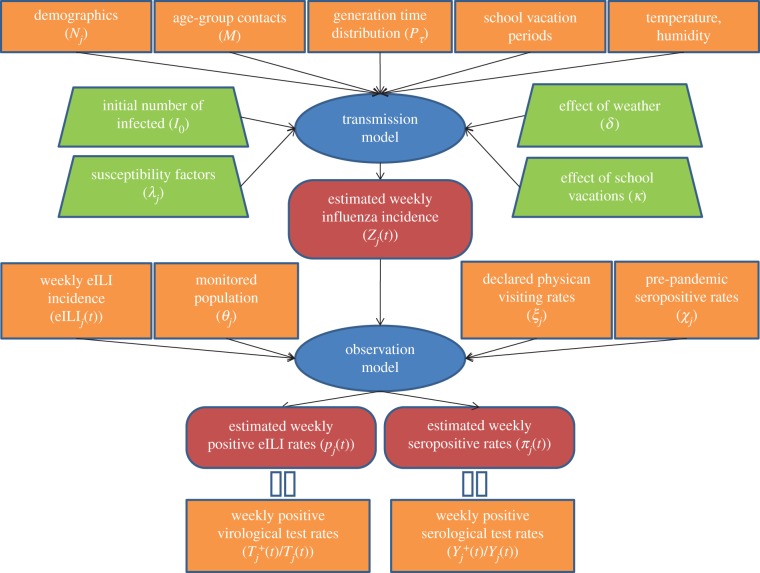
Figure 2 summarizes the modelling scheme used here, which will be described in detail below. In general, the scheme involves a deterministic transmission model that characterizes the transmission of influenza in the Israeli population as a whole. The transmission model estimates the number of new infectives each day over the course of the pandemic. This information is fed into a stochastic observation model that makes predictions about the results of serological and virological tests of the population. The likelihood function is defined based on the probability of obtaining the laboratory results (the serological and virological data at our disposal) given the clinical surveillance data and the transmission model output.
Figure 2.
A diagram describing the components of the modelling scheme. Ovals represent model components. Rectangles represent data used by the model. Trapezoids represent model parameters that are estimated by the model fitting. Rounded rectangles represent model outputs. The transmission model output feeds into the observation model whose two types of output are compared with the available virological and serological data. (Online version in colour.)
2.2.1. Transmission model
For the description of the infection process, we use the deterministic version of a discrete-time, age-of-infection model framework [29]. The model is extended to incorporate multiple age-groups. The model follows the general SIR framework that assumes each individual at any time is in one of three classes: susceptible, infected or recovered. The model equations are
 |
2.1a |
| 2.1b |
Here, ij(t) is the number of newly infected from age-group j (1 ≤ j ≤ m) and Sj(t) is the number of susceptible individuals from age-group j on day t. A detailed description of the model parameters and the derivation of the model equations can be found in the electronic supplementary material (§B). Briefly, the model parameters are (see also tables 2 and 3):
— Nj, the population size of group j.
— Pτ(1 ≤ τ ≤ d), the infectivity profile or generation time distribution.
— Mjk, the contact frequency matrix (§2.1).
— σjk(t), the effect of SVs on the contact structure. For the most part, we assume SVs affect contacts among school children only, with the parameter κ measuring the percentage change in the number of contacts among school children during SV periods (see equation S2 in electronic supplementary material, B). We also explored other options for modelling the effect of SVs (see Discussion and electronic supplementary material, G).
— w(t), the effect of weather conditions on the probability of infection. Two alternative forms are considered using either AH or temperature. In both cases, the parameter δ measures the strength of the seasonal forcing related to AH or temperature (see electronic supplementary material, B equation S3).
— λj, the susceptibility of individuals from age-group j which is proportional to the probability of a susceptible individual from age-group j being infected upon contact with an infected person. The value of each of these terms by itself is not very informative. However, by comparing these values, the relative susceptibility of the different age-groups can be assessed. We define
 as the relative susceptibility of the age-groups.
as the relative susceptibility of the age-groups.—
 the daily number of imported influenza cases of age-group j.
the daily number of imported influenza cases of age-group j.
Given the model formulation, one can obtain the formula for the reproductive number on day t – R(t), measuring the mean number of infections caused by one infected individual throughout the individual's period of infectivity, given the conditions on day t (electronic supplementary material, C). In our case, there are two external factors that cause the value of R to vary in time: weather conditions and the effect of SVs. In addition, R is reduced in time owing to the depletion of susceptible individuals in the population.
Table 2.
List of the fixed model parameters.
| parameter | description | value | source |
|---|---|---|---|
| Pτ (1 ≤ τ ≤ d) | generation time distribution | gamma distribution with d = 7 days, mean of 2.7 days and variance of 1.8 days | based on data collected during the initial stages of the pandemic [2,3] |
| Mij (1 ≤ i, j ≤ 5) | contact frequency matrix for the five modelled age-groups in Israel | see electronic supplementary material, table S4 | generated using the eight POLYMOD matrices (see electronic supplementary material, §A.4) |
| Nj | population size of age-group j at the end of 2009 | see electronic supplementary material, table S6 | Israel Central Bureau of Statistics [30] |
| θj | fraction of monitored population (Maccabi HMO members) in age-group j at the end of 2009 | see electronic supplementary material, table S6 | social security report on HMO membership for 2009 [31] |
| ξj | fraction of individuals of age-group j that declare to would consult their physician when experiencing influenza-like symptoms | see electronic supplementary material, table S6 | results of a survey conducted in Israel in 2011 [28] |
 |
average daily number of imported cases in age group J detected in Israel during June 2009 | see electronic supplementary material, table S6 | data collected during the pandemic's initial stages [2,3] |
| ηj | distribution of confirmed cases among age-groups detected in Israel up to beginning of June 2009 | see electronic supplementary material, table S6 | data collected during the pandemic's initial stages [2,3] |
| χj | baseline pre-pandemic fraction of seropositive individuals in age-group j | see electronic supplementary material, tables S2 and S6 | results of serology tests taken prior to the pandemic [18] |
| S0j | initial fraction of susceptible individuals in age-group j | set to either 1−χj or 1 for all age-groups | according to assumption regarding protection of pre-pandemic seropositives (§§2.2.3, 2.2.4) |
Table 3.
Maximum-likelihood estimates (MLE) and bootstrapped 95% CI for the parameters together with results obtained from fitting the basic model, under two alternative scenarios: assuming pre-pandemic seropositive individuals were protected completely or not at all from the pandemic strain (see §§2.2.2 and 2.2.3 for details). We present here the values of the estimated relative susceptibility of the age-groups  . The values of λj themselves are given in electronic supplementary material, table S8.
. The values of λj themselves are given in electronic supplementary material, table S8.
| parameter/result | description | MLE [95% CI] assuming pre-pandemic seropositives were fully protected | MLE [95% CI] assuming pre-pandemic seropositives were not protected |
|---|---|---|---|
 |
relative susceptibility of individuals of age-group j compared to individuals of the most susceptible age-group |
 = 1.0 [1.0–1.0] = 1.0 [1.0–1.0] = 0.81 [0.71–0.93] = 0.81 [0.71–0.93] = 0.69 [0.58–0.84] = 0.69 [0.58–0.84] = 0.40 [0.31–0.49] = 0.40 [0.31–0.49] = 0.21 [0.11–0.34] = 0.21 [0.11–0.34] |
 = 1.0 [1.0–1.0] = 1.0 [1.0–1.0] = 0.68 [0.58–0.78] = 0.68 [0.58–0.78] = 0.63 [0.52–0.76] = 0.63 [0.52–0.76] = 0.38 [0.30–0.45] = 0.38 [0.30–0.45] = 0.17 [0.09–0.25] = 0.17 [0.09–0.25] |
| δ | strength of seasonal forcing in the transmission rates related to absolute humidity conditions | 0.12 [0.09–0.15] | 0.10 [0.08–0.13] |
| κ | percentage change in number of contacts among school children during school vacations | −0.18 [(−0.28)–(−0.08)] | −0.20 [(−0.33)–(−0.10)] |
| I0 | total number of infected individuals on 1 June 2009 |
3035 [2223–3785] | 3789 [2932–4412] |
| AR | overall attack rates | AR1 = 0.41 [0.35–0.47] AR2 = 0.48 [0.45–0.50] AR3 = 0.30 [0.26–0.33] AR4 = 0.13 [0.10–0.15] AR5 = 0.03 [0.02–0.05] |
AR1 = 0.42 [0.35–0.48] AR2 = 0.54 [0.49–0.56] AR3 = 0.31 [0.28–0.34] AR4 = 0.13 [0.11–0.15] AR5 = 0.03 [0.02–0.04] |
| R | reproductive number at the initiation of the pandemic | 1.43 [1.38–1.47] | 1.40 [1.35–1.44] |
| negative log-likelihood | 731 | 691 |
2.2.2. Observation model
The observation model links the incidence of influenza given by the transmission model to the observed virological and serological data. We start by formulating the relationship between influenza incidence and the virological data. We set Zj(t) to be the weekly incidence of influenza by summing up the daily incidence of newly infected from age-group j—ij(t)—given by the transmission model. Only a proportion of the individuals infected in the full population are reported and assigned with one of the clinical diagnoses in our dataset. This proportion depends on both the fraction of monitored population in each age-group— —which is known (electronic supplementary information, table S6), and a reporting rate—rj(t)—which is the fraction of infected individuals of age-group j who consult their physician on week t and are diagnosed with eILI. The number of infected in age-group j who were monitored and diagnosed with eILI on week t is thus
—which is known (electronic supplementary information, table S6), and a reporting rate—rj(t)—which is the fraction of infected individuals of age-group j who consult their physician on week t and are diagnosed with eILI. The number of infected in age-group j who were monitored and diagnosed with eILI on week t is thus  . For the main part of this paper, we assume constant reporting rates in time rj, setting rj = ξj, where ξj are the fraction of individuals from age-group j that declare they would consult their physician when experiencing influenza-like symptoms, according to the results of a survey conducted in Israel in 2011 [32]. We performed a sensitivity analysis to assess the effect of uncertainty regarding the used reporting rates on the model outcome (electronic supplementary material, F). We also examined several model variants to assess the effect of varying reporting rates in time as well as varying fraction of symptomatic cases in the different age-groups, on the model outcome (electronic supplementary material, G,H).
. For the main part of this paper, we assume constant reporting rates in time rj, setting rj = ξj, where ξj are the fraction of individuals from age-group j that declare they would consult their physician when experiencing influenza-like symptoms, according to the results of a survey conducted in Israel in 2011 [32]. We performed a sensitivity analysis to assess the effect of uncertainty regarding the used reporting rates on the model outcome (electronic supplementary material, F). We also examined several model variants to assess the effect of varying reporting rates in time as well as varying fraction of symptomatic cases in the different age-groups, on the model outcome (electronic supplementary material, G,H).
The virological dataset consists of Tj(t)—the number of tests performed on week t on individuals of age-group j; and  —the number of positive tests for the H1N1pdm influenza strain. These tests were conducted almost exclusively on individuals diagnosed with one of the clinical diagnoses included in our eILI dataset, which is why we need to use this extended dataset and not just the ILI dataset. The probability that an individual of age-group j who is diagnosed with eILI on week t is infected is given by
—the number of positive tests for the H1N1pdm influenza strain. These tests were conducted almost exclusively on individuals diagnosed with one of the clinical diagnoses included in our eILI dataset, which is why we need to use this extended dataset and not just the ILI dataset. The probability that an individual of age-group j who is diagnosed with eILI on week t is infected is given by  . Therefore, the probability of obtaining
. Therefore, the probability of obtaining  positive cases out of the Tj(t) eILI cases which are virologically tested is given by the binomial distribution
positive cases out of the Tj(t) eILI cases which are virologically tested is given by the binomial distribution
| 2.2 |
Next, we formulate the relation between influenza incidence and the serological data. These data consist of Yj(t)—the number of serological tests performed on week t on individuals of age-group j; and  —the number of seropositive tests for H1N1pdm. The state of seropositives in the population on week t is assumed to be represented by the results of the serological tests on week t + 2, as there is a gap of two weeks on average between infection and the rise of antibodies in the serum to a detectable level known as the seroconversion period [33]. The number of seropositives in the population on each week is determined by the cumulative number of infected individuals. In addition, prior to the pandemic, some individuals were already seropositive to the pandemic strain owing to cross reactivity with previously circulating influenza strains. From the serological tests of serums taken between April 2008 and April 2009, we have Y0j and
—the number of seropositive tests for H1N1pdm. The state of seropositives in the population on week t is assumed to be represented by the results of the serological tests on week t + 2, as there is a gap of two weeks on average between infection and the rise of antibodies in the serum to a detectable level known as the seroconversion period [33]. The number of seropositives in the population on each week is determined by the cumulative number of infected individuals. In addition, prior to the pandemic, some individuals were already seropositive to the pandemic strain owing to cross reactivity with previously circulating influenza strains. From the serological tests of serums taken between April 2008 and April 2009, we have Y0j and  , the number of pre-pandemic total/positive tests for each age-group (electronic supplementary material, table S2). The ratio
, the number of pre-pandemic total/positive tests for each age-group (electronic supplementary material, table S2). The ratio  is used as an estimate of the fraction of pre-pandemic seropositives within the population.
is used as an estimate of the fraction of pre-pandemic seropositives within the population.
Because it is difficult to assess to what extent being seropositive prior to the pandemic is a correlate of immunity to the pandemic strain [34], two extreme alternatives were examined: (i) there is complete correlation, so that a pre-pandemic seropositive individual could not be infected by the pandemic strain, and (ii) there is no correlation, so that a pre-pandemic seropositive has the same probability of being infected as a pre-pandemic seronegative individual. In the first case, the probability of obtaining  seropositive tests out of a total of Yj tests on week t + 2 of the epidemic may be modelled as
seropositive tests out of a total of Yj tests on week t + 2 of the epidemic may be modelled as
 |
2.3a |
Here, χj is the probability an individual of age-group j was seropositive prior to the pandemic, and the attack rate  is the probability an individual of age-group j was infected by week t of the pandemic, and so became seropositive by week t + 2. Because, in this case, we assume the two are mutually exclusive their sum πj(t) is the probability an individual is seropositive by week t + 2. In the second case, we model the same probability as follows:
is the probability an individual of age-group j was infected by week t of the pandemic, and so became seropositive by week t + 2. Because, in this case, we assume the two are mutually exclusive their sum πj(t) is the probability an individual is seropositive by week t + 2. In the second case, we model the same probability as follows:
 |
2.3b |
Here, πj(t) (the probability an individual is seropositive by week t + 2) is the sum of χj and the attack rate  multiplied by the factor (1 − χj) to exclude the proportion of infected individuals that were already seropositive before the pandemic started. In addition, the two models differ in their initial conditions regarding the proportion of susceptible individuals at the start of the pandemic (see below).
multiplied by the factor (1 − χj) to exclude the proportion of infected individuals that were already seropositive before the pandemic started. In addition, the two models differ in their initial conditions regarding the proportion of susceptible individuals at the start of the pandemic (see below).
2.2.3. Initial conditions
We estimate I0—the initial total number of infected at the point of time in which the fit to the data begins (1 June). Dividing the I0 infected individuals among the different age-groups was done according to ηj—the distribution of confirmed H1N1pdm cases among the five age-groups up to that point in time [2,3]. However, we also investigated other possibilities to divide I0 (e.g. according to the size of each age-group) and found out that generally the exact distribution of the I0 infected among the age-groups has no major effect on the outcome. When using equation (2.3a) in the observation model, the initial fraction of susceptible individuals in each age-group (S0j) is set to 1 − χj, as it is assumed in this case that the pre-pandemic seropositives are immune to the pandemic strain. When using equation (2.3b) in the observation model, we set S0j = 1, so that all individuals have the potential of being infected by the pandemic strain. We also investigated a model variant in which we estimated S0j, while assuming the same probability of infection upon contact for each age-group (see electronic supplementary material, G).
2.2.4. Model variants
We have examined fitting several model variants, distinguished by the incorporation of different assumptions affecting the number of model parameters being fitted (see electronic supplementary material, G), as well as the type of weather data employed to modulate the transmission rates (AH or temperature). The results of the model variants were compared using Akaike information criterion (AIC) [35].
2.3. Inference
We use a maximum-likelihood approach for estimating model parameters. Our likelihood function is a conditional likelihood: the probability of obtaining the observed results of the virological and serological tests from the transmission and observation models, given the clinical surveillance data. Conditional likelihoods are used when part of the relevant data is not modelled (see [36]). In our case, the eILI surveillance data, which depend also on the circulation of other viruses leading to ILI symptoms, are not modelled, so that the relevant likelihood is that of the virological and serological data conditional on the eILI data, which are used to compute the probability pj(t) of a virological test being positive (see equation (2.2)). Thus, for a set of parameter values  the likelihood function is given by the product of the two binomial probabilities described in equations (2.2) and (2.3a or 2.3b):
the likelihood function is given by the product of the two binomial probabilities described in equations (2.2) and (2.3a or 2.3b):
 |
2.4 |
Our conditional likelihood approach differs from previous works integrating virological, serological and clinical data [19,22,24], which incorporated the probability of observing the clinical data into their likelihood function. The difficulty in incorporating the ILI surveillance data into the likelihood stems from the fact that, because the ILI data include a large proportion of non-influenza cases, the number of ILI cases at each point in time depends also on the current incidence of other viruses, which are likely to follow their own dynamics. This introduces the need to include many extra parameters in the likelihood function, one for each time point: the number of non-influenza ILI cases, or, equivalently, the fraction of ILI cases which are true influenza cases. In [19], the availability of clinical and virological data from other regions of England was used to provide an estimate of the fraction of non-influenza-related cases within the clinical cases in London throughout the pandemic. Such external data are not available to us. In [22,24], the estimation of the many extra parameters was avoided by assuming a prior distribution for these parameters (in the prior distribution, the parameters are independently distributed according to a gamma distribution), and using this prior distribution to integrate out the extra parameters, so that only the two parameters of the gamma prior need to be estimated. In our approach, we opted to avoid the problem of the unknowns introduced by the ILI data by not fitting the clinical data altogether and therefore use the conditional likelihood approach as described above.
In a previous work, we have focused on modelling the stochasticity of the infection process described by the transmission model and the stochasticity in the reporting of clinical diagnoses [8]. Here, for the sake of simplicity, the stochasticities involved in these processes are ignored, and the likelihood function is based on the stochasticity of the sampling processes involved in the laboratory testing, as it is assumed that the latter are dominant, owing to the small populations involved in these data. This assumption was verified by testing the fitting procedure on simulated data incorporating process and observation noise (see electronic supplementary material, D).
An optimization program written in Matlab was used to find the maximum-likelihood parameter estimates. The program used a combination of a nonlinear optimization algorithm (fminunc function) and a simulated annealing algorithm (simulannealbnd function). To ensure that the global maximum is found, the program was run numerous times, each time using a random set of initial parameter values. The program was tested on simulated data to verify the identifiability of the model parameters, and was found to work well with these data (electronic supplementary material, D). 95% CI for the parameter estimates were calculated using parametric bootstrapping (electronic supplementary material, D) and likelihood profiles (electronic supplementary material, E). We also conducted sensitivity analyses to assess the effect of uncertainty related to our fixed parameter values on the outcome of the model fit (electronic supplementary material, F).
2.4. Testing the effect of vaccination
The transmission model used in the data fitting (equation (2.1)) does not include vaccination as the vaccination stocks for the pandemic strain reached Israel late in the epidemic, and the vaccination coverage attained was too small to have any tangible impact on the epidemic (see electronic supplementary material, figure S12). Nevertheless, after fitting the model to the data, we wish to test the hypothetical effect of vaccination using the model. For this purpose, equation (2.1b) of the transmission model was extended to incorporate vaccination in the following manner:
| 2.5 |
Here, vj(t) is the number of vaccinations given to individuals of age-group j on day t and ζ is the efficacy of the vaccine which was set to 70% [37]. In testing the effect of vaccination, the transmission model parameters were set to the maximum-likelihood estimates as obtained by the basic model fit (table 3). Assuming vaccine coverage C in the total population, we searched for the optimized allocation of vaccinations within the different age-groups— —that minimizes the total attack rate, and evaluated the effect of the vaccinations on the pandemic dynamics. We either assumed that the vaccinations were given immediately at the beginning of the pandemic
—that minimizes the total attack rate, and evaluated the effect of the vaccinations on the pandemic dynamics. We either assumed that the vaccinations were given immediately at the beginning of the pandemic  or examined other scenarios in which vaccination is conducted later in the pandemic and the allocation of the vaccines is spread evenly over a period of time (e.g. we set
or examined other scenarios in which vaccination is conducted later in the pandemic and the allocation of the vaccines is spread evenly over a period of time (e.g. we set  to model allocation of vaccines during September 2009). When examining the effect of vaccination during the pandemic we need to revise equation (2.5) further in order to take into consideration ‘wasting’ of vaccines on individuals who are already protected (after being infected) at the time of vaccination (see electronic supplementary material, J).
to model allocation of vaccines during September 2009). When examining the effect of vaccination during the pandemic we need to revise equation (2.5) further in order to take into consideration ‘wasting’ of vaccines on individuals who are already protected (after being infected) at the time of vaccination (see electronic supplementary material, J).
3. Results
3.1. Model fit
In the following, we present the results of fitting our basic model variant, described in the Methods section, to the virological and serological datasets, while using AH as the weather factor modulating the transmission rates. Table 3 gives the maximum-likelihood estimates and 95% confidence intervals (CIs) obtained while assuming pre-pandemic seropositives were fully protected and while assuming they were not protected any more than seronegative individuals (§§2.2.2 and 2.2.3). The results obtained in the two scenarios are very similar, the most notable difference being the attack rate in school children estimated as a little lower in the first scenario. As the differences between the two alternatives are relatively minor, we focus from this point forward on the results of the second scenario (i.e. assuming a pre-pandemic seropositive has the same probability of being infected as a pre-pandemic seronegative individual), which obtained the higher likelihood among the two scenarios. We found that employing AH as the seasonal driver yields better results than using temperature, regardless of the model variant used (see electronic supplementary material, table S10). Models incorporating more parameters, such as reporting rates that vary in time, managed in some cases to obtain a better fit to the data in terms of their AIC score, while not having any major impact on the parameter estimates and the estimated pandemic dynamics obtained from this simpler model (electronic supplementary material, table S10). In addition, sensitivity analyses showed no major impact on the outcome of the model fit resulting from uncertainty in the fixed model parameters (electronic supplementary material, table S9).
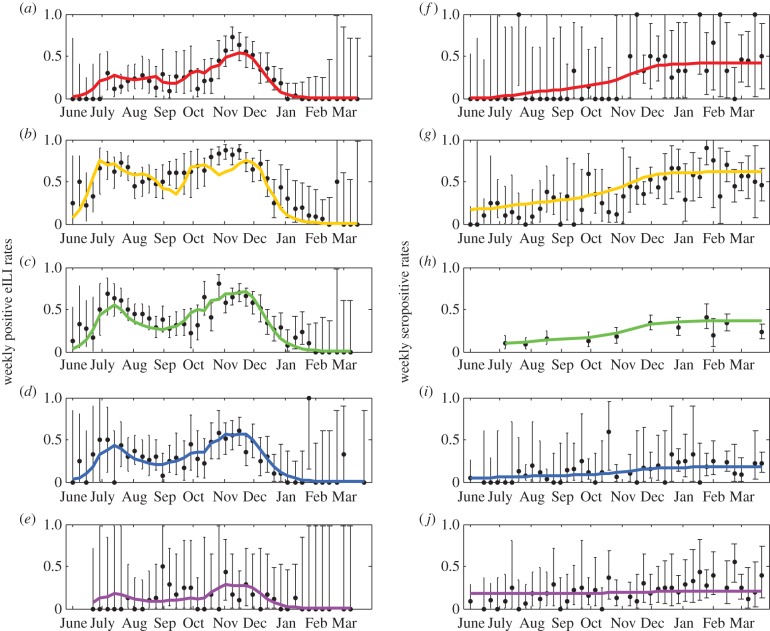
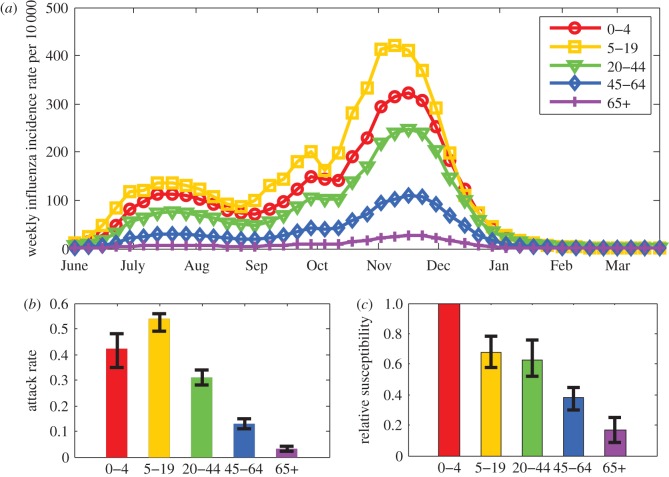
Figure 3a–e shows the fit between the fraction of weekly eILI incidence that is influenza related obtained from the model fit (pj(t) in equation (2.2)) and the proportion of influenza-positive eILI cases in the virological dataset (electronic supplementary material, table S1), including 95% binomial CIs calculated using the Clopper–Pearson method. Figure 3f–j shows the fit between the estimated seroprevalence rates given by the model fit (πj(t) in equation (2.3b)) and the weekly fraction of seropositives in the population according to the serological tests (electronic supplementary material, table S2), including 95% binomial CI. As the figures show, for the most part, the estimates for all age-groups are found within the 95% CI of the observed data. Figure 4a shows the estimated influenza incidence rate given by the transmission model in the five age-groups. The total estimated attack rate is 32% (bootstrapped 95% CI 28–35%), considerably larger than the attack rate of a seasonal influenza epidemic, which is estimated at 5–20% [38]. However, we find large variation in the attack rates between the different age-groups, with the highest attack rates among school children and the lowest among the elderly (figure 4b). We also find that susceptibility to the pandemic strain decreased considerably with age (figure 4c).
Figure 3.
Observed (dots and bars) and estimated (curved lines) weekly rates of influenza-positive eILI cases (a–e) and weekly seroprevalence rates (f–j) in the five age-groups: 0–4 (a,f), 5–19 (b,g), 20–44 (c,h), 45–64 (d,i) and 65+ (e,j). The observed data are the results of the virological and serological tests (given in electronic supplementary material, tables S1 and S2). Binomial 95% CI for the observed data were calculated using the Clopper–Pearson method. The estimated curves were obtained from the model fit using equations (2.2) and (2.3b). (Online version in colour.)
Figure 4.
Results of the model fit (using equations (2.2) and (2.3b), see also table 3). (a) Weekly influenza incidence rate in the five age-groups. (b) Attack rate in the five age groups with bootstrapped 95% CI. (c) Relative susceptibility of individuals in the five age groups with bootstrapped 95% CI. (Online version in colour.)
3.2. Effect of absolute humidity and school vacations
Our basic model employs the effect of both AH and SVs. Results of fitting model variants that employ only AH or only SV reveal that each of these factors by itself is able to produce similar dynamics to the one obtained by the basic model fit (figure 4a), with small but visible differences in the age-group incidence between the three model variants (electronic supplementary material, figure S8). While the two factors present a competing explanation for the obtained pandemic dynamics, the comparison of the three model variants using AIC (electronic supplementary material, table S10) as well as the likelihood surface with respect to the two factors (electronic supplementary material, figure S5) clearly indicates that employing both factors is better in terms of fitting the combination of virological and serological data. Furthermore, tests conducted on simulated data confirmed that the two parameters measuring the effect of AH and SV (δ and κ) are simultaneously identifiable when fitted to the type and amount of data available to us (electronic supplementary material, D).
Given the maximum-likelihood estimates of the basic model employing both factors, we analyse the impact of each of the factors, AH and SV, on the pandemic dynamics. Figure 5a shows the estimated influenza incidence rate for the whole population according to this model with and without the effect of AH and/or SV (i.e. setting the parameter δ and/or κ values to zero). According to this exercise, without the effect of AH or SV, the epidemic would have had a single peak during August. Only the combination of AH and SV has managed to delay the epidemic enough, so that the bulk of the epidemic occurred during November–December. Figure 5b shows the estimated value of the reproductive number R as it changed over the duration of the pandemic (electronic supplementary material, equations S4 and S5), for the four configurations used in figure 5a. At the beginning of the pandemic, R is estimated to be 1.4 (1.35–1.44). Varying AH conditions were responsible for a reduction of up to 20% during summer and an increase of up to 25% during winter in the value of R (dashed line compared with solid line in figure 5b), whereas SVs reduced R by 12% (dotted line compared to solid line in figure 5b). The latter is attributed to an estimated 20% (8–31%) reduction in contacts among school-aged children during SVs. Whenever R declines below the threshold R = 1, the incidence of influenza cases starts to decrease. The declines in the epidemic during the summer and the beginning of October were the result of the combined effect of SV and unfavourable AH conditions, whereas the decline in the third wave during November is attributed by the model to the depletion of the susceptible population.
Figure 5.
(a) The combined and separate effect of absolute humidity (AH) and school vacations (SV) on influenza incidence in the population as a whole. Using the parameter estimates obtained by the model fit incorporating both factors (equations (2.2) and (2.3b)), we project the dynamics of the pandemic without the effect of one or both factors by running the model while setting the relevant parameter(s) (κ and/or δ) to zero. The inset shows the overall attack rates in each of the scenarios. (b) The combined and separate effect of AH and SV on the reproductive number R in the four scenarios described in (a). (Online version in colour.)
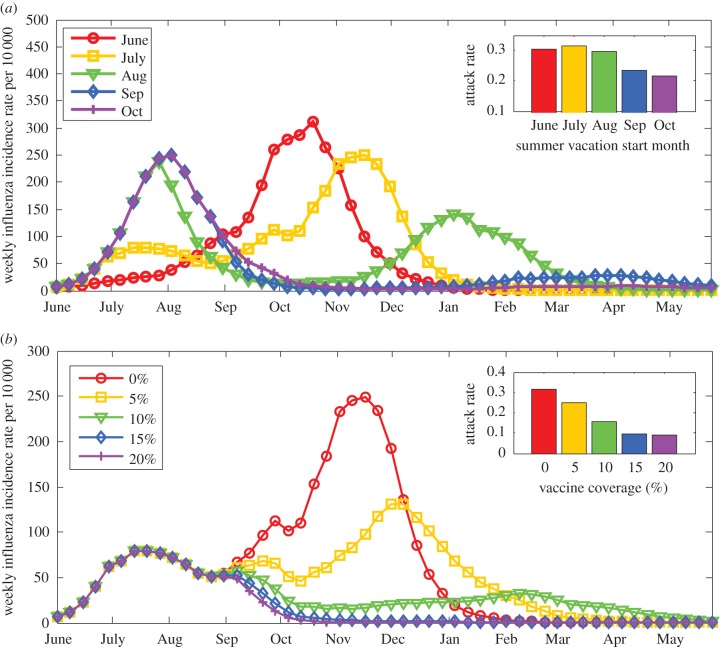
In terms of total attack rates, SV alone (without the effect of AH), would have reduced the attack rates from 34% to 27%. However, with the effect of AH, SV increased the overall attack rates from 24% (had there been no vacations) to 32%. That is, by delaying the epidemic and extending it into winter, the overall epidemic size with SVs turned out to be larger than it would have been otherwise. Similar results can be seen in figure 6a which shows the outcome of the pandemic according to the model if the same SV periods were shifted in time, so that the summer vacation starts either on the first of June, July (the actual dates), August, September or October. When vacations start at June or July, the bulk of the epidemic is postponed to wintertime, and when vacations start at September or later the big epidemic wave occurs in summer. When summer vacations start in August, there are two big waves, in summer and in late winter. Once more, postponing the epidemic to winter increases the overall epidemic size.
Figure 6.
(a) Effect of timing of school vacations on the pandemic dynamics. Using the parameter estimates obtained by the model fit (equations (2.2) and (2.3b)), we project the dynamics of the pandemic if the timing of school vacations were shifted, so that summer vacation would start on 1 June, 1 July (actual), 1 August and so forth. The inset shows the overall attack rates in each of the scenarios. According to the model, postponing the epidemic to winter increased the overall epidemic size. (b) Effect of optimal vaccine allocation on the pandemic dynamics with hypothetical vaccination campaigns during September 2009, after the first wave of the pandemic in Israel. Using the parameter estimates obtained by the model fit (equations (2.2) and (2.3b)), we project the dynamics of the pandemic assuming overall vaccine coverage of 0–20% of the population. The inset shows the overall attack rates in each of the scenarios. According to the model, with a coverage of 15% using optimal allocation consisting of vaccinating almost only school children, the outbreak would have been mitigated (see electronic supplementary material, figure S14 for more details). (Online version in colour.)
3.3. Effect of optimal vaccine allocation
Using the model with the estimated parameters given by the maximum-likelihood estimates (table 3), we tested what would have been the effect of different vaccination campaigns on the pandemic. Figure 6b shows the results of a hypothetical vaccination campaign during September 2009, after the first wave of the pandemic. With up to 20% coverage in the population as a whole, the optimal allocation strategy, which reduces the total attack rate to a minimum, is to vaccinate almost only school children (5–19). Vaccinating just 15% of the population using the optimal allocation strategy (vaccinating 58% of all school children) would have completely mitigated the winter wave of the outbreak.
To illustrate the advantage of vaccinating children over adults, we have compared the outcome of vaccinating 20% of the population, where all the vaccinations are either given to children 0–19, young adults 20–44 or to adults 45 + years old. In each case, we have calculated the overall attack rate and its distribution among all five age-groups (electronic supplementary material, figure S15). Vaccinating the children outperforms vaccinating the adults by a huge margin. An important finding is that vaccinating the children is advantageous not only from an overall perspective, but also from the adults' perspective, as the attack rates in the adult age-groups are smaller when the children are vaccinated compared with when the adults themselves are vaccinated. This finding was obtained while assuming the efficacy of the vaccine is the same for all age-groups, whereas typically the efficacy is smaller in the elderly [23,39], which would only strengthen this result.
4. Discussion
In this study, we reconstructed the dynamics of the 2009 pandemic in Israel in five age-groups by fitting a dynamic model to unique virological and serological datasets. Our model has managed to fit the results of the virological and serological tests in five age-groups well (figure 3), while estimating a relatively small number of parameters. Although the model was not fitted to the ILI data, the reconstructed dynamics follows a similar trend suggested by the ILI data (figure 1a), with two small waves during summer and autumn followed by a large wave during winter (electronic supplementary material, figure S10). Analysis of the model shows that these waves were triggered by the combination of varying weather conditions and SVs. The estimated attack rate for the pandemic in the whole population of Israel is 32%, with a large variation in the attack rates among different age-groups (figure 4b). Our estimation of the attack rate in the different age-groups resembles results of previous studies [19,20,22,33,40]. The distribution of infectives among the age-groups is explained in part by the age-group contact patterns, with school children having the highest number of contacts and the elderly the least. However, the model indicates increasing immunity with age to be another significant factor that is essential in reproducing the observed pattern of attack rates in the different age-groups (figure 4c). This result reaffirms similar conclusions made by previous serological studies [40–42] as well as modelling studies of the pandemic in other countries [20,43–45]. Nonetheless, it should be noted that the differences in susceptibility between individuals in the 0–4 age-group and individuals in the 5–19 and 20–44 age-groups, as estimated by our model, might also be related to behavioural differences that render young children more likely to be infected when contacting an infectious individual.
We obtained an estimate of R = 1.4 at the beginning of the pandemic, which is in the range of other estimates from different parts of the world (1.2 ≤ R ≤ 2.2) [10,12,16,19,20,22,24,44]. In the beginning of July, owing to the combined effect of SVs and increasing AH levels, our estimate for R is just above 1 (figure 5b), which is in line with previous estimates made based on incidence of confirmed cases at the initial phase of the pandemic in Israel [3]. Our formulation of R takes into account the existing prior immunity at the beginning of the pandemic (as given by the estimated parameters λj, which encompass the probability of being infected upon contact). We can calculate the value of the basic reproductive number R0 if we assume young children (0–4) were completely susceptible to the pandemic strain, by plugging λ1 in place of each λj in electronic supplementary material, equation (S1) at time t = 0, leading to an estimate of R0 = 2.2. Notably, this estimate of R0 for the pandemic is considerably lower than an estimate (R0 = 3.25) obtained from fitting 11 years of seasonal influenza epidemics in Israel [8].
Our estimate of the impact of AH on the transmission of influenza is similar to a previous estimate based on modelling 11 years of seasonal influenza epidemics in Israel [8]. As in the seasonal influenza study, we have found that AH better explains the seasonality of influenza in Israel than temperature. The estimated 12% reduction in R during SVs is around the low range of the 14–50% reduction estimated by modelling studies in other countries [10,12,13,15–20]. Obviously, the effect of SVs is a social-related phenomenon that may vary from population to population. In the basic model formulation presented here, SVs can affect contacts among school children only. In the electronic supplementary material, we present a model variant in which contacts of school children with other age-groups could also be affected during SVs. Using this variant, we obtained a larger reduction in the number of contacts among school children during SVs when compared with the reduction estimated in the basic model variant (40% compared with 20%), paired with an increase (of approx. 60%) in the number of contacts between school children and other age-groups during these periods (see electronic supplementary material, G). This increase may be attributed to increasing within-families contacts during SVs. This model variant has managed to improve the overall fit of the model (as seen in its improved AIC score) while not having any major effect on the rest of our results.
Our pandemic model suggests that the interplay of seasonality in the transmission and mitigation efforts can lead to some complex and unexpected outcomes, which should be of consequence when considering the use of school closures as a measure to mitigate an ongoing epidemic. The model demonstrates that having SVs at the time they occurred actually increased the magnitude of the pandemic in Israel, by delaying the epidemic and extending it into winter when better conditions for influenza prevail (figures 5a and 6a). The only scenario in which closing the schools early in the epidemic would have been advantageous in terms of reducing the overall size of the epidemic is if the time gained by delaying the epidemic were used to vaccinate a larger part of the population. This result was not reported in other modelling works that projected the course of the pandemic without the effect of SVs [10,20]. These modelling works either did not model the effect of weather conditions [20] or found the effect of weather conditions in their settings to be minimal [10]. Without the effect of weather, SVs would always reduce the magnitude of the epidemic. Only when combined with seasonality induced by weather (or some other seasonal forcing) can SVs cause an increase in the overall attack rate.
The actual vaccination campaign against the pandemic strain in Israel was ‘too little too late’ owing to the late arrival of the vaccine to Israel to have any effect on the pandemic dynamics (see electronic supplementary material, J). Using the model, we investigated what would have been the effect of vaccinations had they been delivered early enough to influence the epidemic. The model demonstrates that vaccinating just 15% of the population during September 2009, after the first wave of the outbreak, using an optimal vaccine allocation which includes vaccination of mostly school children, would have mitigated the winter wave of the outbreak entirely (figure 6b). In an unrealistic scenario in which a vaccination campaign could have taken place prior to the initiation of the pandemic, a campaign covering 20% of the population with an optimal allocation consisting of mostly school children (78% of all school children) would have prevented the outbreak completely (electronic supplementary material, figure S13). In optimizing the allocation of vaccines, we considered only the effect of vaccination on the total attack rate. However, we have found that vaccinating children is advantageous over vaccinating adults even from the perspective of reducing the infection rates among adults (electronic supplementary material, figure S15). This result is in line with previous modelling studies [23,46–49] and a randomized controlled trial study [50] that have shown that vaccinating children can be an effective method to protect the elderly from influenza. In our case, the combination of higher contact rates in children and lower probability of infection in the elderly means that concentrating on vaccinating the children would have been the optimal strategy in reducing both morbidity and mortality. Because it appears that increased protection for older individuals compared with younger individuals is a hallmark of influenza pandemics [51], vaccinating children to protect the whole population should probably be the preferred course of action in the case of a future pandemic.
Supplementary Material
Acknowledgements
The authors acknowledge three anonymous referees whose helpful review helped to improve this paper. The authors also acknowledge Prof. David Earn, Prof. Pejman Rohani and Prof. Jacob Oleson for their helpful comments and suggestions on a preliminary version of the paper appearing in Dr Yaari's PhD thesis.
Authors' contributions
R.Y., G.K., L.S. and A.H. conceived and designed the study. R.Y. implemented the study and wrote the first draft. G.K., L.S. and A.H. revised and provided critical comments on the draft. E.M. and M.M. collected and analysed the virological and serological data.
Competing interests
We declare we have no competing interests.
Funding
R.Y. and A.H. were supported by the Israel National Institute for Health Policy Research.
References
- 1.Heesterbeek H, et al. 2015. Modeling infectious disease dynamics in the complex landscape of global health. Science 347, aaa4339. ( 10.1126/science.aaa4339) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roll U, Yaari R, Katriel G, Barnea O, Stone L, Mendelson E, Huppert A. 2011. Onset of a pandemic: characterizing the initial phase of the swine flu (H1N1) epidemic in Israel. BMC Infect. Dis. 11, 92 ( 10.1186/1471-2334-11-92) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katriel G, Yaari R, Huppert A, Roll U, Stone L. 2011. Modelling the initial phase of an epidemic using incidence and infection network data: 2009 H1N1 pandemic in Israel as a case study. J. R. Soc. Interface 8, 856–867. ( 10.1098/rsif.2010.0515) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaman J, Kohn M. 2009. Absolute humidity modulates influenza survival, transmission, and seasonality. Proc. Natl Acad. Sci. USA 106, 3243–3248. ( 10.1073/pnas.0806852106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDevitt J, Rudnick S, First M, Spengler J. 2010. Role of absolute humidity in the inactivation of influenza viruses on stainless steel surfaces at elevated temperatures. Appl. Environ. Microbiol. 76, 3943–3947. ( 10.1128/AEM.02674-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaman J, Pitzer VE, Viboud C, Grenfell BT, Lipsitch M. 2010. Absolute humidity and the seasonal onset of influenza in the continental United States. PLoS Biol. 8, e1000316 ( 10.1371/journal.pbio.1000316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barreca AI, Shimshack JP. 2012. Absolute humidity, temperature, and influenza mortality: 30 years of county-level evidence from the United States. Am. J. Epidemiol. 176(Suppl 7), S114–S122. ( 10.1093/aje/kws259) [DOI] [PubMed] [Google Scholar]
- 8.Yaari R, Katriel G, Huppert A, Axelsen JB, Stone L. 2013. Modelling seasonal influenza: the role of weather and punctuated antigenic drift. J. R. Soc. Interface 10, 20130298 ( 10.1098/rsif.2013.0298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaman J, Goldstein E, Lipsitch M. 2011. Absolute humidity and pandemic versus epidemic influenza. Am. J. Epidemiol. 173, 127–135. ( 10.1093/aje/kwq347) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Earn D, He D, Loeb M, Fonseca K. 2012. effects of school closure on incidence of pandemic influenza in Alberta, Canada. Ann. Intern. Med. 173–182. ( 10.7326/0003-4819-156-3-201202070-00005) [DOI] [PubMed] [Google Scholar]
- 11.Chowell G, et al. 2012. The influence of climatic conditions on the transmission dynamics of the 2009 A/H1N1 influenza pandemic in Chile BMC Infect. Dis. 12, 298 ( 10.1186/1471-2334-12-298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamerius J, Viboud C, Shaman J, Chowell G. 2015. Impact of school cycles and environmental forcing on the timing of pandemic influenza activity in Mexican states, May–December 2009. PLoS Comput. Biol. 11, e1004337 ( 10.1371/journal.pcbi.1004337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cauchemez S, Valleron A-J, Boëlle P-Y, Flahault A, Ferguson NM. 2008. Estimating the impact of school closure on influenza transmission from Sentinel data. Nature 452, 750–754. ( 10.1038/nature06732) [DOI] [PubMed] [Google Scholar]
- 14.Heymann AD, Hoch I, Valinsky L, Kokia E, Steinberg DM. 2009. School closure may be effective in reducing transmission of respiratory viruses in the community. Epidemiol. Infect. 137, 1369–1376. ( 10.1017/S0950268809002556) [DOI] [PubMed] [Google Scholar]
- 15.Eames KTD, Tilston NL, Brooks-Pollock E, Edmunds WJ. 2012. Measured dynamic social contact patterns explain the spread of H1N1v influenza. PLoS Comput. Biol. 8, e1002425 ( 10.1371/journal.pcbi.1002425) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chowell G, Echevarría-Zuno S, Viboud C, Simonsen L, Tamerius J, Miller MA, Borja-Aburto VH. 2011. Characterizing the epidemiology of the 2009 influenza A/H1N1 pandemic in Mexico. PLoS Med. 8, e1000436 ( 10.1371/journal.pmed.1000436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali S, Kadi A, Ferguson N. 2013. Transmission dynamics of the 2009 influenza A (H1N1) pandemic in India: the impact of holiday-related school closure. Epidemics 5, 157–163. ( 10.1016/j.epidem.2013.08.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang KE, Lipsitch M, Shaman J, Goldstein E. 2014. The US 2009 A(H1N1) influenza epidemic: quantifying the impact of school openings on the reproductive number. Epidemiology 25, 203–206. ( 10.1097/EDE.0000000000000055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birrell PJ, et al. 2011. Bayesian modeling to unmask and predict influenza A/H1N1pdm dynamics in London. Proc. Natl Acad. Sci. USA 108, 18 238–18 243. ( 10.1073/pnas.1103002108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Te Beest DE, Birrell PJ, Wallinga J, De Angelis D, van Boven M. 2015. Joint modelling of serological and hospitalization data reveals that high levels of pre-existing immunity and school holidays shaped the influenza A pandemic of 2009 in the Netherlands. J. R. Soc. Interface 12, 20141244 ( 10.1098/rsif.2014.1244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weil M, Shohat T, Bromberg M, Bassal R, Dichtiar R, Mandelboim M, Sofer D, Cohen D, Mendelson E. 2013. The dynamics of infection and the persistence of immunity to A(H1N1)pdm09 virus in Israel. Influenza Other Respir. Viruses 7, 838–846. ( 10.1111/irv.12071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorigatti I, Cauchemez S, Pugliese A, Ferguson NM. 2012. A new approach to characterising infectious disease transmission dynamics from sentinel surveillance: application to the Italian 2009–2010 A/H1N1 influenza pandemic. Epidemics 4, 9–21. ( 10.1016/j.epidem.2011.11.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baguelin M, Flasche S, Camacho A, Demiris N, Miller E, Edmunds WJ. 2013. Assessing optimal target populations for influenza vaccination programmes: an evidence synthesis and modelling study. PLoS Med. 10, e1001527 ( 10.1371/journal.pmed.1001527) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorigatti I, Cauchemez S, Ferguson NM. 2013. Increased transmissibility explains the third wave of infection by the 2009 H1N1 pandemic virus in England. Proc. Natl Acad. Sci. USA 110, 13 422–13 427. ( 10.1073/pnas.1303117110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sever L. 2011. Characteristics of the 2009 A/H1N1 influenza patients in the community and hospitals (table 13). Master's thesis, School of Public Health, Tel-Aviv University.
- 26.Mossong J, et al. 2008. Social contacts and mixing patterns relevant to the spread of infectious diseases. PLoS Med. 5, e74 ( 10.1371/journal.pmed.0050074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Israel Meteorological Service (IMS). Meteorological database archive. See https://data.gov.il//ims (accessed 15 March 2016).
- 28.Google Trends: Results of search for ‘Flu’ in Israel between June 2009 and March 2010. See https://www.google.com/trends/explore#q=%D7%A9%D7%A4%D7%A2%D7%AA&geo=IL&date=6%2F200910m&cmpt=geo&tz=Etc%2FGMT-3 (accessed 16 July 2015).
- 29.Katriel G. 2013. Stochastic discrete-time age-of-infection epidemic models. Int. J. Biomath. 6, 1250066 ( 10.1142/S1793524512500660) [DOI] [Google Scholar]
- 30. Israel Central Bureau of Statistics. 2010 Statistical abstract of Israel 2010, Table 2.19. See http://www.cbs.gov.il/shnaton61/st02_19.pdf (accessed 1 March 2016).
- 31.Bendalak J. 2010. Membership in health maintenance organization in 2009. See www.btl.gov.il/Publications/survey/Documents/seker_227.pdf (accessed 1 March 2016).
- 32.Yamin D. 2012. Influenza vaccination policy. PhD thesis, Department of Industrial Engineering and Management, Ben-Gurion University of the Negev.
- 33.Baguelin M, Hoschler K, Stanford E, Waight P, Hardelid P, Andrews N, Miller E. 2011. Age-specific incidence of A/H1N1 2009 influenza infection in England from sequential antibody prevalence data using likelihood-based estimation. PLoS ONE 6, e17074 ( 10.1371/journal.pone.0017074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cox RJ. 2013. Correlates of protection to influenza virus, where do we go from here? Hum. Vacc. Immunother. 9, 405–408. ( 10.4161/hv.22908) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burnham KP, Anderson DR. 2002. Model selection and multimodel inference: a practical information-theoretic approach. Berlin, Germany: Springer. [Google Scholar]
- 36.Pawitan Y. 2001. In all likelihood: statistical modelling and inference using likelihood. Oxford, UK: Oxford University Press. [Google Scholar]
- 37.Osterholm MT, Kelley NS, Sommer A, Belongia EA. 2012. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect. Dis. 12, 36–44. ( 10.1016/S1473-3099(11)70295-X) [DOI] [PubMed] [Google Scholar]
- 38.Huppert A, Barnea O, Katriel G, Yaari R. 2012. Modeling and statistical analysis of the spatio-temporal patterns of seasonal influenza in Israel. PLoS ONE 7, e45107 ( 10.1371/journal.pone.0045107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fleming DM, Andrews NJ, Ellis JS, Bermingham A, Sebastianpillai P, Elliot AJ, Miller E, Zambon M. 2010. Estimating influenza vaccine effectiveness using routinely collected laboratory data. J. Epidemiol. Commun. Health 64, 1062–1067. ( 10.1136/jech.2009.093450) [DOI] [PubMed] [Google Scholar]
- 40.Van Kerkhove MD, Hirve S, Koukounari A, Mounts AW, Allwinn R, Bandaranayake DD. 2013. Estimating age-specific cumulative incidence for the 2009 influenza pandemic: a meta-analysis of A(H1N1)pdm09 serological studies from 19 countries. Influenza Other Respir. Viruses 7, 872–886. ( 10.1111/irv.12074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller E, Hoschler K, Hardelid P, Stanford E, Andrews N, Zambon M. 2010. Incidence of 2009 pandemic influenza A H1N1 infection in England: a cross-sectional serological study. Lancet 375, 1100–1108. ( 10.1016/S0140-6736(09)62126-7) [DOI] [PubMed] [Google Scholar]
- 42.Steens A, et al. 2011. Age-dependent patterns of infection and severity explaining the low impact of 2009 influenza A (H1N1): evidence from serial serologic surveys in the Netherlands. Am. J. Epidemiol. 174, 1307–1315. ( 10.1093/aje/kwr245) [DOI] [PubMed] [Google Scholar]
- 43.Simon C, Yosinao N. 2011. A mathematical model to distinguish sociological and biological susceptibility factors in disease transmission in the context of H1N1/09 influenza. J. Theor. Biol. 286, 50–56. ( 10.1016/j.jtbi.2011.07.008) [DOI] [PubMed] [Google Scholar]
- 44.Opatowski L, Fraser C, Griffin J, de Silva E, van Kerkhove MD, Lyons EJ, Cauchemez S, Ferguson NM. 2011. Transmission characteristics of the 2009 H1N1 influenza pandemic: comparison of 8 southern hemisphere countries. PLoS Pathog. 7, e1002225 ( 10.1371/journal.ppat.1002225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merler S, Ajelli M, Pugliese A, Ferguson NM. 2011. Determinants of the spatiotemporal dynamics of the 2009 h1n1 pandemic in Europe: implications for real-time modelling. PLoS Comput. Biol. 7, e1002205 ( 10.1371/journal.pcbi.1002205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Longini IM, Halloran ME. 2005. Stragegy for distribution of influenza vaccine to high-risk groups and children. Am. J. Epidemiol. 161, 303–306. ( 10.1093/aje/kwi053) [DOI] [PubMed] [Google Scholar]
- 47.Bansal S, Pourbohloul B, Meyers LA. 2006. A comparative analysis of influenza vaccination programs. PLoS Med. 3, 1816–1825. ( 10.1371/journal.pmed.0030387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dushoff J, Plotkin JB, Viboud C, Simonsen L, Miller M, Loeb M, Earn DJD, Lipsitch M. 2007. Vaccinating to protect a vulnerable subpopulation. PLoS Med. 4, e174 ( 10.1371/journal.pmed.0040174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Medlock J, Galvani AP. 2009. Optimizing influenza vaccine distribution. Science 325, 1705–1708. ( 10.1126/science.1175570) [DOI] [PubMed] [Google Scholar]
- 50.Loeb M, et al. 2010. Effect of influenza vaccination of children on infection rates in Hutterite communities: a randomized trial. JAMA 303, 943–950. ( 10.1001/jama.2010.250) [DOI] [PubMed] [Google Scholar]
- 51.Greer AL, Tuite A, Fisman DN. 2010. Age, influenza pandemics and disease dynamics. Epidemiol. Infect. 138, 1542–1549. ( 10.1017/S0950268810000579) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.