SUMMARY
The participation of a specific subset of B cells and how they are regulated in cancer is unclear. Here, we demonstrate that the proportion of CD5+ relative to interleukin-6 receptor α (IL-6Rα) expressing B cells was greatly increased in tumors. CD5+ B cells responded to IL-6 in the absence of IL-6Rα. IL-6 directly bound to CD5, leading to activation of the transcription factor STAT3 via gp130 and its downstream kinase JAK2. STAT3 upregulated CD5 expression, thereby forming a feed-forward loop in the B cells. In mouse tumor models, CD5+ but not CD5−B cells promoted tumor growth. CD5+ B cells also showed activation of STAT3 in multiple types of human tumor tissues. Thus, our findings demonstrate a critical role of CD5+ B cells in promoting cancer.
INTRODUCTION
Extensive prior studies suggest the importance of various immune cells, including those of myeloid lineage and T cells in regulating cancer inflammation and antitumor immunity (Grivennikov et al., 2010; Mantovani et al., 2008; Trinchieri, 2012; Yu et al., 2009). Several seminal studies have also indicated an important role of B cells in promoting cancer progression (Ammirante et al., 2010; de Visser et al., 2005; Mantovani, 2011; Woo et al., 2014). However, there are other reports indicating that B cells can mediate antitumor effects (DiLillo et al., 2010; Li et al., 2009). Our previous study shows that tumor-associated B cells promote tumor invasion by producing multiple pro-angiogenic factors in a STAT3-dependent manner (Yang et al., 2013b). Moreover, B cell infiltration and STAT3 activation in patient tumor-associated tissues correlate negatively with survival, at least in a limited number of ovarian cancer patients examined (Yang et al., 2013a). However, STAT3 is activated only in a subpopulation of B cells in multiple types of human tumor-associated tissues (Yang et al., 2013a; Yang et al., 2013b). The identity of the B cells in tumor and tumor-related tissues that are positive for STAT3 activation remains unknown.
A crucial role of STAT3 in promoting proliferation, survival and invasion in diverse cancers has been established (Bollrath et al., 2009; Grivennikov et al., 2009; Yu and Jove, 2004; Yu et al., 2007; Yu et al., 2009). STAT3 was originally discovered in the context of IL-6-IL-6 receptor signaling (Heinrich et al., 1998; Taga and Kishimoto, 1997). Subsequently, many other cytokines, such as IL-10, as well as growth factors and chemokines, have been identified as STAT3 activators (Donnelly et al., 1999; Kortylewski et al., 2009; Lamprecht et al., 2008; Stout et al., 2004). STAT3 in turn, mediates the expression of some of these activators, forming a feed-forward loop that facilitates persistent STAT3 activation. This occurs not only in tumor cells but also in various types of immune cells in the tumor microenvironment, promoting tumor growth, invasion and suppression of T helper 1 (Th1) cell antitumor immunity in cancer (Kortylewski et al., 2009; Lee et al., 2010; Yu et al., 2009). While many cytokines and other mediators have been shown to contribute to cancer progression through STAT3, IL-6 has been considered by many as the most crucial STAT3 activator for cancer progression (Catlett-Falcone et al., 1999; Grivennikov et al., 2009; Grivennikov and Karin, 2010; Yu et al., 2007; Yu et al., 2009). However, although B cells have now been shown to promote cancer, the expression of IL-6Rα is restricted to a small percentage of normal B cells (Hoge et al., 2013). Therefore, it remains to be investigated whether any other receptor(s) on B cells could contribute to IL-6 signaling and promote cancer progression.
CD5+ B lymphocytes are a relatively minor population of B cells in both human and murine lymphoid organs (Baumgarth, 2011; Berland and Wortis, 2002). However, they display some unique properties in that they are self-renewing and possess a propensity for malignant transformation. CD5+ B lymphocytes are considered to be the normal counterpart of human chronic lymphocytic leukemia (Dong et al., 2003; Zheng et al., 2002). A subset of CD5+ B cells have also been shown to be regulatory B cells, playing an important role in dampening several autoimmune pathologic conditions, such as collagen-induced arthritis, autoimmune encephalitis, chronic colitis among others (Matsushita et al., 2008; Yanaba et al., 2008). The ability of the CD5+ regulatory B cells in modulating immune responses and inflammation in autoimmune diseases is believed to be mediated by IL-10 (Xing et al., 2015; Yoshizaki et al., 2012). However, whether and how CD5+ B cells may dampen antitumor immune responses and/or enhance cancer-promoting inflammation remains to be explored. Furthermore, functional ligands for CD5 are still elusive.
Here we investigated whether and how CD5 contributes to B cell-mediated tumor progression. Our results demonstrate that CD5 bound and responded to IL-6, which activated STAT3 by gp130 and JAK2 in B cells in tumor microenvironment. Furthermore, STAT3 activation elevated expression of CD5, thereby forming a feed-forward loop. Moreover, CD5+ B cells and STAT3 activation and poor patient survival correlated, at least in a limited number of patients. Thus, our results suggest that CD5+ B cells play a critical role in the tumor microenvironment for cancer progression.
RESULTS
CD5+ B Cells Increase and Respond to IL-6 in the Tumor Environment
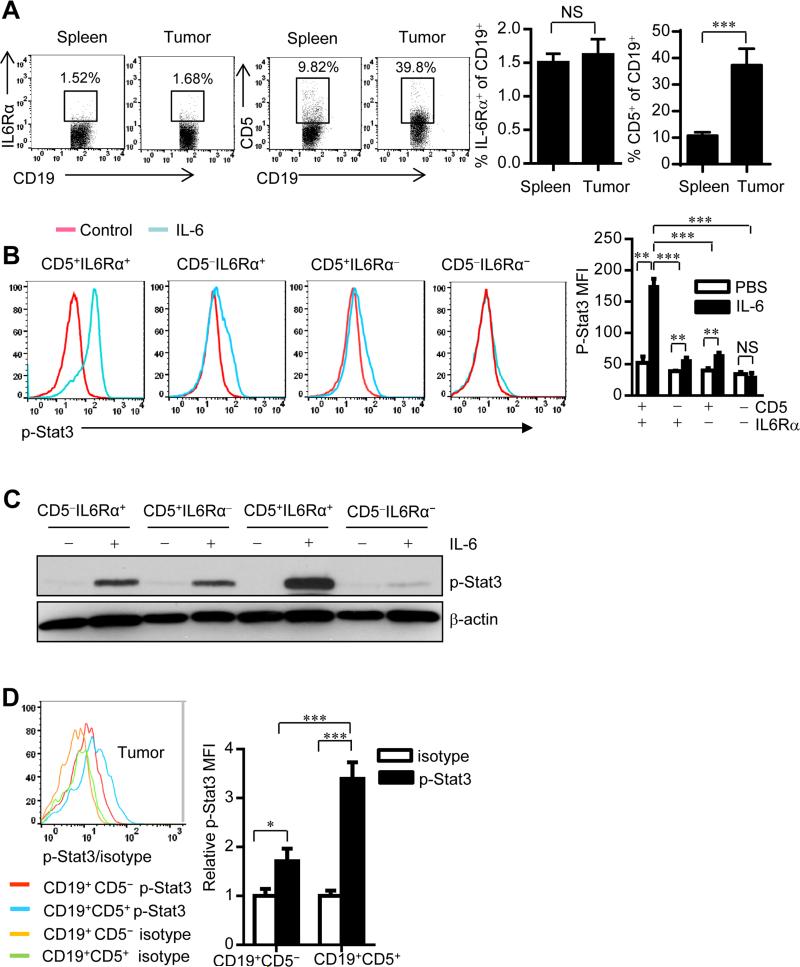
We have shown that B cells and STAT3 activation in omentum could be indicative of poor survival for ovarian cancer patients (Yang et al., 2013a). Because IL-6 is a key activator of STAT3 and is important in inflammation-mediated cancer progression (Bollrath et al., 2009; Grivennikov et al., 2009; Grivennikov and Karin, 2010; Heinrich et al., 2003; Yu and Jove, 2004; Yu et al., 2009), we explored the possibility that IL-6-IL-6 receptor signaling activates STAT3 in tumor-associated B cells. However, flow cytometric analyses of both mouse splenic and human peripheral immune cells indicated that the percentage of IL-6Rα+ B cells was low (Figure S1). The percentage of IL-6Rα+ B cells remained low in the tumor microenvironment (Figure 1A). Because IL-10-producing regulatory B cells are CD5 positive (Yoshizaki et al., 2012) and IL-10 production is STAT3 dependent (Yu et al., 2007), we reasoned that CD5 might aid B cells by responding to IL-6. We found that CD19+ CD5+ B cells were significantly increased, relative to IL-6Rα+ B cells, within tumors compared to periphery (Figure 1A). We found higher percentage of IgMhiIgDlo and CD11b+ B-1 population in CD5+ B cells from naïve mouse peritoneal cavity than spleen (Figure S1B). We further checked IgD, IgM and CD11b expression in CD5+ B cell in the tumor microenvironment, compared to their splenic counterparts from tumor bearing mice (as well as from naïve mice, Figure S1C). Although CD5+ B cell proportion was increased in the tumor, the percentages of IgMhiIgDlo B-1 B cells, IgMloIgDhi B-2 B cells, and CD11b+ population in CD5+ B cells were similar in the tumor and in the spleens of tumor-bearing mice (Figure S1C). These results suggest that tumor-associated CD5+ B cells are not likely converted from B2 cells. Flow cytometric analyses of mouse splenic cells showed that CD5+ B cells within IL-6Rα− B cells responded to IL-6 for Stat3 activation (Figure 1B), which was confirmed by protein blotting analyses (Figure 1C).
Figure 1. CD19+CD5+ B cells significantly increase in tumors and are responsive to IL-6.
(A) Flow cytometric analysis of splenic and tumor IL-6Rα+ and CD5+ frequencies within CD19+ B cells from B16 tumor bearing mouse. Data are shown as means ± SEM (n = 10); ***P < 0.001.
(B) Levels of p-Stat3 in subpopulations of mouse splenic B cells upon IL-6 stimulation. Quantification is shown as means ± SEM (n = 3). **P < 0.01, ***P < 0.001.
(C) Western blotting showing p-Stat3 level in FACS-sorted CD5−IL6Rα+, CD5+IL6Rα−, CD5+IL6Rα+ and CD5−IL6Rα− B cells with or without IL-6 treatment for 15 min.
(D) Levels of p-Stat3 in B16 tumor-infiltrating B cells shown by flow cytometry. Quantification of mean fluorescence intensity (MFI) from one of three independent experiments is shown as means ± SEM (normalized to isotype control; n = 5) *P < 0.05, ***P < 0.001. See also Figure S1 and Figure S2.
Because IL-6 is important in tumors and the tumor microenvironment by activating STAT3, we next examined whether CD5 is required for tumor-derived factor-stimulated STAT3 activation in B cells. Only CD5+ B cells, but not CD5− B cells, responded to tumor-conditioned medium (TCM) in terms of Stat3 activation, which was IL-6-dependent (Figure S2). Furthermore, CD5+ B cells in mouse tumors exhibited significantly higher Stat3 activity than their CD5− counterparts (Figure 1D). Thus, CD5+ B cells respond to IL-6.
Stat3 activation in CD5+IL-6Rα− B cells is not mediated by sIL-6Rα
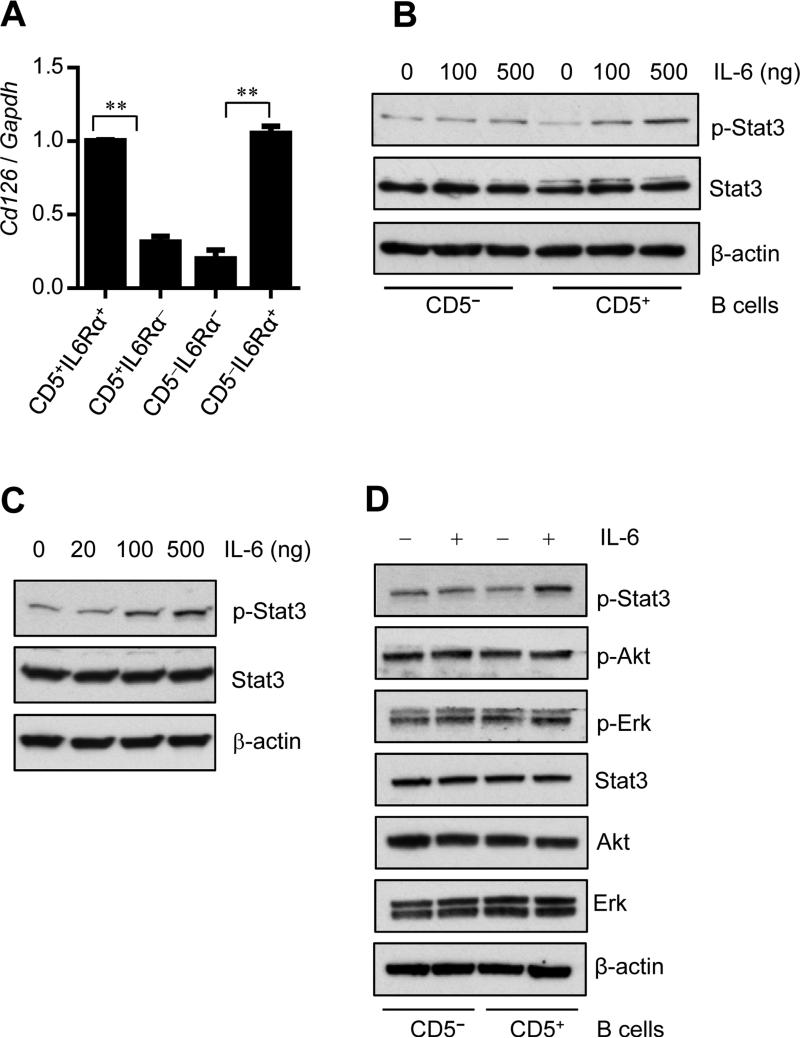
In many inflammatory and tumor microenvironment conditions, IL-6R is known to be cleaved from the surface of the cell to form soluble IL-6Rα (sIL-6Rα) which nevertheless can activate IL-6 signaling and Stat3 by virtue of “IL-6 trans-signaling” (Neurath and Finotto, 2011; Rose-John, 2012; Rose-John and Neurath, 2004). To address whether CD5+IL-6Rα− B cells respond to IL-6 for Stat3 activation through sIL-6Rα, we performed real-time PCR to measure IL-6Rα mRNA level in various subsets of B cells. IL-6Rα (CD126) positive B cells expressed higher amounts of IL-6Rα than IL-6Rα negative B cells (Figure 2A). Although both CD5+ IL-6Rα− and CD5− IL-6Rα−B cells expressed much lower amount of IL-6Rα mRNA than IL-6Rα+B cells, CD5+ IL-6Rα− B cells exhibited higher p-Stat3 than CD5−IL-6Rα− B cells after IL-6 stimulation (Figure 1B and Figure 2A). Furthermore, we measured sIL-6Rα in the tumor microenvironment by ELISA and we found that sIL-6Rα was not detectable in the culture medium containing either mouse IL-6 or TCM (Figure S3). In addition, using B cells derived from IL-6Rα (CD126)-deficient (Il6ra−/−) mice we assessed whether Stat3 activation was contributed by sIL-6-IL-6 trans-signaling. CD5+ and CD5− B cells from Il6ra−/− mice were sorted, then stimulated with IL-6 in HBSS for 15min. Stat3 activation was determined by protein blotting. The results confirmed that IL-6 can stimulate Stat3 activation in Il6ra−/− CD5+ B cells, but not in Il6ra−/− CD5− B cells (Figure 2B). Because T cells express CD5, we also confirmed that IL-6 stimulated Stat3 activation in T cells derived from Il6ra−/− mice (Figure 2C). Thus, IL-6 trans-signaling does not contribute measurably to IL-6-induced, CD5-associated Stat3 activation. In addition, CD5 is necessary for Stat3 activation in Il6ra−/− B and T cells. Interestingly, IL-6 does not increase p-ERK and p-Akt levels in CD5+Il6ra−/− B cells (Figure 2D), suggesting that CD5-mediated STAT3 activation may involve pathways that are different from IL-6R.
Figure 2. Stat3 activation in CD5+IL-6Rα− B cells is not mediated by sIL-6Rα.
(A) Real-time RT-PCR measuring CD126 (IL-6Rα) mRNA level in different subclass CD19+ B cells sorted from splenocytes derived from C57BL/6 mice. Data represent means ± SEM of 3 independent experiments, each involving 3 pooled mice per group done in triplicates.
(B-C) Western blotting showing p-Stat3 level in FACS-sorted CD19+ CD5+, CD19+ CD5− B cells (B), and T cells (C). Both B and T cells used were from IL-6Rα-deficient mice and treated with IL-6 for 15 min.
(D) Western blotting showing p-Stat3, p-Akt and p-Erk levels in CD19+ CD5+, and CD19+ CD5− post IL-6 treatment. CD5+ and CD5− B cells from IL-6Ra KO (Il6ra−/−) mice were sorted by FCS and stimulated with IL-6 for 15 min, followed by Western blotting to detect p-Stat3, p-Akt and p-Erk levels. See also Figure S3.
CD5 Directly Binds to IL-6
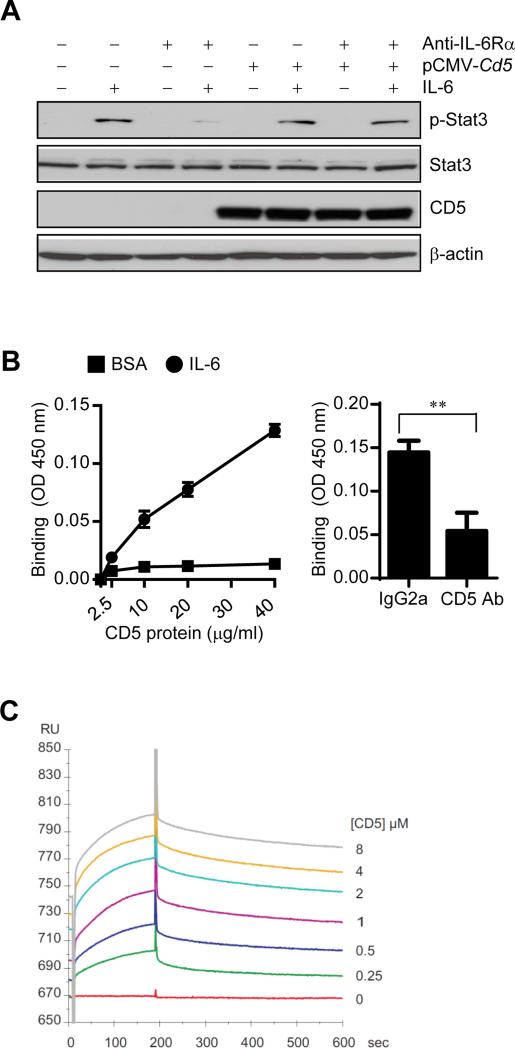
To investigate whether CD5 can directly activate STAT3, we performed experiments to ensure that in the absence of IL-6Rα, IL-6 can induce STAT3 activation by binding to CD5. Blocking IL-6Rα with its antibody has no inhibitory effects on IL-6-stimulated Stat3 activation in B16 melanoma tumor cells overexpressing CD5 (the parental tumor cells are CD5−IL-6Rα+) (Figure 3A).
Figure 3. CD5 directly binds to IL-6.
(A) IL-6-induced Stat3 activation in CD5-overexpressing B16 melanoma cells with or without IL-6Rα blockade was evaluated by Western blotting.
(B) ELISA-type assays showing dose-dependent direct binding of mouse IL-6 to immobilized mouse CD5 (left); and blockade of the IL-6 binding to CD5 by adding an CD5 antibody prior to incubating with IL-6 (right). Data are shown as means ± SEM (n = 3).
(C) Biacore analysis of a serial of increasing concentration of murine CD5 binding to IL-6 immobilized on a CM5 sensor chip.
We next tested whether CD5 can directly bind to IL-6, thereby activate STAT3 in the tumor microenvironment. We performed enzyme-linked immunosorbent (ELISA)-type assays (Ieguchi et al., 2010) to assess if recombinant murine IL-6 binds to immobilized mouse CD5 in a dose-dependent manner. IL-6 bound CD5 (Figure 3B), and in the presence of an antibody against CD5, the binding of soluble IL-6 to CD5 was blocked, suggesting that binding of IL-6 is specific to CD5. The kinetic affinity between murine CD5 and immobilized mIL6 were measured by surface plasmon resonance (Biacore). The KD was approximately 1 ×10−7M (Figure 3C). These results indicated that IL-6 can directly bind to CD5.
CD5-induced STAT3 Activation Requires gp130 and JAK2
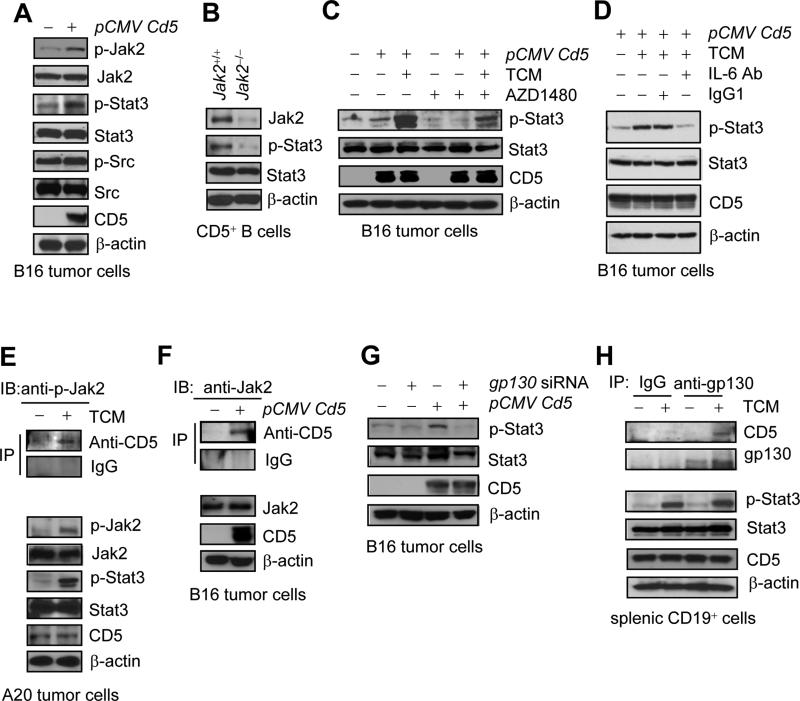
IL-6Rα, along with several other receptors for related cytokines, requires gp130 and JAK for STAT3 activation (Heinrich et al., 1998; Jones et al., 2011; Putoczki and Ernst, 2010). We next investigated the relationship between CD5 and gp130 and Jak2. Overexpression of CD5 activated both Jak2 and Stat3 in mouse B16 tumor cells (Figure 4A). Furthermore, ablation of the Jak2 alleles in CD5+ B cells from tumor bearing mice abrogated tumor-induced Stat3 activation in the CD5+ B cells (Figure 4B). Blocking JAK signaling using a JAK inhibitor, AZD1480 (Hedvat et al., 2009), also reduced Stat3 activation induced by CD5 overexpression and tumor conditioned medium (TCM) (Figure 4C). Neutralizing IL-6 with an anti-IL6 antibody reduced Stat3 activation in the CD5 over-expressing cells cultured with TCM (Figure 4D). In A20 B cell lymphoma cells that express CD5 (although not at a high amount), an interaction between CD5 and Jak2 was detectable upon stimulation of IL-6-containing TCM (Figure 4E). Co-immunoprecipitation with a CD5 antibody followed by protein blotting also showed an interaction between CD5 and Jak2 in CD5-overexpressing B16 tumor cells (Figure 4F). We further explored the effects of silencing gp130 with siRNA on CD5-induced Stat3 activation. Silencing gp130 abrogated Stat3 activation induced by CD5-overexpression (Figure 4G). In addition, gp130 interacted with CD5 in primary mouse B cells, when Stat3 was activated by IL-6-containing tumor-conditioned medium (Figure 4H). Thus, CD5-induced STAT3 activation requires gp130 and JAK2.
Figure 4. CD5-induced Stat3 activation involves Jak2 and gp130.
(A-B) Detection of Stat3 and Jak2 activation by Western blotting in Cd5-overexpressing B16 tumor cells (A), and in splenic Jak2+/+ or Jak2−/− CD5+ B cells from tumor bearing mice (B).
(C) Western blotting showing TCM-induced Stat3 activation in Cd5-overexpression B16 tumor cells is reduced by a Jak inhibitor, AZD1480, in B16 tumor cells.
(D) p-Stat3 levels in CD5-overexpressing B16 tumor cells with or without IL-6 depletion. The tumor cells stimulated for 15 min by TCM, which were pre-treated with either IgG1 isotype control or mouse IL-6 neutralizing antibody for 60 min.
(E) Proteins co-immunoprecipitated by CD5 antibody (top) from TCM-treated or untreated A20 tumor cells or whole lysates (bottom) were immunoblotted for indicated proteins.
(F) Co-immunoprecipitation with CD5 antibody followed by Western blotting showing CD5 and Jak2 in a complex in CD5-overexpressing B16 tumor cells.
(G) Western blotting showing the effect of gp130 silencing on Stat3 activation induced by CD5-overexpression in B16 cells.
(H) Co-immunoprecipitation by gp130 antibody before western blotting, showing CD5-gp130 complexes induced by TCM in naïve splenic CD19+ B cells.
STAT3 Is a Direct Transcriptional Factor for CD5
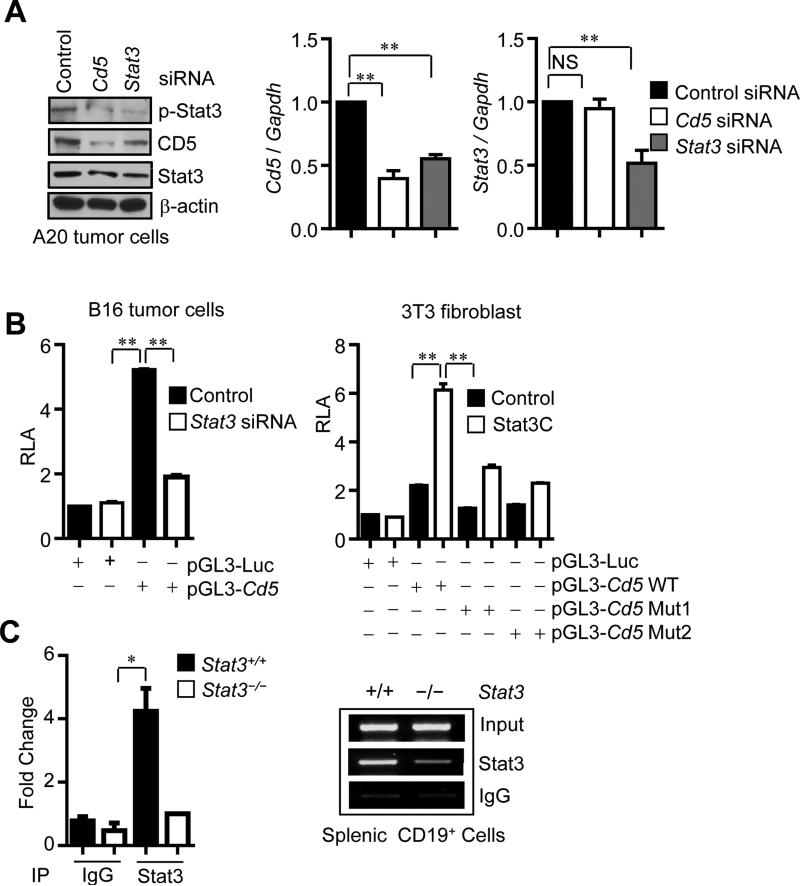
Our data thus far delineated a signaling pathway connecting IL-6 with gp130-Jak2-STAT3 through CD5. However, the percentage of CD5+ B cells in periphery, although approximately 5-fold higher than IL-6Rα+ B cells is still relatively small. Importantly, the percentage of CD5+ B cells was significantly expanded in tumors (Figure 1A). STAT3 is a transcription factor known to upregulate expression of several of its own upstream activators (Herrmann et al., 2014; Kortylewski et al., 2009; Lee et al., 2010; Xin et al., 2013; Yu et al., 2009). We therefore explored the possibility that STAT3 upregulates CD5 expression, thereby forming a feed-forward loop in B cells to promote tumor progression. We showed that ablating Stat3 in splenic and tumor-associated B cells abrogated CD5 expression at the protein and/or RNA levels (Figure S4). Furthermore, silencing Stat3 in A20 B cell lymphoma cells caused a downregulation of CD5 expression, as shown by both protein blotting and quantitative real-time RT-PCR (Figure 5A). These results suggest that Stat3 siRNA knockdown inhibits CD5 at the transcriptional level in normal B cells and malignant B cells. In order to determine whether this transcriptional regulation was direct, we performed luciferase reporter and chromatin immunoprecipitation (ChIP) analyses. Luciferase activity by the CD5 promoter-reporter assessed in both B16 tumor cells and 3T3 fibroblasts indicated that CD5 transcription required Stat3 (Figure 5B). Furthermore, an increase in STAT3 activity through enforced expression of STAT3C, a constitutively-activated mutant form of STAT3, increased CD5 promoter activity (Figure 5B). Mutating the Stat3 DNA-binding sites within the CD5 promoter also abrogated its activity (Figure 5B). A direct binding of Stat3 to the CD5 promoter was also shown by ChIP assay with Stat3 antibody (Figure 5C) or p-Stat3 antibody (Figure S5). Thus, STAT3 is a transcription factor for the CD5 gene.
Figure 5. Stat3 is a direct transcriptional activator of CD5.
(A) Western blotting and real-time RT-PCR showing CD5 and Stat3 at protein and mRNA levels in A20 tumor cells transfected with indicated siRNAs. Results are means ± SEM (normalized to control siRNA; n = 3).
(B) Luciferase assay to analyze the effect of Stat3 activity on Cd5 promoter activity. Relative luciferase activity (RLA) was calculated as the ratio of firefly luciferase activity to Renilla luciferase activity. Results are shown as means ±SEM (n = 3). Data represent one of three experiments.
(C) ChIP assay demonstrating Stat3 binding to the Cd5 promoter. Chromatins were prepared from splenic CD19+ B cells of tumor-bearing mice. Quantitative real-time PCR (left) or regular PCR (right) showing the relative amounts of specific DNA fragments of CD5 promoter after immunoprecipitating by Stat3/IgG and normalizing to the input. Results are means ± SEM (n = 3). See also Figure S4 and Figure S5.
CD5+ but Not CD5− B Cells Promote Tumor Progression
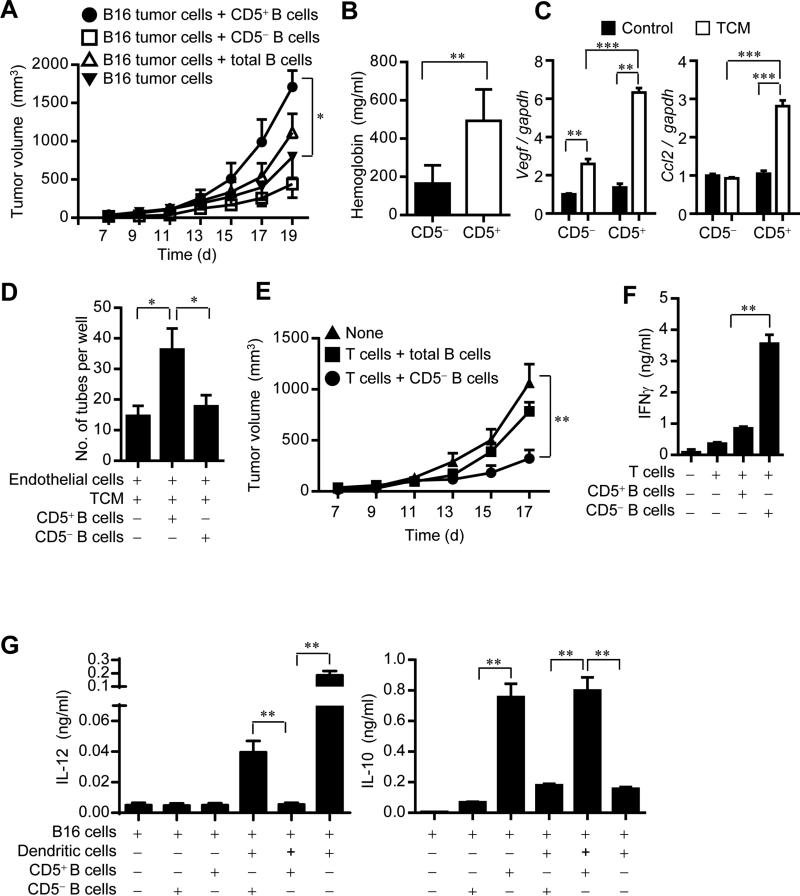
Although several seminal studies have shown a crucial role of B cells in promoting cancer progression (Ammirante et al., 2010; Andreu et al., 2010; de Visser et al., 2005), other reports have indicated that B cells could also exert antitumor effects (DiLillo et al., 2010; Mantovani, 2011), Our recent studies indicated that both B cell tumor-infiltrates and STAT3 activity correlate with poor patient survival, at least in the limited tumor tissues and patients studied. However, only a subpopulation of B cells in the tumor-associated tissues is p-STAT3 positive (Yang et al., 2013a; Yang et al., 2013b). We tested the hypothesis that CD5+ might be the subpopulation of B cells that is STAT3-positive and promotes tumor growth. We purified CD5+ and CD5− B cells, mixed them with B16 tumor cells, and injected the mixtures into Rag1−/− mice. CD5+ B cells, but not CD5− B cells, significantly increased tumor growth (Figure 6A). We also measured p-Stat3 level in tumor cells derived from B16 tumors grown in Rag1−/− mice in the presence of either CD5+ B cells or CD5− B cells. p-Stat3 was higher in B16 tumor cells mixed with CD5+ B cells than those with CD5− B cells (Figure S6A). These observations were confirmed in the MB49 mouse bladder tumor model (Figure S6B, 6C).
Figure 6. CD5+ B cells promote cancer progression in part by inducing angiogenesis and immunosuppression.
(A) Tumor growth curve showing enhanced B16 tumor growth in Rag1−/− mice by adding CD5+ B cells to the tumor cells. Results shown are representative of 3 independent experiments (n = 6). *P < 0.05.
(B) Hemoglobin content measurement from Matrigel plugs containing a mixture of B16 tumor cells and CD5+ or CD5− splenic B cells. Results are means ± SEM (n = 6). *P < 0.05.
(C) Real-time RT-PCR showing mRNA levels of angiogenic factors (VEGF and CCL2) in CD5+ or CD5− splenic B cell incubated with 5% TCM for 16 h. Results are means ± SEM (n = 3).
(D) In vitro tube formation assay using endothelial cells mixed with CD5+ or CD5− B cells at the presence of 2.5% TCM. Results shown are representative of 3 independent experiments, as means ± s.e.m. (n = 6). *P < 0.05.
(E) Tumor growth curve showing inhibition of B16 tumor growth in Rag1−/− mice when CD5− B cells are included in the adoptively transferred T cells. Results shown are representative of 2 independent experiments (n = 6). *P < 0.05.
(F) ELISA assessing IFN-γ production from mouse T cells co-cultured with CD5+ or CD5− splenic B cells in the presence of irradiated B16 tumor cells, means ± SEM (n = 3). **P < 0.01.
(G) Multiplex bead assay detecting IL-10 and IL-12 production from mouse dendritic cells co-cultured with CD5+ or CD5− splenic B cells in the presence of irradiated B16 tumor cells. Data represent means ± SEM (n = 3). **P < 0.01. See also Figure S6 and Figure S7.
We further tested whether CD5+ B cells could contribute to accelerate tumor growth through enhancing tumor angiogenesis. Hemoglobin assays using Matrigel containing tumor cells and CD5-positive or CD5-negative B cells indicated that CD5+ B cells were stronger in inducing tumor angiogenesis than their CD5− counterparts (Figure 6B). CD5+ B cells, especially when stimulated by the IL-6 containing tumor-conditioned medium, secreted more pro-angiogenic factors, including VEGF and CCL2 (Figure 6C), and stimulated angiogenesis in vitro (Figure 6D). We also assessed whether CD5+ B cells could promote tumor growth by inhibiting antitumor immune responses. Adoptively transferring T cells together with CD5− B cells into Rag1−/− mice lacking B and T cells inhibited tumor growth (Figure 6E). Furthermore, in the presence of tumor cells, significantly more IFN-γ was detected in the T cell culture incubated with CD5− B cells than that with CD5+ B cells (Figure 6F). IL-10 production by CD5+ B cells was significantly higher compared to that by CD5− B cells (Figure 6G). In addition, CD5+ B cells were capable of more potently inhibiting IL-12 production by dendritic cells than CD5− B cells. We further showed that CD5+ B cells stimulated Stat3 activation in tumor cells in part by upregulating Stat3-downstream cytokines, including IL-10 and GM-CSF (Figure S7). Thus, CD5+ B cells are the main contributing B cell population for tumor progression. These studies on the roles of CD5+ and CD5− B cells in tumor growth may clarify some controversies surrounding B cells’ roles in cancer.
CD5-STAT3 Feed-forward Loop Correlated with Oncogenic Activity in Human Tumor
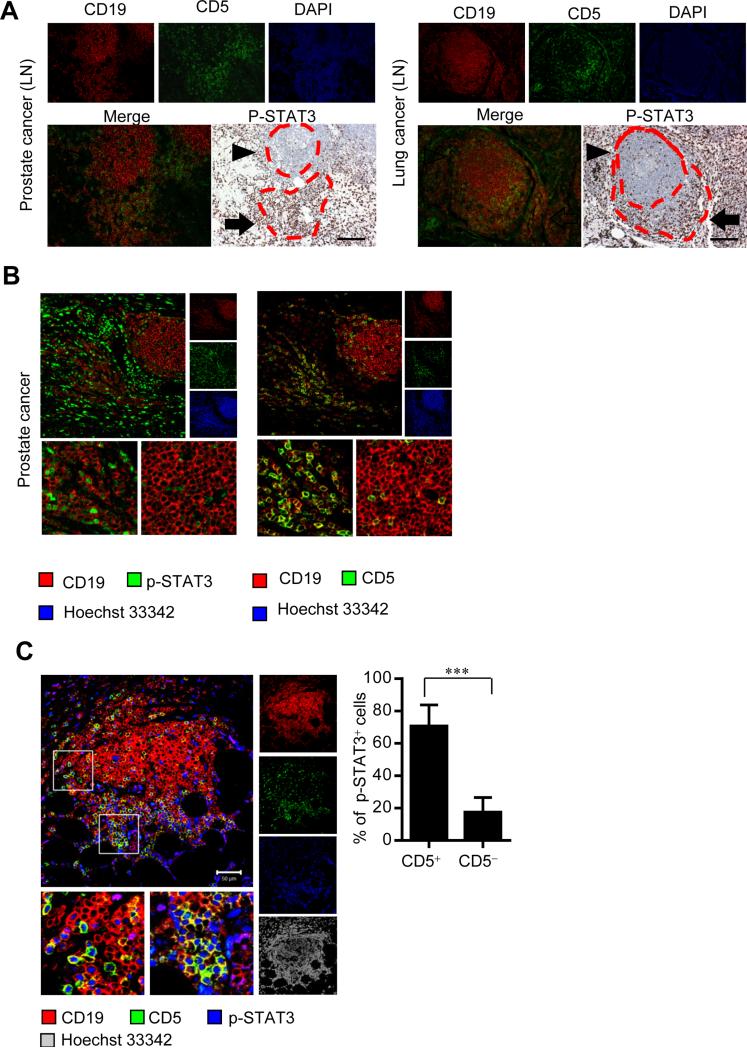
To extend our findings in mice to human cancers, we analyzed CD5 expression and STAT3 activity in tumor draining lymph nodes (LNs) from prostate cancer and lung cancer patients, as well as primary tumors from prostate cancer patients. We were able to detect strong STAT3 activation in the CD19+CD5+ areas but not the CD19+CD5− areas in both primary tumors and tumor draining LNs from prostate and lung cancer patients. Also, in both human primary tumors and tumor LNs, p-STAT3 expression and CD5 positivity in CD19+ B cells strictly correlated (Figure 7A and 7B). We next explored whether the CD5-STAT3 feed-forward loop has any relevance to human cancers. Our previous study, with limited number of patient specimens and corresponding survival data, indicated that the number of B cells in omental tissues of ovarian cancer patients with metastatic diseases correlates with an increase in STAT3 activity and might be a contributing negative factor for patient survival (Yang et al., 2013a). We analyzed these same tissues for CD5 expression and STAT3 activation in B cells. Our results showed that CD5 expression in CD19+ B cells was largely confined to those with elevated STAT3 activity, and STAT3 activated B cells were positive for CD5 (Figure 7C). These results support the findings that CD5 activates STAT3 in B cells, which is consistent with the finding that STAT3 promotes CD5 expression, forming a feed-forward loop that increases of CD5+ B cells and their oncogenic activity in cancer.
Figure 7. p-STAT3 levels and CD5 positivity in CD19+ B cells correlate in human cancer.
(A) Immunofluorescent staining of CD19 and CD5 and immunohistochemical staining of p-STAT3 in consecutive sections of regional lymph nodes (LN) from patients with prostate cancer (left) and non-small cell lung cancer (right). ▶ indicates CD19+CD5− and ➡ shows CD19+CD5+ cells. Scale bars, 100 μm.
(B) Immunofluorescent staining and confocal microscopic images showing co-expression of p-STAT3 (left, green) and CD5 (right, green) in CD19+ B cells (red) in consecutive sections of human prostate cancer tissue. Scale bars, 50 μm.
(C) Immunofluorescent staining and confocal microscopic images (left) showing p-STAT3 (blue) and CD5 (green) in CD19+ B cells (red) in omental tissue sections from ovarian cancer patients. Scale bars: 50 μm. Histograms (right) showing p-STAT3 expressing cell frequencies within CD5+ and CD5− B cells. Results represent means ± SEM, n = 17 (17 images acquired from omental tissue sections from 7 ovarian cancer patients). ***P < 0.001.
DISCUSSION
Although a critical role of IL-6 in regulating the most fundamental functions of B cells has been evident soon after its discovery (Nishimoto et al., 1994), only a small percentage of B cells is IL-6Rα positive in periphery in normal physiology (Hoge et al., 2013). In contrast to B cells, in T cells both CD5 and IL-6Rα are widely expressed. Why only such small percentage of B cells express IL-6Rα requires further investigation. Our findings that CD5 can respond and bind to IL-6 identify a biologically important ligand for CD5. STAT3 signaling as a central axis in both cancer and inflammation is well established. This study suggests that CD5 expression is regulated by STAT3, thereby allowing an increase in percentage of CD5+ B cells in cancer in which STAT3 is activated. While an important role of B cells in promoting as well as inhibiting cancer has been demonstrated (Andreu et al., 2010; de Visser et al., 2005; DiLillo et al., 2010; Mantovani, 2011), our current study has defined CD5+ B cells but not CD5− B cells as tumor promoting. Results from our current study raise the possibility that other cell types may also use CD5 to respond to IL-6, thereby contributing to STAT3 activation.
CD5 is expressed on T cells and a subset of B cells and known to bind to ligand CD72 and modulate lymphocyte activation and differentiation processes (Baumgarth, 2011; Berland and Wortis, 2002). It has also been reported that CD5 interacts with conserved fungal cell wall components and suppresses zymosan-induced septic shock-like syndrome (Vera et al., 2009). Stat3 plays an important role in Th17 cell development. Moreover, CD5 can induce Th17 cell development by promoting Stat3 activation (de Wit et al., 2011). Our findings are consistent with many of these results, and demonstrate that CD5 can bind to IL-6 to induce Stat3 activation. Further studies are required to determine whether at least some of the CD5's biological functions reported (de Wit et al., 2011; Vera et al., 2009) are contributed in part by its ability to bind to IL-6.
Results from our current study also suggest that overexpression of CD5 itself can promote STAT3 activation. Since STAT3 serves as a direct transcriptional factor for CD5, as shown by our data, the increase of CD5+ B cells in the tumor microenvironment is caused by persistent activation of STAT3 in tumor. Activation of STAT3 in tumor cells and in the tumor microenvironment is contributed by many factors, albeit a critical role of IL-6 (Kujawski et al., 2008; Wang et al., 2009). In addition to multiple cytokines and growth factors, chemokines and other ligands of GPCRs including sphingosine-1-phosphate (S1P), have been shown to be critical for persistent activation of STAT3 in tumor, including many types of immune cells in the tumor microenvironment (Deng et al., 2012; Herrmann et al., 2014; Lee et al., 2010; Lee et al., 2009). A common feature of STAT3 activation is that STAT3 regulates expression of their receptors, which in turn activate STAT3, even in the absence of abundant amounts of their ligands. For example, elevated expression of one of the receptors for S1P, S1PR1, enhances STAT3 activity in tumor cells and various immune cells in the tumor microenvironment, promoting cancer progression and facilitating colonization of myeloid cells in the future metastatic sites to provide niches for tumor cells (Deng et al., 2012).
CD5 is overexpressed by several immune cell malignancies, including those of B cells. Our finding that CD5 and STAT3 forming a feed-forward loop provides some mechanistic insights on how CD5 overexpression promotes oncogenic potential of these malignancies. In addition, our analyses of human tumor and tumor-associated tissues suggest that activated STAT3 in B cells is largely confined to CD5+ B cell subpopulation. Since STAT3 drives CD5 expression, and the correlation between CD5 and activated STAT3 could be contributed by upregulation of CD5, suggesting a feed-forward loop that increases of CD5+ B cells and their oncogenic activity in cancer. Furthermore, our results suggest that CD5+ B cells in tumor may be important for both diagnostic and therapeutic purposes. Our findings may also have implications for targeting IL-6 signaling in cancer.
EXPERIMENTAL PROCEDURES
Mice, cells and reagents
Cd19-Cre mice, C57BL/6 mice and Rag1−/− mice were obtained from Jackson Laboratory. Stat3flox/flox mice were kindly provided by Dr. S. Akira (Osaka University) and Jak2flox/flox mice by Dr. K-U Wagner (University of Nebraska Medical Center). IL-6Rα (CD126)-deficient (Il6ra−/−) mice were kindly provided by Dr. Simon A.Jones (School of Medicine, Cardiff University).
B16 mouse melanoma, A20 mouse B cell lymphoma, NIH-3T3 fibroblast cell lines were from ATCC. MB49 mouse bladder carcinoma cells, C4 mouse melanoma cells and mouse endothelial cells were kindly provided by Drs. T. Ratliff (University of Iowa), J. Fidler and S. Huang (M.D. Anderson Cancer Center), respectively. Tumor-conditioned medium was prepared as described (Lee et al., 2009).
Sources of antibodies were: phospho-STAT3 (Y705), phospho-JAK2 (Y1007/1008), phospho-Src (Y416), Cell Signaling Technology; STAT3 (C-20) to detect total STAT3 protein, mouse CD5, Santa Cruz Biotechnology; mouse gp130, Millipore; human CD19, AbD Serotec; human CD5, Thermo Scientific. Antibodies used for flow cytometry: mouse CD5 (53-7.3), mouse CD19 (6D5), mouse IL-6Rα (D7715A7), human CD5 (UCHT2), human CD19 (HIB19), human IL-6Rα (UV4) were from Biolegend; and phospho-Stat3 antibodies were from BD Biosciences. Mouse IL-6 neutralization antibody and isotype control were purchased from eBioscience. Cd5, Stat3 and gp130 siRNAs were obtained from Santa Cruz Biotechnology. IL-6 was purchased from PeproTech and used at 20 ng ml−1. AZD 1480 was from AstraZeneca and used at 1 μM. Recombinant mouse CD5 was obtained from R&D. Biotin anti- mouse IL-6 (MP5-32C11) was purchased from Biolegend.
In vivo mouse experiments
Mouse care and experimental procedures were performed under pathogen-free condition in accordance with established institutional guidance and approved protocols from Institutional Animal Care and Use Committee at City of Hope. Cd19-Cre mice were crossed with Stat3flox/flox to generate mice with Stat3 or Jak2 ablation in B cells. Jak2flox/flox mice (Krempler et al., 2004) were crossed with Mx1-Cre mice which were obtained from the Jackson Laboratory. Mice with Jak2−/− hematopoietic cells were generated by treating Mx1-Cre/Jak2flox/flox mice with poly (I:C) as described previously (Kortylewski et al., 2009). Mouse melanoma B16 or MB49 bladder tumor cells were mixed with different subsets of B cells and then injected subcutaneously into Rag1−/− mice. For adoptive transfer, T cells or B cells were injected intravenously into Rag1−/− mice, and then B16 tumor cells were implanted subcutaneously after 24 h.
IL-6 binding assay
To detect IL-6 binding to CD5, we performed enzyme-linked immunosorbent assay (Ieguchi et al., 2010). Briefly, recombinant mouse CD5 was incubated in a 96-well plate at 4°C for overnight. Unbound CD5 was removed, and 200 μl of blocking buffer (ImmunoChemistry Technologies) was added and incubated at RT for 2 hours. The wells were washed with PBS containing 0.05% Tween-20, and soluble IL-6 in 1% BSA was added to each well and incubated at room temerature for 2 hours. IL-6 was quantified using biotin-avidin assay followed by a colorimetric detection. Anti-mCD5 blocking assay was performed as described above, except that anti-CD5 antibody was added to CD5-coated wells before adding IL-6.
Biacore analysis
To detect IL-6 binding to CD5 affinity, we performed Biacore assay. Binding affinity was measured between recombinant mouse CD5 and recombinant mouse IL6 using a Biacore X100 instrument (GE Healthcare). The IL6 was immobilized on a CM5 sensor chip (800 RU) using amine-coupling chemistry following the manufacture's instructions. Using the kinetic affinity program, serial concentrations of mCD5 (0.25 - 8 μM) were injected at a flow rate of 30 μl/min at 25 °C. In all experiments, data were zero adjusted and the reference cell subtracted. The data were analyzed using the 1:1 model, Biacore X100 BIAevaluation 2.0 software (GE Healthcare) and reported as KD.
Isolation of mouse immune cells
The procedure for isolating immune cells has been described previously (Lee et al., 2010). Mouse splenic CD19+CD5+ and CD19+CD5− B cells were FACS sorted by Aria III Sorter (BD).
Human peripheral blood mononuclear cell (PBMC) preparation
The use of anonymous discarded blood samples was approved by the City of Hope Institutional Review Board and exempt from the informed consent requirement. PBMCs were isolated from citrated human blood by density gradient centrifugation using Ficoll-Paque Plus (GE healthcare biosciences).
Flow cytometry
Data were collected on CyAn and analyzed by FlowJo software (Tree Star). For intracellular p-Stat3 staining, isolated cells were fixed in paraformaldehyde and permeabilized in methanol (Kortylewski et al., 2005).
Immunofluorescent staining and confocal microscopy
Discarded tissue specimens from ovarian cancer, lung cancer and prostate cancer patient were obtained through a City of Hope IRB approved protocol. Tissue section staining using fluorophore-conjugated secondary antibodies and imaging by con-focal microscopy were as previously described (Deng et al., 2012).
Cell transfection and promoter activity assay
For B16 or 3T3 cells, pCMV/Sport6/mCD5, pRC/CMV/STAT3C, the generated luciferase constructs, mouse STAT3 siRNA and gp130 siRNA as well as their controls were transfected into the cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer`s instructions. For A20 cells, mouse Stat3 siRNA and mouse CD5 siRNA as well as their controls were transfected into the cells using Amaxa cell line Nucleofector Kit V (Lonza) according to the manufacturer`s instructions. Putative STAT binding sites in mouse Cd5 gene were identified using TRANSFAC software and 630bp DNA fragment (−558~ +72) containing two putative STAT binding sites was inserted into pGL3-Basic luciferase reporter vector to generate mouse CD5 luciferase construct (pGL3-Cd5 WT). For site-mutation, the putative STAT binding sites, GTGGTTTCACAGG (−175~ −188) and GGAGTTCCCAGC (−329 ~−341) were changed to GTGGTagCACAGG (pGL3-Cd5 Mut2) and GGAG agCCCAGC (pGL3-Cd5 Mut1) by QuickChange® site-directed mutagenesis kit (Stratagene), respectively. Promoter activity was determined as describe (Kortylewski et al., 2009).
Chromatin immunoprecipitation (ChIP) assays
The chromatins were prepared from 5 × 106 splenic CD19+ B cells enriched from tumor bearing mice. The following primers, 5’- CCTTTTACCTCGGGTGAGCT-3’, and 5’- AAGGGGAAGCAGGCAGTGTG -3’, were used to amplify the mouse Cd5 promoter region (−388~−106) spanning the putative STAT-binding sites.
Coimmunoprecipitation and immunoblotting
For the interaction study of CD5 and JAK2, B16 cells were transfected with pCMV/sport6/Cd5 or control vector for 48 hours and lysed in buffer containing 25 mM Tirs-HCl, pH7.4, 1% NP-40, 150 mM NaCl, 5% glycerol and 1 mM EDTA. For interaction study of CD5 and gp130, CD19+ splenic B cells were incubated with 5% TCM; for CD5 and p-JAK2 interaction study, A20 cells were incubated with 10% TCM.
Quantitative Real-Time PCR
Total RNAs from various cell population were purified using the RNeasy system following the manufacturer`s instructions (Qiagen). RNA (0.5 to 1 μg) was reverse-transcribed to cDNA using iScript cDNA Synthesis Kit (Bio-Rad), and real-time PCR reactions were performed as described previously (Lee et al., 2010). Specific primers for mouse Cd5, Stat3, Ccl2, Vegf and Gapdh were purchased from SA Bioscience. Samples were run in triplicate and expressed as means ± SEM.
Tube formation assay and in vivo Matrigel angiogenesis assay
Endothelial cells (ECs) and B cells were co-cultured on neutralized collagen at 1:1 ratio in 1% FBS-RPMI 1640 medium (1.2 mg/ml; BD Biosciences) for 16 h. Cells were fixed in 4% paraformaldehyde for 10 min, washed, and analyzed under an inverted light microscope (Nikon). Closed networks of vessel-like tubes were counted from each well. Mouse splenic CD19+CD5+ or CD19+CD5− B cells were mixed with B16 tumor cells in 600 ul of growth factor-reduced Matrigel (BD Biosciences) and then injected into Rag1−/− mice. Matrigel plugs were dissected 6 d later and analyzed for hemoglobin content as described (Kujawski et al., 2008).
Cytokine measurement
CD5+ or CD5− B cells together with either T cells (1 × 106) or DCs (1 × 105), all from spleens of B16 tumor-bearing mice, were placed into a well of the 48-well plate containing irradiated B16 tumor cells (2 × 105, 7000 rads). Culture medium was collected after 3 days to determine IFN-γ levels by ELISA (eBioscience) and IL-12, IL-10 levels by Multiplex bead assays with LEGEND plex™ Multi-Analyte Flow Assay Kits (Biolegend).
Statistical analysis
Data are shown as means ± SEM. Statistical comparisons between groups were determined using the Student`s t test to calculate the two-tailed p values: *** P < 0.001, **P < 0.01, *P < 0.05.
Supplementary Material
Highlights.
CD5+ B cells increase in tumors and promote tumor progression
CD5 binds to IL-6, leading to STAT3 activation via gp130 and JAK2
STAT3 upregulates CD5 expression, forming a feed-forward loop in B cells
CD5-STAT3 expression correlates with oncogenic activity in human tumor.
ACKNOWLEDGEMENTS
We thank the dedication of staff members at the flow cytometry core, pathology core and light microscopy core at the Beckman Research Institute at City of Hope Comprehensive Cancer Center for their technical assistance. We also acknowledge the contribution of staff members at the animal facilities at City of Hope. This work is funded in part by R01CA122976, R01CA146092, P50 CA107399 and V Foundation Translational Research Grant. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under grant number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
H.Y., C.Z. developed the concept, designed experiments and prepared the manuscript. C.Z and H.X. carried out most of the experiments, the statistical analyses and contributed to manuscript preparation. W.Z. carried out tissue staining and analyses. P.J.Y performed the Biacore assay, analyzed and described the data. Z.Z. and K.L. performed human PBMCs isolation, flow cytometry and analyzed data. W.L. helped siRNA transfection. H.L. contributed to manuscript preparation. R.J. contributed to the manuscript writing. S.F. and L.K. helped discussion of the clinical significance.
REFERENCES
- Ammirante M, Luo JL, Grivennikov S, Nedospasov S, Karin M. B-cell-derived lymphotoxin promotes castration-resistant prostate cancer. Nature. 2010;464:302–305. doi: 10.1038/nature08782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu P, Johansson M, Affara NI, Pucci F, Tan T, Junankar S, Korets L, Lam J, Tawfik D, DeNardo DG, et al. FcRgamma activation regulates inflammation-associated squamous carcinogenesis. Cancer Cell. 2010;17:121–134. doi: 10.1016/j.ccr.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarth N. The double life of a B-1 cell: self-reactivity selects for protective effector functions. Nat Rev Immunol. 2011;11:34–46. doi: 10.1038/nri2901. [DOI] [PubMed] [Google Scholar]
- Berland R, Wortis HH. Origins and functions of B-1 cells with notes on the role of CD5. Annu Rev Immunol. 2002;20:253–300. doi: 10.1146/annurev.immunol.20.100301.064833. [DOI] [PubMed] [Google Scholar]
- Bollrath J, Phesse TJ, von Burstin VA, Putoczki T, Bennecke M, Bateman T, Nebelsiek T, Lundgren-May T, Canli O, Schwitalla S, et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15:91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Catlett-Falcone R, Landowski TH, Oshiro MM, Turkson J, Levitzki A, Savino R, Ciliberto G, Moscinski L, Fernandez-Luna JL, Nunez G, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- de Visser KE, Korets LV, Coussens LM. De novo carcinogenesis promoted by chronic inflammation is B lymphocyte dependent. Cancer Cell. 2005;7:411–423. doi: 10.1016/j.ccr.2005.04.014. [DOI] [PubMed] [Google Scholar]
- de Wit J, Souwer Y, van Beelen AJ, de Groot R, Muller FJ, Klaasse Bos H, Jorritsma T, Kapsenberg ML, de Jong EC, van Ham SM. CD5 costimulation induces stable Th17 development by promoting IL-23R expression and sustained STAT3 activation. Blood. 2011;118:6107–6114. doi: 10.1182/blood-2011-05-352682. [DOI] [PubMed] [Google Scholar]
- Deng J, Liu Y, Lee H, Herrmann A, Zhang W, Zhang C, Shen S, Priceman SJ, Kujawski M, Pal SK, et al. S1PR1-STAT3 signaling is crucial for myeloid cell colonization at future metastatic sites. Cancer Cell. 2012;21:642–654. doi: 10.1016/j.ccr.2012.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo DJ, Yanaba K, Tedder TF. B cells are required for optimal CD4[r]+[/r] and CD8[r]+[/r] T cell tumor immunity: therapeutic B cell depletion enhances B16 melanoma growth in mice. J Immunol. 2010;184:4006–4016. doi: 10.4049/jimmunol.0903009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong HY, Gorczyca W, Liu Z, Tsang P, Wu CD, Cohen P, Weisberger J. B-cell lymphomas with coexpression of CD5 and CD10. Am J Clin Pathol. 2003;119:218–230. doi: 10.1309/u98advkuc26r2rja. [DOI] [PubMed] [Google Scholar]
- Donnelly RP, Dickensheets H, Finbloom DS. The interleukin-10 signal transduction pathway and regulation of gene expression in mononuclear phagocytes. J Interferon Cytokine Res. 1999;19:563–573. doi: 10.1089/107999099313695. [DOI] [PubMed] [Google Scholar]
- Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, Karin M. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Karin M. Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010;21:11–19. doi: 10.1016/j.cytogfr.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedvat M, Huszar D, Herrmann A, Gozgit JM, Schroeder A, Sheehy A, Buettner R, Proia D, Kowolik CM, Xin H, et al. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16:487–497. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich PC, Behrmann I, Muller-Newen G, Schaper F, Graeve L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J. 1998;334(Pt 2):297–314. doi: 10.1042/bj3340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann A, Cherryholmes G, Schroeder A, Phallen J, Alizadeh D, Xin H, Wang T, Lee H, Lahtz C, Swiderski P, et al. TLR9 is critical for glioma stem cell maintenance and targeting. Cancer Res. 2014;74:5218–5228. doi: 10.1158/0008-5472.CAN-14-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoge J, Yan I, Janner N, Schumacher V, Chalaris A, Steinmetz OM, Engel DR, Scheller J, Rose-John S, Mittrucker HW. IL-6 controls the innate immune response against Listeria monocytogenes via classical IL-6 signaling. J Immunol. 2013;190:703–711. doi: 10.4049/jimmunol.1201044. [DOI] [PubMed] [Google Scholar]
- Ieguchi K, Fujita M, Ma Z, Davari P, Taniguchi Y, Sekiguchi K, Wang B, Takada YK, Takada Y. Direct binding of the EGF-like domain of neuregulin-1 to integrins (avb3 and a6b4) is involved in neuregulin-1/ErbB signaling. J Biol Chem. 2010;285:31388–31398. doi: 10.1074/jbc.M110.113878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA, Scheller J, Rose-John S. Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J Clin Invest. 2011;121:3375–3383. doi: 10.1172/JCI57158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortylewski M, Kujawski M, Wang T, Wei S, Zhang S, Pilon-Thomas S, Niu G, Kay H, Mule J, Kerr WG, et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. 2005;11:1314–1321. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- Kortylewski M, Xin H, Kujawski M, Lee H, Liu Y, Harris T, Drake C, Pardoll D, Yu H. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell. 2009;15:114–123. doi: 10.1016/j.ccr.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempler A, Qi Y, Triplett AA, Zhu J, Rui H, Wagner KU. Generation of a conditional knockout allele for the Janus kinase 2 (Jak2) gene in mice. Genesis. 2004;40:52–57. doi: 10.1002/gene.20063. [DOI] [PubMed] [Google Scholar]
- Kujawski M, Kortylewski M, Lee H, Herrmann A, Kay H, Yu H. Stat3 mediates myeloid cell-dependent tumor angiogenesis in mice. J Clin Invest. 2008;118:3367–3377. doi: 10.1172/JCI35213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht B, Kreher S, Anagnostopoulos I, Johrens K, Monteleone G, Jundt F, Stein H, Janz M, Dorken B, Mathas S. Aberrant expression of the Th2 cytokine IL-21 in Hodgkin lymphoma cells regulates STAT3 signaling and attracts Treg cells via regulation of MIP-3alpha. Blood. 2008;112:3339–3347. doi: 10.1182/blood-2008-01-134783. [DOI] [PubMed] [Google Scholar]
- Lee H, Deng J, Kujawski M, Yang C, Liu Y, Herrmann A, Kortylewski M, Horne D, Somlo G, Forman S, et al. STAT3-induced S1PR1 expression is crucial for persistent STAT3 activation in tumors. Nat Med. 2010;16:1421–1428. doi: 10.1038/nm.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Herrmann A, Deng JH, Kujawski M, Niu G, Li Z, Forman S, Jove R, Pardoll DM, Yu H. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15:283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Teitz-Tennenbaum S, Donald EJ, Li M, Chang AE. In vivo sensitized and in vitro activated B cells mediate tumor regression in cancer adoptive immunotherapy. J Immunol. 2009;183:3195–3203. doi: 10.4049/jimmunol.0803773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A. B cells and macrophages in cancer: yin and yang. Nat Med. 2011;17:285–286. doi: 10.1038/nm0311-285. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- Matsushita T, Yanaba K, Bouaziz JD, Fujimoto M, Tedder TF. Regulatory B cells inhibit EAE initiation in mice while other B cells promote disease progression. J Clin Invest. 2008;118:3420–3430. doi: 10.1172/JCI36030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurath MF, Finotto S. IL-6 signaling in autoimmunity, chronic inflammation and inflammation-associated cancer. Cytokine Growth Factor Rev. 2011;22:83–89. doi: 10.1016/j.cytogfr.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Nishimoto N, Ogata A, Shima Y, Tani Y, Ogawa H, Nakagawa M, Sugiyama H, Yoshizaki K, Kishimoto T. Oncostatin M, leukemia inhibitory factor, and interleukin 6 induce the proliferation of human plasmacytoma cells via the common signal transducer, gp130. J Exp Med. 1994;179:1343–1347. doi: 10.1084/jem.179.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putoczki T, Ernst M. More than a sidekick: the IL-6 family cytokine IL-11 links inflammation to cancer. J Leukoc Biol. 2010;88:1109–1117. doi: 10.1189/jlb.0410226. [DOI] [PubMed] [Google Scholar]
- Rose-John S. IL-6 trans-signaling via the soluble IL-6 receptor: importance for the pro-inflammatory activities of IL-6. Int J Biol Sci. 2012;8:1237–1247. doi: 10.7150/ijbs.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose-John S, Neurath MF. IL-6 trans-signaling: the heat is on. Immunity. 2004;20:2–4. doi: 10.1016/s1074-7613(04)00003-2. [DOI] [PubMed] [Google Scholar]
- Stout BA, Bates ME, Liu LY, Farrington NN, Bertics PJ. IL-5 and granulocyte-macrophage colony-stimulating factor activate STAT3 and STAT5 and promote Pim-1 and cyclin D3 protein expression in human eosinophils. J Immunol. 2004;173:6409–6417. doi: 10.4049/jimmunol.173.10.6409. [DOI] [PubMed] [Google Scholar]
- Taga T, Kishimoto T. Gp130 and the interleukin-6 family of cytokines. Annu Rev Immunol. 1997;15:797–819. doi: 10.1146/annurev.immunol.15.1.797. [DOI] [PubMed] [Google Scholar]
- Trinchieri G. Cancer and inflammation: an old intuition with rapidly evolving new concepts. Annu Rev Immunol. 2012;30:677–706. doi: 10.1146/annurev-immunol-020711-075008. [DOI] [PubMed] [Google Scholar]
- Vera J, Fenutria R, Canadas O, Figueras M, Mota R, Sarrias MR, Williams DL, Casals C, Yelamos J, Lozano F. The CD5 ectodomain interacts with conserved fungal cell wall components and protects from zymosan-induced septic shock-like syndrome. Proc Natl Acad Sci U S A. 2009;106:1506–1511. doi: 10.1073/pnas.0805846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yi T, Kortylewski M, Pardoll DM, Zeng D, Yu H. IL-17 can promote tumor growth through an IL-6-Stat3 signaling pathway. J Exp Med. 2009;206:1457–1464. doi: 10.1084/jem.20090207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JR, Liss MA, Muldong MT, Palazzi K, Strasner A, Ammirante M, Varki N, Shabaik A, Howell S, Kane CJ, et al. Tumor infiltrating B-cells are increased in prostate cancer tissue. J Transl Med. 2014;12:30. doi: 10.1186/1479-5876-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H, Lu R, Lee H, Zhang W, Zhang C, Deng J, Liu Y, Shen S, Wagner KU, Forman S, et al. G-protein-coupled receptor agonist BV8/prokineticin-2 and STAT3 protein form a feed-forward loop in both normal and malignant myeloid cells. J Biol Chem. 2013;288:13842–13849. doi: 10.1074/jbc.M113.450049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing C, Ma N, Xiao H, Wang X, Zheng M, Han G, Chen G, Hou C, Shen B, Li Y, Wang R. Critical role for thymic CD19+CD5+CD1dhiIL-10+ regulatory B cells in immune homeostasis. J Leukoc Biol. 2015;97:547–556. doi: 10.1189/jlb.3A0414-213RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaba K, Bouaziz JD, Haas KM, Poe JC, Fujimoto M, Tedder TF. A regulatory B cell subset with a unique CD1d[r]hi[/r]CD5[r]+[/r] phenotype controls T cell-dependent inflammatory responses. Immunity. 2008;28:639–650. doi: 10.1016/j.immuni.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Yang C, Lee H, Jove V, Deng J, Zhang W, Liu X, Forman S, Dellinger TH, Wakabayashi M, Yu H, Pal S. Prognostic significance of B-cells and pSTAT3 in patients with ovarian cancer. PLoS One. 2013a;8:e54029. doi: 10.1371/journal.pone.0054029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Lee H, Pal S, Jove V, Deng J, Zhang W, Hoon DS, Wakabayashi M, Forman S, Yu H. B Cells Promote Tumor Progression via STAT3 Regulated-Angiogenesis. PLoS One. 2013b;8:e64159. doi: 10.1371/journal.pone.0064159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki A, Miyagaki T, DiLillo DJ, Matsushita T, Horikawa M, Kountikov EI, Spolski R, Poe JC, Leonard WJ, Tedder TF. Regulatory B cells control T-cell autoimmunity through IL-21-dependent cognate interactions. Nature. 2012;491:264–268. doi: 10.1038/nature11501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Venkatapathy S, Rao G, Harrington CA. Expression profiling of B cell chronic lymphocytic leukemia suggests deficient CD1-mediated immunity, polarized cytokine response, altered adhesion and increased intracellular protein transport and processing of leukemic cells. Leukemia. 2002;16:2429–2437. doi: 10.1038/sj.leu.2402711. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.