Abstract
The human virome plays important roles in health and immunity. However, current methods for detecting viral infections and antiviral responses have limited throughput and coverage. Here, we present VirScan, a high-throughput method to comprehensively analyze antiviral antibodies using immunoprecipitation and massively parallel DNA sequencing of a bacteriophage library displaying proteome-wide peptides from all human viruses. We assayed over 108 antibody-peptide interactions in 569 humans across four continents, nearly doubling the number of previously established viral epitopes. We detected antibodies to an average of 10 viral species per person and 84 species in at least two individuals. Although rates of specific virus exposure were heterogeneous across populations, antibody responses targeted strikingly conserved “public epitopes” for each virus, suggesting that they may elicit highly similar antibodies. VirScan is a powerful approach for studying interactions between the virome and the immune system.
Introduction
The collection of viruses found to infect humans (the “human virome”) can have profound effects on human health (1). In addition to directly causing acute or chronic illness, viral infection can also alter host immunity in more subtle ways, leaving an indelible footprint on the immune system (2). For example, latent herpesvirus infection has been shown to confer symbiotic protection against bacterial infection in mice through prolonged production of interferon-γ and systemic activation of macrophages (3). This interplay between virome and host immunity has also been implicated in the pathogenesis of complex diseases such as type 1 diabetes, inflammatory bowel disease, and asthma (4). Despite this growing appreciation for the importance of interactions between the virome and host, a comprehensive method to systematically characterize these interactions has yet to be developed (5).
Viral infections can be detected by serological- or nucleic acid-based methods (6). However, nucleic acid tests fail in cases where viruses have already been cleared after causing or initiating tissue damage and can miss viruses of low abundance or viruses not normally present in the sampled fluid or surface. In contrast, humoral responses to infection typically arise within two weeks of initial exposure and can persist over years or decades (7). Tests detecting antiviral antibodies in peripheral blood can therefore identify ongoing and cleared infections. However, current serological methods are predominantly limited to testing one virus at a time and are therefore only employed to address specific clinical hypotheses. Scaling serological analyses to encompass the complete human virome poses significant technical challenges, but would be of great value for better understanding host-virus interactions, and would overcome many of the limitations associated with current clinical technologies. In this work, we present VirScan, a programmable, high-throughput method to comprehensively analyze antiviral antibodies using immunoprecipitation and massively parallel DNA sequencing of a bacteriophage library displaying proteome-wide coverage of peptides from all human viruses.
Results
The VirScan Platform
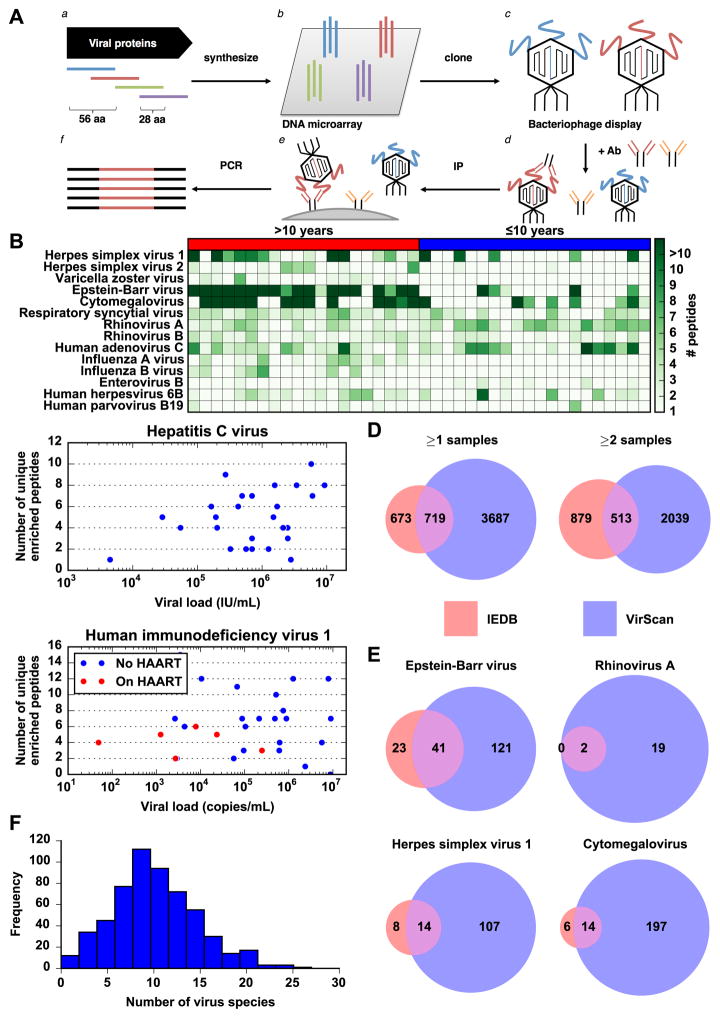
VirScan utilizes the Phage Immunoprecipitation sequencing (PhIP-seq) technology previously developed in our laboratory (8). Briefly, we used a programmable DNA microarray to synthesize 93,904 200-mer oligonucleotides, encoding 56-residue peptide tiles, with 28 residue overlaps, that together span the reference protein sequences (collapsed to 90% identity) of all viruses annotated to have human tropism in the UniProt database (Fig. 1A.a and 1A.b) (9). This library includes peptides from 206 species of virus and over 1,000 different strains. We cloned the library into a T7 bacteriophage display vector for screening (Fig. 1A.c).
Fig. 1.
General VirScan analysis of the human virome. (A) Construction of the virome peptide library and VirScan screening procedure. (a) The virome peptide library consists of 93,904 56 amino acid peptides tiling, with 28 amino acid overlap, across the proteomes of all known human viruses. (b) 200 nt DNA sequences encoding the peptides were printed on a releasable DNA microarray. (c) The released DNA was amplified and cloned into a T7 phage display vector and packaged into virus particles displaying the encoded peptide on its surface. (d) The library is mixed with a sample containing antibodies that bind to their cognate peptide antigen on the phage surface. (e) The antibodies are immobilized and unbound phage are washed away. (f) Finally, amplification of the bound DNA and high throughput sequencing of the insert DNA from bound phage reveals peptides targeted by sample antibodies. Abbreviations: aa, amino acid; Ab, antibody; IP: immunoprecipitation. (B) Antibody profile of randomly chosen group of donors to show typical assay results. Each row is a virus, each column is a sample. The label above each chart indicates whether the donors are over 10 years of age or at most 10 years of age. The color intensity of each cell indicates the number of peptides from the virus that were significantly enriched by antibodies in the sample. (C) Scatter plot of the number of unique enriched peptides (after applying maximum parsimony filtering) detected in each sample against the viral load in that sample. Data are shown for the HCV positive and HIV positive samples for which we were able to obtain viral load data. For the HIV positive samples, red dots indicate samples from donors currently on highly active anti-retroviral therapy at the time the sample was taken, whereas blue dots indicate different donors prior to undergoing therapy. (D) Overlap between enriched peptides detected by VirScan and human B cell epitopes from viruses in IEDB. The entire pink circle represents the 1,392 groups of non-redundant IEDB epitopes that are also present in the VirScan library (out of 1,559 clusters total). The overlap region represents the number of groups with an epitope that is also contained in an enriched peptide detected by VirScan. The purple only region represents the number of non-redundant enriched peptides detected by VirScan that do not contain an IEDB epitope. Data are shown for peptides enriched in at least one (left) or at least two (right) samples. (E) Overlap between enriched peptides detected by VirScan and human B cell epitopes in IEDB from common human viruses. The regions represent the same values as in (D) except only epitopes corresponding to the indicated virus are considered, and only peptides from that virus that were enriched in at least two samples were considered. (F) Distribution of number of viruses detected in each sample. The histogram depicts the frequency of samples binned by the number of virus species detected by VirScan. The mean and median of the distribution are both approximately 10 virus species.
To perform a screen, we incubate the library with a serum sample containing antibodies, recover the antibodies using a mixture of protein A and G coated magnetic beads, and remove unbound phage particles by washing (Fig. 1A.d and 1A.e). Finally, we perform PCR and massively parallel sequencing on the phage DNA to quantify enrichment of each library member due to antibody binding (Fig. 1A.f). Each sample is screened in duplicate to ensure reproducibility. VirScan requires only 2 μg of immunoglobulin (<1 μL of serum) per sample and can be automated on a 96-well liquid handling robot (10). PCR product from 96 immunoprecipitations can be individually barcoded and pooled for sequencing, reducing the cost for a comprehensive viral antibody screen to approximately $25 per sample.
Following sequencing, we tally the read count for each peptide before (“input”) and after (“output”) immunoprecipitation. We then fit a zero-inflated generalized Poisson model to the distribution of output read counts for each input read count and regress the parameters as a function of input read count (fig. S1). Using this model, we calculate a −log10(p-value) for the significance of each peptide’s enrichment. Finally, we call a peptide significantly enriched if its −log10(p-value) is greater than the reproducibility threshold of 2.3 in both replicates (fig. S2).
Characterizing VirScan’s sensitivity and specificity
Fig. 1B shows the antibody profiles of a set of human viruses in sera from a typical group of individuals in a heat map format that illustrates the number of enriched peptides from each virus. We frequently detected antibodies to multiple peptides from common human viruses, such as Epstein-Barr virus (EBV), Cytomegalovirus (CMV), and rhinovirus. As expected, we observed more peptides to be enriched from viruses with larger proteomes, such as EBV and CMV, likely because there are more epitopes available for recognition. We noticed fewer enriched peptides in samples from individuals less than ten years of age compared to their geographically matched controls, in line with an accumulation of viral infections throughout adolescence and adulthood. However, there were occasional samples from young donors with very strong responses to viruses that cause childhood illness, such as Parvovirus B19 and Herpesvirus 6B, which cause the “fifth disease” and “sixth disease” of the classical infectious childhood rashes, respectively (11). These observations are examined in greater detail in Fig. 2.
Fig. 2.
Population stratification of the human virome immune response. The bar graphs depict the differences in exposure to viruses between donors who are (A) less than ten years of age versus over ten years of age, (B) HIV positive versus HIV negative, (C) residing in Peru versus residing in the United States, (D) residing in South Africa versus residing in the United States, and (E) residing in Thailand versus residing in the United States. Asterisks indicate false discovery rate < 0.05.
We developed a computational method to identify the set of viruses to which an individual has been exposed, based on the number of enriched peptides identified per virus. Briefly, we set a threshold number of significant non-overlapping enriched peptides for each virus. We empirically determined that a threshold of three non-overlapping enriched peptides gave the best performance for detecting Herpes simplex virus 1 compared to a commercial serologic test, described below (Table 1). For other viruses, we adjusted the threshold to account for the size of the viral proteome (fig. S3). Next, we tally the number of enriched peptides from each virus. Antibodies generated against a specific virus can cross-react with similar peptides from a related virus. This would lead to false positives because an antibody targeted to an epitope from one virus to which a donor was exposed would also enrich a homologous peptide from a related virus to which the donor may not have been exposed. In order to address this issue, we adopted a maximum parsimony approach to infer the fewest number of virus exposures that could elicit the observed spectrum of antiviral peptide antibodies. For groups of enriched peptides that share a 7 amino acid subsequence and may be recognized by a single specific antibody, we only count it as one epitope for the virus that has the greatest number of other enriched peptides. If this adjusted peptide count is greater than the threshold for that virus, the sample is considered positive for the virus. For this analysis, we also filtered out peptides that were enriched in only one of the 569 samples to avoid spurious hits.
Table 1.
Virscan’s sensitivity and specificity on samples with known viral infections. Sensitivity is the percentage of samples positive for the virus as determined by VirScan out of all n known positives. Specificity is the percentage of samples negative for the virus by VirScan out of all n known negatives.
| Virus | Sensitivity (n) | Specificity (n) |
|---|---|---|
| Hepatitis C virus | 92% (26) * | 97%** (34) |
| Human immunodeficiency virus 1 | 95% (61) * | 100% (33) |
| Herpes simplex virus 1 | 97% (38) | 100% (6) |
| Herpes simplex virus 2 | 90% (20) | 100% (24) |
We found that although the false negative samples did not meet our stringent cut-off for enriching multiple unique peptides, they had detectable antibodies to a recurrent epitope. By modifying the criterion to allow for samples that enrich multiple homologous peptides that share a recurrent epitope as described in the text, the sensitivity of detecting Hepatitis C virus increases to 100% and the sensitivity for detecting HIV increases to 95%. This modified criterion does not significantly affect specificity (fig. S13).
The one false positive was from an individual whose HCV-negative status was self-reported, but had antibodies to as many HCV peptides as 23% of the true HCV positive individuals and is likely to be HCV positive now or in the past. It is possible that this individual was exposed to HCV but cleared the infection. If true, the observed specificity for HCV is 100%.
Using this analytical framework, we measured the performance of VirScan using serum samples from patients known to be infected or not infected with human immunodeficiency virus (HIV) and Hepatitis C virus (HCV), based on commercial ELISA and Western blot assays. For both viruses, VirScan achieves very high sensitivities and specificities of ~95% or higher (Table 1) over a wide range of viral loads (Fig. 1C). The viral genotype was also known for the HCV positive samples. Despite the over 70% amino acid sequence conservation among HCV genotypes (12), which poses a problem for all antibody-based detection methods, VirScan correctly reported the HCV genotype in 69% of the samples. We also compared VirScan to a commercially available serology test that is type specific for the highly related Herpes simplex viruses 1 and 2 (HSV1 and HSV2) (Table 1). These results demonstrate that VirScan performs well in distinguishing between closely related viruses and viruses that range in size from small (HIV and HCV) to very large (HSV1 and HSV2) with high sensitivity and specificity.
Population-level analysis of viral exposures
After ascertaining the performance of VirScan for a panel of viruses, we undertook a large-scale screening of samples with unknown exposure history. Using our multiplex approach, we assayed over 106 million antibody-peptide interactions using samples from 569 human donors in duplicate. We detected antibody responses to an average of 10 species of virus per sample (Fig. 1F). Each person is likely exposed to multiple distinct strains of some viral species. We detected antibody responses to 62 of the 206 species of virus in our library in at least 5 individuals, and 84 species in at least 2 individuals. The most frequently detected viruses are generally those known to commonly infect humans (Table 2, table S1). We occasionally detected what appear to be false positives that may be due to antibodies that cross react with non-viral peptides. For example, 29% of the samples positive for Cowpox virus were right at the threshold of detection and had antibodies against a peptide from the C4L gene that shares an eight amino acid sequence (‘SESDSDSD’) with the Clumping Factor B protein from Staphylococcus aureus, against which humans are known to generate antibodies (13). This will become less of an issue when we test more examples of sera from patients with known infections to determine the set of likely antigenic peptides for a given virus. However, the fact that we do not detect high rates of very rare or virulent viruses strengthens our confidence in VirScan’s specificity (see Supplementary Discussion).
Table 2.
Frequently detected viruses. The ‘%’ column indicates the percentage of samples that were positive for the virus by VirScan. Known HIV and HCV positive samples were excluded when performing this analysis.
| Virus Species | % |
|---|---|
| Human herpesvirus 4 | 87.1% |
| Rhinovirus B | 71.8% |
| Human adenovirus C | 71.8% |
| Rhinovirus A | 67.3% |
| Human respiratory syncytial virus | 65.7% |
| Human herpesvirus 1 | 54.4% |
| Influenza A virus | 53.4% |
| Human herpesvirus 6B | 52.8% |
| Human herpesvirus 5 | 48.5% |
| Influenza B virus | 40.5% |
| Poliovirus | 33.7% |
| Human herpesvirus 3 | 24.3% |
| Human adenovirus F | 20.4% |
| Human adenovirus B | 16.8% |
| Human herpesvirus 2 | 15.5% |
| Enterovirus A | 15.2% |
| Enterovirus B | 13.3% |
We frequently detected antibodies to rhinovirus and respiratory syncytial virus, which are normally found only in the respiratory tract, indicating that VirScan using blood samples is still able to detect viruses that do not cause viremia. We also detected antibodies to influenza, which is normally cleared, and poliovirus, to which most people in modern times generate antibodies through vaccination. Since the original antigen is no longer present, we are likely detecting antibodies secreted by long-lived memory B cells (14).
We detected antibodies to certain viruses less frequently than expected based on previous seroprevalence studies using optimized serum ELISA assays. For example, the frequency at which we detect influenza (53.4%) and poliovirus (33.7%) is lower than expected given that the majority of the population has been exposed to or vaccinated against these viruses. This may be due to reduced sensitivity because of a gradual narrowing and decrease of the long-lived B cell response in the absence of persistent antigen. We also rarely detected antibody responses to small viruses such as JC virus and Torque Teno virus, which are frequently detected using specific tests. We believe that the disparity is due to low titers of antibodies to unmodified, linear epitopes from these viruses. For example, serum antibodies against the major capsid protein of JCV are reported to only recognize conformational epitopes (15). Finally, the frequency of detecting varicella zoster virus (chicken pox) antibodies is also lower than expected (24.3%), even though the frequency of detecting other latent herpesviruses, such as Epstein-Barr virus (87.1%) and cytomegalovirus (48.5%), is similar to the prevalence reported in epidemiological studies (16–18). This may reflect differences in how frequently these viruses shed antigens that stimulate B cell responses or a more limited humoral response that relies on epitopes that cannot be detected in a 56-residue peptide. It might also be possible to increase the sensitivity of detection of these viral antibodies by stimulating memory B cells in vitro to probe the history of infection more deeply.
To assess differences in viral exposure between populations, we split the samples into different groups based on age, HIV status, and geography. We first compared results from children under the age of ten to adults within the United States (HIV-positive individuals were excluded from this analysis) (Fig. 2A). Fewer children were positive for most viruses, including Epstein-Barr virus, HSV1, HSV2, and influenza virus, which is consistent with our preliminary observations comparing the number of enriched peptides (Fig. 1B). In addition to the fact that children may generate lower antibody titers in general, these younger donors probably have not yet been exposed to certain viruses, for example HSV2 which is sexually transmitted (19).
When comparing results from HIV positive to HIV negative samples, we found more of the HIV positive samples to also be seropositive for additional viruses, including HSV2, CMV, and Kaposi’s sarcoma-associated herpesvirus (KSHV) (false discovery rate q < 0.05, Fig. 2B). These results are consistent with prior studies indicating higher risk of these co-infections in HIV positive patients (20–22). Patients with HIV may engage in activities that put them at higher risk for exposure to these viruses. Alternatively, these viruses may increase the risk of HIV infection. HIV infection may reduce the immune system’s ability to control reactivation of normally dormant resident viruses or to prevent opportunistic infections from taking hold and triggering a strong adaptive immune response.
Finally, we compared the evidence of viral exposure between samples taken from adult HIV-negative donors residing in countries from four different continents (the United States, Peru, Thailand, and South Africa). In general, donors outside the United States had higher frequencies of seropositivity (Fig. 2C–E). For example, cytomegalovirus antibodies were found in significantly higher frequencies in samples from Peru, Thailand, and South Africa. Other viruses, such as Kaposi’s sarcoma-associated herpesvirus and HSV1 were detected more frequently in donors from Peru and South Africa, but not Thailand. The observed detection frequency of different adenovirus species varies across populations. Adenovirus C seropositivity was found at similar frequencies in all regions, but Adenovirus D seropositivity was generally higher outside the United States, while Adenovirus B seropositivity was higher in Peru and South Africa, but not in Thailand. The higher rates of virus exposure outside the United States could be due to differences in population density, cultural practices, sanitation, or genetic susceptibility. Interestingly, Influenza B seropositivity was more common in the United States compared to other countries, especially Peru and Thailand. The global incidence of Influenza B is much lower than Influenza A but the standard influenza vaccination contains both Influenza A and B strains, so the elevated frequency of individuals with seroreactivity may be due to higher rates of influenza vaccination in the United States. Other viruses, such as Rhinovirus and Epstein-Barr virus, were detected at very similar frequencies in all the geographic regions.
Analysis of viral epitope determinants
After analyzing responses on the whole virus level, we focused our attention on the specific peptides targeted by these antibodies. We detected antibodies to a total of 8,425 peptides in at least 2 samples, and 15,052 in at least 1 sample. Because of the presence of many related peptides in our library and the Immune Epitope Database (IEDB), for the following analysis we consider a peptide unique only if it does not contain a continuous 7-residue subsequence, the estimated size of a linear epitope, in common with another peptide. Analyzed as such, our VirScan database nearly doubles the 1,559 unique human B cell epitopes from human viruses in the IEDB (23). The epitopes identified in our unbiased analysis demonstrate a significant overlap with those contained in the IEDB (p < 10−30, Fisher’s exact text, Fig. 1D). The amount of overlap is even greater for epitopes from viruses that commonly cause infection (Fig. 1E). We would likely have detected even more antigenic peptides in common with the IEDB if we had tested more samples from individuals infected with rare viruses. We next analyzed the amino acid composition of recurrently enriched peptides. Enriched peptides tend to have more proline and charged amino acids and fewer hydrophobic amino acids, which is consistent with a previous analysis of B cell epitopes in the IEDB (fig. S4) (24). This trend likely reflects enrichment for amino acids that are surface exposed or can form stronger interactions with antibodies.
B cell responses target highly similar viral epitopes across individuals
We compared the profile of peptides recognized by the antibody response in different individuals. We found that for a given protein, each sample generally only had strong responses against one to three immunodominant peptides (Fig. 3). Surprisingly, we found that the vast majority of seropositive samples for a given virus recognized the same immunodominant peptides, suggesting that the antiviral B cell response is highly stereotyped across individuals. For example, in glycoprotein G from respiratory syncytial virus, there is only a single immunodominant peptide comprising positions 141–196 that is targeted by all samples with detectable antibodies to the protein, regardless of the country of origin (Fig. 3A).
Fig. 3.
The human anti-virome response recognizes a similar spectrum of peptides among infected individuals. In the heatmap charts, each row is a peptide tiling across the indicated protein and each column is a sample. The colored bar above each column, labeled at the top of the figure, indicates the country of origin for that sample. The samples shown are a subset of individuals with antibodies to at least one peptide from the protein. The color intensity of each cell corresponds to the −log10(p-value) measure of significance of enrichment for a peptide in a sample (greater values indicates stronger antibody response). Data are shown for (A) Human respiratory syncytial virus Attachment Glycoprotein G (G), (B) Human adenovirus C penton protein (L2), and (C) Epstein-Barr virus nuclear antigen 1 (EBNA1). Data shown are the mean of two replicates.
For other antigens, we observed inter-population serological differences. For example, two overlapping peptides from position 309–364 and 337–392 of the penton base protein from Adenovirus C frequently elicited antibody responses (Fig. 3B). However, donors from the United States and South Africa had much stronger responses to peptide 309–364 (p < 10−6, t-test) relative to donors from Thailand and Peru. We observed that for the EBNA1 protein from Epstein Barr virus, donors from all four countries frequently had strong responses to peptide 393–448 and occasionally to peptide 589–644. However, donors from Thailand and Peru had much stronger responses to peptide 57–112 (p < 10−6, t-test) (Fig. 3C). These differences may reflect variation in the strains endemic in each region. In addition, polymorphism of MHC class II alleles, immunoglobulin genes and other modifiers that shape immune responses in each population likely play a role in defining the relative immunodominance of antigenic peptides.
To determine whether the humoral responses that target an immunodominant peptide are actually targeting precisely the same epitope, we constructed single-, double-, and triple-alanine scanning mutagenesis libraries for 8 commonly recognized peptides. These were introduced into the same T7 bacteriophage display vector and subjected to the same immunoprecipitation and sequencing protocol using samples from the United States. Mutants that disrupt the epitope diminish antibody binding affinity and peptide enrichment. We found that for all 8 peptides tested, there was a single, largely contiguous subsequence in which mutations disrupted binding for the majority of samples. As expected, the triple-mutants abolished antibody binding to a greater extent, and the enrichment patterns were similar among single-, double- and triple-mutants of the same peptide (Fig. 4, figures S5–S11). For 4 of the 8 peptides, a 9 to 15 amino acid region was critical for antibody recognition in >90% of samples (Fig. 4, Figures S5–S7). One other peptide had a region of similar size that was critical in about half of the samples (fig. S8). In another peptide, a single region was important for antibody recognition in the majority of the samples, but the extents of the critical region varied slightly for different samples and occasionally there are donors that recognize a completely separate epitope (fig. S9). The remaining two peptides contained a single triple mutant that abolished binding in the majority of samples, but the critical region also extended further to different extents depending on the sample (figs. S10–S11). Surprisingly, in one of these peptides, in addition to the main region surrounding positions 13–14 that is critical for binding, a single G36A mutation disrupted binding in almost half of the samples whereas none of the double- or triple-alanine mutants that also included the adjacent positions (L35, G37) affected binding (fig. S11). It is possible that G36 plays a role in helping the peptide adopt an antigenic conformation and multiple-mutants containing the adjacent Leu or Gly residues rescue this ability. We occasionally saw other examples of mutations that resulted in patterns of disrupted binding with no simple explanation, illustrating the complexity of antibody-antigen interaction.
Fig. 4.
Recognition of common epitopes within an antigenic peptide from human adenovirus C penton protein (L2) across individuals. Each row is a sample. Each column denotes the first mutated position for the (A) single-, (B) double-, and (C) triple-alanine mutant peptide starting with the N-terminus on the left. Each double- and triple-alanine mutant contains two or three adjacent mutations, respectively, extending towards the C-terminus from the colored cell. The color intensity of each cell indicates the enrichment of the mutant peptide relative to the wild-type. For double-mutants, the last position is blank. The same is true for the last two positions for triple-mutants. Data shown are the mean of two replicates.
The discovery of recurring targeted epitopes led us to ask whether we could apply this knowledge to improve the sensitivity of viral detection with VirScan. We hypothesized that samples showing a strong response to a recurrently targeted “diagnostic” peptide, which we defined as a peptide enriched in at least 30% of known positive samples, are likely to be seropositive even if they do not meet our stringent cutoff requiring at least two non-overlapping enriched peptides. We tested how this modified criterion affected our sensitivity and specificity in detecting HIV and HCV and found that it reduced the number of false negatives without affecting the specificity of the assay (fig. S13). We next turned our attention to respiratory syncytial virus (RSV), a virus for which our detected seroprevalence was lower than reported epidemiological rates, suggesting imperfect sensitivity of our assay. We tested 60 patient sera for antibodies to RSV by ELISA and found 95% were positive, above the reported sensitivity of the assay and consistent with near-universal exposure to this pathogen. Applying the modified criterion to these samples increased our rate of detection by VirScan from 63% to 97% (table S2). These data suggest that assigning more weight to recurrently targeted epitopes can enhance the sensitivity of VirScan and that the performance of the assay can be improved by screening known positives for a particular virus.
Discussion
We have developed VirScan, a technology for identifying viral exposure and B cell epitopes across the entire known human virome in a single, multiplex reaction using less than a drop of blood. VirScan uses DNA microarray synthesis and bacteriophage display to create a uniform, synthetic representation of peptide epitopes comprising the human virome. Immunoprecipitation and high-throughput DNA sequencing reveals the peptides recognized by antibodies in the sample. VirScan is easily automated in 96-well format to enable high throughput sample processing. Barcoding of samples during PCR enables pooled analysis that can dramatically reduce the per-sample cost. The VirScan approach has several advantages for studying the effect of viruses on the host immune system. By detecting antibody responses, it can identify infectious agents that have been cleared after an effective host response. Current serological methods of antiviral antibody detection typically employ the selection of a single optimized antigen in order to achieve high accuracy. In contrast, VirScan’s unique approach does not require such optimization in order to obtain similar performance. VirScan achieves sensitive detection by assaying each virus’s complete proteome to detect any antibodies directed to epitopes that can be captured in a 56-residue fragment and specificity by computationally eliminating cross-reactive antibodies. This unbiased approach identifies exposure to less well-studied viruses for which optimal serological antigens are not known and can be rapidly extended to include new viruses as they are discovered (25).
While sensitive and selective, VirScan also has a few limitations. First, it cannot detect epitopes that require post-translational modifications. Secondly, it cannot detect epitopes that involve discontinuous sequences on protein fragments greater than 56 residues. In principle, the latter can be overcome by using alternative technologies that allow for the display of full-length proteins such as Parallel Analysis of Translated ORFs (PLATO) (26). Third, VirScan is likely to be less specific compared with certain nucleic acid tests that discern highly related virus strains. However, VirScan demonstrates excellent serological discrimination among similar virus species, such as HSV1 and HSV2 and can even distinguish the genotype of HCV 69% of the time. We envision VirScan will become an important tool for first-pass unbiased serologic screening applications. Individual viruses or viral proteins uncovered in this way can subsequently be analyzed in further detail using more focused assays, as we have demonstrated for a panel of immunodominant epitopes.
We have demonstrated that VirScan is a sensitive and specific assay for detecting exposure to viruses across the human virome. Because it can be performed in high-throughput and requires minimal sample and cost, VirScan enables rapid and cost-effective screening of large numbers of samples to identify population-level differences in virus exposure across the human virome. In this work, we analyzed over 106 million antibody-viral peptide interactions in a comprehensive study of pan-virus serology in a large, diverse population. In doing so we detected 84 different viral species in 2 or more individuals. This is likely to be an underestimate of the history of viral infection as only low levels of circulating antibodies may remain from infections that were cleared in the distant past. In addition, an individual could be infected by multiple distinct strains of each viral species. We identified known and novel differences in virus exposure between groups differing in age, HIV status, and geographic location across four different continents. Our results are largely consistent with previous studies, validating the effectiveness of VirScan. For example, cytomegalovirus antibodies were found in significantly higher frequencies in Peru, Thailand, and South Africa whereas Kaposi’s sarcoma-associated herpesvirus and HSV1 antibodies were detected more frequently in Peru and South Africa, but not in Thailand (16, 27–31). We also uncovered previously undocumented serological differences, such as an increased rate of antibodies against Adenovirus B and respiratory syncytial virus in HIV positive individuals compared to HIV negative individuals. These differences may provide insight into how HIV co-infection alters the balance between host immunity and resident viruses, as well as help to identify pathogens that may increase susceptibility to HIV and other heterologous infections. HIV infection may reduce the immune system’s ability to control reactivation of normally dormant resident viruses or to prevent opportunistic infections from taking hold and triggering a strong adaptive immune response. Beyond the epidemiological applications demonstrated here, VirScan could also be applied to identify viral exposures that correlate with disease or other phenotypes in virome-wide association studies.
Our results identified a large number of novel B cell epitopes, cumulatively nearly doubling the number of all previously identified viral epitopes. We have utilized our data to identify globally immunodominant and commonly recognized “public” epitopes. For most species of viruses, one or more peptides are individually recognized in over 70% to 95% of samples positive for that species (table S3). We identified a set of two peptides that together are recognized by >95% of all screened samples and a set of five peptides that together are recognized in >99% of screened samples. These public epitopes could be used to improve vaccine design by piggybacking on the existing antibody response against them. Fusing a public B cell epitope to a protein in a vaccine to which we hope to induce an immune response may increase a vaccine’s efficacy among a broad population by improving presentation of that protein and aiding affinity maturation. Pre-existing B cells recognizing the public epitope can act as antigen presenting cells to process and present T cell epitopes of the fused vaccine target on MHC class I and II (32). Antibodies secreted by these B cells can also participate in immune complexes with the fused vaccine target, which are critical for follicular dendritic cells to prime class switching and affinity maturation of B cells recognizing other epitopes on the fused antigen (33). Finally, we demonstrated that applying more weight to these public epitopes increases the sensitivity of VirScan without significantly affecting specificity, suggesting that this limited subset of peptides can serve as the basis for the next generation of our assay or for other novel diagnostics.
We also found that the precise epitopes recognized by the B cell response are highly similar among individuals across many viral proteins. One possible model for this striking similarity is that these regions possess properties favorable for antigenicity, such as accessibility. Another model is that the same or highly similar B cell receptor sequences that recognize these epitopes are commonly generated. Identical T cell receptor sequences (“public” clonotypes) have been found in multiple individuals and are thought to be the result of biases during the recombination process that favor certain amino acid sequences (34). V(D)J recombination of the immunoglobulin heavy and light chain loci is also heavily biased (35). Highly similar or even identical complementarity determining region 3 (CDR3) sequences have been observed in dengue virus specific antibodies from different individuals (36). It is possible that, rather than being an exception for dengue specific antibodies, this represents a general phenomenon: inherent biases in V(D)J recombination generate the same or similar antibodies in multiple individuals that recognize highly similar epitopes. Slight differences in the antibody CDR3 sequence may subtly alter antibody-antigen interaction, leading to the slight variations observed in the extent of critical epitope regions. Sequencing of antigen specific antibody genes will be required to investigate these possibilities. The same principle may also apply to T cell epitopes and their cognate TCRs.
In conclusion, VirScan is a new method that enables human virome-wide exploration - at the epitope level - of immune responses in large numbers of individuals. We have demonstrated its effectiveness for determining viral exposure and characterizing viral B cell epitopes in high throughput and at high resolution. Our preliminary studies have revealed intriguing general properties of the human immune system, both at the individual and population scale. VirScan will be an important tool in uncovering the effect of host-virome interactions on human health and disease and could easily be expanded to include other human pathogens such as bacteria, fungi and protozoa.
Materials and Methods
Patient samples
Specimens originating from human donors were collected after informed written consent was obtained and under a protocol approved by the local governing human research protection committee. Secondary use of all samples for the purposes of this work was exempted by the Brigham and Women’s Hospital Institutional Review Board (Protocol #: 2013P001337). Samples included donors residing in Thailand (n=48), donors residing in Peru (n=48), donors residing in South Africa (n=48), and the remaining donors residing in the Unites States including HIV+ donors (n=61) and HCV+ donors (n=26). All serum and plasma samples were stored in aliquots at −80°C until use.
Design and cloning of viral peptide and scanning mutagenesis library sequences
For the virome peptide library, we first downloaded all protein sequences in the UniProt database from viruses with human host and collapsed on 90% sequence identity (http://www.uniprot.org/uniref/?query=uniprot: (host:“Human+[9606]”)+identity:0.9). The clustering algorithm UniProt represents each group of protein sequences sharing at least 90% sequence similarity with a single representative sequence. Then, we created 56 aa peptide sequences tiling through all the proteins with 28 aa overlap. We reverse translated these peptide sequences into DNA codons optimized for expression in E. coli, making synonymous mutations when necessary to avoid restriction sites used in subsequent cloning steps (EcoRI and XhoI). Finally, we added the adapter sequence “aGGAATTCCGCTGCGT” to the 5′ end and “CAGGgaagagctcgaa” to the 3′ end to form the 200 nt oligonucleotide sequences.
For the scanning mutagenesis library, we first took the sequences of the peptides to be mutagenized. For each peptide, we made all single-mutants, and consecutive double- and triple-mutants sequences scanning through the whole peptide. Non-alanine amino acids were mutated to alanine and alanines were mutated to glycine. We reverse translated these peptide sequences into DNA codons, making synonymous mutations when necessary to avoid restriction sites used in subsequent cloning steps (EcoRI and XhoI). We also made synonymous mutations to ensure that the 50 nt at the 5′ end of peptide sequence is unique to allow unambiguous mapping of the sequencing results. Finally, we added the adapter sequence “aGGAATTCCGCTGCGT” to the 5′ end and “CAGGgaagagctcgaa” to the 3′ end to form the 200 nt oligonucleotide sequences.
The 200 nt oligonucleotide sequences were synthesized on a releasable DNA microarray. We PCR amplified the DNA using the primers T7-PFA (aatgatacggcggGAATTCCGCTGCGT) and T7-PRA (caagcagaagACTCGAGCTCTTCCCTG), digested the product with EcoRI and XhoI, and cloned the fragment into the EcoRI/SalI site of the T7FNS2 vector (8). The resulting library was packaged into T7 bacteriophage using the T7 Select Packaging Kit (EMD Millipore) and amplified using the manufacturer suggested protocol.
Phage immunoprecipitation and sequencing
We performed phage immunoprecipitation and sequencing using a slightly modified version of previously published PhIP-Seq protocols (8, 10). First, we blocked each well of a 96 deep-well plate with 1 mL of 3% BSA in TBST overnight on a rotator at 4°C. To each pre-blocked well, we added sera or plasma containing approximately 2 μg of IgG (quantified using a Human IgG ELISA Quantitation Set (Bethyl Laboratories)) and 1 mL of the bacteriophage library diluted to approximately 2×105 fold representation (2×1010 pfu for a library of 105 clones) in phage extraction buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 6 mM MgSO4). We performed two technical replicates for each sample. We allowed the antibodies to bind the phage overnight on a rotator at 4°C. The next day, we added 20 μL each of magnetic Protein A and Protein G Dynabeads (Invitrogen) to each well and allowed immunoprecipitation to occur for 4 h on a rotator at 4°C. Using a 96-well magnetic stand, we then washed the beads three times with 400 μL of PhIP-Seq wash buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% NP-40). After the final wash, we resuspended the beads in 40 μL of water and lysed the phage at 95 °C for 10 m. We also lysed phage from the library before immunoprecipitation (“input”) and after immunoprecipitation with beads alone.
We prepared the DNA for multiplexed Illumina sequencing using a slightly modified version of a previously published protocol (36). We performed two rounds of PCR amplification on the lysed phage material using hot start Q5 polymerase according to the manufacturer suggested protocol (NEB). The first round of PCR used the primers IS7_HsORF5_2 (ACACTCTTTCCCTACACGACTCCAGTCAGGTGTGATGCTC) and IS8_HsORF3_2 (GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCCGAGCTTATCGTCGTCATCC). The second round of PCR used 1 μL of the first round product and the primers IS4_HsORF5_2 (AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACTCCAGT) and a different unique indexing primer for each sample to be multiplexed for sequencing (CAAGCAGAAGACGGCATACGAGATxxxxxxxGTGACTGGAGTTCAGACGTGT, where “xxxxxxx” denotes a unique 7 nt indexing sequence). After the second round of PCR, we determined the DNA concentration of each sample by qPCR and pooled equimolar amounts of all samples for gel extraction. Following gel extraction, the pooled DNA was sequenced by the Harvard Medical School Biopolymers Facility using a 50 bp read cycle on an Illumina HiSeq 2000 or 2500. We pooled up to 192 samples for sequencing on each lane and generally obtained approximately 100 – 200 million reads per lane (500,000 to 1,000,000 reads per sample).
Informatics and statistical analysis
We performed the initial informatics and statistical analysis using a slightly modified version of the previously published technique (8, 10). We first mapped the sequencing reads to the original library sequences using Bowtie and counted the frequency of each clone in the “input” and each sample “output” (37). Since the majority of clones are not enriched we use the observed distribution of output counts as a null distribution. We found that a zero-inflated generalized poisson distribution fits our output counts well. We use this null distribution to calculate a p-value for the likelihood of enrichment for each clone. The probability mass function for the zero-inflated generalized poisson distribution is
We used maximum likelihood estimation to regress the parameters π, θ, and λ to fit the distribution of counts after immunoprecipitation for all clones present at a particular frequency count in the input. We repeated this procedure for all of the observed input counts and found that θ and λ are well fit by linear regression and π by an exponential regression as a function of input count (fig. S1). Finally, for each clone we used its input count and the regression results to determine the null distribution based on the zero-inflated generalized poisson model, which we used to calculate the −log10(p-value) of obtaining the observed count.
To call hits, we determined the threshold for reproducibility between technical replicates based on a previously published method (10). Briefly, we made scatter plots of the log10 of the −log10 (p-values) and used a sliding window of width 0.005 from 0 to 2 across the axis of one replicate. For all the clones that fell within each window, we calculated the median and median absolute deviation of the log10 of the −log10 (p-values) in the other replicate and plotted it against the window location (fig. S2). We called the threshold for reproducibility the first window in which the median was greater than the median absolute deviation. We found that the distribution of the threshold −log10 (p-value) was centered around a mean of approximately 2.3 (fig. S12). So we called a peptide a hit if the −log10 (p-value) was at least 2.3 in both replicates. We eliminated the 593 hits that came up in at least three of the twenty-two immunoprecipitations with beads alone (negative control for non-specific binding). We also filtered out any peptides that were not enriched in at least two of the samples.
To call virus exposures, we grouped peptides according to the virus the peptide is derived from. We grouped all peptides from individual viral strains for which we had complete proteomes. The sample was counted as positive for a species if it was positive for any strain from that species. For viral strains that had partial proteomes, we grouped them with other strains from the same species to form a complete set and bioinformatically eliminated homologous peptides (see next paragraph). We set a threshold number of hits per virus based on the size of the virus. We found that there is approximately a power-law relationship between size of the virus and the average number of hits per sample (fig. S3). In comparing results from VirScan to samples with known infection, we empirically determined that a threshold of 3 hits for herpes simplex virus 1 worked the best. We used this value and the slope of the best fit line to scale the threshold for other viruses. We also set a minimum threshold of at least 2 hits in order to avoid false positives from single spurious hits.
To bioinformatically remove cross-reactive antibodies, we first sorted the viruses by total number of hits in descending order. We then iterated through each virus in this order. For each virus, we iterated through each peptide hit. If the hit shared a subsequence of at least 7 aa with any hit previously observed in any of the viruses from that sample, that hit was considered to be from a cross-reactive antibody and would be ignored for that virus. Otherwise, the hit is considered to be specific and the score for that virus is incremented by one. In this way, we summed only the peptide hits that do not share any linear epitopes. We compared the final score for each virus to the threshold for that virus to determine whether the sample is positive for exposure to that virus
To identify differences between populations, we first used Fisher’s exact test to calculate a p-value for the significance of association of virus exposure with one population versus another. Then, we constructed a null-distribution of Fisher’s exact p-values by randomly permuting the sample labels 1000 times and re-calculating the Fisher’s exact p-value for each virus. Using this null-distribution, we calculated the false discovery rate by dividing the number of permutation p-values more extreme than the one observed by the total number of permutations.
IEDB epitope overlap analysis
We downloaded data for all continuous human B cell epitopes from IEDB and filtered out all non-viral epitopes (22). To avoid redundancy in these 4,549 viral epitopes, we grouped together epitopes that are 100% identical or share a 7 aa subsequence, giving us 1,559 non-redundant epitope groups. Of these groups, 1,392 contain a member epitope that is also a subsequence of a peptide in the VirScan library. This represents the total number of epitopes we could detect by VirScan. To determine the number of epitopes we detected, we tallied the number of epitope groups with at least one member that is contained in a peptide that was enriched in one or two samples. Finally, to determine the number of non-redundant new epitopes we detected, we grouped non-IEDB epitopes containing peptides that share a 7 residues subsequence and counted the number of these non-redundant peptide groups.
Scanning mutagenesis data analysis
First, we estimated the fractional abundance of each peptide by dividing the number of reads for that peptide by the total number of reads for the sample. Then, we divided the fractional abundance of each peptide after immunoprecipitation by the fractional abundance before immunoprecipitation to get the enrichment. To calculate relative enrichment, we divided enrichment of the mutated peptide by enrichment of the wild-type peptide. Since most of the single-mutant peptides had wild-type levels of enrichment, we averaged enrichment of the wild-type peptide enrichment with the middle two quartiles of enrichment of single-mutant peptides to get a better estimate of the wild-type peptide enrichment.
Respiratory syncytial virus and Herpesvirus 1 and 2 serology
Serum from 44 donors was tested for Herpesvirus 1 and Herpesvirus 2 antibodies using the HerpeSelect® 1 and 2 Immunoblot IgG kit (Focus Diagnostics) according to manufacturer’s protocol. Serum from 60 donors was tested for Respiratory syncytial virus antibodies using Anti-Respiratory syncytial virus (RSV) IgG Human ELISA Kit (ab108765) according to manufacturer’s protocol.
Supplementary Material
Acknowledgments
We thank Elizabeth Unger and Supranee Buranapraditkun for providing reagents and Kai Wucherpfennig (Harvard) and Hidde Ploegh (MIT) for critical reading of the manuscript, and TWIST Biosciences for providing access to their advanced oligonucleotide synthesis technology. The cohort in Durban, South Africa was funded by the NIH (R37AI067073) and the International AIDS Vaccine Initiative (UKZNRSA1001). T.N. received additional funding from the South African Research Chairs Initiative, the Victor Daitz Foundation and an International Early Career Scientist Award from the Howard Hughes Medical Institute. RTC was funded by grants NIH DA033541 and AI082630. C.B and J.S. were supported by NIH N01-AI-30024 and N01-AI-15422, NIH-NIDCR R01 DE018925-04, the HIVACAT program and CUTHIVAC 241904. K.R. is supported by TRF Senior Research Scholar, the Thailand Research Fund; and the Chulalongkorn University Research Professor Program, Thailand and NIH grant N01-AI-30024. G.J.X. and T.K. were supported by the NSF Graduate Research Fellowships Program. S.J.E. and B.W. are Investigators with the Howard Hughes Medical Institute. G.J.X., T.K., H.B.L., and S.J.E. are inventors on a patent application (PCT Application No. PCT/US14/70902) filed by The Brigham and Women’s Hospital, Inc. that covers the use of phage display libraries to detect antiviral antibodies.
Footnotes
References and Notes
- 1.Wylie KM, Weinstock GM, Storch GA. Emerging view of the human virome. Transl Res. 2012;160:283–290. doi: 10.1016/j.trsl.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duerkop Ba, Hooper LV. Resident viruses and their interactions with the immune system. Nat Immunol. 2013;14:654–659. doi: 10.1038/ni.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton ES, et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 4.Foxman EF, Iwasaki A. Genome-virome interactions: examining the role of common viral infections in complex disease. Nat Rev Microbiol. 2011;9:254–264. doi: 10.1038/nrmicro2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lecuit M, Eloit M. The human virome: New tools and concepts. Trends Microbiol. 2013;21:510–515. doi: 10.1016/j.tim.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Vlaminck I, et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell. 2013;155:1178–1187. doi: 10.1016/j.cell.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammarlund E, et al. Duration of antiviral immunity after smallpox vaccination. Nat Med. 2003;9:1131–1137. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- 8.Larman HB, et al. Autoantigen discovery with a synthetic human peptidome. Nat Biotechnol. 2011;29:535–541. doi: 10.1038/nbt.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The UniProt Consortium. Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42:D191–198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larman HB, et al. PhIP-Seq characterization of autoantibodies from patients with multiple sclerosis, type 1 diabetes and rheumatoid arthritis. J Autoimmun. 2013;43:1–9. doi: 10.1016/j.jaut.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bialecki C, Feder HM, Grant-Kels JM. The six classic childhood exanthems: a review and update. J Am Acad Dermatol. 1989;21:891–903. doi: 10.1016/s0190-9622(89)70275-9. [DOI] [PubMed] [Google Scholar]
- 12.Lee JH, Roth WK, Zeuzem S. Evaluation and comparison of different hepatitis C virus genotyping and serotyping assays. J Hepatol. 1997;26:1001–1009. doi: 10.1016/s0168-8278(97)80108-0. [DOI] [PubMed] [Google Scholar]
- 13.Wertheim HFL, et al. Key role for clumping factor B in Staphylococcus aureus nasal colonization of humans. PLoS Med. 2008;5:0104–0112. doi: 10.1371/journal.pmed.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manz RA, Hauser AE, Hiepe F, Radbruch A. Maintenance of serum antibody levels. Annu Rev Immunol. 2005;23:367–386. doi: 10.1146/annurev.immunol.23.021704.115723. [DOI] [PubMed] [Google Scholar]
- 15.Wang M, et al. Human anti-JC virus serum reacts with native but not denatured JC virus major capsid protein VP1. J Virol Methods. 1999;78:171–176. doi: 10.1016/s0166-0934(98)00180-3. [DOI] [PubMed] [Google Scholar]
- 16.Staras SAS, et al. Seroprevalence of cytomegalovirus infection in the United States, 1988–1994. Clin Infect Dis. 2006;43:1143–1151. doi: 10.1086/508173. [DOI] [PubMed] [Google Scholar]
- 17.Reynolds MA, Kruszon-Moran D, Jumaan A, Schmid DS, McQuillan GM. Varicella seroprevalence in the U.S.: data from the National Health and Nutrition Examination Survey, 1999–2004. Public Health Rep. 125:860–869. doi: 10.1177/003335491012500613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen JI. Epstein–Barr virus infection. N Engl J Med. 2000;343:481–492. doi: 10.1056/NEJM200008173430707. [DOI] [PubMed] [Google Scholar]
- 19.Dong L, et al. A combination of serological assays to detect human antibodies to the avian influenza a H7N9 virus. PLoS One. 2014;9:e95612. doi: 10.1371/journal.pone.0095612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel P, et al. Prevalence and Risk Factors Associated With Herpes Simplex Virus-2 Infection in a Contemporary Cohort of HIV-Infected Persons in the United States. Sex Transm Dis. 2012;39:154–160. doi: 10.1097/OLQ.0b013e318239d7fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stover CT, et al. Prevalence of and risk factors for viral infections among human immunodeficiency virus (HIV)-infected and high-risk HIV-uninfected women. J Infect Dis. 2003;187:1388–1396. doi: 10.1086/374649. [DOI] [PubMed] [Google Scholar]
- 22.Engels EA, et al. Risk factors for human herpesvirus 8 infection among adults in the United States and evidence for sexual transmission. J Infect Dis. 2007;196:199–207. doi: 10.1086/518791. [DOI] [PubMed] [Google Scholar]
- 23.Vita R, et al. The Immune Epitope Database 2.0. Nucleic Acids Res. 2009;38:D854–862. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh H, Ansari HR, Raghava GPS. Improved Method for Linear B-Cell Epitope Prediction Using Antigen’s Primary Sequence. PLoS One. 2013;8:e62216. doi: 10.1371/journal.pone.0062216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mokili JL, Rohwer F, Dutilh BE. Metagenomics and future perspectives in virus discovery. Curr Opin Virol. 2012;2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J, et al. Protein interaction discovery using parallel analysis of translated ORFs (PLATO) Nat Biotechnol. 2013;31:331–4. doi: 10.1038/nbt.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urwijitaroon Y, Teawpatanataworn S, Kitjareontarm A. Prevalence of cytomegalovirus antibody in Thai-northeastern blood donors. Southeast Asian J Trop Med Public Health. 1993;24(Suppl 1):180–182. [PubMed] [Google Scholar]
- 28.Cannon MJ, Schmid DS, Hyde TB. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol. 2010;20:202–213. doi: 10.1002/rmv.655. [DOI] [PubMed] [Google Scholar]
- 29.Mohanna S, et al. Human herpesvirus-8 in Peruvian blood donors: a population with hyperendemic disease? Clin Infect Dis. 2007;44:558–561. doi: 10.1086/511044. [DOI] [PubMed] [Google Scholar]
- 30.Ablashi D, et al. Seroprevalence of human herpesvirus-8 (HHV-8) in countries of Southeast Asia compared to the USA, the Caribbean and Africa. Br J Cancer. 1999;81:893–897. doi: 10.1038/sj.bjc.6690782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith JS, Robinson NJ. Age-specific prevalence of infection with herpes simplex virus types 2 and 1: a global review. J Infect Dis. 2002;186(Suppl):S3–S28. doi: 10.1086/343739. [DOI] [PubMed] [Google Scholar]
- 32.Heit A, et al. CpG-DNA aided cross-priming by cross-presenting B cells. J Immunol. 2004;172:1501–1507. doi: 10.4049/jimmunol.172.3.1501. [DOI] [PubMed] [Google Scholar]
- 33.Aydar Y, Sukumar S, Szakal AK, Tew JG. The influence of immune complex-bearing follicular dendritic cells on the IgM response, Ig class switching, and production of high affinity IgG. J Immunol. 2005;174:5358–5366. doi: 10.4049/jimmunol.174.9.5358. [DOI] [PubMed] [Google Scholar]
- 34.Quigley MF, et al. Convergent recombination shapes the clonotypic landscape of the naive T-cell repertoire. Proc Natl Acad Sci U S A. 2010;107:19414–19419. doi: 10.1073/pnas.1010586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson KJL, Kidd MJ, Wang Y, Collins AM. The shape of the lymphocyte receptor repertoire: lessons from the B cell receptor. Front Immunol. 2013;4:263. doi: 10.3389/fimmu.2013.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parameswaran P, et al. Convergent antibody signatures in human dengue. Cell Host Microbe. 2013;13:691–700. doi: 10.1016/j.chom.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.