Abstract
The posttranslational modification of proteins by the ubiquitin-like small molecule NEDD8 has previously been shown to be vital in a number of cell signaling pathways. In particular, conjugation of NEDD8 (neddylation) serves to regulate protein ubiquitination through modifications to E3 ubiquitin ligases. Despite the prevalence of NEDD8 in neurons, very little work has been done to characterize the role of this modifier in these cells. Here, we use the recently developed NEDD8 Activating Enzyme (NAE) inhibitor MLN4924 and report evidence of a role for NEDD8 in regulating mammalian excitatory synapses. Application of this drug to dissociated rat hippocampal neurons caused reductions in synaptic strength, surface glutamate receptor levels, dendritic spine width, and spine density, suggesting that neddylation is involved in the maintenance of synapses.
Keywords: Ubiquitin, NEDD8, MLN4924, synapse, hippocampus
Introduction
Control of synaptic strength is achieved in part through posttranslational modifications to synaptic proteins (Lu and Roche, 2012). In addition to the well-studied role of phosphorylation at the synapse, ubiquitination has recently emerged as an important regulatory mechanism for a number of proteins located both pre- and postsynaptically (Mabb and Ehlers, 2010; Yamada et al., 2013). Ubiquitination controls the targeting of specific substrates to 26S proteasomes and lysosomes for degradation and can also mediate subcellular distribution and function. Substrate specificity is accomplished by E3 ubiquitin ligases, which are responsible for the final step in the ubiquitination process. These enzymes are divided into a number of different families based on their structure and function, and their activity can be regulated through posttranslational modifications.
The conjugation of the ubiquitin-like protein NEDD8, termed neddylation, has been identified as a regulatory mechanism for a number of E3 ligases and components of E3 complexes (Rabut and Peter, 2008). Notably, all members of the cullin family of proteins, which comprise multi-subunit E3 ligase complexes, are known to be neddylated (Jones et al., 2008). While the exact biological effects of neddylation are still unclear, ample evidence suggests that the conjugation of NEDD8 to cullin proteins stimulates the ligase activity of cullin-based E3 complexes (Bennett et al., 2010; Podust et al., 2000; Read et al., 2000). Members of other E3 ligase families have also been shown to be substrates for neddylation, including the RING finger ubiquitin ligase Mdm2 and the HECT ligase Smurf1 (Xie et al., 2014; Xirodimas et al., 2004). This suggests that neddylation plays a critical role in regulating ubiquitination. Interestingly, many ubiquitin ligases known to be neddylated have also been demonstrated to regulate neuronal function and synaptic strength through the ubiquitination of synaptic proteins (Chen and Matesic, 2007; Cheng et al., 2011; Choo et al., 2012; Colledge et al., 2003; Zhu et al., 2005). However, little work has been done to directly explore the role of neddylation in neurons.
The recent generation of the NEDD8 activating enzyme (NAE) inhibitor MLN4924 has enabled a number of studies exploring the roles of neddylation in various cell types (Fig. 1A) (Bennett et al., 2010; Gu et al., 2014; Soucy et al., 2009). Though initially created as a potential tumor inhibitor, this drug allows for the pharmacological exploration of NEDD8 function. Here, we demonstrate that inhibition of neddylation in cultured hippocampal rat neurons affects synaptic strength and dendritic spine morphology. Our results provide evidence that neddylation plays a role in the maintenance of excitatory synapses.
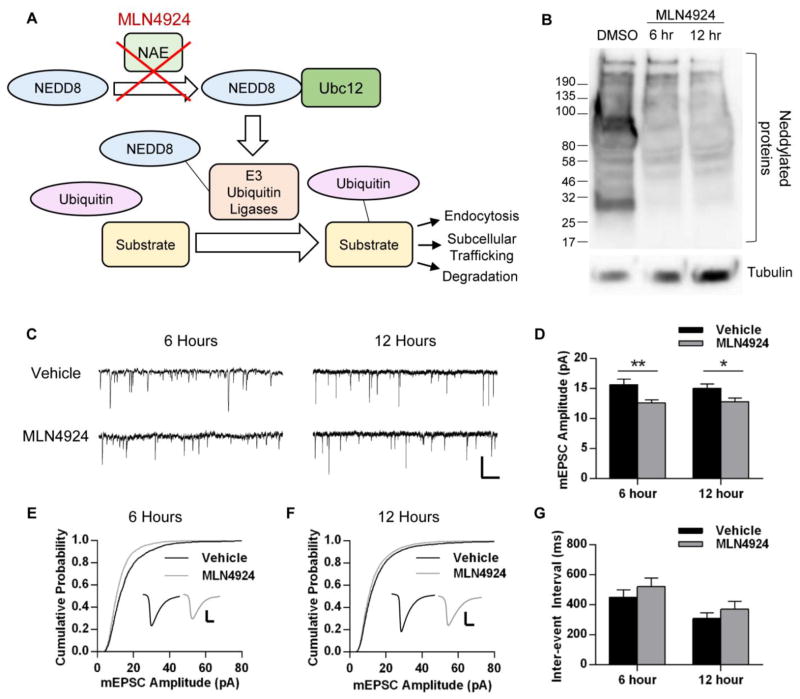
Fig. 1.
Inhibition of neddylation with MLN4924 reduces mEPSC amplitude. A, Diagram depicting process of neddylation and action of the drug MLN4924. Under normal conditions, NEDD8 activating enzyme (NAE) permits NEDD8 to be conjugated to its intermediate E2 enzyme (Ubc12) and then conjugated to its target substrates, which include a number of E3 ubiquitin ligases. The stabilizing and activating effects of NEDD8 conjugation allow the E3 ligases to conjugate ubiquitin to their specific protein substrates, which serves as a signal for surface protein endocytosis, trafficking between subcellular compartments, or degradation via proteasomes or lysosomes. MLN4924 inhibits this regulatory pathway through direct inhibition of NAE. B, Western blot depicting inhibition of NEDD8 conjugation in neurons after treatment with MLN4924. Dissociated cortical rat neurons (DIV 19–25) were treated with DMSO (vehicle) for 12 hours or with 1 μM MLN4924 for 6 or 12 hours. Cells were lysed in the presence of 1,10-orthophenathroline, then lysates were resolved on 4–20% SDS-PAGE and probed for NEDD8 and tubulin (n = 2 independent experiments). C–G, Dissociated hippocampal rat neurons (DIV 19–25) were infected with 2-gene GFP for 16–20 hours. Neurons were treated with 1 μM MLN4924 or DMSO (vehicle) for the last 6 or 12 hours of infection and mEPSCs were recorded from GFP-positive pyramidal-like neurons. C, Representative mEPSC traces recorded from neurons in each condition. Scale bar represents 500 ms, 20 pA. D, Average mEPSC amplitudes of neurons recorded from the four conditions; n > 19 cells per condition across 3 independent experiments. E, Cumulative probability plot of all mEPSC events recorded from cells treated with MLN4924 for 6 hours; p < 0.001, Kolmogorov-Smirnov test; n = 2003, 2224 events. F, Cumulative probability plot from mEPSCs in 12-hour conditions; p < 0.001, K-S test; n = 5781, 5350 events. Insets depict averaged waveform from all events in each condition, scale bar represents 5 ms, 5 pA. G, Average inter-event intervals of neurons in each condition. Graphs depict mean ± SEM. *p < 0.05, **p < 0.01, Student’s t test.
Materials and Methods
Antibodies and Reagents
Antibodies purchased from: pAb surface (N-terminal) GluA1, pAb (C-terminal) GluA1 (Millipore); mAb GFP (Neuromab, UC Davis); pAb GFP (Invitrogen); mAb tubulin (Sigma-Aldrich); pAb NEDD8 (generous gift from Dr. Eric Bennett). Reagents purchased from: tetrodotoxin (TTX), bicuculline (Tocris Bioscience); MLN4924 (Active Biochem, Maplewood, NJ).
Infections and Drug Treatments
Hippocampal cultures were infected with double subgenomic Sindbis virion (2-gene GFP) at 19–25 DIV and allowed to express for 14–22 hours to fill cells for visualization. All drug treatments were conducted within cell media at indicated concentrations for indicated amounts of time, with DMSO of equal volume used as control.
Neuronal cultures
Dissociated hippocampal or cortical neurons from postnatal day 1 rat pups of either sex were plated onto poly-D-lysine-coated glass coverslips at a density of 45,000 cells/cm2 (hippocampal) or onto coated 6-well plastic dishes at ~500,000 cells per well (cortical) and were maintained in B27-supplemented Neurobasal media (Invitrogen) until ≥ 14 days in vitro (DIV), as previously described (Schwarz et al., 2010; Scudder et al., 2014).
Western blots
To test the efficacy of MLN4924, cultured cortical neurons were lysed in a lysis buffer containing 8 M urea, 75 mM NaCl, 50 mM Tris (pH 8.2), 2 mM 1,10-orthophenathroline, 1 mM NaF, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, and 1x protease inhibitor cocktail (Roche). For GluA1 experiments, neurons were lysed in RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate, and 0.1% SDS) with protease inhibitor cocktail. Protein concentrations were determined by BCA protein assay (Pierce) and samples containing equal amounts of total protein were boiled with sample buffer, resolved on 10% or 4–20% SDS-PAGE as specified, and probed with primary antibodies overnight at 4° C. Following incubation with HRP-conjugated secondary antibodies (1 hr, room temperature), blots were developed digitally and band intensities were analyzed using NIH ImageJ. Significance was determined through a paired t-test.
Immunostaining
Following infections and drug treatments, dissociated hippocampal neurons were washed with PBS-MC and fixed with a solution containing 4% paraformaldehyde and 4% sucrose for 10 minutes. Cells were permeabilized with 0.2% Triton X-100 and 2% BSA in PBS-MC for 20 minutes followed by a 1 hour incubation with 5% BSA in PBS-MC for blocking. Primary and secondary antibodies were diluted in 2% BSA in PBS-MC and applied to neurons for 1 hour at room temperature. Coverslips were transferred onto glass slides for confocal imaging. For surface immunostaining, neurons were washed, fixed, blocked in 5% BSA for 12–20 h at 4° C, and incubated with surface GluA1 antibody in 2% BSA for 12–20 h at 4° C. Cells were then permeabilized, blocked, incubated with anti-GFP antibody, and incubated with secondary antibodies.
Confocal Microscopy
Immunofluorescent images were acquired with a Leica DMI6000 inverted microscope equipped with a Yokogawa Nipkon spinning disk confocal head, an Orca ER high-resolution black and white cooled CCD camera (6.45 μm/pixel at 1×), Plan Apochromat 63×/1.4 numerical aperture objective, and an argon/krypton 100 mW air-cooled laser for 488/568/647 nm excitations. Maximum projected confocal Z-stacks were generated and images were analyzed with macros written for NIH ImageJ. Statistical significance was determined using unpaired t-tests or the Kolmogorov-Smirnov (K-S) test for cumulative distributions, conducted in Prism (GraphPad). Images were acquired and analyzed with experimenter blind to conditions.
Whole-cell electrophysiology
For whole-cell patch-clamp recordings of miniature excitatory postsynaptic currents (mEPSCs), dissociated rat hippocampal neurons were bathed at room temperature in an extracellular recording solution containing the following: 119 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 30 mM glucose, 10 mM HEPES (pH 7.2), 1 μM TTX, and 10 μM bicuculline. Glass pipettes were filled with an intracellular solution containing the following (in mM): 10 CsCl, 105 CsMeSO3, 0.5 ATP, 0.3 GTP, 10 HEPES, 5 glucose, 2 MgCl2, and 1 EGTA (pH 7.2). Electrode resistances ranged from 2.5 to 4.5 MΩ and access resistances ranged from 10 to 25 MΩ. Cells were held at −70 mV. Signals were amplified, filtered to 2 or 5 kHz, and digitized at 10 kHz sampling frequency. Infected cells were identified via GFP fluorescence and the experimenter was blind to condition. mEPSCs were analyzed using ClampFit 10.3 (Molecular Devices) to determine amplitude and inter-event interval. Statistical differences between conditions were analyzed through unpaired t-tests for averages and through the Kolmogorov-Smirnov (K-S) test for cumulative distributions, conducted in Prism (GraphPad).
Results
Pharmacological inhibition of the NEDD8 Activating Enzyme reduces synaptic strength
Neddylation is known to be a regulator of a number of functions in diverse cell types, though little work has been done to explore its role in neurons. Using the NEDD8 Activating Enzyme (NAE) inhibitor MLN4924, we investigated the role of this posttranslational modification in the maintenance of neuronal synapses (Fig. 1A). First, we sought to verify that this drug effectively blocks neddylation in cultured neurons. Dissociated cortical neurons were treated with 1 μM MLN4924 or vehicle (DMSO) for 6 or 12 hours. Cells were lysed in the presence of the zinc chelator and COPS5 inhibitor 1,10-orthophenathroline (OPT), which has been shown to allow for detection of neddylated proteins (Bennett et al., 2010). In the control condition, probing the neuronal lysates for NEDD8 yields a number of dark bands which are indicative of neddylated proteins of various molecular weights. Treatment with MLN4924, however, abolishes such bands, demonstrating that this drug prevents the conjugation of NEDD8 to substrates within neurons (Fig. 1B).
We next determined whether inhibition of neddylation would affect synaptic strength. Dissociated hippocampal rat neurons were first infected with 2-gene GFP for visualization of cell morphology (16–20 hours total), then treated with 1 μM MLN4924 or vehicle (DMSO) for 6 hours. Miniature excitatory postsynaptic currents (mEPSCs) were recorded from GFP-positive pyramidal-like cells to determine the effect on synaptic strength. We observed that six hours of MLN4924 treatment caused a significant decrease in the amplitude of mEPSCs (DMSO, amplitude: 15.67 ± 0.88 pA, n = 17 cells; MLN4924: 12.60 ± 0.52 pA, n = 19 cells; p < 0.01; Fig. 1C–E). We observed no changes to inter-event interval (DMSO, IEI: 450.9 ± 49.24 ms; MLN4924: 521.4 ± 58.19 ms; p = 0.37; Fig. 1C, G), suggesting a postsynaptic alteration. However, since the entire mEPSC amplitude distribution does not fully shift to the left (Fig 1E, F), it is also possible that we are observing preferential selection of weaker synapses over stronger ones, rather than a reduction in the strength of all synapses.
To determine whether a longer treatment with MLN4924 would enhance this reduction, we increased the treatment time to 12 hours. We observed a decrease in mEPSC amplitude comparable to that of the shorter treatment (DMSO, amplitude: 15.05 ± 0.71 pA, n = 46 cells; MLN4924: 12.83 ± 0.59 pA, n = 43; p < 0.05; Fig. 1C, F) and no change in inter-event interval (DMSO, IEI: 309 ± 38.35 ms; MLN4924: 371.3 ± 50.99 ms; p = 0.33; Fig. 1C, G), demonstrating that no further reduction in synaptic strength occurs with longer inhibition of neddylation.
Application of MLN4924 reduces surface GluA1 expression
Changes in mEPSC amplitude are often accompanied by changes in the levels of surface glutamate receptors, particularly AMPA receptors (AMPARs). To assess whether these observed changes in mEPSC amplitude are the result of altered postsynaptic surface AMPAR expression, we infected hippocampal neurons with GFP to visualize morphology, treated with MLN4924 or vehicle for 6 hours, and stained for surface GluA1 (an AMPAR subunit) using immunofluorescent antibodies. We observed reduced surface GluA1 fluorescence in MLN4924-treated neurons (DMSO, normalized surface signal: 1.00 ± 0.04, n = 40 cells; MLN4924: 0.88 ± 0.03, n = 40 cells; p < 0.05; Fig. 2A, B).
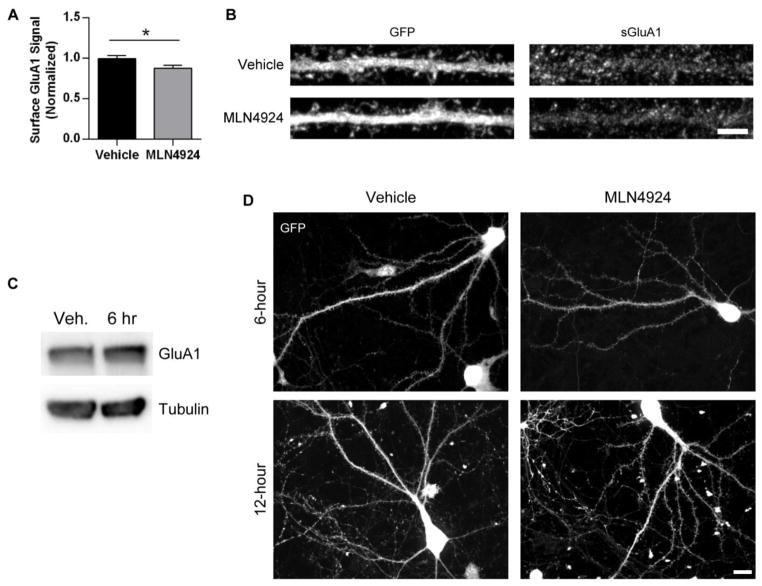
Fig. 2.
MLN4924 reduces surface AMPAR expression. A, Cultured hippocampal neurons (DIV 19–25) were infected with 2-gene GFP for 16–20 hours and treated with 1 μM MLN4924 or DMSO (vehicle) for the last 6 hours of infection. Cells were then fixed and stained for surface GluA1 using immunofluorescent antibodies. Images of GFP-positive neurons were acquired with a confocal microscope and straightened dendrites were analyzed for GluA1 expression. Values were normalized to the vehicle-treated condition; n = 40 neurons per condition across two independent experiments; p < 0.05, Student’s t test. Graph depicts mean ± SEM. B, Representative z-stacked immunofluorescent images of dendrites from vehicle- and MLN4924-treated neurons, stained for GFP and GluA1. Scale bar represents 5 μm. C, Representative Western blot depicting total GluA1 after MLN4924 treatment. Dissociated rat cortical neurons (DIV19–25) were treated with 1 μM MLN4924 or vehicle for 6 hours, then lysed. Lysates were resolved on 10% SDS-PAGE and probed with GluA1 and tubulin antibodies. D, Representative z-stacked images of dissociated hippocampal neurons expressing GFP after 6 and 12 hours of vehicle or MLN4924 treatment. Scale bar represents 10 μm.
To determine whether total GluA1 protein levels are altered after inhibition of neddylation, dissociated cortical neurons were treated with 1 μM MLN4924 or DMSO for 6 hours and lysed. Lysates were resolved on SDS-PAGE and probed with GluA1 antibodies. We did not observe a significant change in total GluA1 levels after MLN4924 treatment (MLN4924, normalized level: 1.26, n = 3 independent experiments; p = 0.58; Fig. 2C). This may indicate that receptors have been removed from synapses but not yet degraded, or that synapses have been dismantled but the component proteins are still present.
Our observations of reduced surface GluA1 and mEPSC amplitude do not appear to be the result of poor neuronal health, as neurons in all conditions appeared healthy and with comparable morphologies (Fig. 2D). Taken together, these data indicate that the inhibition of neddylation in hippocampal neurons leads to functional changes that reduce synaptic strength.
Inhibition of neddylation leads to alterations to dendritic spines
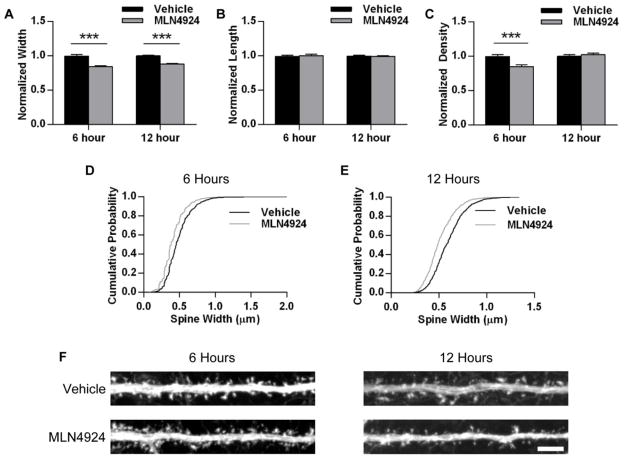
The majority of excitatory synapses are located on dendritic spines. In hippocampal pyramidal neurons, there is a strong correlation between the numbers of dendritic spines and excitatory synapses (Nimchinsky et al., 2004). Additionally, spine morphology is closely related to synaptic function (Hering and Sheng, 2001; Matsuzaki et al., 2001). To determine whether the observed functional changes caused by neddylation inhibition are accompanied by structural changes in neurons, we examined the size and density of dendritic spines in dissociated hippocampal neurons treated with 1 μM MLN4924 for 6 hours. MLN4924 treatment produced a significant decrease in spine width (DMSO, normalized width: 1.00 ± 0.02, n = 64 dendrites; MLN4924: 0.84 ± 0.01, n = 64 dendrites; p < 0.001; Fig. 3A, D) and density (DMSO, normalized density: 1.00 ± 0.03; MLN4924: 0.85 ± 0.02; p < 0.001; Fig. 3C), but did not affect spine length (p = 0.93; Fig. 3B). We extended our treatment time to 12 hours and observed a similar reduction in width (DMSO, normalized width: 1.00 ± 0.01, n = 70 dendrites; MLN4924: 0.88 ± 0.01, n = 79; p < 0.001; Fig. 3A, E) and no reduction in density (p = 0.52; Fig. 3C) or length (p = 0.67; Fig. 3B), demonstrating that spines do not continue shrinking after the initial reduction in width. These data indicate that the functional reduction in synaptic strength caused by inhibition of neddylation is accompanied by structural changes to dendritic spines.
Fig. 3.
Inhibition of neddylation affects dendritic spines. Cultured hippocampal neurons (DIV 19–25) were infected with 2-gene GFP for 16–20 hours and treated with 1 μM MLN4924 or DMSO (vehicle) for the last 6 or 12 hours. Cells were fixed and stained for GFP. GFP-positive cells with pyramidal-like morphology were chosen for analysis. A, Average dendritic spine width (across widest point) for dendrites from vehicle- and MLN4924-treated neurons, normalized to vehicle condition. B, Average dendritic spine length (from shaft to tip) for dendrites from each condition, normalized to vehicle. C, Average spine density for dendrites in each condition, normalized to vehicle. D, Cumulative probability plot of width measurements from all spines analyzed from neurons treated with vehicle or MLN4924 for 6 hours; p < 0.001, K-S test; n = 2630, 2244 spines. E, Cumulative probability plot for all spine widths from 12-hour conditions; p < 0.001, K-S test; n = 1274, 1257 spines. F, Representative images of dendrites straightened from GFP-expressing neurons treated with 6 or 12 hours of vehicle or MLN4924. Scale bar represents 5 μm. ***p < 0.001, Student’s t-test; n > 60 dendrites per condition across two independent experiments. Graphs depict mean ± SEM.
Discussion
Synaptic strength is tightly controlled by posttranslational modifications to synaptic proteins, including surface glutamate receptors such as AMPA and NMDA receptors and scaffolding proteins such as PSD-95. In addition to the relatively well-characterized phosphorylation of such proteins, research in recent years has highlighted a critical role of ubiquitination in the control of synaptic strength (Schwarz and Patrick, 2012; Yamada et al., 2013). Neddylation is known to be an important regulator of a number of ubiquitin E3 ligases and components of ligase complexes that also have known roles in neurons, including Smurf1 (Cheng et al., 2011; Xie et al., 2014), Cul4b (Chen and Matesic, 2007; Liu et al., 2012), Cul3 (Zhu et al., 2005), parkin (Choo et al., 2012; Um et al., 2012), and Mdm2 (Colledge et al., 2003; Xirodimas et al., 2004). Therefore, the disruption of this process in neurons would likely affect some aspects of neuronal function.
The generation of MLN4924 as a specific inhibitor of NEDD8 Activating Enzyme allowed us to determine whether neddylation plays a role in neuronal function. We observed that dissociated hippocampal neurons treated with MLN4924 displayed reduced synaptic strength, as measured by mEPSC amplitude and surface GluA1 staining. This effect appears to be maximal at 6 hours, as no further changes were observed with longer treatments (Fig. 1, Fig. 2). In addition to these functional reductions, we also observed structural changes in these neurons. Specifically, inhibition of NEDD8 conjugation caused a reduction of dendritic spine width and density (Fig. 3). Importantly, these results did not appear to be due to reduced cell health, as neuronal morphology remained normal after a 12-hour treatment and no signs of poor health were observed (Fig. 2D). Interestingly, spine density was only reduced at the 6-hour time-point and appears normal at 12 hours. It is possible that excitatory neurons engage in some sort of homeostatic mechanism of spine outgrowth to make up for the reductions in synaptic strength that initially occur. Additionally, formation of new synapses on existing spines or a compensatory change in release probability could also explain why there is no observed decrease in mEPSC inter-event interval at 6 hours despite the spine density reduction.
These results suggest that neddylation plays a role in the maintenance of excitatory synapses. Since NEDD8 is able to modify a number of different E3 ligases and components of ligase complexes that are known to be involved in the development and maintenance of synapses, there are a number of substrates that could be responsible for the observed changes in synaptic strength. One major role of ubiquitination at synapses is in the regulation of AMPA receptor trafficking, which is accomplished through the direct ubiquitination of receptor subunits by the E3 ligases RNF167 and Nedd4-1 (Lin et al., 2011; Lussier et al., 2012; Schwarz et al., 2010; Scudder et al., 2014). However, in these studies ubiquitination serves as a negative regulator of synaptic strength through the reduction of surface AMPAR levels. While the possibility that these ligases are regulated by neddylation has not yet been investigated, it appears unlikely that such a mechanism would explain our results given that inhibition of these ligases would either increase synaptic strength or have no effect. Thus, the observed reduction in surface AMPAR expression is likely not directly related to ubiquitin-dependent AMPAR trafficking.
As our study uses only a pharmacological manipulation to explore the role of neddylation in neurons, further research using more precise interventions will be instrumental in determining the exact targets of neddylation at the synapse and how this regulatory mechanism affects neuronal function. Additionally, future work can explore whether this modification can be regulated in an activity-dependent manner to provide dynamic control of synaptic strength.
There is evidence suggesting that NEDD8 plays a role in neurodegenerative disorders such as Alzheimer’s and Parkinson’s Disease, based on the high levels of NEDD8 present in Lewy Bodies and neurofibrillary tangles in neural tissue collected from patients with these disorders (Dil Kuazi et al., 2003; Mori et al., 2005). Additionally, some of the known NEDD8 substrates have been linked to mental retardation and autism (Tsai et al., 2012; Zou et al., 2007). A greater understanding of the role this posttranslational modification plays in neurons could allow for mechanistic insight into these diseases and potentially pave the way for therapeutic interventions.
Highlights.
The neddylation inhibitor MLN4924 affects synaptic strength in hippocampal neurons
MLN4924 reduced mEPSC amplitude and surface AMPAR expression
MLN4924 reduced dendritic spine width and density
MLN4924 did not negatively impact neuronal health
Acknowledgments
We thank Eric Bennett and members of the Patrick laboratory for helpful discussion and Lara Dozier for technical assistance. This work was supported by an NSF Graduate Research Fellowship (S.L.S), NIH Grant NS060847 (G.N.P.), NIH Training Grant T32 AG00216 (S.L.S.), and Grant P50-GMO85764 from the Center for Systems Biology.
Abbreviations
- NEDD8
neural precursor cell expressed, developmentally down-regulated 8
- AMPAR
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor
- Ubc12
ubiquitin carrier protein 12
- RING
really interesting new gene
- HECT
homologous to the E6-AP carboxyl terminus
- Mdm2
mouse double minute 2 homolog
- GFP
green fluorescent protein
- DMSO
dimethyl sulfoxide
- BSA
bovine serum albumin
- mEPSC
miniature excitatory postsynaptic current
Footnotes
Conflict of Interest: The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Samantha L. Scudder, Email: sscudder@ucsd.edu.
Gentry N. Patrick, Email: gpatrick@ucsd.edu.
References
- Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell. 2010;143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Matesic LE. The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev. 2007;26:587–604. doi: 10.1007/s10555-007-9091-x. [DOI] [PubMed] [Google Scholar]
- Cheng PL, Lu H, Shelly M, Gao H, Poo MM. Phosphorylation of E3 ligase Smurf1 switches its substrate preference in support of axon development. Neuron. 2011;69:231–243. doi: 10.1016/j.neuron.2010.12.021. [DOI] [PubMed] [Google Scholar]
- Choo YS, Vogler G, Wang D, Kalvakuri S, Iliuk A, Tao WA, Bodmer R, Zhang Z. Regulation of parkin and PINK1 by neddylation. Human molecular genetics. 2012;21:2514–2523. doi: 10.1093/hmg/dds070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, Langeberg LK, Lu H, Bear MF, Scott JD. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40:595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dil Kuazi A, Kito K, Abe Y, Shin RW, Kamitani T, Ueda N. NEDD8 protein is involved in ubiquitinated inclusion bodies. The Journal of pathology. 2003;199:259–266. doi: 10.1002/path.1283. [DOI] [PubMed] [Google Scholar]
- Gu Y, Kaufman JL, Bernal L, Torre C, Matulis SM, Harvey RD, Chen J, Sun SY, Boise LH, Lonial S. MLN4924, an NAE inhibitor, suppresses AKT and mTOR signaling via upregulation of REDD1 in human myeloma cells. Blood. 2014;123:3269–3276. doi: 10.1182/blood-2013-08-521914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering H, Sheng M. Dendritic spines: structure, dynamics and regulation. Nat Rev Neurosci. 2001;2:880–888. doi: 10.1038/35104061. [DOI] [PubMed] [Google Scholar]
- Jones J, Wu K, Yang Y, Guerrero C, Nillegoda N, Pan ZQ, Huang L. A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. Journal of proteome research. 2008;7:1274–1287. doi: 10.1021/pr700749v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Hou Q, Jarzylo L, Amato S, Gilbert J, Shang F, Man HY. Nedd4-mediated AMPA receptor ubiquitination regulates receptor turnover and trafficking. Journal of neurochemistry. 2011;119:27–39. doi: 10.1111/j.1471-4159.2011.07221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HC, Enikolopov G, Chen Y. Cul4B regulates neural progenitor cell growth. BMC neuroscience. 2012;13:112. doi: 10.1186/1471-2202-13-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Roche KW. Posttranslational regulation of AMPA receptor trafficking and function. Current opinion in neurobiology. 2012;22:470–479. doi: 10.1016/j.conb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier MP, Herring BE, Nasu-Nishimura Y, Neutzner A, Karbowski M, Youle RJ, Nicoll RA, Roche KW. Ubiquitin ligase RNF167 regulates AMPA receptor-mediated synaptic transmission. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19426–19431. doi: 10.1073/pnas.1217477109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabb AM, Ehlers MD. Ubiquitination in postsynaptic function and plasticity. Annual review of cell and developmental biology. 2010;26:179–210. doi: 10.1146/annurev-cellbio-100109-104129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Ellis-Davies GC, Nemoto T, Miyashita Y, Iino M, Kasai H. Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nature neuroscience. 2001;4:1086–1092. doi: 10.1038/nn736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori F, Nishie M, Piao YS, Kito K, Kamitani T, Takahashi H, Wakabayashi K. Accumulation of NEDD8 in neuronal and glial inclusions of neurodegenerative disorders. Neuropathology and applied neurobiology. 2005;31:53–61. doi: 10.1111/j.1365-2990.2004.00603.x. [DOI] [PubMed] [Google Scholar]
- Nimchinsky EA, Yasuda R, Oertner TG, Svoboda K. The number of glutamate receptors opened by synaptic stimulation in single hippocampal spines. J Neurosci. 2004;24:2054–2064. doi: 10.1523/JNEUROSCI.5066-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust VN, Brownell JE, Gladysheva TB, Luo RS, Wang C, Coggins MB, Pierce JW, Lightcap ES, Chau V. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4579–4584. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Peter M. Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects’ review series. EMBO reports. 2008;9:969–976. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB, Pierce JW, Podust VN, Luo RS, Chau V, et al. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Molecular and cellular biology. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz LA, Hall BJ, Patrick GN. Activity-dependent ubiquitination of GluA1 mediates a distinct AMPA receptor endocytosis and sorting pathway. J Neurosci. 2010;30:16718–16729. doi: 10.1523/JNEUROSCI.3686-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz LA, Patrick GN. Ubiquitin-dependent endocytosis, trafficking and turnover of neuronal membrane proteins. Molecular and cellular neurosciences. 2012;49:387–393. doi: 10.1016/j.mcn.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scudder SL, Goo MS, Cartier AE, Molteni A, Schwarz LA, Wright R, Patrick GN. Synaptic Strength Is Bidirectionally Controlled by Opposing Activity-Dependent Regulation of Nedd4-1 and USP8. J Neurosci. 2014;34:16637–16649. doi: 10.1523/JNEUROSCI.2452-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Tsai NP, Wilkerson JR, Guo W, Maksimova MA, DeMartino GN, Cowan CW, Huber KM. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell. 2012;151:1581–1594. doi: 10.1016/j.cell.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um JW, Han KA, Im E, Oh Y, Lee K, Chung KC. Neddylation positively regulates the ubiquitin E3 ligase activity of parkin. Journal of neuroscience research. 2012;90:1030–1042. doi: 10.1002/jnr.22828. [DOI] [PubMed] [Google Scholar]
- Xie P, Zhang M, He S, Lu K, Chen Y, Xing G, Lu Y, Liu P, Li Y, Wang S, et al. The covalent modifier Nedd8 is critical for the activation of Smurf1 ubiquitin ligase in tumorigenesis. Nature communications. 2014;5:3733. doi: 10.1038/ncomms4733. [DOI] [PubMed] [Google Scholar]
- Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Yamada T, Yang Y, Bonni A. Spatial organization of ubiquitin ligase pathways orchestrates neuronal connectivity. Trends in neurosciences. 2013;36:218–226. doi: 10.1016/j.tins.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Perez R, Pan M, Lee T. Requirement of Cul3 for axonal arborization and dendritic elaboration in Drosophila mushroom body neurons. J Neurosci. 2005;25:4189–4197. doi: 10.1523/JNEUROSCI.0149-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Liu Q, Chen B, Zhang X, Guo C, Zhou H, Li J, Gao G, Guo Y, Yan C, et al. Mutation in CUL4B, which encodes a member of cullin-RING ubiquitin ligase complex, causes X-linked mental retardation. American journal of human genetics. 2007;80:561–566. doi: 10.1086/512489. [DOI] [PMC free article] [PubMed] [Google Scholar]