Abstract
The PR-domain containing 16 (Prdm16) protein is a powerful inducer of the thermogenic phenotype in fat cells. In both developmental (brown) and induced (beige) thermogenic adipose tissue, Prdm16 has a critical role in maintaining proper tissue structure and function. It has roles throughout the course of differentiation, beginning with lineage determination activity in precursor cells, and continuing with coactivator functions that enable and maintain thermogenic gene expression. These abilities are primarily mediated by interactions with other adipogenic factors, suggesting that Prdm16 acts to coordinate the overall brown adipose phenotype. Mouse models have confirmed that thermogenic adipose depends upon Prdm16, and that this type of fat tissue provides substantial metabolic protection against the harmful effects of a high fat/high energy diet. Activation of Prdm16, therefore, holds promise for stimulating thermogenesis in fat cells to reduce human obesity and its complications.
Keywords: adipogenesis, BAT, beige/brown/white adipose tissue, obesity, Prdm16, thermogenesis, WAT
Abbreviation
- AD
activation domain
- BAT
brown adipose tissue
- HKMT
histone lysine methyltransferase
- KO
knockout
- MiR
micro-RNA
- Prdm16
PR domain containing 16
- PRR
proximal regulatory region
- RD
repression domain
- WAT
white adipose tissue
- ZF
zinc-finger
Introduction
The identification of Prdm16 (PR domain containing 16) as a key transcriptional regulator of brown adipose identity1 led to important advances in our understanding of brown and beige fat biology. Previously, no other transcription factors were known to provide proper specification of the brown fat lineage. The ability of Prdm16 to activate the full thermogenic genetic program in white adipose and even non-adipose cells opened up opportunities to explore the molecular development of brown adipose tissue. This also revitalized hope that methods could be developed to activate thermogenic fat in people as a means to reduce obesity and associated metabolic dysfunction. Over the past several years, the roles of Prdm16 and its mechanisms of action in fat cells have been studied in greater detail. While the biology has (predictably) not been quite so straightforward, Prdm16 continues to occupy a central role in regulating the development and function of thermogenic adipose tissue. Here, we discuss our current understanding of Prdm16 function and the insights into thermogenic fat biology that have resulted.
Prdm16 in Brown Adipogenesis
At least three major types of adipose cells are commonly considered. White adipocytes represent the majority of the adult adipose mass and are vastly expanded (in size and number) in obesity. The cells are typified by a single large unilocular lipid droplet, minimal cytoplasm, and are specialized for efficiently storing energy reserves as lipid until required. Brown adipocytes, in contrast, have many small (multi-)locular lipid droplets, cytoplasm densely packed with mitochondria, and express the protein Uncoupling protein-1 (Ucp1) in their mitochondrial inner membranes (reviewed by Cannon and Nedergaard2). Ucp1 is the definitive marker of thermogenic fat cells, as it is physically responsible for leaking protons across the inner mitochondrial membrane to disconnect the electron transport chain from ATP synthesis, with heat as the product. Whereas brown adipose tissue is derived embryonically and maintains a relatively consistent phenotype under different conditions, the third class of fat cells (variably entitled beige, brite, or iBAT) are inducible and exhibit thermogenic characteristics when they appear within white adipose tissue following challenges such as cold exposure. These 3 types of fat cells (white, brown and beige) appear to have distinct developmental origins, and each expresses certain “signature” genes that distinguish it from the others.3-10
Brown and beige fat activity can act in rodents to limit the weight (fat) gain caused by overeating.11-13 This apparent counter-regulatory mechanism, known as diet-induced thermogenesis, is somehow activated by nutritional status or specific nutrients.11 In addition to this natural defense mechanism, engineering animals to have more thermogenic fat mass also staves off obesity (reviewed by Harms and Seale14). Importantly, the thermogenic fat mass in adult humans is positively correlated with leanness and metabolic health15,16 and can be activated.17 It therefore seems reasonable to believe that the ongoing studies of thermogenesis in animal models will have eventual clinical application.
The molecular basis for the phenotypic differences between white adipose and brown adipose cells has been a long-standing question. The transcription factor Pparγ (Peroxisome-proliferator activator receptor gamma) is the master regulator of adipogenesis in all types of fat cells.18-20 Other DNA-binding factors and co-regulators provide for certain thermogenic-specific functions that distinguish brown from white fat. For instance, CCAAT/enhancer binding protein (C/ebp)-α and -β activate expression of Pparγ to regulate adipose differentiation21 and were known to regulate Ucp1 expression as early as 1994.22 Pparα is highly expressed in brown adipose and participates in the expression of many thermogenic genes, including Pgc1α and Prdm16.23 Pparγ coactivator 1α (Pgc1α), stimulates mitochondrial biogenesis, and its increased expression and activity provides for the very high density of mitochondria that are found in brown and beige fat cells.24,25 The winged-helix protein forkhead box C2 (Foxc2) can also induce parts of a brown fat phenotype.26 These factors, however, are insufficient on their own to coordinate all aspects of the brown adipose phenotype.
In a search for determinants of brown fat cells, Prdm16 was identified as a transcriptional factor having highly enriched expression in brown fat relative to visceral white fat tissue.1 Mochizuki et al.27 and Nishikata et al.28 had previously isolated Prdm16 in searches for a gene affected by the reciprocal translocation t(1;3)(p36,q21) linked to myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML). In an adipose context, Prdm16 was robustly expressed in BAT (versus WAT) but was not induced by cold exposure of mice, suggesting a role controlling rather participating in the thermogenic program.1 Strikingly, ectopic expression of Prdm16 induced a comprehensive brown fat program in adipocytes including mitochondrial biogenesis via Pgc1a and cyclic-AMP signal responsiveness; but also repressed WAT-selective markers.1 Conversely, knockdown of Prdm16 prevented expression of the thermogenic program without affecting overt adipogenesis.1 Many of these activities were DNA-binding independent in mutation studies.1 This pattern of expression and activities positioned Prdm16 as a determinant of brown adipose identity.
BAT Determination vs. Skeletal Muscle and WAT
Two of the most striking observations in subsequent experiments with Prdm16 in adipogenesis were that Prdm16 could convert determined myoblasts into brown adipocytes, and that loss of Prdm16 in brown preadipocytes resulted in a myogenic phenotype.4 A common developmental precursor for BAT, dermis, and skeletal muscle had been first suggested by En1 fate mapping experiments.29 Lineage tracing using genetic tracing with markers from the skeletal myogenesis field (Myf5Cre; Pax7CreER) produced labeling of BAT and skeletal muscle.4,30 These results suggested that there was a potential bipotent precursor during embryogenesis that gave rise to both BAT and skeletal muscle, and that Prdm16 acted as a master switch between the two lineages (reviewed by Sanchez-Gurmaches and Guertin31).
Additional evidence has reinforced the notion that BAT and muscle share common origins and traits. For instance, global gene expression analyses showed that brown fat precursors and skeletal muscle cells express many common mRNA and miRNAs.32,33 Consistent with this, mitochondria from BAT were substantially more similar to those from skeletal muscle than those from WAT.34 In mice, inactivation of Myogenin, an essential driver of muscle cell differentiation, resulted in a rather dramatic expansion of BAT.35 Conversely, loss of several genes, including Ewing sarcoma (EWS)36 and Ehmt1,37 results in a loss of BAT phenotype and an upregulation of muscle genes.
Several specific MiRs tied to Prdm16 have also been implicated in enforcing a fate choice between brown adipose and myogenic lineages. MiR-193b and -635 were found to be upregulated by Prdm16, acting through Pparα, and their expression could repress muscle gene expression in myoblasts.38 Conversely, loss of MiR-193b/635 impaired adipogenesis while increasing myogenic RNA levels.38 In contrast, MiR-133 expression is enriched in myoblasts and is a repressor of brown adipose fate, directly targeting the 3’ untranslated region of the Prdm16 transcript.39,40 MiR-133 is temperature-regulated, and acute cold exposure of animals decreases its expression as Prdm16 and Ucp1 levels increase.39,40 This is consistent with β-adrenergic signaling inhibiting expression of the Mef2 transcription factors upstream of MiR-133.39,41 In vivo antagonism of MiR-133 in myogenic satellite cells resulted in the appearance of muscle-derived Prdm16-positive brown adipocytes embedded between muscle fibers.40 Collectively, therefore, MiRs regulating and regulated by Prdm16 can function to enforce negative and positive BAT lineage choices.
While it remains clear that there is a common progenitor for the two lineages in early development, the relationship between them may not be quite as simple as a binary fate switch hinged on Prdm16 expression. Deletion of Prdm16 in vivo produces a loss of BAT phenotype, but approaches WAT in appearance and does not show induction of myogenic markers.42 Expression of the Myf5Cre and Pax7CreER transgenes occurs in the embryo prior to myogenic determination and may not necessarily reflect the endogenous gene expression pattern. Therefore, the common precursor may be further upstream and multipotent rather than bipotent. Indeed, Myf5Cre tracing studies show that dermis is also labeled along with BAT and muscle.6 Nonetheless, manipulations around Prdm16 suggest that adult progenitors for skeletal muscle and brown fat retain at least some degree of intrinsic lineage bipotency.37,40
In vivo, there appears to be biological redundancy at work that is absent in cell culture models (Fig. 1). A Prdm16 BAT-selective knockout (KO) model, using the Myf5Cre driver to delete Prdm16 loxP-conditional alleles, showed a delayed rather than complete loss of thermogenic function that became apparent by three months of age.42 Concurrent loss of the closely related gene Prdm3 (also known as MECOM for Mds1/Evi1-COMplex) accelerated the deterioration, with defects appearing earlier between one and two months of age.42 However, BAT initially formed, suggesting that there is additional redundancy during embryogenesis and neonatally to allow for BAT formation. Loss of euchromatic histone-lysine-N-methyltransferase 1 (Ehmt1), a Prdm16-interacting partner, is associated with a dramatic block in brown adipose development which includes elevated skeletal muscle gene transcripts.37 Therefore, Ehmt1 may be a common component able to interact with several Prdm-family factors, including Prdm16 and Prdm3.42 Nonetheless, loss of Ehmt1 does not cause ectopic skeletal muscle differentiation in BAT,37 indicating that the cells had been committed to the adipose lineage.
Figure 1.
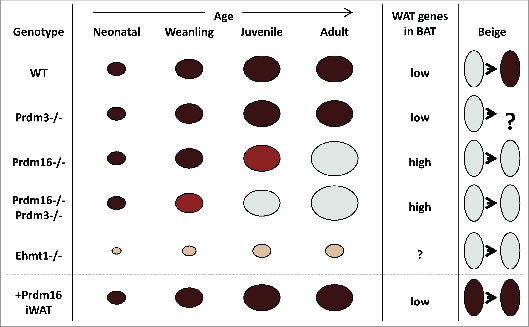
Prdm16 in BAT determination. Prdm16 is essential for maintaining BAT in adult mice. However, redundancy exists for BAT determination at earlier stages of development, although not for suppression of WAT-selective genes. The redundant factor during juvenile and early adult periods is Prdm3. The basis for compensation neonatally remains unknown. In contrast, the Prdm16-interacting protein Ehmt1 is required at all stages, suggesting that it may act as a common partner to several Prdm family members to allow for BATogenesis. Ectopic expression of Ap2-Prdm16 in WAT results in results in a beige fat pad.
In cell culture, genetic loss of Prdm16 prevents expression of the thermogenic phenotype but does not inhibit general adipogenesis,42 showing that the isolated cells have either already been determined to become adipocytes, or that they possess redundancy for the adipogenic aspects of Prdm16 (such as the induction and coactivation of Pparγ). The thermogenic deficiency due to the absence of Prdm16 is apparent in isolated precursors from all ages (embryonic, neonatal, adult) however, and is therefore cell autonomous and contrasts with the delayed tissue phenotype in vivo.42 In neither case is there significant upregulation of myogenic factors, suggesting that the shRNA knockdown of Prdm16 may have additional effects (such as impairment of Pparγ activity) that are necessary for myogenesis to ensue. Indeed, in vitro CRISPR-based knockdown of Prdm16 (in lieu of shRNA) produced results that agree with the genetic knockout, yielding adipogenesis without thermogenic gene expression (JI, unpublished).
The model that emerges, then, is that Prdm16 is sufficient for generating a thermogenic adipocyte but is context-dependent in its necessity. Through its ability to induce Pparγ expression, repress muscle transcripts, and activate thermogenic genes, Prdm16 can reprogram an amenable cell such as a myoblast. However, loss of Prdm16 is inadequate to convert an in vivo brown adipocyte into a myoblast, likely because genes such as Pparγ and C/ebpα/β have been activated by other means and are active in maintaining adipose identity. Embryonically, BAT still appears in the absence of both Prdm16 and Prdm3, suggesting that there remains an unidentified participant early in development.
Prdm16 in Beige Adipose
Some of the thermogenic adipose depots that have been detected in adult humans resemble mouse beige or “inducible” brown fat rather than developmental BAT.3,43 While the distribution of classic brown vs. beige type fat cells in various human depots is not completely settled,43-46 there is unquestionably a therapeutic allure to the idea of inducing a thermogenic phenotype in white fat. This concept has been explored in several mouse models, showing that Prdm16 is an essential factor for making beige fat.
Wildtype subcutaneous depots have low but detectable levels of Prdm16 and acquire a thermogenic phenotype in response to environmental (cold) or pharmacological (CL316243, B3-adrenergic agonist; thiazolidinedione, Pparγ ligand) challenge. Beige induction by rosiglitizone (thiazolidinedione) is thought to be partly mediated by stabilization of Prdm16 protein.47 Additionally, rosiglitazone promotes the SirT1-mediated deacetylation of Pparγ which, in turn, increases its interaction with Prdm16 to drive tissue beigeing.48 Adipose-specific expression of Prdm16 in mice was sufficient to induce beigeing in subcutaneous fat pads, which provided the animals with increased insulin sensitivity and resistance to high fat diet-provoked weight gain.49 There were notable differences in response to ectopic Prdm16 between different depots that correlated with their pre-existing susceptibility to beigeing. In particular, Prdm16 induced a strong thermogenic phenotype in the inguinal subcutaneous depot whereas visceral depots were largely unresponsive.49 BAT, having high levels of endogenous Prdm16, was neither helped nor hindered by having more Prdm16. Prdm16 protein levels followed the phenotypes despite robust RNA expression in all depots, suggesting that visceral depots translationally or post-translationally down-regulate Prdm16 protein in a way that others do not.49 These results showed that Prdm16 was sufficient to produce beigeing in WAT that was metabolically beneficial for the animals.
The requirement for Prdm16 in beige adipose function was demonstrated by Cohen et al. using a tissue-selective knockout mouse.50 With an Adiponectin-Cre driver, Prdm16 was inactivated specifically in adipose tissues. BAT was largely unaffected by the deletion; in contrast to the results of Harms et al.,42 this may be due to the use of a different Cre-expressing transgene that is expressed in differentiating cells rather than in precursors. Deletion of Prdm16 from WAT, however, resulted in a loss of beigeing ability within the subcutaneous depots after cold exposure or CL316243 injection.50 The mice were able to maintain a normal body temperature in cold conditions, likely due to their intact BAT depots and intact shivering response. However, when subjected to a high-fat diet, the knockout animals gained slightly more weight than their controls. Interestingly, despite a relatively subtle increase in fat mass, the Prdm16 adipose-knockout mice developed quite a profound degree of insulin resistance caused predominantly by inefficient suppression of hepatic glucose production. Prdm16-null subcutaneous WAT assumed a molecular profile similar to visceral WAT, including an inflammatory signature linked to Wilm's tumor-1 (Wt1) expression.50 Taken together, these data illustrate that beige adipose (as defined by the proportion of Ucp1+ cells) affects overall metabolism, and requires Prdm16 to do so.
Prdm16 Structure and Function
The Prdm16 protein can be divided into six major domains (Fig. 2). At the N-terminus is the PR(/SET) domain, the namesake feature of the Prdm family for homology to similar domains in Prdm1 (PRD1-BF1/Blimp1) and Prdm2 (Riz1), and similar to the SET domain in histone lysine methyltransferase (HKMT) proteins. Following the PR domain is a cluster of seven C2H2-type zinc-finger motifs that constitute zinc-finger domain-1 (ZF1), which is capable of sequence-specific DNA binding and through which many protein-protein interactions occur. An ill-defined proximal regulatory region (PRR) then occurs prior to a repression domain (RD) containing several critical and well-defined interaction motifs. A subsequent second zinc-finger domain (ZF2) is composed of three additional C2H2-type zinc-fingers and participates in DNA binding and protein interactions. Finally, an acidic activation domain (AD) is designated toward the C-terminus of the protein. Before, between, and after these domains are several additional stretches of between 80 and 150 amino acids for which clear purposes have not been defined.
Figure 2.
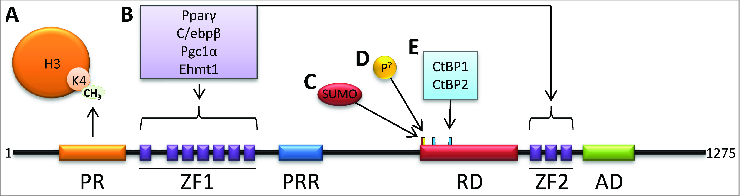
Prdm16 domain structure. Prdm16 can be partitioned into six major regions, including a PR/SET domain (PR); two zinc-finger domain containing seven or three C2H2 zinc finger motifs (ZF1/ZF2); a proximal regulatory region (PRR); a repression domain (RD); a second zinc-finger domain containing three C2H2 zinc finger motifs (ZF2); and an acidic activation domain (AD). (A) The PR domain has Histone-H3 lysine-4 (H3K4) monomethylase activity. (B) ZF1 and ZF2 have protein-protein interactions with a variety of factors. (C–E) Specific sequence motifs regulate Prdm16-mediated gene repression. (C) Sumoylation enhances interactions with CtBP. (D) By homology to MECOM, Prdm16 is potentially phosphorylated at Ser758. (E) Of two consensus CtBP motifs, the second is functional and required for interaction with CtBP1 and CtBP2.
The PR domain spans approximately 125 amino acids and, despite being a prominent feature in the protein, has been dispensable in deletion experiments for adipogenic and thermogenic induction by Prdm16.1,51 The PR domain has HKMT activity and was seen to provide the initial histone-3/lysine-9 (H3K9) mono-methylation event required for subsequent methylations to dimethyl and trimethyl status by other factors.52 Curiously, this function was demonstrated to occur cytoplasmically, which contrasts with our observations of strict nuclear localization for Prdm16 in the context of brown adipose cells (PS, unpublished). That the PR domain is not essential in vitro for brown adipogenesis51 may indicate that PR domain function and cellular localization is lineage-specific. Alternatively, a low level of Prdm16 protein may be cytoplasmic in brown adipose yet remain unremarked compared to the very high levels in the nucleus. Additionally, redundant HKMTs52 could obscure this Prdm16 function if the residual HKMT activity is sufficient to allow the specific brown-adipogenic properties of Prdm16(ΔPR) to emerge. Therefore, expression of Prdm16(ΔPR) in a cell system disabled for H3K9 monomethylation could help to demonstrate a requirement for the PR domain in thermogenic differentiation.
The Prdm16 ZF1 is an essential part of the protein.51 ZF1 has sequence-specific DNA binding activity,28 but it is unclear if direct DNA binding by Prdm16 is occurring at critical target genes in the context of brown adipogenesis.1 Despite extensive ChIP-seq studies of Prdm16 in brown adipose, the in vitro Prdm16 binding site has not emerged as a consensus motif from the statistical analysis of the enriched loci (M. Harms & PS, unpublished). Several explanations could cooperate to explain this. Firstly, Prdm16 may directly bind DNA but only at a subset of loci that are hidden among other events. Secondly, the binding sites may be degenerate and therefore difficult to detect. In this case, different DNA binding modes between ZF1, ZF2, or ZF1+ZF2 would add additional complexity. Thirdly, and particularly relevant at key thermogenic genes, Prdm16 has key protein-protein interactions through its ZF domains with DNA-binding factors such as Pparγ/α4 and C/ebpα/β/δ51. Indeed, Prdm16 has been shown to enhance the transcriptional function of C/ebp and Ppar factors as well as the coactivators Pgc1α and Pgc1β, though the mechanism by which this occurs is not yet clear.1,4,51 Together, the data suggest that Prdm16 is a key component of an activating transcriptional complex at thermogenic genes, with DNA binding provided by one or more other complex members.
A repressive function for Prdm16 in brown adipocytes has been mapped to the RD domain, and specifically to a C-terminal binding protein (CtBP) interaction within this region.53 Expression of Prdm16 in white adipocytes strongly represses white-specific genes such as Angiotensinogen (Agt) and Resistin (Retn), and BAT tissue from Prdm16-KO animals exhibits strongly upregulated expression of these genes.42 This pattern of Prdm16 regulation is absolutely dependent on its interaction with CtBP1 or CtBP2 through the second of two 5-amino acid CtBP-interaction motifs in Prdm16.53 Interestingly, Prdm16 can be sumoylated at a four amino acid motif at the N-terminal end of the RD and just before the first CtBP motif. In the context of myeloid differentiation, sumoylation of Prdm16 at this site enhances its interaction with CtBP, and repression of target genes is impaired if sumoylation is prevented.54 By homology to Prdm3, there is also a potential phosphorylatable residue (Ser758) for casein kinase II/protein phosphatase-1α between the SUMO and CtBP sites that could participate in this regulation.55
The relative roles of Ehmt1 and CtBP in mediating the repressive functions of Prdm16 are unclear. Both Ehmt1 and CtBP directly interact with Prdm16 to repress non-BAT gene expression.37,53 However, Ehmt1-linked repression was largely described for myogenic genes,37 whereas CtBP was linked to repression of the WAT-selective gene program. It has not been clearly established if these are actual separable functions of Prdm16, or if both muscle and WAT tissue signatures are being suppressed simultaneously, and it remains to be tested if the two activities cooperate or if they act independently. Use of the Prdm16(ΔCtBP) mutants in conjunction with Ehmt1 disruption could provide insight into their respective mechanisms of action.
Relatively little is specifically known about the PRR, ZF2, or AD domains. The ZF2 domain has been shown in vitro to provide DNA binding activity.28 For the same reasons as for ZF1 in brown adipogenesis, however, it is not obvious if or how DNA binding by ZF2 is required. Proteins that interact with ZF2 also appear to interact with ZF1, albeit independently since the binding still occurs when isolated domains are expressed in vitro.4,51 Further work will need to be done to discover how these domains contribute to Prdm16 function.
Within the Prdm family, the most closely related protein to Prdm16 is Prdm3, with which it shares a nearly identical exon/intron organization and domain structure, and overall 52% amino acid identity and 79% conservation. Notably, the conserved domains show even higher similarity - for instance, the ZF1 domain is 92% similar between proteins. Both PR domains of have H3K9 monomethylation activity52; and the PR-domain-lacking isoform of Prdm3 (also known as Evi1) is also adipogenic like Prdm16(ΔPR).56 The two proteins have very similar consensus DNA binding sites for both ZF1 and ZF2.28,57,58 Prdm3 has similar interactions to Prdm16, including with C/ebpβ56 and CtBP.59,60 Indeed, Prdm3 provides some redundancy for Prdm16 in brown adipogenesis, as its loss in a double-knockout (Myf5Cre;Prdm16;Prdm3) mouse drastically accelerates the BAT deterioration phenotype from loss of Prdm16.42 Nonetheless, the individual loss of Prdm16 in BAT produces a loss-of-function with age that does not occur without Prdm3 and in vitro expression of Prdm16 produces a more robust browning effect than overexpression of Prdm3.42 Repression of white adipose genes (e.g., Agt, Retn) is also a distinct capability of Prdm16 that is absent from Prdm3.42 These latter observations imply that, despite qualitative overlap in many functions, Prdm16 has specialized brown adipogenic properties for which Prdm3 cannot substitute.
Prdm16 in Non-Adipose Systems
Expression of Prdm16 is observed in a variety of tissues with little obvious phenotypic similarity to BAT, including brain, heart, kidney, lung, uterus, and testis.1,27 We do not yet understand how Prdm16 is modified by context to provide such a variety of functions. One can envision mechanisms including physical interactions with various tissue-specific factors, differential signaling and protein modifications, unique chromatin landscapes, and combinations of these influences that make Prdm16 multifunctional. In some cases, however, there may be a common underlying pattern that is not yet clear, or that has remained unremarked next to more prominent features. Studies from other systems, therefore, will be informative for ruling in or out specific Prdm16 mechanisms in BAT.
Prdm16 is essential for maintaining certain stem cell populations in the haematopoietic and nervous systems.61 In those tissues, it was found to modulate oxidative stress, in part by regulating expression of Hgf.61 Loss of Prdm16 increased reactive oxygen species, cell death, and depletion of stem cell numbers.61 Whether Prdm16 has a similar role in adipose progenitors has not been described; however, regulation of protective activity against ROS in an intensely energetic tissue such as BAT would not be unexpected.
The Drosophila gene Hamlet is homologous to mammalian Prdm16, and several Hamlet interactions may potentially be informative of Prdm16 function. Induction of Hamlet expression in response to activity of the Swi/Snf chromatin remodelling complex was shown to be important for regulating the progressive differentiation of neural progenitors and preventing dedifferentiation.62 Hamlet also participates in Notch signaling to regulate fate choice in olfactory neurogenesis.63 The relevance of the Hamlet work to mammals is supported by a recent study showing that inhibition of Notch signaling in mouse WAT resulted in upregulation of Prdm16 and adipose tissue beigeing.64 Whether there are conserved mechanisms and protein domains underpinning these functions remains to be determined.
Questions and Directions
Prdm16 is a multifunctional protein having effects in brown/beige fat cell determination and differentiation, and in inhibiting gene expression of alternative lineages (Fig. 3). For instance, Prdm16 acts with C/ebpβ as an inducer of Pparγ and adipogenesis,51 but with Ehmt1 to inhibit myogenic gene expression.37 However, the known constellation factors are presently insufficient to explain the specific BAT effects of Prdm16. Identification of molecular markers for BAT that are unaffected by Pparγ disruption6 will provide leverage for refining our understanding of the how Prdm16 induces the earliest events adipogenesis. As well, the relationships between Prdm16 and other brown-adipose inducing factors, particularly the required lineage commitment factor Early B-cell factor 2 (Ebf2),65 remain poorly understood. Assembling these parts into a coherent mechanism will also give us a better grasp of the events involved in inducing and maintaining a thermogenic phenotype.
Figure 3.
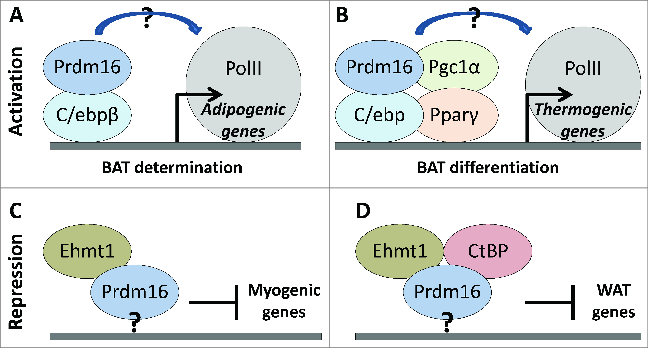
Model of gene regulation by Prdm16. Questions remain to be answered regarding the specific mechanisms by which Prdm16 selects between target gene (A and B) activation or (C and D) repression; recognizes target gene regulatory elements; and interacts with co-regulatory proteins. (A) Prdm16 and C/ebpβ cooperate to activate adipogenic gene expression. (B) In differentiation, some (or all) of Prdm16, C/ebp, Pparγ, and Pgc1α assemble a transactivating complex at thermogenic genes. How either complex (A) or (B) interacts with RNA polymerase II (PolII) to stimulate transcription remains to be described. (C) Myogenic genes are repressed by Prdm16 and Ehmt1. (D) WAT genes are repressed by Prdm16+Ehmt1 and Prdm16+CtBP; it is unclear if the complex involves Prdm16+Ehmt1+CtBP. It is unclear how sequence-specific DNA binding of the Prdm16 complex occurs at repressive gene elements (C and D).
Stabilization and maintenance of thermogenic genes in an accessible and prepared state might be one purpose for Prdm16 in differentiated brown adipocytes. Even in the absence of a continuous inductive signal (e.g., cold exposure), BAT remains phenotypically brown and physiologically poised in an inactive-but-ready state. Cold, however, is an acute and deadly threat, and having a reserve of primed BAT may be essential for survival until additional thermoprotective mechanisms can be recruited. Without Prdm16 and Prdm3, this stability is lost in adult animals and the BAT becomes dysfunctional and WAT-like.42 Ectopic expression of Prdm16 in subcutaneous WAT results in beigeing,49 although it is not yet clear if this conversion requires an initiating thermogenic stimulation to occur. This possible maintenance function of Prdm16 should be explored, being relevant to therapeutic applications in which a stable thermogenic fat pad would be preferable to transient beigeing.
The thermogenic gene program is intact in newborn animals lacking both Prdm16 and Prdm3, suggesting that redundant or alternative mechanisms exist to defend BAT mass against Prdm16 and Prdm3 loss when mice are most vulnerable to cold.42 Nonetheless, Prdm16 has (in our hands) a stronger pro-thermogenic activity, and whatever alternatives exist then fail or are lost after several months. Interaction with Ehmt137 may be a common feature of the Prdm16-redundant factors allowing for pro-thermogenic differentiation. The lack of an overt phenotype in Prdm16/Prdm3 double-null embryonic BAT is highly suggestive of an unknown third factor that is active or available only in early development.
An alternative hypothesis could involve redundancy that is not intracellular but rather reflects the presence of multiple progenitor cell populations. This would invoke distinct Prdm-independent embryonic (“developmental”) BAT stem cells that are progressively supplanted post-weaning by Prdm-dependent adult adipose stem cells. This model would be reminscent of the waves of embryonic, fetal, and later myoblasts that populate developing skeletal muscle (reviewed in Hollway & Curry66). Notably, however, cultured embryonic brown preadipocytes exhibited similar Prdm16-dependent defects to those observed from adults, suggesting that in vivo environment may be modifying a cell-autonomous phenotype.42
Repression of typical white-selective adipose genes, however, is a purely Prdm16-dependent function.42 Prdm3 has primarily white-adipose inducing activities,56 consistent with robust expression of the white-selective markers repressed by Prdm16 in BAT. Considering the domain-level structural similarity between the two proteins (Fig. 1), including the presence of identical CtBP-interaction in both, this contrast suggests that there are other unique molecular properties that could involve selective protein-protein or protein-DNA interactions. Direct comparison of these factors to tease apart their molecular similarities and differences will be insightful for constructing a model of brown adipose differentiation. Furthermore, the robustness of this repressive Prdm16 function suggests that the expression of these white-selective genes is purposeful, although we do not yet know why.
Conclusion
Our improved understanding of how Prdm16 acts in thermogenic fat cells is providing new insight into the origins, differentiation, and maintenance of brown and beige adipose. Ongoing investigations of Prdm16's molecular mechanisms and its collaborations with interacting factors will deepen our abilities to manipulate this tissue. In the face of alarming obesity-related challenges, the idea of using thermogenic fat therapeutically remains promising, feasible, and necessary.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Matthew Harms and Suzanne Shapira for assistance with review and editing of the text.
Funding
PS is supported by NIGMS/NIH award DP2OD007288 and by a Searle Scholars Award.
References
- 1. Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell Metab 2007; 6:38-54; PMID:17618855; http://dx.doi.org/ 10.1016/j.cmet.2007.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev 2004; 84:277-359; PMID:14715917; http://dx.doi.org/ 10.1152/physrev.00015.2003 [DOI] [PubMed] [Google Scholar]
- 3. Wu J, Boström P, Sparks LM, Ye L, Choi JH, Giang A-H, Khandekar M, Virtanen KA, Nuutila P, Schaart G, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 2012; 150:366-76; PMID:22796012; http://dx.doi.org/ 10.1016/j.cell.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scimè A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fatskeletal muscle switch. Nature 2008; 454:961-7; PMID:18719582; http://dx.doi.org/ 10.1038/nature07182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Long JZ, Svensson KJ, Tsai L, Zeng X, Roh HC, Kong X, Rao RR, Lou J, Lokurkar I, Baur W, et al. A smooth muscle-like origin for beige adipocytes. Cell Metab 2014; 19:810-20; PMID:24709624; http://dx.doi.org/ 10.1016/j.cmet.2014.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang W, Kissig M, Rajakumari S, Huang L, Lim H-W, Won K-J, Seale P. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc Natl Acad Sci U S A 2014; 111:14466-71; PMID:25197048; http://dx.doi.org/ 10.1073/pnas.1412685111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chau Y-Y, Bandiera R, Serrels A, Martínez-Estrada OM, Qing W, Lee M, Slight J, Thornburn A, Berry R, McHaffie S, et al. Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat Cell Biol 2014; 16:367-75; PMID:24609269; http://dx.doi.org/ 10.1038/ncb2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu W, Shan T, Yang X, Liang S, Zhang P, Liu Y, Liu X, Kuang S. A heterogeneous lineage origin underlies the phenotypic and molecular differences of white and beige adipocytes. J Cell Sci 2013; 126:3527-32; PMID:23781029; http://dx.doi.org/ 10.1242/jcs.124321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gupta RK, Mepani RJ, Kleiner S, Lo JC, Khandekar MJ, Cohen P, Frontini A, Bhowmick DC, Ye L, Cinti S, et al. Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell Metab 2012; 15:230-9; PMID:22326224; http://dx.doi.org/ 10.1016/j.cmet.2012.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tran K-V, Gealekman O, Frontini A, Zingaretti MC, Morroni M, Giordano A, Smorlesi A, Perugini J, De Matteis R, Sbarbati A, et al. The vascular endothelium of the adipose tissue gives rise to both white and brown fat cells. Cell Metab 2012; 15:222-9; PMID:22326223; http://dx.doi.org/ 10.1016/j.cmet.2012.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rothwell NJ, Stock MJ. A role for brown adipose tissue in diet-induced thermogenesis. Nature 1979; 281:31-5; PMID:551265; http://dx.doi.org/ 10.1038/281031a0 [DOI] [PubMed] [Google Scholar]
- 12. Lowell BB, V SS, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, Kozak LP, Flier JS. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature 1993; 366:740-2; PMID:8264795; http://dx.doi.org/ 10.1038/366740a0 [DOI] [PubMed] [Google Scholar]
- 13. Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab 2009; 9:203-9; PMID:19187776; http://dx.doi.org/ 10.1016/j.cmet.2008.12.014 [DOI] [PubMed] [Google Scholar]
- 14. Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med 2013; 19:1252-63; PMID:24100998; http://dx.doi.org/ 10.1038/nm.3361 [DOI] [PubMed] [Google Scholar]
- 15. Van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JMAFL, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJJ. Cold-activated brown adipose tissue in healthy men. N Engl J Med 2009; 360:1500-8; PMID:19357405; http://dx.doi.org/ 10.1056/NEJMoa0808718 [DOI] [PubMed] [Google Scholar]
- 16. Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med 2009; 360:1509-17; PMID:19357406; http://dx.doi.org/ 10.1056/NEJMoa0810780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cypess AM, Chen Y-C, Sze C, Wang K, English J, Chan O, Holman AR, Tal I, Palmer MR, Kolodny GM, et al. Cold but not sympathomimetics activates human brown adipose tissue in vivo. Proc Natl Acad Sci U S A 2012; 109:10001-5; PMID:22665804; http://dx.doi.org/ 10.1073/pnas.1207911109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev 1994; 8:1224-34; PMID:7926726; http://dx.doi.org/ 10.1101/gad.8.10.1224 [DOI] [PubMed] [Google Scholar]
- 19. Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell 1999; 4:585-95; PMID:10549290; http://dx.doi.org/ 10.1016/S1097-2765(00)80209-9 [DOI] [PubMed] [Google Scholar]
- 20. Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Spiegelman BM, Mortensen RM. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell 1999; 4:611-7; PMID:10549292; http://dx.doi.org/ 10.1016/S1097-2765(00)80211-7 [DOI] [PubMed] [Google Scholar]
- 21. Wu Z, Xie Y, Bucher NL, Farmer SR. Conditional ectopic expression of CEBP beta in NIH-3T3 cells induces PPAR gamma and stimulates adipogenesis. Genes Dev 1995; 9:2350-63; PMID:7557387; http://dx.doi.org/ 10.1101/gad.9.19.2350 [DOI] [PubMed] [Google Scholar]
- 22. Yubero P, Manchado C, Cassard-Doulcier AM, Mampel T, Viñas O, Iglesias R, Giralt M, Villarroya F. CCAATenhancer binding proteins alpha and beta are transcriptional activators of the brown fat uncoupling protein gene promoter. Biochem Biophys Res Commun 1994; 198:653-9; PMID:8297376; http://dx.doi.org/ 10.1006/bbrc.1994.1095 [DOI] [PubMed] [Google Scholar]
- 23. Hondares E, Rosell M, Díaz-Delfín J, Olmos Y, Monsalve M, Iglesias R, Villarroya F, Giralt M. Peroxisome proliferator-activated receptor α (PPARα) induces PPARγ coactivator 1α (PGC-1α) gene expression and contributes to thermogenic activation of brown fat: involvement of PRDM16. J Biol Chem 2011; 286:43112-22; PMID:22033933; http://dx.doi.org/ 10.1074/jbc.M111.252775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 1998; 92:829-39; PMID:9529258; http://dx.doi.org/ 10.1016/S0092-8674(00)81410-5 [DOI] [PubMed] [Google Scholar]
- 25. Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab 2006; 3:333-41; PMID:16679291; http://dx.doi.org/ 10.1016/j.cmet.2006.04.002 [DOI] [PubMed] [Google Scholar]
- 26. Cederberg A, Gronning LM, Ahren B, Tasken K, Carlsson P, Enerback S. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell 2001; 106:563-73; PMID:11551504; http://dx.doi.org/ 10.1016/S0092-8674(01)00474-3 [DOI] [PubMed] [Google Scholar]
- 27. Mochizuki N, Shimizu S, Nagasawa T, Tanaka H, Taniwaki M, Yokota J, Morishita K. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood 2000; 96:3209-14; PMID:11050005 [PubMed] [Google Scholar]
- 28. Nishikata I, Sasaki H, Iga M, Tateno Y, Imayoshi S, Asou N, Nakamura T, Morishita K. A novel EVI1 gene family, MEL1, lacking a PR domain (MEL1S) is expressed mainly in t(1;3)(p36;q21)-positive AML and blocks G-CSF-induced myeloid differentiation. Blood 2003; 102:3323-32; PMID:12816872; http://dx.doi.org/ 10.1182/blood-2002-12-3944 [DOI] [PubMed] [Google Scholar]
- 29. Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev Biol 2006; 296:164-76; PMID:16730693; http://dx.doi.org/ 10.1016/j.ydbio.2006.04.449 [DOI] [PubMed] [Google Scholar]
- 30. Lepper C, Fan C-M. Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genes N Y N 2000 2010; 48:424-36; PMID:20641127; http://dx.doi.org/ 10.1002/dvg.20630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sanchez-Gurmaches J, Guertin DA. Adipocyte lineages: tracing back the origins of fat. Biochim Biophys Acta 2014; 1842:340-51; PMID:23747579; http://dx.doi.org/ 10.1016/j.bbadis.2013.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Walden TB, Timmons JA, Keller P, Nedergaard J, Cannon B. Distinct expression of muscle-specific microRNAs (myomirs) in brown adipocytes. J Cell Physiol 2009; 218:444-9; PMID:18937285; http://dx.doi.org/ 10.1002/jcp.21621 [DOI] [PubMed] [Google Scholar]
- 33. Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, Hamilton DL, Gimeno RE, Wahlestedt C, Baar K, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci U S A 2007; 104:4401-6; PMID:17360536; http://dx.doi.org/ 10.1073/pnas.0610615104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Forner F, Kumar C, Luber CA, Fromme T, Klingenspor M, Mann M. Proteome differences between brown and white fat mitochondria reveal specialized metabolic functions. Cell Metab 2009; 10:324-35; PMID:19808025; http://dx.doi.org/ 10.1016/j.cmet.2009.08.014 [DOI] [PubMed] [Google Scholar]
- 35. Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 1993; 364:501-6; PMID:8393145; http://dx.doi.org/ 10.1038/364501a0 [DOI] [PubMed] [Google Scholar]
- 36. Park JH, Kang HJ, Kang SI, Lee JE, Hur J, Ge K, Mueller E, Li H, Lee B-C, Lee SB. A multifunctional protein, EWS, is essential for early brown fat lineage determination. Dev Cell 2013; 26:393-404; PMID:23987512; http://dx.doi.org/ 10.1016/j.devcel.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature 2013; 504:163-7; PMID:24196706; http://dx.doi.org/ 10.1038/nature12652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sun L, Xie H, Mori MA, Alexander R, Yuan B, Hattangadi SM, Liu Q, Kahn CR, Lodish HF. Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol 2011; 13:958-65; PMID:21743466; http://dx.doi.org/ 10.1038/ncb2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Trajkovski M, Ahmed K, Esau CC, Stoffel M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat Cell Biol 2012; 14:1330-5; PMID:23143398; http://dx.doi.org/ 10.1038/ncb2612 [DOI] [PubMed] [Google Scholar]
- 40. Yin H, Pasut A, Soleimani VD, Bentzinger CF, Antoun G, Thorn S, Seale P, Fernando P, van Ijcken W, Grosveld F, et al. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab 2013; 17:210-24; PMID:23395168; http://dx.doi.org/ 10.1016/j.cmet.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci U S A 2007; 104:20844-9; PMID:18093911; http://dx.doi.org/ 10.1073/pnas.0710558105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Harms MJ, Ishibashi J, Wang W, Lim H-W, Goyama S, Sato T, Kurokawa M, Won K-J, Seale P. Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metab 2014; 19:593-604; PMID:24703692; http://dx.doi.org/ 10.1016/j.cmet.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sharp LZ, Shinoda K, Ohno H, Scheel DW, Tomoda E, Ruiz L, Hu H, Wang L, Pavlova Z, Gilsanz V, et al. Human BAT possesses molecular signatures that resemble beigebrite cells. PloS One 2012; 7:e49452; PMID:23166672; http://dx.doi.org/ 10.1371/journal.pone.0049452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee P, Swarbrick MM, Zhao JT, Ho KKY. Inducible brown adipogenesis of supraclavicular fat in adult humans. Endocrinology 2011; 152:3597-602; PMID:21791556; http://dx.doi.org/ 10.1210/en.2011-1349 [DOI] [PubMed] [Google Scholar]
- 45. Cypess AM, White AP, Vernochet C, Schulz TJ, Xue R, Sass CA, Huang TL, Roberts-Toler C, Weiner LS, Sze C, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat Med 2013; 19:635-9; PMID:23603815; http://dx.doi.org/ 10.1038/nm.3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jespersen NZ, Larsen TJ, Peijs L, Daugaard S, Homøe P, Loft A, de Jong J, Mathur N, Cannon B, Nedergaard J, et al. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supraclavicular region of adult humans. Cell Metab 2013; 17:798-805; PMID:23663743; http://dx.doi.org/ 10.1016/j.cmet.2013.04.011 [DOI] [PubMed] [Google Scholar]
- 47. Ohno H, Shinoda K, Spiegelman BM, Kajimura S. PPARγ agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab 2012; 15:395-404; PMID:22405074; http://dx.doi.org/ 10.1016/j.cmet.2012.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, Rosenbaum M, Zhao Y, Gu W, Farmer SR, et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell 2012; 150:620-32; PMID:22863012; http://dx.doi.org/ 10.1016/j.cell.2012.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest 2011; 121:96-105; PMID:21123942; http://dx.doi.org/ 10.1172/JCI44271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cohen P, Levy JD, Zhang Y, Frontini A, Kolodin DP, Svensson KJ, Lo JC, Zeng X, Ye L, Khandekar MJ, et al. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell 2014; 156:304-16; PMID:24439384; http://dx.doi.org/ 10.1016/j.cell.2013.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-CEBP-beta transcriptional complex. Nature 2009; 460:1154-8; PMID:19641492; http://dx.doi.org/ 10.1038/nature08262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pinheiro I, Margueron R, Shukeir N, Eisold M, Fritzsch C, Richter FM, Mittler G, Genoud C, Goyama S, Kurokawa M, et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 2012; 150:948-60; PMID:22939622; http://dx.doi.org/ 10.1016/j.cell.2012.06.048 [DOI] [PubMed] [Google Scholar]
- 53. Kajimura S, Seale P, Tomaru T, Erdjument-Bromage H, Cooper MP, Ruas JL, Chin S, Tempst P, Lazar MA, Spiegelman BM. Regulation of the brown and white fat gene programs through a PRDM16CtBP transcriptional complex. Genes Dev 2008; 22:1397-409; PMID:18483224; http://dx.doi.org/ 10.1101/gad.1666108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nishikata I, Nakahata S, Saito Y, Kaneda K, Ichihara E, Yamakawa N, Morishita K. Sumoylation of MEL1S at lysine 568 and its interaction with CtBP facilitates its repressor activity and the blockade of G-CSF-induced myeloid differentiation. Oncogene 2011; 30:4194-207; PMID:21516122; http://dx.doi.org/ 10.1038/onc.2011.132 [DOI] [PubMed] [Google Scholar]
- 55. Bard-Chapeau EA, Gunaratne J, Kumar P, Chua BQ, Muller J, Bard FA, Blackstock W, Copeland NG, Jenkins NA. EVI1 oncoprotein interacts with a large and complex network of proteins and integrates signals through protein phosphorylation. Proc Natl Acad Sci U S A 2013; 110:E2885-94; PMID:23858473; http://dx.doi.org/ 10.1073/pnas.1309310110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ishibashi J, Firtina Z, Rajakumari S, Wood KH, Conroe HM, Steger DJ, Seale P. An Evi1-CEBPβ complex controls peroxisome proliferator-activated receptor γ2 gene expression to initiate white fat cell differentiation. Mol Cell Biol 2012; 32:2289-99; PMID:22473998; http://dx.doi.org/ 10.1128/MCB.06529-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Delwel R, Funabiki T, Kreider BL, Morishita K, Ihle JN. Four of the seven zinc fingers of the Evi-1 myeloid-transforming gene are required for sequence-specific binding to GA(CT)AAGA(TC)AAGATAA. Mol Cell Biol 1993; 13:4291-300; PMID:8321231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Funabiki T, Kreider BL, Ihle JN. The carboxyl domain of zinc fingers of the Evi-1 myeloid transforming gene binds a consensus sequence of GAAGATGAG. Oncogene 1994; 9:1575-81; PMID:8183551 [PubMed] [Google Scholar]
- 59. Izutsu K, Kurokawa M, Imai Y, Maki K, Mitani K, Hirai H. The corepressor CtBP interacts with Evi-1 to repress transforming growth factor beta signaling. Blood 2001; 97:2815-22; PMID:11313276; http://dx.doi.org/ 10.1182/blood.V97.9.2815 [DOI] [PubMed] [Google Scholar]
- 60. Palmer S, Brouillet JP, Kilbey A, Fulton R, Walker M, Crossley M, Bartholomew C. Evi-1 transforming and repressor activities are mediated by CtBP co-repressor proteins. J Biol Chem 2001; 276:25834-40; PMID:11328817; http://dx.doi.org/ 10.1074/jbc.M102343200 [DOI] [PubMed] [Google Scholar]
- 61. Chuikov S, Levi BP, Smith ML, Morrison SJ. Prdm16 promotes stem cell maintenance in multiple tissues, partly by regulating oxidative stress. Nat Cell Biol 2010; 12:999-1006; PMID:20835244; http://dx.doi.org/ 10.1038/ncb2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Eroglu E, Burkard TR, Jiang Y, Saini N, Homem CCF, Reichert H, Knoblich JA. SWISNF complex prevents lineage reversion and induces temporal patterning in neural stem cells. Cell 2014; 156:1259-73; PMID:24630726; http://dx.doi.org/ 10.1016/j.cell.2014.01.053 [DOI] [PubMed] [Google Scholar]
- 63. Endo K, Karim MR, Taniguchi H, Krejci A, Kinameri E, Siebert M, Ito K, Bray SJ, Moore AW. Chromatin modification of Notch targets in olfactory receptor neuron diversification. Nat Neurosci 2012; 15:224-33; PMID:22197833; http://dx.doi.org/ 10.1038/nn.2998 [DOI] [PubMed] [Google Scholar]
- 64. Bi P, Shan T, Liu W, Yue F, Yang X, Liang X-R, Wang J, Li J, Carlesso N, Liu X, et al. Inhibition of Notch signaling promotes browning of white adipose tissue and ameliorates obesity. Nat Med 2014; 20:911-8; PMID:25038826; http://dx.doi.org/ 10.1038/nm.3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rajakumari S, Wu J, Ishibashi J, Lim H-W, Giang A-H, Won K-J, Reed RR, Seale P. EBF2 determines and maintains brown adipocyte identity. Cell Metab 2013; 17:562-74; PMID:23499423; http://dx.doi.org/ 10.1016/j.cmet.2013.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hollway G, Currie P. Vertebrate myotome development. Birth Defects Res Part C Embryo Today Rev 2005; 75:172-9; PMID:16187310; http://dx.doi.org/ 10.1002/bdrc.20046 [DOI] [PubMed] [Google Scholar]


