Abstract
The dynamic motions of protein structural elements, particularly flexible loops, are intimately linked with diverse aspects of enzyme catalysis. Engineering of these loop regions can alter protein stability, substrate binding, and even dramatically impact enzyme function. When these flexible regions are structurally unresolvable, computational reconstruction in combination with large-scale molecular dynamics simulations can be used to guide the engineering strategy. Here, we present a collaborative approach consisting of both experiment and computation that led to the discovery of a single mutation in the F/G loop of the nitrating cytochrome P450 TxtE that simultaneously controls loop dynamics and completely shifts the enzyme's regioselectivity from the C4 to the C5 position of L-tryptophan. Furthermore, we find that this loop mutation is naturally present in a subset of homologous nitrating P450s and confirm that these uncharacterized enzymes exclusively produce 5-nitro-L-tryptophan, a previously unknown biosynthetic intermediate.
Enzymes rely on finely tuned molecular recognition events to selectively install chemical functionalities. This is especially true for reactions with highly reactive intermediates such as those catalyzed by enzymes of the cytochrome P450 superfamily, which activate molecular oxygen at the heme cofactor to access regio- and stereoselective oxygenation.1-7 While the precise positioning of catalytic groups in the active site is essential for such modifications, dynamic aspects of secondary structural elements, in particular flexible loops, are also integral to P450 catalysis.8-10 The functional dynamics of these enzymes are characterized by open and closed arrangements of the B/C and F/G loops that act like lids upon substrate binding.8-12 Not only do these loops seal the active site from bulk solvent to produce a catalytically productive microenvironment, they are also believed to have a direct role in substrate recognition and alignment during catalysis.8
This picture is supported by a host of crystallographic studies that have assigned structural significance to various positions along the B/C loop as well as the B and F helices.8 Although the F/G loop has been identified as a specificity-determining region (SDR) on the basis of mutational studies,8,13 there is only limited structural evidence of its direct interaction with substrates.14-16 This observation raises the question of whether these contacts are too transient to be captured by crystallography, or whether it is indeed rare that residues in these loops can extend sufficiently deep into the active site of P450s to contribute to substrate positioning during catalysis.
Here we present the discovery of such a functional position in the F/G loop of the nitrating P450 TxtE from Streptomyces scabies and demonstrate its control over regioselectivity and loop dynamics. TxtE is a rare example of an enzyme that catalyzes a direct and regioselective aromatic nitration.17-19 It differs from other known P450s by combining nitric oxide, usually an inhibitor of heme cofactors, with molecular oxygen to generate a putative ferric peroxynitrite intermediate which then disproportionates to nitrate L-tryptophan, producing 4-nitro-L-tryptophan in the thaxtomin A pathway.20
In previous work, we obtained high-resolution structures of TxtE and described a key rearrangement of the B/C loop that anchors the substrate proximal to the heme, but we were unable to determine whether such specific interactions extend to the disordered F/G loop.21 Motivated by this knowledge gap and the unique chemistry of TxtE together with its potential application as a nitration biocatalyst, we set out to study the role of the F/G loop in the recognition and positioning of the L-tryptophan substrate. We turned to computational methods to reconstruct the unresolved loop region (residues 176–183) and characterize its functional dynamics on a biologically relevant timescale through GPU-accelerated simulations and modern statistical analysis approaches. In silico methods of this type are particularly powerful when combined with laboratory techniques to overcome limitations in the experimental data. The sparse structural information on P450 F/G loops, and that of TxtE in particular, makes TxtE engineering a natural application of this paradigm. Our discovery process combined these large-scale simulations with mutagenesis and X-ray crystallography to identify a single position in the F/G loop that interacts directly with the substrate to control regioselectivity not only in TxtE, but also across other nitrating P450s.
Results and Discussion
To better understand the functional significance of the F/G loop, it was necessary to map out the dynamics of TxtE by sampling timescales 200–2000x longer than previous reports on cytochrome P450 enzymes,22-35 i.e. a time scale relevant to these motions and in a format that allows for their repeated observation. To achieve this, we performed Markov state model (MSM)36 molecular dynamics (MD) simulations on the 100 μs timescale. The substrate co-crystal structure of TxtE (PDB ID: 4TPO)21 served as the structural basis for the simulations after homology-based reconstruction and equilibration of the missing F/G loop (Fig. 1a, Supplementary Fig. 1). We carried out several rounds of adaptive sampling37 and monitored the progress through hidden Markov models (HMMs).38 Based on what is known about the structural plasticity in P450s8, we chose to define the conformational space of TxtE in terms of the F/G and B/C loop motions. Clustering of the dataset revealed that the F/G loop obeys bimodal dynamics that do not appear to be coupled with the motions of the B/C loop. The F/G loop of TxtE transitions between an unstructured set of open-lid ensembles similar to the MD starting structures that feature a solvent-accessible active site and a narrow set of closed-lid conformations that cut off the active site from bulk water (Fig. 1b). We achieved convergence within the conformational space of the F/G and B/C loops and observed on the order of 100 transitions between the open- and closed-lid conformations. Throughout, the active site was represented as the substrate-bound ferric peroxynitrite intermediate, which allowed us to investigate the catalytic step just prior to nitration (Supplementary Fig. S2).
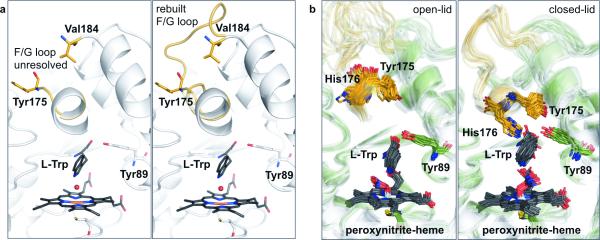
Figure 1. Dynamic range of the TxtE F/G loop.
a, The missing density between residues 175 and 184 of the wild-type crystal structure (PDB ID: 4TPO) indicates a disordered F/G loop (left panel), which was rebuilt through homology modeling (right panel). b, Subsequent large-scale MD simulations show that the F/G loop transitions between a set of disordered open-lid (left panel) and a set structured closed-lid conformations in which the structurally unresolved His176 engages in a direct interaction with the substrate l-Trp (right panel). Hydrogen atoms are omitted for the sake of clarity. The substrate and heme are in black stick rendition and the F/G loop is shown in orange. A PyMOL session file of (b) is available as part of the Supplementary Information.
While in line with the general picture of P450 F/G loop dynamics, our simulation data afforded details that had been inaccessible using structural methods alone. The closed-lid state is characterized by a tightly packed active site in which a small number of water molecules occupy discrete positions, as previously reported.39 The active site is sealed off from bulk solvent by a network of hydrogen bonds that involves Tyr89 of the B’2/C loop, Tyr175 of the F/G loop, the amino acid moiety of the substrate, and a water molecule (Fig. 2a). Strikingly, His176 of the F/G loop is in direct contact with the substrate through a tightly packed edge-to-face interaction (Supplementary Fig. S3). In contrast, none of the open-lid structural ensembles shows Tyr89 and Tyr175 in contact with one another, and His176 points away from the active site (Fig. 2b). The set of open-lid TxtE conformations is instead characterized by an influx of disordered water molecules into the active site. While the open-lid F/G loop is necessary for substrate binding and product release, the exclusion of solvent is necessary for a catalytically productive active site. Its hydration instead frequently results in the separation of the substrate and peroxynitrite-heme by layers of water molecules, which allows the substrate to rearrange and adopt a variety of loosely-bound poses at the opening of the active site (Supplementary Fig. S4).
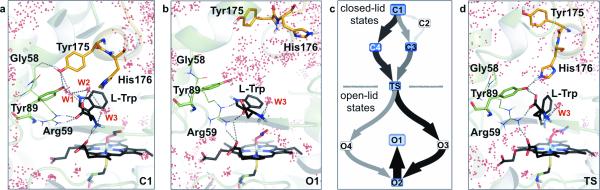
Figure 2. Productive and non-productive active site arrangements are separated by a single conformational transition state and depend on whether the F/G loop is closed or open.
a, In the closed-lid state, the F/G loop (orange) promotes a set of productive and tightly-packed active site conformations that support a small number of enclosed structural water molecules (W1–W3). These form a hydrogen-bond network with the substrate's amino acid moiety whereas the hydrophobic substrate indole ring is ‘de-wetted’ and rests proximal to the ferric peroxynitrite-species. b, The open-lid state enables substrate binding and product release. It also renders the active site accessible to bulk solvent in the presence of substrate, which can interfere with its productive alignment in the active site. c, The productive (C1–C4) and non-productive (O1–O4) active site states are connected by a single conformational transition state (TS). The Supplementary Information contains additional detail on the diagram. d, Analysis of the TS reveals that the Gly58-Tyr175-Tyr89 contacts in (a) are replaced by a set of His176-Tyr89 interactions, which identifies His176 as a mutational target. The substrate and heme are in black and represent the geometric mean of the corresponding state. Water molecules (red spheres) were drawn from 20 random structures across the corresponding state.
While the amino acid moiety of the substrate is bound to the protein in a manner consistent with the co-crystal structure,21 the substrate indole ring can occupy the active site pocket in two distinct orientations. One shows the indole-NH in contact with the heme-bound peroxynitrite oxygen (Supplementary Fig. S5) and resembles the orientation evident in the co-crystal structure (‘unflipped’). The other has the indole ring rotated by ~180° (‘flipped’), with its C4 closest to the nitrogen atom of the heme-bound peroxynitrite (Supplementary Fig. S5). The flipped orientation accounts for the overwhelming majority of observed closed-lid states (78%, Table 1). However, substrate poses with a flipped indole ring are the exception in the open-lid state and account for a mere 7%. Instead, the predominant substrate orientation is that of the unflipped indole ring (81%), with the remaining 12% accounting for substrate positions that are loosely bound and distant from the heme center.
Table 1.
Comparison of nitration activities (TTN, total turnover number), regioselectivities, dissociation constants, and open- and closed-lid populations of TxtE variants.
| ||||||
|---|---|---|---|---|---|---|
| Entry | Enzyme | TTNa | 5NT:4NT | Kd (μM)b | % Openc (f;u;l)e | % Closed (f;u;l) |
| 1 | TxtE | 96 ± 5 | 1:57 | 39 ± 2 | 56 (7|81|12) | 44 (78|22|0) |
| 2 | TxtE_His176Phe | 38 ± 0.3 | >99:1 | 2.6 ± 0.1 | 11 (10|82|8) | 89 (66|34|0) |
| 3 | TxtE_His176Tyr | 17 ± 0.9 | >99:1 | 4.9 ± 0.3 | 8 (9|77|17) | 92 (63|37|0) |
| 4 | TxtE_His176Trp | 5.9 ± 0.5 | >99:1 | 1.3 ± 0.1 | 9 (12|79|9) | 91 (59|41|0) |
| 5 | Virginiae_TxtE | 57 ± 7 | >99:1 | 140 ± 6 | N/Dd | N/D |
Total turnover numbers (TTN) with standard deviations are reported for the major product. Reaction conditions and data analysis are described in the Methods section. TTNs and regioselectivities were determined by HPLC.
Dissociation constants (Kd) with standard deviations are reported. Differential UV-vis spectra and Kd plots can be found in Supplementary Figure S49.
Open- and closed-lid populations were determined by MD. Simulation methods are described in the Supplementary Information.
N/D: not determined. Abbreviations: spinach ferredoxin (Fd), spinach ferredoxin NADP+ reductase (Fr), 5-nitrotryptophan (5NT), and 4-nitrotryptophan (4NT).
f: flipped, u: unflipped, and l: loosely bound substrate orientations are listed in % of the corresponding open/closed population.
Decomposition of these two macroscopic states, open- and closed-lid, into their underlying micro-states allowed us to further subdivide and discretize the free energy landscape. Rather than being defined by a large-scale conformational change, the geometric differences between these micro-states are on the order of side chain reorganizations and lend themselves to a more comprehensive analysis of the closed-to-open dynamics. We determined the connectivity between these sub-states and employed transition path theory (TPT)40 to determine the flux between states and to map out the kinetic rates of transitions across the network (Fig. 2c). This statistical framework allowed us to outline the steps separating the closed- from the open-lid conformations and to identify the interactions that gate their transitions. In particular, we characterized a single intermediate state central to all observed closed-to-open transitions in our dataset (Fig. 2d). Unsurprisingly, this state shares attributes of both the open- and the closed-lid conformations, featuring an interrupted Tyr175-Tyr89 contact, a new His176-Tyr89 interaction, and a partial opening of the F/G loop. This result suggests a transition whereby His176 substitutes for Tyr175, producing a significantly weakened interaction between the F/G loop and Tyr89.
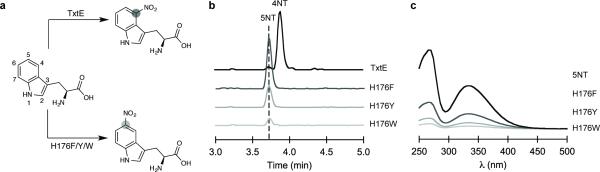
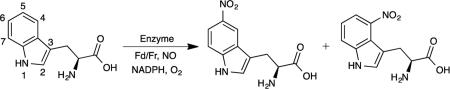
Based on these observations, we hypothesized that mutations at His176 could shift the loop equilibrium to the catalytically competent closed state when substrate is bound, with the goal of improving binding and thereby enhancing catalytic efficiency, particularly on non-natural substrates. We selected two relatively conservative mutations, intended either to provide a stronger contact to Tyr89 as a preemptive measure against lid opening (His176Tyr) or raise the free energy barrier of the closed-to-open transition through increased steric demand (His176Phe), which were tested with L-tryptophan. Consistent with our hypothesis, the binding affinity of the His176Phe and His176Tyr variants for L-tryptophan increased 15- and 8-fold, respectively, compared with the wild-type enzyme (Table 1). However, instead of the anticipated 4-nitro-L-tryptophan, we detected a new product at a shorter retention time but with a mass spectrum that nevertheless corresponds to a nitrated tryptophan (Fig. 3, Supplementary Fig. S8–S10). Derivatives of nitrotryptophan have characteristic spectral signatures that can be used to determine the connectivity of the nitro group to the tryptophan core.41-43 The wavelength of maximum absorption for the new nitro product (λmax = 330 nm) was consistent with that reported for 5-nitro-L-tryptophan (λmax = 330 nm)41 and was further confirmed by HPLC co-injection with an authentic standard (Fig. 3, Supplementary Fig. S9–S10, Supplementary Fig. S31–S32). No other naturally occurring or engineered enzymes have been reported to exclusively nitrate L-tryptophan at the C5 position.
Figure 3. Discovery of a nitration regioselectivity switch at His176.
a, Schematic of the regiospecific nitration catalyzed by wild-type TxtE and the TxtE_His176Phe/Tyr/Trp variants. b, LC-MS chromatograms of the nitrated products synthesized by wild-type TxtE and the TxtE_His176Phe/Tyr/Trp variants. c, Diode array detector (DAD) UV-visible absorption spectra to detect the 5-nitrotryptophan (5NT) standard and the nitrated products produced by the TxtE_His176Phe/Tyr/Trp variants. Abbreviation: 4-nitrotryptophan (4NT).
Motivated by these results, we performed site-saturation mutagenesis at His176 to explore the effect any of the remaining amino acid substitutions might have on regioselectivity. While most mutations significantly reduced or completely abolished activity, several variants generated detectable levels of nitrated product(s). Mutation from His176 to asparagine, glycine, serine, cysteine, or methionine resulted in a mixture of 4- and 5-nitro-L-tryptophan (Supplementary Fig. S12–S16), while substitution of His176 with phenylalanine, tyrosine, and tryptophan all resulted in nitration exclusively at the C5-position.
To better understand how the substitutions at residue 176 give rise to improved binding of L-tryptophan and a complete shift in regioselectivity we characterized the His176Phe/Tyr/Trp variants with MD and the His176Phe/Tyr variants with X-ray crystallography. Compared with the nearly 50:50 ratio of closed vs. open states observed in the wild-type TxtE simulations, the computational data of the His176Phe/Tyr/Trp mutants showed a substantial increase in their relative closed state populations with ratios around 90:10 (Table 1). HMM-state decomposition and TPT mapping reveal significant kinetic and thermodynamic effects for each of the C5-selective variants. The larger aromatic side chains of these amino acids allow for tighter packing of the closed-lid F/G loop and provide thermodynamic stabilization compared to the open-lid conformation. The added bulk further increases the contact surface of the F/G loop with neighboring residues, particularly Tyr89, during the closed-to-open transition, leading to an increased kinetic barrier and slowed conformational exchange. Thermodynamic stabilization of the closed-lid conformation and slowed closed-to-open transitions are consistent with the tighter binding constants and lower total turnover numbers (TTNs) determined for the C5-selective variants relative to wild-type TxtE (Table 1). As with His176 in the wild-type closed-lid conformation, the MD simulations show the side chains of the residues at position 176 in the C5-selective variants engaged in edge-to-face interactions with the substrate indole moiety (Fig. 4b–d) and the substrate adopting one of two orientations: a minor conformation in which the indole nitrogen faces the iron center and a major one in which it is flipped. In the former orientation, the substrate alignment does not change relative to the wild-type enzyme.21 The slowed closed-to open transitions also limit the partial substrate release necessary for the flipping of the indole moiety (Supplementary Fig. S4). Further analysis of the substrate populations suggests that the Phe/Tyr/Trp variants favor the catalytically relevant flipped orientation of the substrate indole moiety in the closed-lid conformation to a lesser extent than wild-type TxtE (Table 1). Interestingly, in the flipped scenario, the increased steric demand of Phe/Tyr/Trp at position 176 shifts the substrate alignment relative to the heme co-factor, placing the indole C5 closest to the nitrogen of the ferric peroxynitrite (Fig. 4b–d, 5). In contrast, the wild-type simulations show the C4 position closest to the peroxynitrite, corresponding with the observed regioselectivity (Fig. 5).
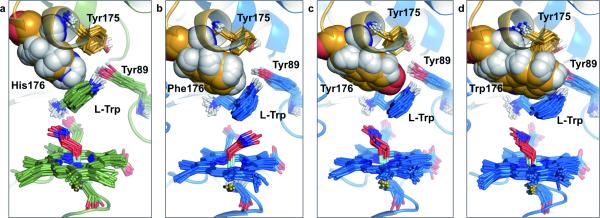
Figure 4. MD simulations show the 5-selective variants in similar edge-to-face interactions with the l-Trp substrate indole-moiety than what is observed with wild-type TxtE.
Compared to the wild-type His176 (a), the increased steric demand of Phe176 (b), Tyr176 (c), and Trp176 (d) causes the substrate indole moiety to adopt a shifted binding orientation that is packed more tightly against the back of the active site, which further determines the orientation of the heme-Fe-bound peroxynitrite (Fig. 5). The substrate indole responds first by adopting a retreated conformation (compare (a) with (b)) and then by assuming an increasingly parallel orientation relative to the plane of the heme cofactor (compare (b) with (c) and (d)). The F/G loop is shown in orange.
Figure 5. Mutation of His176 to Phe fine-tunes substrate-to-peroxynitrite alignment and active site water network.
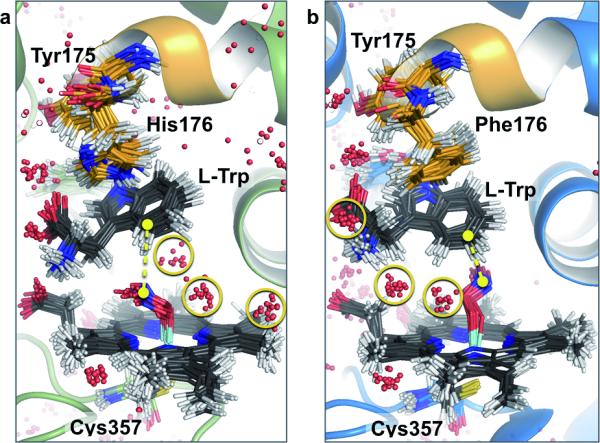
TxtE wild-type simulations predominantly show the C4-carbon of l-Trp proximal to the peroxynitrite-nitrogen, indicated by yellow distance marker (a), while simulations of the His176Phe mutant predominantly show the C5-carbon closest to the peroxynitrite-N (b). The two active sites support a distinct set of structured water molecules (highlighted by yellow circles).
We also obtained high-resolution X-ray crystal structures of the His176Phe and His176Tyr variants bound to L-tryptophan. In contrast to the wild-type TxtE, both of these structures show fully-resolved closed-lid conformation F/G loops, lending support to our hypothesis that these mutations stabilize the closed-lid conformation. The conformation of the F/G loop in these structures shows remarkable agreement with that predicted by our reconstruction and simulation procedures, and both Phe176 and Tyr176 are shown to interact directly with the co-crystallized substrate as predicted (Fig. 6). Other than the F/G loop, no significant structural deviation was observed from the previous co-crystal structure.21 As with the wild-type structure, the co-crystallized tryptophan was found to occupy only the unflipped orientation, most likely due to inherent differences between the crystalline and solution state, as well as the absence of the ferric peroxynitrite intermediate included in our simulations.
Figure 6. Crystal structures of His176Phe/Tyr variants have a resolved closed-lid F/G loop that aligns with the predicted closed-lid MD geometries.
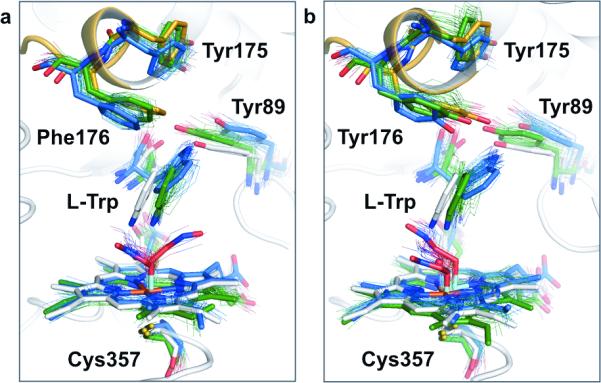
a, His176Phe (crystal structure in white and orange, MD geometries in blue and green). b, His176Tyr (crystal structure in white and orange, MD geometries in blue and green). The crystal structures in (a) and (b) (white) show the F/G loop (orange) in the closed-lid state and further show an active site arrangement that supports the predicted closed-lid MD geometries - for both, the flipped (blue) and unflipped (green) indole-group ensembles. The range of dynamics is indicated through the overlaid hair-lines, corresponding to 20 structures randomly drawn from each of the MD states.
Given the demonstrated control over regioselectivity of the residue at position 176 in the S. scabies TxtE enzyme, we investigated the natural diversity at this position in related enzymes. Seven unique homologs of TxtE were identified using the BLAST search algorithm (sequence identity > 65%), and multiple sequence alignment revealed a natural subpopulation with a tryptophan (but not phenylalanine or tyrosine) at the position corresponding to His176 in four annotated P450s of unknown function from: Streptomyces virginiae (Uniprot ID K9MYX3), Streptomyces sp. Mg1 (B4VER3), Streptomyces lavendulae (I2FGE0), and Saccharomonospora marina XMU15 (H5X7S6) (Fig. 7a, Supplementary Fig. S50–S51). All of the other residues that were identified to bind L-tryptophan in the wild-type structure (PDB ID: 4TPO) are conserved in these P450s, suggesting that they would also accept L-tryptophan as a substrate (Supplementary Fig. S50). We synthesized the genes for these enzymes and expressed them in E. coli. LC-MS analysis and HPLC co-injection demonstrated that, as with the C5-selective His176Trp TxtE variant, all four of the homologous enzymes with tryptophan at the amino acid corresponding to His176 exclusively nitrate L-tryptophan at the C5 position (Fig. 7b and c, Supplementary Fig. S17–S20, Supplementary Fig. S34–S37). It is interesting to note that no natural products have yet been identified with a 5-nitro-L-tryptophan core and that this nitrated building block may be a component of undiscovered natural products. For a point of comparison to S. scabies TxtE_His176Trp, we further characterized the most stable and highly expressed homolog from S. virginiae (Virginiae_TxtE). Virginiae_TxtE produces exclusively 5-nitro-L-tryptophan with a higher turnover and lower affinity than the corresponding His176Trp mutant of S. scabies TxtE (Table 1). Upon mutation of Virginiae_TxtE Trp176 to His176, the nitration activity is significantly attenuated, but the product distribution shifts to favor C4 nitration (Supplementary Fig. S21). These data demonstrate that the relationship between residue 176 and regioselective nitration of tryptophan is preserved across the range of homologous enzymes despite the subtlety of the rearrangement that gives rise to it and the modest overall sequence identity (67–77%).
Figure 7. Identification of naturally-occurring TxtE homologs that catalyze the production of 5-nitrotryptophan.
a, Sequence sub-alignment of the F/G loops of TxtE and seven homologs (sequence identity: 67–77%) reveals natural occurrence of Trp176 in several homologs. Colors in the sequence logo refer to the hydrophobic (black), basic (blue), acidic (red), polar (green), and other (purple) residues. b, LC-MS chromatograms of the nitrated products synthesized by the Trp176-containing TxtE homologs. c, DAD UV-visible absorption spectra to detect the 5-nitrotryptophan (5NT) standard and the nitrated products produced by the Trp176-containing TxtE homologs from S. virginiae, S. sp. Mg1, S. lavendulae, and S. marina XMU15.
Conclusions
In order to create a catalytically productive microenvironment, P450s undergo open-to-closed transitions that involve large, dynamic motions of the F/G loop and adjoining F and G helices.8,11,12 Through a collaborative approach between experiment and simulation, we have identified a single F/G loop residue in the nitrating P450 TxtE that not only plays a key role in gating these transitions, but also acts to control regioselectivity and can be used as a predictor of regioselectivity in homologous nitrating P450s. This approach also showcases how a thorough understanding of structure-function relationships, as obtained through protein engineering efforts, can be a catalyst for the discovery of previously uncharacterized enzymes.44
Taken together, our results demonstrate that TxtE largely controls regioselectivity through precise positioning of the substrate with respect to the ferric peroxynitrite intermediate. TxtE chemistry stands in contrast to peroxidase or globin catalyzed tryptophan nitration, wherein 5-nitrotryptophan is typically observed as the minor product or not at all.41-43,45-48 It has been suggested that the 5-nitrotryptophan regioisomer is produced only in the presence of an electrophilic nitronium ion.42,48 Notably, the production of 5-nitrotryptophan by the His176Phe/Tyr/Trp mutants and the TxtE homologs having tryptophan at the corresponding position makes an electrophilic substitution mechanism a possibility, though a caged radical mechanism could also give rise to the observed behavior.20 The mechanism of TxtE-catalyzed nitration, how TxtE has evolved from its P450 counterparts, and what natural products contain 5-nitro-L-tryptophan are still outstanding questions.
Our study showcases a rare example of how a single residue in the cytochrome P450 F/G loop can interact directly with the substrate to contribute to active site organization.14-16 We speculate that direct interactions between the F/G loop and the substrate could exist, albeit transiently, in other P450s as well. A more rigorous and extensive understanding of the dynamics and stabilization of flexible loops in P450s and other enzymes could be useful towards the engineering of new substrate contacts to alter active site chemistry or even engineer novel enzymatic function.49,50
Methods
Computational details
The high-resolution TxtE co-crystal structure with PDB accession ID 4TPO served as the structural template. Missing residues of the TxtE F/G loop were reconstructed with Schrödinger Inc.'s structure prediction tool Prime. The heme-iron(III)-peroxynitrite complex involved in the TxtE catalyzed aromatic nitration cycle was used to model the active form of TxtE in the presence of L-Trp substrate. Force field parameters were generated with the antechamber module of AMBER12, and production MD simulations were carried out utilizing the GPU-accelerated pmemd.cuda code. One hundred independent production runs were performed for 0.5 microseconds each, utilizing the GPU nodes of NCSA's Blue Waters supercomputer. Mixtape v.0.2.2 (zenodo.org/record/12638#.Vjtef7Rx1N0), which is now part of MSMBuilder 3 (msmbuilder.org), was used to construct a 10 state hidden Markov model (HMMs) of this dataset. 100 new trajectories (10 from each state) were seeded from this state decomposition and were run for 0.2 microseconds each. This equal-weighted adaptive sampling process was performed a total of three times, resulting in an aggregate sampling of 110 microseconds. The final dataset was used to generate a 9 state HMM and its connectivity was examined with transition path theory (TPT). A more detailed description of the computational protocol is available as part of the Supplementary Information.
Purified enzyme reactions
A solution of purified enzyme (150 μL, 2.5 μM in 25 mM Tris pH 8) was aliquoted in quadruplicate into a 96-well plate (Evergreen Scientific). A solution (50 μL) containing the following was added to each well: L-tryptophan (5 μL, 20 mM in 25 mM Tris pH 8, 500 μM final concentration), spinach ferredoxin NADP+ reductase (2 μL, 17 U/mL in 1 M Tris pH 8, 0.17 U/mL final concentration), spinach ferredoxin (2 μL, 1 mg/mL in Tris pH 8, 0.01 mg/mL final concentration), nicotinamide adenine dinucleotide phosphate (NADPH, Codexis, Inc.) (2 μL, 89 mM in 25 mM Tris pH 8, 890 μM final concentration), and diethylamine NONOate sodium salt hydrate (DEANO, 1 μL, 100 mM in 10 mM sodium hydroxide) in 38 μL of 25 mM Tris pH 8. The plate was covered, wrapped in foil, and the reactions proceeded overnight with shaking. To each well, a solution of the internal standard p-toluenesulfonamide (25 μL, 4 mM stock in 0.15 M HCl with 0.4% DMSO, 444 μM final concentration) was added with shaking for 5 min. Each reaction (200 μL) was applied to a 0.5-mL 3-kDA MWCO centrifugal filter (Millipore) and centrifuged at 20,817g for 1 h at room temperature. The filtrate was transferred to a 96-well assay plate (Agilent Technologies) and analyzed by HPLC for total turnover numbers or transferred to a vial insert (Agilent Technologies) and analyzed by liquid chromatography mass spectrometry (LC-MS). The wavelength of maximum absorption for the new nitro product (λmax = 330 nm) was consistent with that reported for 5-nitro-L-tryptophan (λmax = 330 nm)41 and 1-nitro-N-α-acetyl-L-tryptophan amide (λmax = 332 nm).42,43 Since the latter compound is known to be unstable in acidic conditions, treatment of the enzyme-generated products with hydrochloric acid (10 μL, 6M) allowed us to distinguish between 1- and 5-nitro-L-tryptophan.42,43
Dissociation constants
A solution of purified enzyme (100 μL, 4.3–5 μM) in 25 mM Tris buffer (pH 8) was aliquoted in a half area 96 well plate (Greiner Bio One) and L-tryptophan prepared in 25 mM Tris (pH 8) was titrated at varying concentrations. For TxtE_His176Phe and TxtE_His176Trp the following final L-tryptophan concentrations were used: 0, 1, 3, 6, 12, 24, 47, 94, 189, 377, 566, 755, and 1132 μM. For TxtE_His176Tyr the following final L-tryptophan concentrations were used: 0, 3, 6, 12, 24, 47, 94, 189, 377, 566, 755, and 1132 μM. For Virginiae_TxtE the following final L-tryptophan concentrations were used: 0, 24, 47, 189, 377, 566, 755, and 1132 μM. The volume change (6 μL) was the same for each well. After at least 10 min of shaking, spectra were recorded from 350 to 500 nm with a 5-nm step size on a plate reader (Infinite M200, Tecan) at least in triplicate. For each substrate concentration, differential UV-visible spectra were determined by subtracting the enzyme alone control spectrum from the enzyme with substrate. The difference in the absorbance of each spectrum at the λmax and λmin was calculated in at least triplicate and the average with standard deviation was plotted versus the substrate concentration. Each data set was fitted to a binding isotherm model using KaleidaGraph as follows ΔΔA = (ΔΔAmax[L-tryptophan]) / (Kd + [L-tryptophan]) (Supplementary Fig. S49).
We note that we are characterizing the recombinant TxtE outside its native context and without its native reductase or nitric oxide source. Instead a surrogate spinach reductase system and an artificial nitric oxide donor with a short half-life are used as previously reported.20 Under these reaction conditions, TxtE activity is low and measurements of kcat, KM, and specific activity are not possible. Instead, as is common for cytochrome P450s, we have reported total turnover numbers and dissociation constants, which are useful for assessing relative activities.
Additional materials and procedures are described in the Supplementary Information.
Supplementary Material
Acknowledgments
We thank Dr. Jens Kaiser and Pavle Nikolovski of the Beckman Molecular Observatory (Caltech) for assistance with crystallography and Dr. Scott Virgil and the 3CS Center for Catalysis and Chemical Synthesis (Caltech) for assistance with LC-MS analyses. This work was funded by the Gordon and Betty Moore Foundation through grant GBMF2809 to the Caltech Programmable Molecular Technology Initiative (to F.H.A). S.C.D is supported by Ruth L. Kirschstein NRSA postdoctoral fellowship from the National Institutes of Health (5F32GM106618). G.K. acknowledges support from the Lawrence Scholars Program, the NIH Simbios Program (U54 GM072970), and the Center for Molecular Analysis and Design (Stanford). J.K.B.C. acknowledges the support of the Resnick Sustainability Institute (Caltech). The Beckman Molecular Observatory is supported by the Gordon and Betty Moore Foundation, the Beckman Institute, and the Sanofi-Aventis Bioengineering Research Program (Caltech). The authors thank Dr. Sabine Brinkmann-Chen, Dr. Todd K. Hyster, Dr. John A. McIntosh, Dr. Christopher K. Prier, Robert T. McGibbon, and Mohammad M. Sultan for helpful discussions. This research is part of the Blue Waters sustained-petascale computing project, which is supported by the National Science Foundation (awards OCI-0725070 and ACI-1238993) and the state of Illinois. Correspondence and material requests should be addressed to F.H.A. for experimental work and V.S.P. for computational work. The content of this paper is solely the responsibility of the authors and does not represent the official views of any of the funding agencies.
Footnotes
Author contributions
S.C.D. and G.K. contributed equally to this work. S.C.D. and G.K. designed the research. S.C.D., G.K., J.K.B.C., and Y.S. performed research. F.H.A and V.S.P. supervised and provided advice. S.C.D., G.K., and J.K.B.C. analyzed the data. S.C.D., G.K., J.K.B.C., and F.H.A wrote the text and conceived the figures with input from all of the authors.
References
- 1.Meunier B, de Visser SP, Shaik S. Mechanism of oxidation reactions catalyzed by cytochrome P450 enzymes. Chem. Rev. 2004;104:3947–3980. doi: 10.1021/cr020443g. [DOI] [PubMed] [Google Scholar]
- 2.Shaik S, de Visser SP. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. Ortiz de Montellano PR, editor. Vol. 2. Plenum; New York: 2005. pp. 45–85. [Google Scholar]
- 3.Whitehouse CJ, Bell SG, Wong LL. P450(BM3) (CYP102A1): connecting the dots. Chem. Soc. Rev. 2012;41:1218–1260. doi: 10.1039/c1cs15192d. [DOI] [PubMed] [Google Scholar]
- 4.Podust LM, Sherman DH. Diversity of P450 enzymes in the biosynthesis of natural products. Nat. Prod. Rep. 2012;29:1251–1266. doi: 10.1039/c2np20020a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S, et al. Substrate recognition by the multifunctional cytochrome P450 MycG in mycinamicin hydroxylation and epoxidation reactions. J. Biol. Chem. 2012;287:37880–37890. doi: 10.1074/jbc.M112.410340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denisov IG, Sligar SG. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. Ortiz de Montellano PR, editor. Springer; Cham: 2015. pp. 69–109. Ch. 3. [Google Scholar]
- 7.Ortiz de Montellano PR. In: Cytochrome P450 Structure, Mechanism, and Biochemistry. Ortiz de Montellano PR, editor. Springer; Cham: 2015. pp. 111–176. Ch. 4. [Google Scholar]
- 8.Pochapsky TC, Kazanis S, Dang M. Conformational plasticity and structure/function relationships in cytochromes P450. Antiox. Redox Signal. 2010;13:1273–1296. doi: 10.1089/ars.2010.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poulos TL. In: Fifty years of Cytochrome P450 Research. Yamazaki H, editor. Springer; 2014. pp. 75–94. [Google Scholar]
- 10.Poulos TL. In: Fifty years of Cytochrome P450 Research. Yamazaki H, editor. Springer; Tokyo: 2014. pp. 75–94. Ch. 4. [Google Scholar]
- 11.Hasemann CA, Kurumbali RG, Boddupalli SS, Peterson JA, Diesenhofer J. Structure and function of cytochromes P450: a comparitive analysis of three crystal structures. Structure. 1995;2:41–62. doi: 10.1016/s0969-2126(01)00134-4. [DOI] [PubMed] [Google Scholar]
- 12.Poulos TL. Cytochrome P450 flexibility. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13121–13122. doi: 10.1073/pnas.2336095100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fasan R, Chen MM, Crook NC, Arnold FH. Engineered alkane-hydroxylating cytochrome P450(BM3) exhibiting nativelike catalytic properties. Angew. Chem. 2007;46:8414–8418. doi: 10.1002/anie.200702616. [DOI] [PubMed] [Google Scholar]
- 14.Yano JK, et al. Crystal structure of a thermophilic cytochrome P450 from the archaeon Sulfolobus solfataricus. J. Biol. Chem. 2000;275:31086–31092. doi: 10.1074/jbc.M004281200. [DOI] [PubMed] [Google Scholar]
- 15.Kells PM, Ouellet H, Santos-Aberturas J, Aparicio JF, Podust LM. Structure of cytochrome P450 PimD suggests epoxidation of the polyene macrolide pimaricin occurs via a hydroperoxoferric intermediate. Chem. Biol. 2010;17:841–851. doi: 10.1016/j.chembiol.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, et al. Structural analysis of HmtT and HmtN involved in the tailoring steps of himastatin biosynthesis. FEBS Lett. 2013;587:1675–1680. doi: 10.1016/j.febslet.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 17.Buddha MR, Tao T, Parry RJ, Crane BR. Regioselective nitration of tryptophan by a complex between bacterial nitric-oxide synthase and tryptophanyl-tRNA synthetase. J. Biol. Chem. 2004;279:49567–49570. doi: 10.1074/jbc.C400418200. [DOI] [PubMed] [Google Scholar]
- 18.Winkler R, Hertweck C. Biosynthesis of nitro compounds. Chembiochem. 2007;8:973–977. doi: 10.1002/cbic.200700042. [DOI] [PubMed] [Google Scholar]
- 19.Ju KS, Parales RE. Nitroaromatic compounds, from synthesis to biodegradation. Microbiol. Mol. Bio. Rev. 2010;74:250–272. doi: 10.1128/MMBR.00006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barry SM, et al. Cytochrome P450-catalyzed L-tryptophan nitration in thaxtomin phytotoxin biosynthesis. Nat. Chem. Biol. 2012;8:814–816. doi: 10.1038/nchembio.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dodani SC, et al. Structural, functional, and spectroscopic characterization of the substrate scope of the novel nitrating cytochrome P450 TxtE. Chembiochem. 2014;15:2259–2267. doi: 10.1002/cbic.201402241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Kenaan C, Hamdane D, Hoa GH, Hollenberg PF. Effect of conformational dynamics on substrate recognition and specificity as probed by the introduction of a de novo disulfide bond into cytochrome P450 2B1. J. Biol. Chem. 2009;284:25678–25686. doi: 10.1074/jbc.M109.032748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sano E, et al. Mechanism of the decrease in catalytic activity of human cytochrome P450 2C9 polymorphic variants investigated by computational analysis. J Comput. Chem. 2010;31:2746–2758. doi: 10.1002/jcc.21568. [DOI] [PubMed] [Google Scholar]
- 24.Hendrychova T, et al. Flexibility of human cytochrome P450 enzymes: molecular dynamics and spectroscopy reveal important function-related variations. Biochim. Biophys. Acta. 2011;1814:58–68. doi: 10.1016/j.bbapap.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 25.Hendrychova T, Berka K, Navratilova V, Anzenbacher P, Otyepka M. Dynamics and hydration of the active sites of mammalian cytochromes P450 probed by molecular dynamics simulations. Curr. Drug Metab. 2012;13:177–189. doi: 10.2174/138920012798918408. [DOI] [PubMed] [Google Scholar]
- 26.Miao Y, et al. Coupled flexibility change in cytochrome P450cam substrate binding determined by neutron scattering, NMR, and molecular dynamics simulation. Biophys. J. 2012;103:2167–2176. doi: 10.1016/j.bpj.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pulawski W, et al. Low-temperature molecular dynamics simulations of horse heart cytochrome c and comparison with inelastic neutron scattering data. Eur. Biophys. J. 2013;42:291–300. doi: 10.1007/s00249-012-0874-9. [DOI] [PubMed] [Google Scholar]
- 28.Cong S, Ma XT, Li YX, Wang JF. Structural basis for the mutation-induced dysfunction of human CYP2J2: a computational study. J. Chem. Inf. Model. 2013;53:1350–1357. doi: 10.1021/ci400003p. [DOI] [PubMed] [Google Scholar]
- 29.Cui YL, et al. Molecular dynamic investigations of the mutational effects on structural characteristics and tunnel geometry in CYP17A1. J. Chem. Inf. Model. 2013;53:3308–3317. doi: 10.1021/ci400553w. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi K, et al. Evaluation of influence of single nucleotide polymorphisms in cytochrome P450 2B6 on substrate recognition using computational docking and molecular dynamics simulation. PLoS One. 2014;9:e96789. doi: 10.1371/journal.pone.0096789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hollingsworth SA, Poulos TL. Molecular dynamics of the P450cam-Pdx complex reveals complex stability and novel interface contacts. Protein Sci. 2015;24:49–57. doi: 10.1002/pro.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan JR, Zheng QC, Cui YL, Li WK, Zhang HX. Investigation of ligand selectivity in CYP3A7 by molecular dynamics simulations. J. Biomol. Struct. Dyn. 2015:1–8. doi: 10.1080/07391102.2015.1054884. [DOI] [PubMed] [Google Scholar]
- 33.Roccatano D. Structure, dynamics, and function of the monooxygenase P450 BM-3: insights from computer simulations studies. J. Phys. Condens. Matter. 2015;27:273102. doi: 10.1088/0953-8984/27/27/273102. [DOI] [PubMed] [Google Scholar]
- 34.Narayan AR, et al. Enzymatic hydroxylation of an unactivated methylene C-H bond guided by molecular dynamics simulations. Nat. Chem. 2015;7:653–660. doi: 10.1038/nchem.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui YL, et al. Structural features and dynamic investigations of the membrane-bound cytochrome P450 17A1. Biochim. Biophys. Acta. 2015;1848:2013–2021. doi: 10.1016/j.bbamem.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 36.Noé F, Prinz JH. In: An Introduction to Markov State Models and Their Application to Long Timescale Molecular Simulation Vol. 797 Advances In Experimental Medicine and Biology. Pande Vijay S., Bowman Gregory R., Noé Frank., editors. Springer; Dordrecht: 2014. pp. 75–90. Ch. 6. [Google Scholar]
- 37.Bowman GR, Ensign DL, Pande VS. Enhanced modeling via network theory: Adaptive sampling of Markov state models. J. Chem. Theory Comput. 2010;6:787–794. doi: 10.1021/ct900620b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGibbon RT, Ramsundar B, Sultan MM, Kiss G, Pande VS. Understanding protein dynamics with L1-regularzied reversible hiddne markov models. arXiv. 2014 [Google Scholar]
- 39.Yu F, et al. Structural insights into the mechanism for recognizing substrate of the cytochrome P450 enzyme TxtE. PLoS One. 2013;8:e81526. doi: 10.1371/journal.pone.0081526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanden-Eijnden E. In: Computer Simulations in Condensed Matter Systems: From Materials to Chemical Biology Vol. 703 Lecture Notes in Physics. Ferrario M, Ciccotti G, Binder K, editors. Springer; Verlag: 2006. pp. 453–493. [Google Scholar]
- 41.Herold S, Shivashankar K, Mehl M. Myoglobin scavenges peroxynitrite without being significantly nitrated. Biochemistry. 2002;41:13460–13472. doi: 10.1021/bi026046h. [DOI] [PubMed] [Google Scholar]
- 42.Sala A, Nicolis S, Roncone R, Casella L, Monzani E. Peroxidase catalyzed nitration of tryptophan derivatives. Mechanism, products and comparison with chemical nitrating agents. Eur. J. Biochem. 2004;271:2841–2852. doi: 10.1111/j.1432-1033.2004.04219.x. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki T, et al. Nitration and nitrosation of N-acetyl-L-tryptophan and tryptophan residues in proteins by various reactive nitrogen species. Free Radical Biol. Med. 2004;37:671–681. doi: 10.1016/j.freeradbiomed.2004.05.030. [DOI] [PubMed] [Google Scholar]
- 44.Brinkmann-Chen S, Cahn JK, Arnold FH. Uncovering rare NADH-preferring ketol-acid reductoisomerases. Metab. Eng. 2014;26c:17–22. doi: 10.1016/j.ymben.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Padmaja S, Ramazenian MS, Bounds PL, Koppenol WH. Reaction of peroxynitrite with L-tryptophan. Redox Rep. 1996;2:173–177. doi: 10.1080/13510002.1996.11747045. [DOI] [PubMed] [Google Scholar]
- 46.Herold S. Nitrotyrosine, dityrosine, and nitrotryptophan formation from metmyoglobin, hydrogen peroxide, and nitrite. Free Radical Biol. Med. 2004;36:565–579. doi: 10.1016/j.freeradbiomed.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 47.Roncone R, Barbieri M, Monzani E, Casella L. Reactive nitrogen species generated by heme proteins: mechanism of formation and targets. Coord. Chem. Rev. 2006;250:1286–1293. [Google Scholar]
- 48.Nuriel T, Hansler A, Gross SS. Protein nitrotryptophan: formation, significance and identification. J. Proteomics. 2011;74:2300–2312. doi: 10.1016/j.jprot.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 50.Nestl BM, Hauer B. Engineering of flexible loops in enzymes. ACS Catal. 2014;4:3201–3211. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.