Abstract
We have previously developed the enabling techniques for sulfoglycomics based on mass spectrometry (MS) analysis of permethylated glycans, which preserves the attractive features of more reliable MS/MS sequencing compared with that performed on native glycans, while providing an easy way to separate and hence enrich the sulfated glycans. Unlike LC-MS/MS analysis of native glycans in negative ion mode that has been more widely in use, the characteristics and potential benefits of similar applications based on permethylated sulfated glycans have not been fully investigated. We report here the important features of reverse phase-based nanoLC-MS/MS analysis of permethylated sulfated glycans in negative ion mode and demonstrate that complementary sets of diagnostic fragment ions afforded can allow rapid identification of various fucosylated, sialylated sulfated glycotopes and definitive determination of the location of sulfate in a way difficult to achieve by other means. A parallel acquisition of both higher collision energy and trap-based MS2 coupled with a product dependent 1 is conceivably the most productive sulfoglycomic workflow currently possible and the manually curated fragmentation characteristics presented here will allow future developments in automating data analysis.
Graphical abstract

INTRODUCTION
Myriad studies have implicated the importance of additional sulfation on the non-reducing terminal epitopes of N- and O-glycans, which alters their physicochemical properties and modulates their functioning as cognate ligands of endogenous glycan binding proteins and those of pathogens1. Among the most well established cases are GlcNAc-6-O-sulfate (GlcNAc6S) on α2-3-sialyl Lewis X, contributing to the preferred ligand for L-selectin in mediating lymphocyte homing to peripheral lymph nodes2,3, and that on α2-6-sialyl LacNAc, making it a higher affinity ligand than the non-sulfated counterpart for human CD22, an inhibitory receptor for B cell activation4. Gal-6-O-sulfate (Gal6S) has also been considered as an essential modification on α2-3-sialyl LacNAc to make it a high affinity ligand for Siglec 8/F based on glycan array studies5 but in this6 and most other cases, the occurrence of sulfated glycotopes in their physiologically relevant settings remains largely unsubstantiated, especially when well-defined monoclonal antibody is not available.
An inherent technical problem in detecting sulfated glycans by high sensitivity mass spectrometry (MS)-based glycomic mapping is the extra negative charge imparted and their generally lower abundance relative to sialylated ones. Thus, other than those derived from secreted and epithelial mucins7–11, sulfated counterparts of N- and O-glycans are generally not detected in most of the current glycomic analyses, with a few notable cases of exceptions12–14. Recognizing this limitation, we have previously developed the enabling sample preparation techniques for sulfoglycomics based on MALDI-MS analysis of permethylated glycans15,16. Since the sulfate would remain the only substituent carrying negative charge, fully methylated sulfated glycans can be selectively detected by MS in negative ion mode and readily separated from the more abundant non-sulfated ones in a way that is difficult to achieve with native glycans. More recently in several immediate applications6,17,18, we have shown that a C18 reverse phase (RP)-based nanoLC-nanoESI-MS/MS analysis of permethylated sulfated glycans in negative ion mode is possible and that diagnostic fragment ions afforded can allow determination of the location of sulfate. Compared with our initial MALDI-MS/MS approach, the most important advantage is higher sensitivity particularly in analyzing disulfated glycans intact. It also allows more comprehensive data dependent MS2 acquisition in an automated fashion with potentials for incorporating multimode and/or multistage fragmentations.
We have now systematically investigated the most useful fragmentation characteristics afforded by higher energy collision dissociation (HCD) versus ion trap CID, based on a panel of synthetic standards and a complex pool of sulfated glycans derived from cultured human bronchial epithelial cells. We show that an efficient mapping of various isomeric fucosylated, sialylated, sulfated glycotopes by negative ion mode nanoLC-MS/MS analysis of permethylated glycans can benefit from data dependent parallel acquisition of both HCD and ion trap CID MS2, supplemented further by a product ion dependent MS3 scan function, and how the generated data can be productively utilized. The manually verified dataset of over hundred glycan entries would additionally serve to guide current development of much needed computational tools for sulfoglycomic data analysis.
EXPERIMENTAL PROCEDURES
Sulfated glycan standards and samples
Synthesis of non-sialylated sulfated Galβ-3/4GlcNAc standards with azide linker has been previously described19. Synthesis of the sialylated sulfated LacNAc standards with either ethyl amine or ethyl azide linkers, and preparation of sulfated sialic acid carrying glucosyl ceramide20 from sea urchin are described in the Supporting Information, which also provides more details on the release of N- and O-glycans from mucin sample secreted by cultured human bronchial epithelial cells via standard procedures16. All glycan samples and the synthetic sulfated glycan standards were separately permethylated and fractionated by Oasis MAX (Waters) cartridge into non-, mono- and disulfated samples exactly as described previously21. Sulfated glycolipids were extracted into chloroform layer after neutralization and quenched with water. Prior to MS analysis, aliquots from all fractions were additionally cleaned up by applying to ZipTip C18 (Millipore) in 5 % acetonitrile/0.1 % formic acid and eluted by 50% acetonitrile/0.1% formic acid.
NanoESI-MS and MS/MS analysis
The permethylated glycan standards dissolved in 50 μl of 50% acetonitrile/0.1% formic acid were infused offline into an LTQ-Orbitrap Velos hybrid mass spectrometer (Thermo Scientific), equipped with a PicoView nanosprayer (New Objective) applied at −2.0 kV. The isolation width for both MS2 and MS3 was set to 3 Da and the normalized collision energies varied from 30 to 100% at 10% increments for CID and 50 to 130% at 10% increments for HCD for offline optimization studies. Online nanoLC-MS/MS analyses were performed on the same instrument fitted with a nanoACQUITY UPLC system (Waters). The permethylated glycan samples were dissolved in 5% acetonitrile/0.1% formic acid, loaded onto a nanoACQUITY UPLC 2G C18 trap column (180-μm × 20-mm, 100Å, 5 μm particle size), and separated on a nanoACQUITY UPLC BEH130 C18 column (75-μm × 250-mm, 130Å, 1.7 μm particle size), at a constant flow rate of 300 nl/min, with a column temperature of 35°C and a linear gradient of 10 to 70% acetonitrile/0.1% formic acid in 22 min, followed by a sharp increase to 95% acetonitrile in 17 min and then held isocratically for another 10 min.
For each data dependent acquisition cycle, the full-scan MS spectrum was acquired in the Orbitrap at 60,000 resolution (at m/z 400) with automatic gain control (AGC) target value of 5 × 106, followed by parallel HCD/CID MS2 on five most intense ions with a 90 sec dynamic exclusion interval. The AGC target value and normalized collision energy applied for CID experiments in the ion trap were set as 5,000 and 50%, respectively. For HCD to be detected in the Orbitrap analyzer at 30,000 resolution, the respective values were 50,000 and 80% (N-glycans) or 100% (O-glycans). Alternatively for product dependent MS3 data acquisition, each CID MS2 was followed by CID MS3 scans on the 2 most intense targeted MS2 product ions (m/z 528.18, m/z 702.26, m/z 1063.44). The AGC target value and normalized collision energy applied were set as 1000, 45% and 50, 35%, for MS2 and MS3, respectively. Data were processed by Xcalibur software v2.1 and all MS/MS data were interpreted manually.
RESULTS and DISCUSSION
NanoLC-MS characteristics of permethylated sulfated glycans
Using an acidic acetonitrile/water solvent system, the permethylated monosulfated glycans were found to elute earlier than their disulfated counterparts on a nanoACQUITY UPLC BEH130 column (Fig. 1) but it should be noted that the reverse is true using other C18 stationary phase such as the Acclaim PepMap RSLC column. As expected, permethylated glycans were not sufficiently well separated on a C18 column. The extracted ion chromatogram (XIC) peak widths tend to be split and/or broad primarily due to many unresolved structural and stereoisomers. The isomeric permutation increases with each increment of a LacNAc unit, Fuc, or sialic acid, which also resulted in expected shifts to later elution times (Fig. S-1). We have strategically opted for nanoLC conditions that would elute all commonly found monosulfated O-glycans within an effective window of about 15–20 min. In essence, this mimics a long lasting direct infusion analysis to allow complementary HCD and ion trap CID MS/MS scans repeatedly acquired for a majority of peaks detected above preset intensity threshold.
Figure 1. Extracted ion chromatograms of representative permethylated, sialylated O-glycans (A) and N-glycans (B) differing in degree of sulfation.

The normalized extracted ion chromatograms from 2 separate LC-MS/MS runs for the mono-sulfated (lower trace) and disulfated or monosulfated but with one under-methylated carboxylic group (upper trace) were aligned and partially superimposed for comparison. In the case of N-glycans, the monosulfated fraction also contained non-sulfated, under-methylated species.
Under these conditions, the fully methylated sulfated glycans could be efficiently separated from those with one sulfate less but carrying instead a sialic acid with free non-methylated carboxylic group (Fig. 1). Our sample pre-fractionation scheme therefore enables separate nanoLC-MS/MS analyses of enriched, permethylated mono- and disulfated glycans free from non-sulfated ones and the interfering under-methylated species. The sulfated glycans were invariably detected in negative ion mode as [M-H]− or [M-2H]2−, with the number of negative charge corresponds to the number of sulfate carried plus any sialic acid with a free carboxylic group. The m/z values of the monosulfated glycans are therefore the same for the MALDI- and nanoESI-MS signals since both would be detected as [M-H]− (Fig. S-2-1). On the other hand, the disulfated glycans were detected mostly as [M-2H]2−, with a small amount of [M+Na-2H]− in nanoESI-MS, but as [M-SO3-H]− in MALDI-MS. On this basis, a careful data analysis showed that all major sulfated O-glycan signals afforded by MALDI-MS could be detected by nanoLC-MS/MS at better signal-to-noise ratio, particularly for the disulfated O-glycans (Fig. S-2-2). At MS level, these signals can each be assigned a glycosyl composition corresponding to a permutation of Hex, HexNAc, Fuc and NeuAc (Tables S-1, S-2), from which core types along with how many LacNAc (Gal-GlcNAc) units and occurrence of terminal sialylated and/or fucosylated epitopes can be further inferred. Supportive evidence for this presumptive assignment would, however, need to be sought from MS/MS data.
Characteristic MS2 ions afforded by permethylated sulfated glycans
An important feature of negative ion mode MS/MS analysis of permethylated sulfated glycans is that only fragment ions retaining the sulfate and hence the negative charge will be observed. Typically among the most useful are the diagnostic non-reducing terminal B ions informing the presence of terminal sulfo HexNAc (m/z 324), sulfo NeuAc (m/z 440), sulfo LacNAc (m/z 528), sulfo Lewis (m/z 702), sulfo sialyl LacNAc (m/z 889), and sulfo sialyl Lewis (m/z 1063). In addition, we have previously identified several low mass ions afforded by HCD MS2 that are indicative of the specific location of sulfate on the terminal sialic acid, LacNAc or LacdiNAc units6,18,22. To more systematically define the diagnostic ions for this purpose, we have now examined a panel of 8 synthetic sulfated type 1 and 2 chain (Gal-3/4GlcNAc) standards, with and without terminal sialylation (Fig. 2).
Figure 2. Characteristic low mass HCD fragment ions for determination of the location of sulfate on terminal glycotopes.
Useful and diagnostic ions are marked with *. Several ions can be assigned as the expected glycosidic and ring cleavage A ions and denoted according to the established nomenclature29 while additional type of cleavages have previously been observed in MALDI-based high energy cleavages and negative ion mode cleavages24,28, e.g. the E and D ions, for the permethylated glycans. A few others including m/z 181, 195, 234 and 264 are highly reproducible and diagnostic but their origins are uncertain, with probable structures proposed as drawn without implying any mechanistic basis for their formation. The standards were synthesized with linker as drawn here but the respective parent ions and reducing end fragment ions are not considered for current purposes. All gave an abundant ion at m/z 97 corresponding to HSO4− (not shown) but only those sialylated ones additionally produced a high intensity ion at m/z 111, possibly resulting from the sulfate acquiring a methyl group from the carboxylic group.
A 6-sulfo on GlcNAc of a terminal LacNAc unit (Gal-4GlcNAc) would afford a prominent ion at m/z 195, along with other diagnostic ions at m/z 234 and 264 (Fig. 2A), These 3 ions were also given by the α2-3 and α2-6-sialylated, 6-sulfo LacNAc (Fig. 2B,C) but m/z 195 and 264 were not detected if the Gal was 3-linked to the 6-sulfo-GlcNAc in a type 1 chain (Gal-3GlcNAc) (Fig. 2D), suggesting that their formation required concomitant elimination of the 4-O-glycosyl substituent on the GlcNAc but preserving the 3-O-Me. Conversely, the 3,5A ion at m/z 167, which requires an O-Me on the 4-position to couple with 6-O-sulfo, was only afforded by 6-sulfo-GlcNAc of a type 1 (Fig. 2D) and not type 2 (Fig. 2A) chain. The O,4A ion at m/z 139 was also much more prominent in 6-sulfo type 1 chain although it can, in principle, be formed from any 6-sulfo residue. Detection of m/z 195 along with m/z 234 and 264 is therefore diagnostic of a 6-sulfo-GlcNAc on LacNAc or sialyl LacNAc, whereas detection of m/z 234 without m/z 195 but accompanied instead by strong O,4A ion at m/z 139 and 3,5A ion at m/z 167 is indicative of 6-sulfo-GlcNAc on type 1 chain. Although the mechanistic basis underlying the production of the ions at m/z 195/234/264 is unknown, plausible structures consistent with the interpretation above are shown as insets in Fig. 2. Notably, since the fragment ion at m/z 195 lacks the N-acetylated C2, it can also be derived from a terminal 6-sulfo-Gal (Fig. 2E and G). In general, a non-reducing terminal sulfated Gal is identified by ions at m/z 253, 283 and 301, corresponding to E1, B1 and C1 ions, respectively (Fig. 2E, G, and H). A pair of ions at m/z 153 (2,4A) and 181 is diagnostic of a terminal 3-O-sulfated Gal (Fig. 2H). If the sulfate is on the 6-position of Gal, the 3,5A ion at m/z 167 would be the base peak below m/z 400, accompanied by a rather prominent ion at m/z 269 (Fig. 2E, G). In that respect, 6-sulfo Gal of a type 2 chain may be marginally distinguishable from type 1 chain by additional presence of the 3,5A ion at m/z 371. The formation of m/z 167 and 269 was not affected by presence of a sialic acid at the 3-position (Fig. 2F) and this is important to identify terminal α2-3-sialyl 6′-sulfo Gal as the putative ligand for Siglec 8/F. Additional evidence can be sought from the weak signals corresponding to E2 and B2 ions of NeuAc-6-sulfo-Gal at m/z 614 and 644, respectively. Finally, sulfate on the side chain of NeuAc can be distinguished by the diagnostic ion at m/z 296 (m/z 326 for NeuGc) to corroborate the B1 ion at m/z 440 (m/z 470 for NeuGc), as shown by the HCD MS2 data on a sulfated sialic acid-carrying glucosylceramide standard isolated from sea urchin (Fig. 2I).
From these characteristic fragment ions, it is clear that each of the sulfated glycotopes can be unambiguously defined. However, the common presence of structural isomers, particularly with respect to location of sulfates, implies that glycomic MS2 is rarely performed on precursors representing a single pure structure. As shown by this work (compiled in Table S-1 and S-2), the characteristic low mass HCD MS2 ions at m/z 167, 195/234/264, 153/181/253 were detected for most of the identified sulfated O-glycan structures, indicative of no strict preference for 6-sulfo on either the Gal or GlcNAc of a LacNAc, or 3-sulfo on terminal Gal that is not further extended. Such a heterogeneity and spectral complexity can, however, be significantly reduced in cases when one or more of the GlcNAc6S-, Gal3S- and Gal6S-sulfotransferase activities are not expressed in the biological sources examined. We have previously demonstrated that double and triple knockout of the responsible sulfotransferases would indeed result in a more restricted set of detectable MS2 ions accurately reflecting the allowed sulfation pattern18.
Parallel HCD and CID MS2 with product dependent MS3
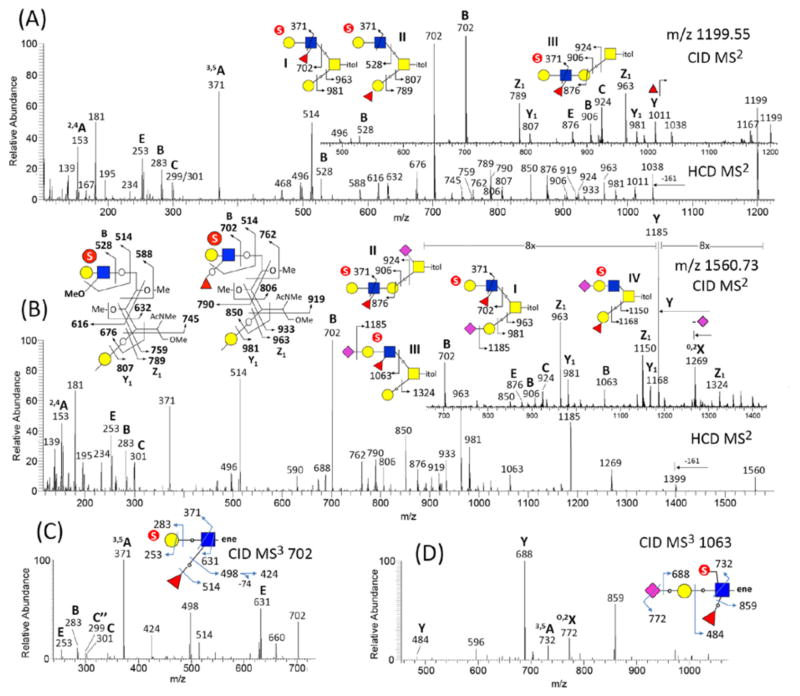
Other than the characteristic low mass ions that were produced only by HCD and not trap CID, the two fragmentation modes otherwise afforded similar cleavage patterns for the monosulfated O-glycans. In the case of structures represented by the precursor at m/z 1199 (Fig. 3A), the dominant B ion at m/z 702 indicates that a sulfated Fuc(Hex-HexNAc) is the major terminal epitope carried on the 6-arm of a core 2 structure I, as supported by i) a series of satellite ions at m/z 762, 790, 806, 850, 919, and 933, leading to ii) the Z1 and Y1 ions at m/z 963 and 981, respectively. The former set of ions resulting from concerted cleavages at the GalNAcitol (see schematic drawing in Fig. 2) has been previously observed under negative ion mode MALDI-MS/MS of sulfated O-glycans21,23, which firmly establishes the 6-arm substituents. It is clear that an isomeric structure II carrying instead a non-fucosylated sulfated LacNAc on the 6-arm also existed, as indicated by the B ions at m/z 528, its analogous satellite ions at m/z 588, 616, 632, 676, 745, and 759, and the Z1 and Y1 ions at m/z 789 and 807. Finally, a trio of E, B and C ions at m/z 876, 906 and 924, derived from cleavages at the Gal 3-linked to GalNAcitol reveal a third isomer corresponding to an extended core 1 structure III. In the case of m/z 1560 (Fig. 3B), the same set of ions at m/z 702, 762, 790, 806, 850, 919, 933, 963, 981, which were more prominent in the HCD than trap CID spectrum, implicates an isomer carrying a sulfated Fuc(Hex-HexNAc) as 6-arm substituent, and hence a sialylated Gal at the 3-arm for a core 2 structure I. The presence of m/z 876, 906 and 924 would imply that this terminal sulfated epitope could also be carried on the 3-arm of an extended core 1 structure II additionally sialylated at the C6 of the GalNAcitol. In addition, the B ion at m/z 1063 suggests that a terminal sulfated sialylated Fuc(Hex-HexNAc) exists along with a non-extended Gal at the 3-arm, as supported by the Z1 ion at m/z 1324 for a core 2 structure III. Finally, the Z1 and Y1 ions at m/z 1150 and 1168 implicate a fourth isomeric structure IV despite the absence of m/z 889 to corroborate the presence of a sulfated, sialylated LacNAc substituent on the 6-arm.
Figure 3. Exemplary HCD/CID MS2 of permethylated, monosulfated O-glycans (A–B) and CID MS3 of monosulfated terminal glycotopes (C–D).
Each of the existing isomeric structures for the parent O-glycans as can be deduced from their respective HCD and CID MS2 spectra, are shown here as structures I–III or IV with associated key fragmentation patterns. Schematic drawings are also provided for the origins of satellite ions associated with the B ions for sulfated lacNAc (m/z 528) and sulfated fucosylated lacNAc (m/z 702) extending from the 6-arm of reducing end GalNAcitol. Loss of 161 u from the parent is consistent with cleavage between C2-C3 of the reducing end GalNAcitol concomitant with elimination of the OMe on the C4. CID MS3 of the sulfated fucosylated lacNAc in (C) clearly confirms location of sulfate on the terminal Hex, but does not rule out sulfate on internal HexNAc. Further loss of 74 u can be assigned as losing the C5-C6 substituents analogous to an O,4A cleavage and can only be produced if the C6 is not sulfated.
From these exemplary assignments, it is clear that each of the precursors represented by a unique glycosyl composition comprises many structural isomers, which can be delineated by identifying the constituent core types and non-reducing terminal sulfated glycotope ensemble via the characteristic fragment ions. Going through the whole set of HCD/CID MS2 data for the monosulfated O-glycans (Table S-1) afforded the following summary with respect to deriving useful fragmentation pattern:
The low mass HCD MS2 ions allow determination of the location of sulfate but cannot define that for each of the co-existing individual sulfated glycotopes;
The diagnostic B ions define the complement of non-reducing terminal sulfated glycotopes collectively presented by the isomeric constituents of a unique precursor mass; further detection of associated satellite ions would confirm that a particular sulfated glycotope is carried on the 6-arm of an O-glycan, or extends from the 3-arm via another Hex residue at the reducing end;
Y1 ions resulting from loss of the 6-arm substituents and both Y1 and Z1 ions produced at the 3-arm of the reducing end GalNAcitol are usually quite distinctive to support assignment of the arm extension via the diagnostic fragment ions described above;
Y ion (−375 u) and 0,2X ion (−375+84 u) arising through loss of terminal sialic acids are very prominent particularly in trap CID MS2; loss of terminal Fuc (−188 u) or elimination of it (−206 u) are also common for fucosylated structures; these are, however, not informative of the location of Fuc/sialic acid
For a more definitive assignment of the identified terminal sulfated glycotopes, a further MS3 is necessary, which can be performed in subsequent target analysis or programmed as product dependent MS3 within the initial LC-MS/MS run. The B ions such as m/z 528, 702, 1063 can be targeted, which will thus be automatically selected for MS3 if detected in the trap CID MS2 spectrum. For example, the MS3 spectra of m/z 702 (Fig. 3C) and 1063 (Fig. 3D) both showed a prominent elimination of Fuc to give m/z 498 and 859, respectively. In the former, the MS3 ions at m/z 253/283/299/301 identify a terminal sulfated Hex, whereas in the latter, an internal sulfated HexNAc is identified by loss of non-sulfated sialyl Hex to give the ion at m/z 484. The 3,5A ions at m/z 371 and 732 define a type 2 LacNAc chain for sulfated, non-sialylated and sialylated LeX type glycotopes, respectively. Unfortunately, due to the low mass cutoff, the MS3 analysis by ion trap did not afford the highly informative low mass ions indicative of the precise location of sulfate. This obvious limitation can, however, be remedied by using HCD for MS3, a feature now made available in Orbitrap Fusion24.
Additional MS2 features for doubly charged disulfated O-glycans
For doubly charged disulfated O-glycans, the trap CID spectrum contained both singly and doubly charged fragment ions. The former set arose through loss of a sulfated moiety, thus converting a doubly charged disulfated precursor ion to singly charged monosulfated fragment ion. This included a very informative set of ions arising through loss of terminal sulfated Hex (−284 u), HexNAc (−325 u), Fuc-Hex (−458 u) and Fuc-HexNAc (−499 u), which unambiguously inform their presence (see Table S-2). The latter set was derived mostly from neutral loss of non-sulfated moiety, often corresponding to loss or elimination of Fuc and sialic acids, similar to those afforded by monosulfated glycans. More useful doubly charged fragment ions are those corresponding to larger disulfated B ions carrying 2 or more LacNAc repeats (see below and Table S-2), which were hardly observed in HCD MS2. As in the case for the monosulfated O-glycans, the heterogeneity in sulfation pattern renders the HCD MS2 data less useful since all diagnostic ions were commonly observed (Table S-2), resulting in essentially very similar spectra for the low mass region (below m/z 400). The slight variation in relative abundance of these diagnostic ions is indicative of preferred location of sulfate but insufficient to provide meaningful quantification. Occasionally, the HCD MS2 data afforded more distinctive singly charged B ion and associated satellite ion signals than trap CID MS2, particularly when these overlap with doubly charged fragment ions within the noisy m/z region of the trap CID spectra (e.g. Fig. 4C).
Figure 4. Representative CID MS2 spectra of doubly charged permethylated disulfated O-glycans.
The corresponding HCD MS2 afforded very similar sets of low mass ions not observed in CID. Only the mass region of HCD MS2 useful for structural assignments are additionally shown here (D, F). Loss of 98 and 112 u corresponds to loss of sulfate and −48 u corresponds to further concerted loss of the OMe from C3 of the HexNAc, as described in the text.
As shown in Fig. 4A and B for the exemplary CID MS2 spectra of 2 disulfated O-glycans differing in only one extra Hex substituting for Fuc residue, the common diagnostic losses observed include elimination of one sulfate (−98 u), loss of sulfated Hex (−284 u), and sulfated Fuc-Hex (−458 u), which implicate not only presence of terminal sulfated Hex but also a terminal sulfated Fuc-Hex. A concerted cleavages involving elimination of the sulfated terminal residue(s) and an OMe from adjacent carbon, reminiscent of previously observed G ion25, would result in further loss of 48 u. Thus the observed losses of 284 u and 284+48 u are indicative of a HexS-HexNAc glycotope. Both (Fuc)HexS-HexNAc or HexS-(Fuc)HexNAc- would allow a concerted loss of 458+48 u but only the former can additionally lose the (Fuc)HexS moiety directly via a single glycosidic cleavage (−458 u). Similar loss of terminal sulfated Fuc0-2(Hex-HexNAc) moiety yielded the respective Y ions, each accompanied by weak 1,5X ion at 28 u higher, and further supported by the corresponding B ions at m/z 528, 702 and 876. Importantly, a doubly charged disulfated B ion at m/z 695.5 was detected, which implicates an extended core 3 structure with a disulfated, difucosylated dilacNAc epitope directly attached to C3 of the GalNAcitol (Fig. 4A). On the other hand, a doubly charged B ion at m/z 608.4, corresponding to a disulfated, monofucosylated dilacNAc epitope, was detected along with the doubly charged trio at m/z 695/710/719, which collectively identified an extended core 1 structure (Fig. 4B). It can be further observed that, in general, the C ion defining the entire C3 substituent of the reducing end GalNAcitol of an extended core 1 structure was rather prominent, whereas such a C ion was absent in extended core 3 structures when a GlcNAc was directly attached to the GalNAcitol.
In addition to m/z 608 and 695, further examples of the doubly charged disulfated B ions include m/z 521, 731, 833, 920 and higher (Table S-2), corresponding to disulfated (Hex-HexNAc)2, Fuc1HexNAc1(Hex-HexNAc)2, Fuc1(Hex-HexNAc)3 and Fuc2(Hex-HexNAc)3, respectively. In the example shown in Fig. 4C, the B ions defining terminal monosulfated Hex-HexNAc, Fuc1(Hex-HexNAc), Fuc1(Hex-HexNAc)2 and Fuc2(Hex-HexNAc)2 at m/z 528, 702, 1151 and 1325 were corroborated respectively by the Y ions at m/z 1808, 1634, 1185 and 1011. The corresponding C ions carrying an extra Hex at m/z 1373 and 1547, and the respective Z1 ions at m/z 963 and 789 are indicative of direct attachment of the monosulfated Fuc1-2(Hex-HexNAc)2 chain to C3 of GalNAcitol. In addition, the doubly charged B ions for disulfated Fuc1-2(Hex-HexNAc)2 at m/z 608 and 695 were clearly detected and corroborated by the doubly charged Y ions at m/z 850 and 937 arising through loss of a non-sulfated Fuc1(Hex-HexNAc) and Hex-HexNAc, respectively. Finally, the presence of an extended core 1 isomer with a disulfated Fuc2(Hex-HexNAc)3-Hex- attached to the GalNAcitol was identified by the doubly charged B ion at m/z 920 and C ion at m/z 1031.
The pairwise detection of B ions and their corresponding Y ions not only allows identification of mono- and disulfated, fucosylated LacNAc repeats extending variably from the core in all possible permutations but also any unexpected sulfated terminal epitope. In addition to the presence of sulfated Fuc-Hex and Fuc-HexNAc not hitherto reported, which are clearly identified by aforementioned loss of 458 u and 499 u, another unusual sulfated epitope identified is sulfated Fuc2HexNAc1(Hex-HexNAc) as represented by the B ion at m/z 1121 (Table S-1 and S-2). In the example shown in Fig. 4D, this B ion was paired with the Y ion at m/z 981. Intriguingly, there was no terminal sulfated HexNAc or sulfated Fuc-HexNAc, the B and Y ions of which would have been readily detected. Instead it implicates a sulfated, difucosylated LacNAc unit further extended by a terminal HexNAc, possibly a sulfated blood group A-Ley. Its exact sequence would require further target MS3 not previously pre-programmed in the run since this is not an expected glycosyl composition. As a final note, sialylation was found to induce not only a very prominent neutral loss of sialic acid from the doubly charged disulfated glycans via trap CID MS2 but also a very characteristic loss of 112 u accompanying the loss of 98 u (Fig. S-3), both attributable to elimination of sulfate and thus converting the precursor to singly charged species. It is common also to find further loss of terminal sialic acid and sialyl Hex after the initial elimination of the sulfate (−98 u).
Overall sulfoglycomic features
In summary, the sulfated O-glycans derived from the mucins secreted by cultured human bronchial epithelial cells analyzed in this work represent one of the most complex sulfoglycomic entities primarily because it includes all 3 common modes of sulfation, namely Gal3S, Gal6S and GlcNAc6S. This renders all possible sulfation patterns on LacNAc units that can be sialylated and/or fully fucosylated on both Hex and HexNAc, as well as further extension by additional LacNAc units, to be variably distributed on different arms of isomeric cores 1-4 structures. Such complexity defies resolution of individual isomers. Here, we focus primarily on the kind of terminal sulfated glycotopes that can be identified based on characteristic fragmentation pattern. We showed that, in negative ion mode, the diagnostic losses of sulfated glycotopes from disulfated O-glycans can complement detection of key B ions, which can be mono- or disulfated. The additional satellite ions associated with cleavages at GalNAcitol, when detected, are useful to locate the sulfated glycotopes on the 6-arm, whereas a direct conjugation to the 3-arm of core 2 can often be identified by the corresponding C ions. We have also identified a series of monosulfated O-glycans with reducing end Hexitol instead of GalNAcitol, likely to be an O-mannosylated series (Table S-1, Fig. S-4). These carry the same sets of sulfated glycotopes defined by the key B ions at m/z 528, 702, and 889 but apparently more restricted in size, with no disulfated counterparts. In contrast, most monosulfated GalNAcitol based O-glycans identified have the disulfated counterparts although not necessarily for each possible isomeric structure. Discounting some odd glycosyl compositions, the range of cores 1+2 and 3+4 based mono- and disulfated O-glycans detected are additionally presented in Table S-3, arranged to show the number of LacNAc repeats and Fuc residues.
Finally, MALDI-MS screening of the permethylated N-glycan fractions revealed the presence of sulfated N-glycans, a majority of which were disulfated (Fig. S-5). Upon trap CID MS2, it is clear that most of the sulfated terminal epitopes are based on HexNAc-HexNAc unit instead of Hex-HexNAc. Consistent with it being a lacdiNAc, i.e. GalNAc-GlcNAc, the sulfate was found to localize on the terminal HexNAc, giving a strong B1 ion at m/z 324, and a 3,5A ion at m/z 167 (Fig. S-5B, C). Additional 3,5A ion at m/z 412 is indicative of 4-linked HexNAc, which can be further fucosylated to give the B2 ion at m/z 743, along with the common B2 ion for nonfucosylated sulfated lacdiNAc at m/z 569. Sulfated sialylated and non-sialylated LacNAc, as well as fucosylated LacNAc epitopes, were also present as can be inferred from the composition (Fig. S-5A) and supported by B ions at m/z 528, 702 and 889 produced by respective LC-MS/MS analysis. The dominance of sulfated lacdiNAc is striking when lacdiNAc was hardly present in the corresponding non-sulfated N-glycans (Fig. S-5A). This phenomenon is, however, reminiscent of our previous sulfoglycomic analysis of N-glycans from an ovarian cell line22.
CONCLUSION
We demonstrated how negative ion mode nanoLC-MS/MS can be efficiently applied to permethylated sulfated N- and O-glycans, their elution orders in accordance with the degree of sulfation or negative charge on a reverse phase C18 nanoAcquity column using acidic solvent, and their ionization and fragmentation characteristics. The feasibility of applying this simple solvent conditions without adducts would allow a rapid switch between positive and negative ion modes and thus enable a dual mode analysis in one LC-MS/MS run. We showed that HCD-MS2 is extremely useful in producing low mass ions, which can be reliably used to confidently assign location of sulfate on terminal type 1 and 2 Gal-GlcNAc unit. For glycomic applications when extreme heterogeneity is normal, these will be most useful if applied at MS3 level targeting the B ions corresponding to sulfated glycotopes. At MS2 level, rapid data dependent acquisition in later generations of LTQ-Oritrap MS systems allows repetitive parallel HCD and CID MS2 triggering without compromising MS1 survey and can further accommodate a product dependent MS3 targeting the few most common sulfated glycotopes of specific interest. For monosulfated glycans, the fragmentation afforded by HCD and CID is similar but the latter suffers from low mass cut-off. In the case of disulfated glycans, CID provides doubly charged disulfated fragment ions in addition to loss of monosulfated moiety to give singly charged fragment ions. These extremely useful doubly charged fragment ions and those singly charged ions of larger m/z are commonly absent in HCD MS2 optimized to ensure a good yield of low mass diagnostic ions. It is therefore advantageous to acquire both HCD and CID, coupled with product dependent MS3.
Supplementary Material
Acknowledgments
This work was supported by Academia Sinica and National Science Council grants (NSC 99-2311-B-001-021-MY3; NSC 102-2311-B-001-026-MY3) to KKH. We gratefully acknowledge Dr. Lan-Yi Chang (Institute of Biological Chemistry, Academia Sinica) for preparing and providing the crude glycolipid extracts of sea urchin. The LC-MS/MS data were acquired at the Core Facilities for Protein Structural Analysis at Academia Sinica, supported under the Taiwan National Core Facility Program for Biotechnology, grant no. NSC101-2319-B-001-003, NSC102-2319-B-001-003.
Footnotes
Notes: The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information. Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Fukuda M, Hiraoka N, Akama TO, Fukuda MN. J Biol Chem. 2001;276:47747–47750. doi: 10.1074/jbc.R100049200. [DOI] [PubMed] [Google Scholar]
- 2.Rosen SD. Annu Rev Immunol. 2004;22:129–156. doi: 10.1146/annurev.immunol.21.090501.080131. [DOI] [PubMed] [Google Scholar]
- 3.Kawashima H. Biol Pharm Bull. 2006;29:2343–2349. doi: 10.1248/bpb.29.2343. [DOI] [PubMed] [Google Scholar]
- 4.Kimura N, Ohmori K, Miyazaki K, Izawa M, Matsuzaki Y, Yasuda Y, Takematsu H, Kozutsumi Y, Moriyama A, Kannagi R. J Biol Chem. 2007;282:32200–32207. doi: 10.1074/jbc.M702341200. [DOI] [PubMed] [Google Scholar]
- 5.Tateno H, Crocker PR, Paulson JC. Glycobiology. 2005;15:1125–1135. doi: 10.1093/glycob/cwi097. [DOI] [PubMed] [Google Scholar]
- 6.Patnode ML, Cheng CW, Chou CC, Singer MS, Elin MS, Uchimura K, Crocker PR, Khoo KH, Rosen SD. J Biol Chem. 2013;288:26533–26545. doi: 10.1074/jbc.M113.485409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karlsson NG, Thomsson KA. Glycobiology. 2009;19:288–300. doi: 10.1093/glycob/cwn136. [DOI] [PubMed] [Google Scholar]
- 8.Thomsson KA, Holmen-Larsson JM, Angstrom J, Johansson ME, Xia L, Hansson GC. Glycobiology. 2012;22:1128–1139. doi: 10.1093/glycob/cws083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joncquel Chevalier Curt M, Lecointe K, Mihalache A, Rossez Y, Gosset P, Leonard R, Robbe-Masselot C. Glycobiology. 2015 doi: 10.1093/glycob/cwv004. [DOI] [PubMed] [Google Scholar]
- 10.Struwe WB, Gough R, Gallagher ME, Kenny DT, Carrington SD, Karlsson NG, Rudd PM. Mol Cell Proteomics. 2015 doi: 10.1074/mcp.M114.044867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiwamoto T, Katoh T, Evans CM, Janssen WJ, Brummet ME, Hudson SA, Zhu Z, Tiemeyer M, Bochner BS. J Allergy Clin Immunol. 2014 doi: 10.1016/j.jaci.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balog CI, Stavenhagen K, Fung WL, Koeleman CA, McDonnell LA, Verhoeven A, Mesker WE, Tollenaar RA, Deelder AM, Wuhrer M. Mol Cell Proteomics. 2012;11:571–585. doi: 10.1074/mcp.M111.011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dwyer CA, Katoh T, Tiemeyer M, Matthews RT. J Biol Chem. 2015 doi: 10.1074/jbc.M114.614099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji IJ, Hua S, Shin DH, Seo N, Hwang JY, Jang IS, Kang MG, Choi JS, An HJ. Anal Chem. 2015;87:2869–2877. doi: 10.1021/ac504339t. [DOI] [PubMed] [Google Scholar]
- 15.Yu SY, Wu SW, Hsiao HH, Khoo KH. Glycobiology. 2009;19:1136–1149. doi: 10.1093/glycob/cwp113. [DOI] [PubMed] [Google Scholar]
- 16.Khoo KH, Yu SY. Methods Enzymol. 2010;478:3–26. doi: 10.1016/S0076-6879(10)78001-0. [DOI] [PubMed] [Google Scholar]
- 17.Shibata TK, Matsumura F, Wang P, Yu S, Chou CC, Khoo KH, Kitayama K, Akama TO, Sugihara K, Kanayama N, Kojima-Aikawa K, Seeberger PH, Fukuda M, Suzuki A, Aoki D, Fukuda MN. J Biol Chem. 2012;287:6592–6602. doi: 10.1074/jbc.M111.305334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patnode ML, Yu SY, Cheng CW, Ho MY, Tegesjo L, Sakuma K, Uchimura K, Khoo KH, Kannagi R, Rosen SD. Glycobiology. 2013;23:381–394. doi: 10.1093/glycob/cws166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tu Z, Hsieh HW, Tsai CM, Hsu CW, Wang SG, Wu KJ, Lin KI, Lin CH. Chem Asian J. 2013;8:1536–1550. doi: 10.1002/asia.201201204. [DOI] [PubMed] [Google Scholar]
- 20.Klein A, Diaz S, Ferreira I, Lamblin G, Roussel P, Manzi AE. Glycobiology. 1997;7:421–432. doi: 10.1093/glycob/7.3.421. [DOI] [PubMed] [Google Scholar]
- 21.Cheng PF, Snovida S, Ho MY, Cheng CW, Wu AM, Khoo KH. Anal Bioanal Chem. 2013;405:6683–6695. doi: 10.1007/s00216-013-7128-2. [DOI] [PubMed] [Google Scholar]
- 22.Yu SY, Chang LY, Cheng CW, Chou CC, Fukuda MN, Khoo KH. Glycoconj J. 2013;30:183–194. doi: 10.1007/s10719-012-9396-z. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi M, Mitoma J, Hoshino H, Yu SY, Shimojo Y, Suzawa K, Khoo KH, Fukuda M, Nakayama J. J Pathol. 2011;224:67–77. doi: 10.1002/path.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Senko MW, Remes PM, Canterbury JD, Mathur R, Song Q, Eliuk SM, Mullen C, Earley L, Hardman M, Blethrow JD, Bui H, Specht A, Lange O, Denisov E, Makarov A, Horning S, Zabrouskov V. Anal Chem. 2013;85:11710–11714. doi: 10.1021/ac403115c. [DOI] [PubMed] [Google Scholar]
- 25.Yu SY, Wu SW, Khoo KH. Glycoconj J. 2006;23:355–369. doi: 10.1007/s10719-006-8492-3. [DOI] [PubMed] [Google Scholar]
- 26.Domon B, Costello CE. Glycoconj J. 1988;5:397–409. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.