Abstract
Human cytochrome P450 enzymes (P450s, CYPs) are major oxidative catalysts that metabolize various xenobiotic and endogenous compounds. Many carcinogens induce cancer only after metabolic activation and P450 enzymes play an important role in this phenomenon. P450 1B1 mediates bioactivation of many procarcinogenic chemicals and carcinogenic estrogen. It catalyzes the oxidation reaction of polycyclic aromatic carbons, heterocyclic and aromatic amines, and the 4-hydroxylation reaction of 17β-estradiol. Enhanced expression of P450 1B1 promotes cancer cell proliferation and metastasis. There are at least 25 polymorphic variants of P450 1B1 and some of these have been reported to be associated with eye diseases. In addition, P450 1B1 polymorphisms can greatly affect the metabolic activation of many procarcinogenic compounds. It is necessary to understand the relationship between metabolic activation of such substances and P450 1B1 polymorphisms in order to develop rational strategies for the prevention of its toxic effect on human health.
Keywords: Cytochrome P450 1B1, Cancer activation, Polymorphism
INTRODUCTION
Cytochrome P450 enzymes (P450s, CYPs) are heme-thiolate monooxygense enzymes found in a variety of living organisms including animals, fungi, bacteria, and plants (1). These are the major catalysts of oxidative metabolism of xenobiotic chemicals, and therefore have been a subject of numerous toxicological, pharmacological, and drug metabolism studies (2,3).
In 1958, Klingenberg first discovered a pigment in the liver microsomes that binds CO and demonstrates strong absorbance at 450 nm (4,5). Four years later, Omura and Sato reported additional properties of this biochemical system and coined the name “cytochrome P450” for “Pigment 450” (5,6). Early work on P450 proteins was carried out with human tissue samples (7,8). For the several human P450 enzymes, cDNA molecules were cloned in 1980s and the recombinant P450 enzymes were heterologously expressed and purified in mammalian, yeast, and bacterial systems in late 1980s and early 1990s (9).
The human genome includes 57 P450 genes (http://drnelson.uthsc.edu/cytochromeP450.html) and among them, about 15 are considered to be primary catalysts of xenobiotic metabolism (10). There are three enzymes in the human P450 1 family including P450 1A1, 1A2, and 1B1. This review will focus on metabolic importance of human P450 1B1 and the role of its polymorphisms in cancer development.
HUMAN CYTOCHROME P450 1B1
P450 1B1 was originally found in cultures of keratinocytes as a new dioxin-inducible gene (11). In humans, P450 1B1 is expressed primarily in the kidney, spleen, thymus, prostate, lung, intestine, and colon but barely detectable in liver (12,13). Moreover, high P450 1B1 expression has been observed in various hormone-mediated cancers, such as breast, ovarian, endometrial, and prostate cancers (14).
P450 1B1 has never been purified from human tissues (12), but Guengerich and colleagues were able to express and characterize the recombinant P450 1B1 enzyme (15). P450 1A1, 1A2, and 1B1 share a common transcriptional regulation through the aryl hydrocarbon (Ah) system, which is comprised of the aryl hydrocarbon receptor (AhR) and AhR nuclear translocator (ARNT) (16). The P450 1B1 gene is transcriptionally activated when a ligand binds to the cytoplasmic AhR complex (17). The ligand bound AhR translocates to the nucleus, where it forms a heterodimer complex with ARNT (17). This tertiary complex of the ligand (e.g., dioxin), AhR and ARNT subsequently binds to enhancer sites of the P450 1B1 gene and opens up the chromatin structure, so that transcription factors can bind to the promoter and cause more rapid transcription (17).
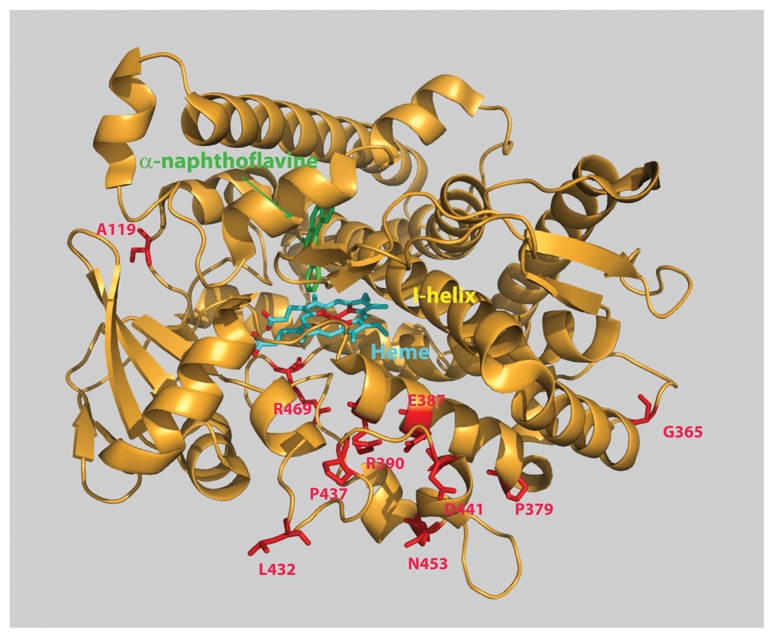
The X-ray crystal structure of P450 1B1 in the complex with α-naphthoflavone was determined by Eric Johnson’s group (Fig. 1) (18). The narrow substrate binding cavity of P450 1B1 contains a slot-like active site, which is well adapted to bind to its characteristic substrates that possess hydrophobic and planar aromatic ring structures (18). The Phe231 residue of P450 1B1 induces a distortion of the F-helix accommodating for π-π stacking with α-naphthoflavone, a feature also present in the structure of P450 1A2 (Fig. 1) (18).
Fig. 1.
The X-ray crystal structure of P450 1B1 in the complex with α-naphthoflavone (18). Polymorphic residues are indicated in red color.
O-deethylation of 7-ethoxyresorufin (EROD) is used as a model reaction catalyzed by P450 1B1. The catalytic activity P450 1B1 is similar to that of P450 1A1 and 1A2 (12). The roles of P450 1B1 in the bioactivation of a very broad spectrum of chemical carcinogens including polycyclic hydrocarbons, heterocyclic amines, aromatic amines, and nitropolycyclic hydrocarbons have been well documented (12,19) (Table 1). An E. coli strain overexpressing P450 and N-acetyltransferase has been used to characterize the chemical genotoxicity of P450 enzymes (20,21). A P450 1B1-based genotoxicity system has been utilized to characterize potent inhibition of the enzyme by tetramethylstilbene (22).
Table 1.
Chemical carcinogens metabolically activated by human P450 1B1a
| Polycyclic aromatic carbons | Heterocyclic amines | Aromatic amines | Nitropolycyclic hydrocarbons | Estrogens |
|---|---|---|---|---|
| Benzo[a]pyrene | MeIQ | 2-Aminoanthracene | 1-Nitropyrene | 17β-Estradiol |
| Dibenzo[a,l]pyrene | MeIQx | 2-Aminofluorene | 2-Nitropyrene | Estrone |
| Benzo[a]anthracene | IQ | 4-Aminobiphyenyl | 6-Nitrochryrene | |
| Dimethylbenz[a]anthtracene | Trp-P1 | O-Aminoazotoluene | 2-Nitrofluoranthene | |
| Benzo[c]phenanthrene-3,4-diol | Trp-P2 | 6-Aminochrysene | 1,8-Dinitropyrene | |
| 5-Methylchrysene | PhIP | 1-Aminopyrene |
(12)
ASSOCIATIONS OF CANCER AND DISEASES
P450 1B1 activity has been under a close experimental scrutiny because this enzyme mediates metabolic bioactivation of many chemical carcinogens (Table 1). This functional feature of P450 1B1 helps to understand its role in cancer initiation and progression. As a recognized driver of cancer development, P450 1B1 has been considered a promising cancer biomarker and a potential target for anticancer therapy. In humans, P450 1A1 and 1A2 are believed to be the major enzymes that catalyze the activation of procarcinogenic polycyclic aromatic hydrocarbons (PAHs). However, several studies have established that P450 1B1 also plays a very important role in metabolic activation of PAHs (13,23,24). There have been several reports about the effects of the P450 1B1 gene knockout in vivo. In mice with a knockout of the P450 1B1 gene, lower rates of tumor growth and elevated protection against DNA adduct formation were observed when carcinogenic agents such as 7, 12-dimethylbenz[a]anthracene and dibenzo[a,l]pyrene were administered (23,25,26). In addition, P450 1B1 gene-knockout mice displayed attenuated tumor tissue metastasis induced by benzo[a]pyrene (27). These reports strongly imply that the mechanisms of P450 1B1-induced cell proliferation, migration, and invasion may have a considerable preclinical and clinical significance.
P450 1B1 is an efficient catalyst for estrogen hydroxylation. It catalyzes the 4-hydroxylation reaction of 17β-estradiol (E2) that produces the less active metabolite, 4-hydroxyestradiol (12,17,28,29). P450 1A2 and 3A4 can hydroxylate 17β-estradiol but the major hydroxylated product is 2-hydroxyestradiol (30). 4-Hydroxyestradiol is believed to cause estrogen-dependent tumors (12). It can be converted to chemically more reactive species, quinones and semiquinones, which covalently bind DNA (31). Ortiz de Montellano’s group reported that the mutation of Val395 to Leu in human P450 1B1 changed the specificity of the 17β-estradiol reaction from 4-hydroxylation to 2-hydroxylation (32), which suggested that estradiol carcinogenicity of P450 1B1 depends on that single amino acid residue (32).
Chun and colleagues previously described a selective and potent inhibitor of P450 1B1 and its utility in preventing cancer development (22). Presence of 2,4,3′,5′-tetramethoxystilbene (TMS) inhibited EROD activity of P450 1B1 with an IC50 value of 6 nM, demonstrating a 500-fold selectivity over P450 1A2. TMS also strongly and selectively inhibited 4-hydroxylation of 17β-estradiol by P450 1B1-expressing membranes (IC50 90 nM) or purified P450 1B1 (IC50 390 nM) (22,33). These studies showed that TMS is a selective and potent competitive inhibitor of P450 1B1 and can be considered as a prospective preventive agent for estrogen-dependent tumors. The molecular mechanism of cancer cell proliferation by P450 1B1 has been revealed recently. The transcription factor Sp1, involved in cell growth and metastasis, was found to be positively regulated by P450 1B1 (unpublished data). It is likely that P450 1B1 promotes cell proliferation and metastasis by inducing the epithelial-mesenchymal transition (EMT) and Wnt/β-catenin signaling via Sp1 induction (unpublished data). This study suggests that Sp1 acts as a key mediator in the promotion of cancer cell proliferation and metastasis stimulated by P450 1B1.
P450 1B1 POLYMORPHISMS
Genetic polymorphisms can dramatically alter specific actions of affected proteins. In particular, polymorphisms in genes encoding P450 enzymes have a considerable impact on the fate of xenobiotics, since these subtle DNA changes represent the most frequent cause of variations in oxidative metabolism of drugs or bioactivation of toxicants (5).
To date, at least 25 allelic variants of P450 1B1 have been identified, all of which are located in the coding region (Table 2) (http://www.cypalleles.ki.se/). Stoilov et al. first identified 19 allelic variants of P450 1B1 during the search for genetic variations associated with primary congenital glaucoma (34,35). P450 1B1-deficient mice exhibit abnormalities in their ocular drainage structure and trabecular meshwork that are similar to those reported in human primary congenital glaucoma patients (36). The mechanism of these impairments is still unclear but P450 1B1 is possibly involved in the metabolism of steroids, retinol and retinal, arachidonate, and melatonin (12,36).
Table 2.
Allelic variants of CYP1B1 with polymorphisms located in the coding region
| Allelic variants | Nucleotide changes | Amino acid changes | References |
|---|---|---|---|
| CYP1B1*2 | 142C>G; 255G>T | R48G; A119S | (35,37) |
| CYP1B1*3 | 4326C>G | L432V | (35) |
| CYP1B1*4 | 4390A>G | N453S | (35) |
| CYP1B1*5 | 142C>G; 4326C>G | R48G; L432V | (38) |
| CYP1B1*6 | 142C>G; 355G>T; 4326C>G | R48G; A119S; L432V | (38) |
| CYP1B1*7 | 142C>G; 355G>T; 4326C>G; 4360C>G | R48G; A119S; L432V; A443G | (38) |
| CYP1B1*8 | 4326C>G; 4353G>C; 4379C>T | L432V; D441H | Rahman et al., unpublished |
| CYP1B1*9 | |||
| CYP1B1*10 | |||
| CYP1B1*11 | 171G>C | W57C | (35) |
| CYP1B1*12 | 182G>A | G61E | (35) |
| CYP1B1*13 | 501_502insT | 167Frameshift | (35) |
| CYP1B1*14 | 841G>T | E281X | (35) |
| CYP1B1*15 | 863_864insC | 288Frameshift | (34) |
| CYP1B1*16 | Large deletion | Splicing defect | (34) |
| CYP1B1*17 | 4096_4108del | 355Frameshift | (34) |
| CYP1B1*18 | 4125G>T | G365W | (35) |
| CYP1B1*19 | 4168C>T | P379L | (35) |
| CYP1B1*20 | 4191G>A | E387K | (35) |
| CYP1B1*21 | 4201G>A | R390H | (35) |
| CYP1B1*22 | 4232_4241dup | 404Frameshift | (35) |
| CYP1B1*23 | 4342C>T | P437L | (35) |
| CYP1B1*24 | 4377delG | 449Frameshift | (35) |
| CYP1B1*25 | 4437C>T | R469W | (35) |
| CYP1B1*26 | 4435_4461dup | 477Frameshift | (35) |
Adopted from “http://www.cypalleles.ki.se/”.
Ingelman-Sundberg and colleagues characterized two common P450 1B1 mutations, R48G and A119S (37). The steady-state kinetic analysis showed no differences in 17β-estradiol hydroxylation activities in this P450 1B1*2 mutant protein and only a minor increase in the apparent Km for EROD was observed (37). In another study, three novel allelic variants, P450 1B1*5 (R48G/L432V), *6 (R48G/A119S/L432V), and *7 (R48G/A119S/L432V/A443G) have been identified in Ethiopian population and functional consequences of these mutations have been analyzed (38). The frequencies of P450 1B1*5, *6, and *7 were 0.7, 6, and 7%, respectively (38). Recombinant P450 1B1*6 and *7 exhibited altered kinetics with a significantly high apparent Km and low kcat values for the hydroxylation of 17β-estradiol (38).
Recently, five non-synonymous SNP allelic variants (W57X, 290Frameshift, Y81N, E229K, and R368H) were detected in coloboma/microphthalmia patients (http://www.cypalleles.ki.se/) (39). It is known that P450 1B1 can contribute to retinoic acid synthesis during embryonic development and, at the same time, retinoic acid receptor signaling regulates choroid fissure closure. Functional consequences of expression of these novel P450 1B1 variants observed in that study suggest that P450 1B1 may regulate proper optic fissure closure by affecting retinoic acid signaling (39).
CONCLUSIONS
In this review, we considered mechanisms of metabolic activation of chemical carcinogens by P450 1B1. Development of selective P405 1B1 inhibitors with increased therapeutic effectiveness is a promising avenue to control cancer growth and metastasis. Pharmacological roles of P450 1B1 in metabolism of clinical drugs have not been intensively studied.
The risks of metabolic activation of chemical carcinogens in different individuals vary because of polymorphisms in genes encoding metabolic enzymes. P450 1B1 polymorphisms have been implicated as risk factors in various diseases, which may arise from impaired P450 1B1 enzymatic activity. It is therefore extremely important to carefully consider the functional significance of genetic variability of P450 enzymes in the initiation and progression of cancer.
ACKNOWLEDGEMENT
This paper was supported by Konkuk University in 2013.
REFERENCES
- 1.Ortiz de Montellano PR. Cytochrome P450: Structure, Mechanism, and Biochemistry. Plenum Press; New York: 2005. [DOI] [Google Scholar]
- 2.Guengerich FP. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol. 2001;14:611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- 3.Porter TD, Coon MJ. Cytochrome P-450. Multiplicity of isoforms, substrates, and catalytic and regulatory mechanisms. J Biol Chem. 1991;266:13469–13472. [PubMed] [Google Scholar]
- 4.Klingenberg M. Pigments of rat liver microsomes. Arch Biochem Biophys. 1958;75:376–386. doi: 10.1016/0003-9861(58)90436-3. [DOI] [PubMed] [Google Scholar]
- 5.Lee IS, Kim D. Polymorphic metabolism by functional alterations of human cytochrome P450 enzymes. Arch Pharm Res. 2011;34:1799–1816. doi: 10.1007/s12272-011-1103-2. [DOI] [PubMed] [Google Scholar]
- 6.Omura T, Sato R. A new cytochrome in liver microsomes. J Biol Chem. 1962;237:1375–1376. [PubMed] [Google Scholar]
- 7.Beaune P, Dansette P, Flinois JP, Columelli S, Mansuy D, Leroux JP. Partial purification of human liver cytochrome P-450. Biochem Biophys Res Commun. 1979;88:826–832. doi: 10.1016/0006-291X(79)91482-7. [DOI] [PubMed] [Google Scholar]
- 8.Wang PP, Beaune P, Kaminsky LS, Dannan GA, Kadlubar FF, Larrey D, Guengerich FP. Purification and characterization of six cytochrome P-450 isozymes from human liver microsomes. Biochemistry. 1983;22:5375–5383. doi: 10.1021/bi00292a019. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez FJ. The molecular biology of cytochrome P450s. Pharmacol Rev. 1988;40:243–288. [PubMed] [Google Scholar]
- 10.Guengerich FP. Cytochrome P450: what have we learned and what are the future issues? Drug Metab Rev. 2004;36:159–197. doi: 10.1081/DMR-120033996. [DOI] [PubMed] [Google Scholar]
- 11.Sutter TR, Guzman K, Dold KM, Greenlee WF. Targets for dioxin: genes for plasminogen activator inhibitor-2 and interleukin-1 beta. Science. 1991;254:415–418. doi: 10.1126/science.1925598. [DOI] [PubMed] [Google Scholar]
- 12.Guengerich FP. In: Human cytochrome P450 enzymes in Cytochrome P450: Structure, Mechanism, and Biochemistry. Ortiz de Montellano PR, editor. Plenum Press; New York: 2005. pp. 377–530. [Google Scholar]
- 13.Shimada T, Hayes CL, Yamazaki H, Amin S, Hecht SS, Guengerich FP, Sutter TR. Activation of chemically diverse procarcinogens by human cytochrome P-450 1B1. Cancer Res. 1996;56:2979–2984. [PubMed] [Google Scholar]
- 14.Chun YJ, Oh YK, Kim BJ, Kim D, Kim SS, Choi HK, Kim MY. Potent inhibition of human cytochrome P450 1B1 by tetramethoxystilbene. Toxicol Lett. 2009;189:84–89. doi: 10.1016/j.toxlet.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Shimada T, Gillam EM, Sutter TR, Strickland PT, Guengerich FP, Yamazaki H. Oxidation of xenobiotics by recombinant human cytochrome P450 1B1. Drug Metab Dispos. 1997;25:617–622. [PubMed] [Google Scholar]
- 16.Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J Biol Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- 17.Sissung TM, Price DK, Sparreboom A, Figg WD. Pharmacogenetics and regulation of human cytochrome P450 1B1: implications in hormone-mediated tumor metabolism and a novel target for therapeutic intervention. Mol Cancer Res. 2006;4:135–150. doi: 10.1158/1541-7786.MCR-05-0101. [DOI] [PubMed] [Google Scholar]
- 18.Wang A, Savas U, Stout CD, Johnson EF. Structural characterization of the complex between alpha-naphthoflavone and human cytochrome P450 1B1. J Biol Chem. 2011;286:5736–5743. doi: 10.1074/jbc.M110.204420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim D, Guengerich FP. Cytochrome P450 activation of arylamines and heterocyclic amines. Annu Rev Pharmacol Toxicol. 2005;45:27–49. doi: 10.1146/annurev.pharmtox.45.120403.100010. [DOI] [PubMed] [Google Scholar]
- 20.Parikh A, Gillam EM, Guengerich FP. Drug metabolism by Escherichia coli expressing human cytochromes P450. Nat Biotechnol. 1997;15:784–788. doi: 10.1038/nbt0897-784. [DOI] [PubMed] [Google Scholar]
- 21.Kim D, Guengerich FP. Selection of human cytochrome P450 1A2 mutants with enhanced catalytic activity for heterocyclic amine N-hydroxylation. Biochemistry. 2004;43:981–988. doi: 10.1021/bi035593f. [DOI] [PubMed] [Google Scholar]
- 22.Chun YJ, Kim S, Kim D, Lee SK, Guengerich FP. A new selective and potent inhibitor of human cytochrome P450 1B1 and its application to antimutagenesis. Cancer Res. 2001;61:8164–8170. [PubMed] [Google Scholar]
- 23.Shimada T, Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004;95:1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rendic S, Guengerich FP. Contributions of human enzymes in carcinogen metabolism. Chem Res Toxicol. 2012;25:1316–1383. doi: 10.1021/tx300132k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buters J, Quintanilla-Martinez L, Schober W, Soballa VJ, Hintermair J, Wolff T, Gonzalez FJ, Greim H. CYP1B1 determines susceptibility to low doses of 7,12-dimethylbenz[a]anthracene-induced ovarian cancers in mice: correlation of CYP1B1-mediated DNA adducts with carcinogenicity. Carcinogenesis. 2003;24:327–334. doi: 10.1093/carcin/24.2.327. [DOI] [PubMed] [Google Scholar]
- 26.Buters JT, Sakai S, Richter T, Pineau T, Alexander DL, Savas U, Doehmer J, Ward JM, Jefcoate CR, Gonzalez FJ. Cytochrome P450 CYP1B1 determines susceptibility to 7, 12-dimethylbenz[a]anthracene-induced lymphomas. Proc Natl Acad Sci USA. 1999;96:1977–1982. doi: 10.1073/pnas.96.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galvez-Peralta M, Shi Z, Chen J, Miller ML, Nebert DW. Oral benzo[a]pyrene in CYP1A1/1B1(−/−) double-knockout mice: Microarray analysis during squamous cell carcinoma formation in preputial gland duct. Int J Cancer. 2013;132:2065–2075. doi: 10.1002/ijc.27897. [DOI] [PubMed] [Google Scholar]
- 28.Hayes CL, Spink DC, Spink BC, Cao JQ, Walker NJ, Sutter TR. 17 beta-estradiol hydroxylation catalyzed by human cytochrome P450 1B1. Proc Natl Acad Sci USA. 1996;93:9776–9781. doi: 10.1073/pnas.93.18.9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanna IH, Dawling S, Roodi N, Guengerich FP, Parl FF. Cytochrome P450 1B1 (CYP1B1) pharmacogenetics: association of polymorphisms with functional differences in estrogen hydroxylation activity. Cancer Res. 2000;60:3440–3444. [PubMed] [Google Scholar]
- 30.Yamazaki H, Shaw PM, Guengerich FP, Shimada T. Roles of cytochromes P450 1A2 and 3A4 in the oxidation of estradiol and estrone in human liver microsomes. Chem Res Toxicol. 1998;11:659–665. doi: 10.1021/tx970217f. [DOI] [PubMed] [Google Scholar]
- 31.Bolton JL, Thatcher GR. Potential mechanisms of estrogen quinone carcinogenesis. Chem Res Toxicol. 2008;21:93–101. doi: 10.1021/tx700191p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishida CR, Everett S, Ortiz de Montellano PR. Specificity determinants of CYP1B1 estradiol hydroxylation. Mol Pharmacol. 2013;84:451–458. doi: 10.1124/mol.113.087700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chun YJ, Lee SK, Kim MY. Modulation of human cytochrome P450 1B1 expression by 2,4,3′,5′-tetramethoxystilbene. Drug Metab Dispos. 2005;33:1771–1776. doi: 10.1124/dmd.105.006502. [DOI] [PubMed] [Google Scholar]
- 34.Stoilov I, Akarsu AN, Sarfarazi M. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum Mol Genet. 1997;6:641–647. doi: 10.1093/hmg/6.4.641. [DOI] [PubMed] [Google Scholar]
- 35.Stoilov I, Akarsu AN, Alozie I, Child A, Barsoum-Homsy M, Turacli ME, Or M, Lewis RA, Ozdemir N, Brice G, Aktan SG, Chevrette L, Coca-Prados M, Sarfarazi M. Sequence analysis and homology modeling suggest that primary congenital glaucoma on 2p21 results from mutations disrupting either the hinge region or the conserved core structures of cytochrome P4501B1. Am J Hum Genet. 1998;62:573–584. doi: 10.1086/301764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vasiliou V, Gonzalez FJ. Role of CYP1B1 in glaucoma. Annu Rev Pharmacol Toxicol. 2008;48:333–358. doi: 10.1146/annurev.pharmtox.48.061807.154729. [DOI] [PubMed] [Google Scholar]
- 37.McLellan RA, Oscarson M, Hidestrand M, Leidvik B, Jonsson E, Otter C, Ingelman-Sundberg M. Characterization and functional analysis of two common human cytochrome P450 1B1 variants. Arch Biochem Biophys. 2000;378:175–181. doi: 10.1006/abbi.2000.1808. [DOI] [PubMed] [Google Scholar]
- 38.Aklillu E, Oscarson M, Hidestrand M, Leidvik B, Otter C, Ingelman-Sundberg M. Functional analysis of six different polymorphic CYP1B1 enzyme variants found in an Ethiopian population. Mol Pharmacol. 2002;61:586–594. doi: 10.1124/mol.61.3.586. [DOI] [PubMed] [Google Scholar]
- 39.Prokudin I, Simons C, Grigg JR, Storen R, Kumar V, Phua ZY, Smith J, Flaherty M, Davila S, Jamieson RV. Exome sequencing in developmental eye disease leads to identification of causal variants in GJA8, CRYGC, PAX6 and CYP1B1. Eur J Hum Genet. 2014;22:907–915. doi: 10.1038/ejhg.2013.268. [DOI] [PMC free article] [PubMed] [Google Scholar]