Abstract
Humans are exposed to thousands of chemicals with inadequate toxicological data. Advances in computational toxicology, robotic high throughput screening (HTS), and genome-wide expression have been integrated into the Tox21 program to better predict the toxicological effects of chemicals. Tox21 is a collaboration among US government agencies initiated in 2008 that aims to shift chemical hazard assessment from traditional animal toxicology to target-specific, mechanism-based, biological observations using in vitro assays and lower organism models. HTS uses biocomputational methods for probing thousands of chemicals in in vitro assays for gene-pathway response patterns predictive of adverse human health outcomes. In 1999, NIEHS began exploring the application of toxicogenomics to toxicology and recent advances in NextGen sequencing should greatly enhance the biological content obtained from HTS platforms. We foresee an intersection of new technologies in toxicogenomics and HTS as an innovative development in Tox21. Tox21 goals, priorities, progress, and challenges will be reviewed.
Keywords: Tox21, high throughput screening, toxicogenomics, toxicology, robotics, pathways, in vitro, NextGen sequencing
1 Introduction
There are more than 100,000 chemicals being used in commerce (Zeiger and Margolin, 2000) and only a small fraction of these have undergone adequate toxicological evaluation. There is a need to develop alternative approaches for screening and prioritising these chemicals for more extensive toxicological studies in animals. The purpose of this review is to:
present high throughput screening (HTS) technologies and the Tox21 concept
discuss Tox21 Phase I and Phase II efforts
describe our interest in Next Generation (NextGen) sequencing and related sequencing technologies for improving toxicogenomics
describe the vision of incorporating HTS toxicogenomics as part of Tox21 Phase III.
2 The beginnings of Tox21
2.1 Tox21 and HTS Technologies
Tox21 was organised in response to the NTP Roadmap (NTP, 2004) and the US National Academy of Science report on ‘Toxicology in the 21st Century, A Vision and Strategy’ (NRC, 2007) and was initiated by the signing of a 5-year memorandum of understanding (MoU) on ‘High-Throughput Screening, Toxicity Pathway Profiling and Biological Interpretation of Findings’. The MoU was released in 2008 and signed by representatives of the National Human Genome Research Institute (NHGRI), the National Institute for Environmental Health Sciences (NIEHS), and the US Environmental Protection Agency (EPA). In 2010, this MoU was revised to include the US Food and Drug Administration (US FDA).
The goals of the Tox21 program are to:
identify patterns of compound-induced biological response in order to characterise toxicity and disease pathways, facilitate cross-species extrapolation, and model low-dose extrapolation
prioritise compounds for more extensive toxicological evaluation either in vitro or in vivo
develop predictive models for biological response in humans (Tice et al., 2013).
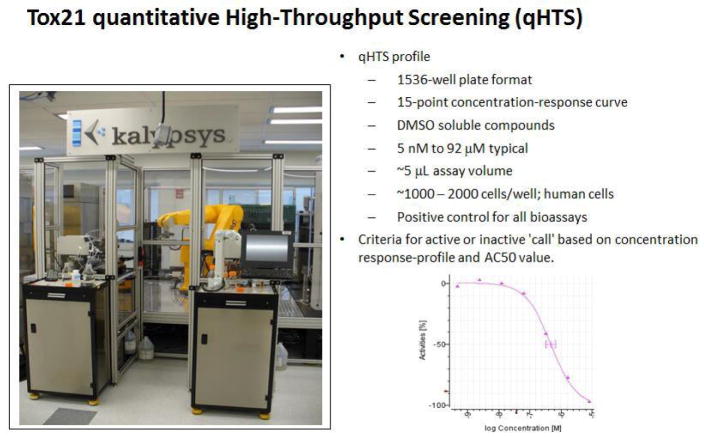
To accomplish Tox21 goals, compound libraries have been established for testing in in vitro quantitative high throughput screening (qHTS) assays in much the same way pharmaceutical companies perform drug discovery by using high precision robots to do many of the assay manipulations more accurately and more quickly (Attene-Ramos et al., 2013). The use of 1,536 well plates allows many thousands of chemical compounds to be screened at the same time using relatively short-term incubation durations of up to 48 hours depending on the needs of the assay (Figure 1). Compounds must be soluble in the solvent, dimethylsulfoxide (DMSO), volatile compounds are avoided, and a positive control must be included in each experiment. Each 1,536 well contains ~5 μL volume and a few thousand cells; reagents can be added but, due to the small volume, media removal/wash steps cannot be part of the experimental protocol. Tox21 assays are conducted primarily in human cells to limit the need for cross-species extrapolation. The quantitative nature of the HTS assay involves generating a 15-point concentration-response curve (repeated in triplicate); the Hill equation or other curve-fitting algorithm is used to calculate an AC50 – an ‘active concentration’ that produces a half-maximal response. For each assay, each compound can be classified as ‘active’, ‘inconclusive’, or ‘inactive’. For active compounds, the AC50 or other measure of activity can be used to rank compounds by relative potency.
Figure 1.
Tox21 qHTS (see online version for colours)
Notes: (a) Automated robotic dispenser and plate holders containing 1,536 well plates for specific assays after short-term chemical exposure. (b) qHTS profile criteria. (c) Concentration-response curve on x and y axes, respectively. DMSO – dimethylsulfoxide; AC50 – concentration in assay that produces half maximal response.
A few words should be included about the development of the Tox21 qHTS effort at the NIH (Shukla et al., 2010). Funding to build the Tox21 program represents a multi-year pooling of federal resources and expertise from NIEHS/NTP, EPA, and NCATS with later participation by the FDA. For example, the significant costs in building a 10,000 compound library (Tox21 10K library) were shared by NIEHS/NTP, EPA, and NCATS and involved the compound selection process; acquisition of the compounds from commercial sources; an independent chemical analysis for identity, purity, and stability in DMSO; and the process of arraying the chemical stock solutions into formats compatible for robotic qHTS (Tice et al., 2013). Screening began in 2005 at the NIH Chemical Genomics Center (NCGC), whose goal is to translate the discoveries of the Human Genome Project into biological and disease insights and ultimately develop new therapeutics for human disease. This goal was to be accomplished by drug discovery efforts similar to those conducted by pharmaceutical companies by using small molecule assay development, high-throughput screening, chemoinformatics, and chemistry (Thomas et al., 2009). The NCGC initiated several programs including Assay Development and High-Throughput Screening, Chemistry Technology, RNAi (interference RNA technology), and toxicology in the 21st century (Tox21). In 2012, the NCGC became part of a new institute called NCATS that was established to develop innovations to reduce, remove, or bypass costly and time-consuming bottlenecks in the translational research pipeline in an effort to speed the delivery of new drugs, diagnostics, and medical devices to patients.
Each of the consortia partners in Tox21 contributes important sets of technologies and expertise (Figure 2), which are distributed across various working groups, including chemical selection, assays and pathways, informatics, and targeted testing (Tice et al., 2013). A major task of the chemical selection working group has been to consider the nearly 100,000 chemicals in commerce and select representative agents to comprise a 10,000 (10K) compound library for qHTS assays. Once the compounds were identified, a major effort was made to procure and analytically determine the identity, purity and stability of each compound. A requirement for the 10K library was that the compound must be soluble in DMSO, be non-volatile, and be expected to be stable under normal analysis conditions. In addition, libraries of defined chemical mixtures and aqueous soluble compounds have been established for use in the Tox21 program. The study of volatile organic compounds (VOC’s) in high throughput mode to screen for the toxicity of these important environmental contaminants awaits more sophisticated incubation chambers compatible with robotic equipment and HTS protocols. The assays and pathways group considers nominations for new assays for HTS. Initially, the focus for assays has been on those that interrogate agonism or antagonism of nuclear receptors and also those assays measuring up-regulation of cellular ‘stress response’ pathways. This working group continues to identify toxicity pathways and corresponding assays that measure their activation, and to prioritise HTS assays for inclusion in Tox21 at NCATS. The informatics working group evaluates HTS assay performance and develops prioritisation schemes and toxicity prediction models. Importantly, this working group helps prepare HTS data for public release, that is accessible at various public URL sites including PubChem, CEBS, and ACToR as described in the following section. The Targeted Testing group has been formed to prioritise ‘active’ compounds from the HTS assays for further evaluation in other cell models (e.g., stem cells), alternative animal models (e.g., zebrafish), and to a very limited extent in acute rodent studies. For example, a number of mitochondrial toxicants have been identified by HTS screening. Prioritisation criteria for future studies by the targeted testing group include consideration of chemical potency (e.g., AC50) and efficacy in HTS screens, cytotoxicity, NTP nomination status, in silico toxicity prediction data, and any additional toxicology data found in various databases (e.g., Leadscope, Chemtrack).
Figure 2.
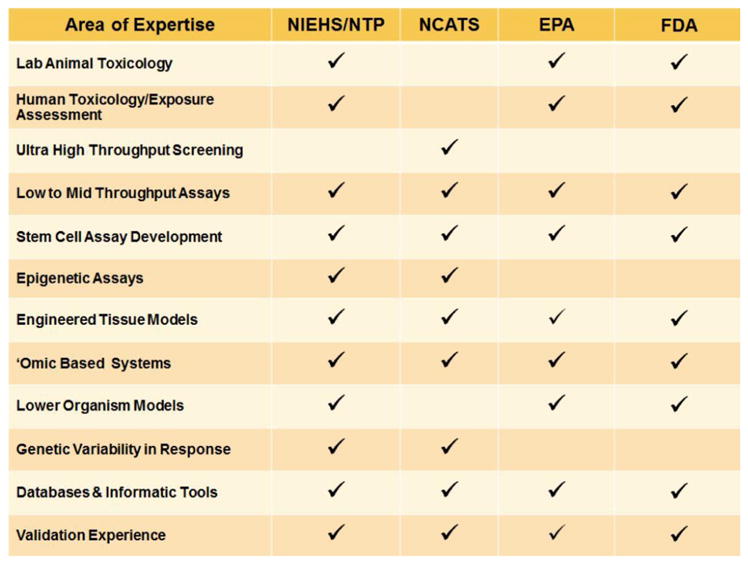
Tox21 partner capabilities (see online version for colours)
Notes: Areas of expertise of Tox21 consortia members are shown. Unique areas of expertise at NIEHS, EPA, and FDA are in human toxicology, preclinical testing, and alternative model species while qHTS robotics and high content screening technologies are housed at the NCATS facility.
2.2 Tox21 data repositories
Chemical effects on biological systems (CEBS) is a publically available resource supported by the NTP and available at http://www.niehs.nih.gov/research/resources/databases/cebs/). CEBS houses data from academic, industrial, and governmental laboratories and is designed to display data in the context of biology and study design and to permit data integration across studies for novel meta-analysis. PubChem is a database of chemical molecules (http://pubchem.ncbi.nlm.nih.gov/) of < 1,000 atoms and their activities in biological assays, including Tox21 HTS data. PubChem is maintained by the National Center for Biotechnology Information (NCBI) as part of NIH. ACToR (http://epa.gov/actor) is the Aggregated Computational Toxicology Online Resource created by the EPA’s NCCT. ACToR serves as EPA’s online compilation of all the public sources of chemical toxicity data, aggregating data from over 1,000 public sources on over 500,000 chemicals. The database is searchable by chemical name, structure, and other identifiers and can be used to find all publicly available information on hazards, exposure, and risk assessment.
2.3 Tox21 in phases
The concept of selecting, acquiring, chemically analysing, and assembling large relevant libraries of chemicals for eventual testing in HTS assays has been a collaborative undertaking that occurred in phases (Tice et al., 2013). The NTP at NIEHS and EPA began a pilot program with the NCGC in 2005 termed ‘Tox21 Phase I’, that involved screening a library of 1,408 compounds (1,353 unique chemicals) of interest to NTP and another library of 1,462 compounds (1,384 unique chemicals) of interest to EPA. The NTP library was screened in 140 qHTS assays that primarily represented 77 cell-based reporter gene endpoints including nuclear receptor transactivation assays (e.g., estrogen receptor alpha, androgen receptor) and those that measured effects on stress response pathways (e.g., p53, caspase, hypoxia inducible factor, heat shock protein), as well as others in various cell types [see Tice et al. (2013) for the complete list]. The EPA library was scored in a subset of these assays.
In addition, the EPA has conducted its own screening program known as ‘ToxCast™’ (Sipes et al., 2013). ToxCast™ focuses on smaller chemical libraries (i.e., hundreds of compounds) but a much wider breadth of biology that covers many different cellular phenotypes. ToxCast™ Phase I screened 320 compounds (309 unique) that were primarily pesticides and some endocrine active compounds in approximately 550 assays. In Phase II, the ToxCast™ effort screened about 700 compounds in more than 700 assays and nearly 1,000 compounds were screened in several endocrine activity assays.
During Tox21 Phase I, the chemical selection working group had been diligently working to produce the much larger ‘10K Compound Library’ for qHTS screening (Attene-Ramos et al., 2013; Huang et al., 2014). The library consists of about 10,500 compounds and of those, 8,307 are unique substances. To evaluate technical variability, the 10K library contains 88 single-sourced compounds included in duplicate on each 1,536 well plate. Multiple wells of a positive control compound that was specific to each assay are also included in each plate. Nominations of specific chemicals to the 10K library were roughly one-third each from the NTP, EPA, and NCATS. The types of compounds from each agency reflected each program’s primary interest. For example, NCATS contributed drugs and drug-like compounds. The NTP contributed NTP-studied chemicals, nominated and related compounds obtained from external collaborators, chemicals tested in various in vivo regulatory assays, and formulated compound mixtures. The EPA provided the compounds in ToxCast™ Phases I and II as well as compounds from the EPA antimicrobial registration and endocrine disruptor screening programs and the FDA drug-induced liver injury program. Compound identities and structures can be found at http://www.epa.gov/ncct/dsstox/sdf_tox21s.html. The 10K compound library is screened in each qHTS assay three separate times at 15 concentrations per qHTS run to ensure reproducibility and to evaluate quality control. In Phase II, much effort has been devoted to screening the 10K library in numerous reporter-driven assays for nuclear receptors and cell stress pathways as described in the next section, ‘Tox21 assays’. Screening the 10K library has occurred over a period of more than 5 years and is still continuing as part of Phase II since new HTS assays continue to be developed and validated for HTS screening against large chemical library sets like the 10K library.
2.4 Tox21 assays
The assays for Tox21 are usually florescence or luminescence driven assays that must be sufficiently sensitive and reproducible to work in a 1536-well plate format. For assays that involve the use of reporter constructs, the detection vector is stably transfected into a compatible human cell type such as HEK293 embryonic kidney cells or HepG2 liver cells. Every 1,536 well plate in each assay contains cells exposed to the solvent control DMSO only and one or two known positive control substances. The assay is evaluated for its ability to detect known active compounds; other desirable features are a signal to noise-background ratio (S/NB) above 2.0, a ‘Z-factor’ at least > 0.5 that reflects both the assay signal dynamic range and the data variation associated with the signal measurements, and a coefficient of variation of less than 10%.
Suitability of each assay is evaluated for compatibility with qHTS screening by the assays and pathways working group prior to being used to screen the 10K library. The focus in Tox21 Phase II is on nuclear receptor (Huang et al., 2011) and stress response pathway assays (Attene-Ramos et al., 2013). Figure 3 shows some representative Phase II assays. Fluorescence or luminescence data are scaled to the positive control data to generate concentration response curves. In order to derive consistent curve features, positive data are modelled to mathematical equations such the Hill equation to determine the AC50 concentration in order to compare results from one chemical to another. The relative biological activity of compounds is based on the maximal magnitude of the response, the shape of their response curves, and concentration compared to known active compounds.
Figure 3.
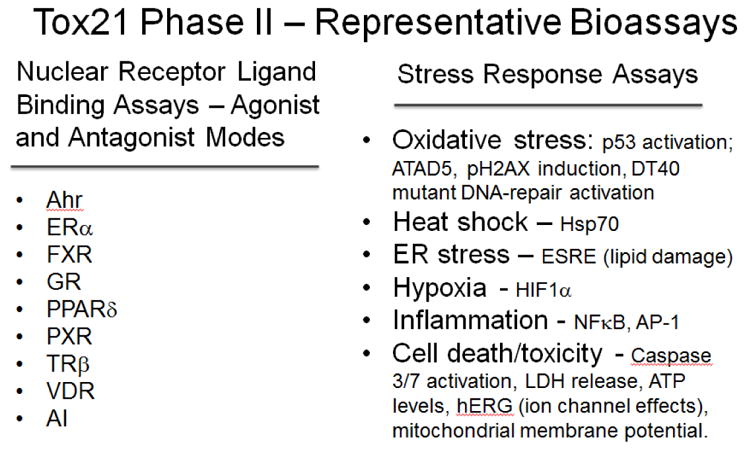
qHTS assays in Tox21 Phase II
Notes: Two categories of assays were evaluated in the second phase of Tox21 that included nuclear ligand receptor binding assays and another set of cellular biology assays relevant to toxicity including oxidative stress, heat shock, ER (endoplasmic reticulum), stress, hypoxia and cytotoxicity. (Ahr, aryl hydrocarbon receptor; ERα, estrogen receptor alpha; FXR, farnesoid X receptor; GR, glucocorticoid receptor; PPARδ, peroxisome proliferator-activated receptor gamma receptor; PXR, pregnane X receptor; TRβ, thyroid hormone receptor beta; VDR, vitamin D receptor; AI, apolipoprotein A-1 receptor; ATAD5, ATPase family, AAA domain containing 5; pH2AX, phosphorylation of histone 2A; DT40, chicken bursal lymphoma cell line; ER stress, endoplasmic reticulum stress; ESRE, ER stress response element; hERG, potassium ion channel called, ether-a-go-go-related gene; HIF1α, hypoxia inducible factor 1-alpha; Hsp70, heat shock protein 70kD; LDH, lactate dehydrogenase; NFκB, nuclear factor of kappa light chain polypeptide gene enhancer in B-cells).
The limits of Tox21 Phase II results are important to recognise. The extent of biological pathways that are currently interrogated are limited. The primary focus of Phase II has been upon the use of reporter gene assays using immortalised human cell lines (compared to terminally differentiated cells or pluripotent cell types). Even though thousands of chemicals are being screened, volatile chemicals or those not soluble in DMSO are excluded. There is a general focus on single compounds but people are more generally exposed to mixtures. A major challenge is the limited capability of xenobiotic metabolism in these cells, since many compounds are metabolically activated or deactivated in vivo. Also, the use of single cell systems does not allow for the multicellular and organisational complexity of tissues and organs in a whole organism. Finally, qHTS assays are limited to acute exposure scenarios of no more than two days while many diseases result from low dose, chronic exposure conditions. Equally daunting are the somewhat limited availability of computational platforms for analysing the very large datasets (i.e., ‘big data’) generated in Tox21.
Another challenge presented by screening thousands of compounds in a single or dual measure assay (e.g., reporter, cytotoxicity) per well is the limited amount of biological information that comes from the discrete cellular biology being measured (e.g., p53 activation only) or the chemical ligand interaction being assayed (e.g., thyroid receptor-beta). Certainly, there is high interest in endocrine disruptor compounds in the environment that antagonise or activate certain nuclear receptor targets (e.g., estrogen and androgen receptors). However, there are a multitude of other ligand-receptor interactions or cellular biology assays that might be altered by chemical exposure in a way that adversely affects human health. It is also recognised that cellular context plays a major role in assay response such that different responses in liver, kidney, and breast epithelial cells are very possible if not likely. Further, use of established cell lines that are immortalised or neoplastically transformed would be expected to have different cellular physiology and receptor status compared to terminally differentiated, normal human cells from different tissues and organs. Some of these limitations are being addressed within the Tox21 program by investigating several different genomic-based platforms that would allow for the monitoring of about 1,500 genes in order to create unique gene expression patterns or ‘gene signatures’ upon exposure to each chemical (i.e.,) in a variety of cell types, including human primary and stem cells (undifferentiated and differentiated) in various high throughput formats.
3 Tox 21: the future
3.1 Introduction
The beginnings of Tox21 were initially conceived around reporter-based, single-measure assays that measured chemical interactions with nuclear receptors or showed activity upon discrete cellular functions in stress response assays. The concept of exploring how chemicals perturb ‘biological space’ is a concept that has grown out of HTS screening of thousands of chemicals for pharmaceutical drug discovery (Nettles et al., 2006; Petit-Zeman, 2004) and is now being applied to toxicology. High throughput screens for drugs have traditionally provided readouts based on fluorescence, luminescence, and absorption (Petit-Zeman, 2004) and like pharma, Tox21 scientists are now looking to future technologies that will allow the capture of more information across biological space. High content screening, involving the use of automated microscopy and image analysis to cultured cells exposed to toxicants (Tolosa et al., 2015) is one way to capture more information from a single assay. The Tox21 community is actively engaged in this research area.
Another way to increase biological space is through the use of gene expression technologies. Rather than observe the expression change of just one gene transcript in cells from each sample well (i.e., a single measure), researchers have considered multiplexing or measuring many transcripts at once to extend their knowledge of a chemical’s effect upon cellular pathways. At the beginning of the new millennium, microarrays printed on a glass slide were hailed as a new high content technology that could measure all genes in each sample; for toxicology, the term ‘toxicogenomics’ represented an exciting new tool for toxicology initiated at NIEHS (Nuwaysir et al., 1999). As powerful as microarrays are, microarray platforms are not high throughput, cost-efficient, or sensitive enough to analyse multiple transcripts for the thousands of samples envisioned for the Tox21 program.
Next generation sequencing (NGS) is a relatively new technology that has been rapidly maturing since 2008 and has provided increased transcriptional information content while declining in cost. NGS technologies simultaneously measure tens of thousands of sequencing reactions simultaneously and can convey base-pair resolution for all cellular transcripts (Koboldt et al., 2013). The NTP wanted to understand the NGS technology, termed ‘RNA-Seq’, that is designed to measure the transcriptome, in order to gain experience and to determine its possible Tox21 applications. Since microarrays have been the gold standard for measuring the transcriptome over the past decade, the NTP compared how well microarray and RNA-Seq performed in analysing control and chemically-treated samples.
Beyond a comparison of microarray and RNA-Seq platforms, the NTP is looking toward future use of NGS platforms that could be adaptable to high throughput needs of the Tox21 program. Those needs involve study of transcript expression responses of different cell types to thousands of environmental chemicals in a rapid and cost-effective manner. The following section will describe in some detail an initial NTP study with RNA-Seq analysis. The data and bioinformatics experience have contributed toward the future directions for Tox21 in widening an understanding of a chemical effects upon biological space.
3.2 Toxicogenomics: microarray and NextGen sequencing
Toxicogenomics is an enabling set of gene expression technologies applied to the field of toxicology that allows researchers to essentially query an entire genome for responses to compound exposure (Khan et al., 2014). Ideally, toxicogenomics would measure or ‘profile’ all gene expression changes at the transcript and protein levels in biological samples after toxicant exposure. Gene and protein profiling from toxicogenomic studies is providing new information from those portions of the genome responding to toxicant injury rather than examining gene readouts on a one-by-one basis. A major goal for toxicogenomics is to define new biomarkers and ‘gene signatures’ of chemical toxicity that provide much greater definition than current indicators for classifying toxicants for health risk and for monitoring toxicity. New toxicity biomarkers and signatures should:
reflect the diverse structure of the thousands of chemicals and other stressors in the environment
show toxicity according to dose and time
incorporate the genetic diversity of cell types, tissues, organs, or surrogate organisms that serve as models for human toxic response and disease.
When human cells or tissues can be utilised, this can often provide one less level of extrapolation of results.
Microarray technologies have been developed into stable platforms (e.g., Affymetrix, Agilent, Illumina) for sensitively measuring about 40,000 genomic elements in humans and rodent species. However, as detailed as microarray analysis has been in defining gene expression changes, newer ‘NextGen’ sequencing technologies such as RNA-Seq can better define responses of the transcriptome at an unprecedented depth of gene expression coverage and sensitivity in transcript detection (Ning et al., 2014). NextGen sequencing represents a series of new technologies for massive parallel DNA sequencing that provides more information about the transcriptome based on alignment and counting of millions of short sequences called ‘reads’ (e.g., 100 base pairs per read) with an improved accuracy compared to hybridisation-based microarrays, the current standard for expression analysis (Ding et al., 2010).
The NTP at NIEHS wanted to explore the potentially increased value of NextGen sequencing for gene expression in order to consider using this powerful technology in the Tox21 HTS program. A recently published study (Merrick et al., 2013) reports on a comparison of microarray (Agilent) and RNA-Seq, using the NextGen sequencing platform by Illumina. In this study, a set of exposure conditions was purposely chosen that would produce a robust gene expression response. Aflatoxin B1 (AFB1) is still an important environmental contaminant and hepatic carcinogen. At 1 ppm exposure, rats will uniformly develop liver tumours after 12–24 months of exposure. Prior work had shown that rats evaluated at the end of a 90-day exposure to AFB1 at 1 ppm did not show any sign of tumours but did reveal a significant gene expression response. This exposure scenario was selected to compare microarray and NextGen sequencing (RNA-Seq) platforms.
RNA was extracted from male F344/N rat liver after animals were exposed for 90 days to normal feed (control) or 1 ppm AFB1 in feed. RNA from four control and four AFB1 treated rats were individually analysed. After isolation and fragmentation of mRNA, a cDNA library of nucleotide sequences was created that has millions of short DNA reads generated in 75-base paired-end orientation by RNA-Seq analysis. The goals of the current study were to:
to more precisely define gene expression changes related to carcinogenesis produced by AFB1 exposure prior to onset of malignancy
establish a high resolution map of the F344/N rat liver transcriptome.
In this particular study, Figure 4 shows more than 60% of differentially expressed genes (DEGs) were found by RNA-Seq using the DESeq algorithm compared to standard analysis using a microarray platform (or other specialised algorithms like Cuffdiff), suggesting RNA-Seq analysis was equal to if not greater in sensitivity to microarray detection of gene expression changes. In addition, this study described the rat liver transcriptome in greater detail than before as shown in Figure 5 with the AFB1-responsive gene, Akr7a3. Reads were assigned to each exon and aligned well with the RefSeq model for Akr7a3. The number of reads across the entire gene transcript are summed for each animal and the comparison of means between control and AFB1 are believed to be more accurate than a fluorescence value from a single microarray probe hybridising to the 3′-end of the gene. In addition, several splice variants, isoforms, and novel transcripts were also described that were not found by microarray analysis. A follow-up study was used to compare these RNA-Seq data from frozen liver with RNA-Seq analysis after RNA was extracted from fixed liver in paraffin blocks from the same animals (Auerbach et al., 2014). Differentially expressed transcripts and activated canonical pathways were highly similar between fresh and paraffin blocked liver, suggesting archival tissues might be suitable for deriving mechanistic gene signatures with corresponding pathophysiology.
Figure 4.
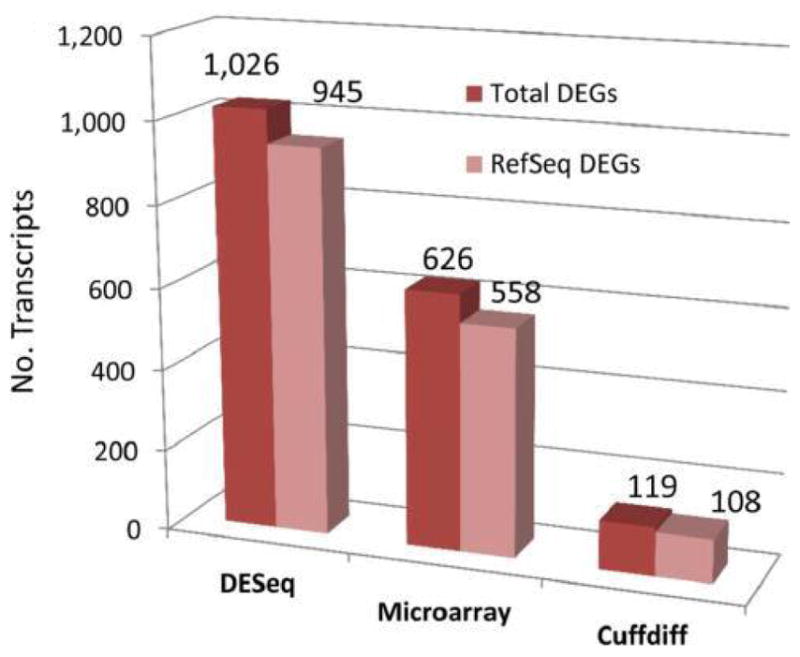
Differentially expressed genes (DEGs) identified from differential expression algorithms, DESeq and Cuffdiff, using RNA-Seq sequencing data compared to DEGs using the same RNA by microarray analysis (see online version for colours)
Notes: Male rats were exposed to control feed or 1 ppm aflatoxin B1 (AFB1) in feed for 90 days, then sacrificed and liver RNA isolated. The number of DEGs that were observed with a two-fold change p < 0.005 of genes by DESeq compared to microarray and Cuffdiff analyses for all eight animals (n = 4 control; n = 4 AFB1) is shown. Total number DEGs and those that match an existing Ref-Seq annotation are displayed in the bar chart.
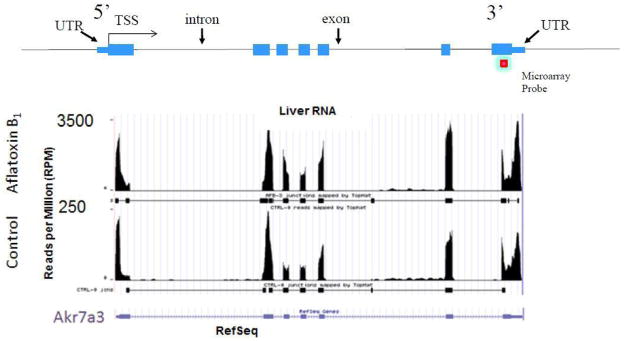
Figure 5.
Comparison of RNA-Seq and microarray data (see online version for colours)
Notes: Akr7a3 – aflatoxin aldehyde reductase – is a 7 exon containing, known response gene induced by AFB1. The number of reads per million is on the ‘y’ axis and the rat genomic sequence that span the Akr7a3 gene is on the ‘x’ axis. A model gene is shown at the top of the figure including the UTR (untranslated region), TSS (transcriptional start site), introns and exons. Only one microarray probe hybridises with the 3′-portion of the Akr7a3 transcript compared to thousands of short reads over each exon, that are summed for each rat RNA sample.
RNA-Seq analysis of many other rodent tissues has been published. A recent study involving 27 chemical treatments affecting the rat transcriptome reported a concordance between RNA-Seq and microarray data (Wang et al., 2014). They found the concordance between microarray and RNA-Seq was affected by transcript abundance and biological complexity of the mechanism of action. RNA-Seq found more differentially expressed genes than microarray as validated by qPCR due to more accurate detection of low level transcripts but both RNA-Seq and microarray platforms similarly could develop classifiers for mechanistic prediction of chemical toxicity. Nevertheless, the discovery potential, sensitivity for low abundance transcripts and more complete description the transcriptome of coding and non-coding genes by RNA-Seq analysis suggests this platform will shape the future of gene expression studies (Li et al., 2015; Yu et al., 2014).
The gains by RNA-Seq over microarrays are promising but it should acknowledged that the time and complexity of bioinformatic analysis of RNA-Seq data is a limiting factor and the cost of performing analysis is still higher when compared to microarrays. Despite these challenges, the expense of sequencing is dropping so that cost equivalency with microarrays is likely in the near future. In addition, RNA-Seq samples can be individually tagged with specific ‘barcodes’ so that a high degree of sample multiplexing is possible. These capabilities of NextGen sequencing are likely to play a significant role for the field of toxicogenomics in the future.
3.3 Tox21 phase III
The Tox21 program is moving towards its third phase of development (Tice et al., 2013), to address a number of issues and challenges that are in the planning or pilot stages. The ability to use human cell lines (e.g., HepaRG) that have metabolic competency greater than conventional cell lines (e.g., HepG2) is viewed as a priority for the next phase of Tox21 qHTS testing of the 10K library and other compounds of interest. For example, the HepaRG liver cell line can be terminally differentiated into hepatocytes that exhibit a wide complement of xenobiotic metabolism enzymes, nuclear receptors such as PXR (pregnane X receptor) and CAR (constitutive androstane receptor), and ion/drug transporters. This is important since many compounds require biotransformation (metabolic activation) to create reactive chemical intermediates that bind to DNA or other macromolecules to mediate their toxicity.
Furthermore, the acute exposure periods (up to a few days) in short-term in vitro assays may not adequately allow enough time for the development of toxicity when repeated or long-term exposure is needed. Researchers are starting to expand the limits of incubation times and conditions. Use of defined, supplemented media and long-term cultures are being studied for their use in chemical exposed conditions. In one such study (Klein et al., 2014), HepaRG cells were successfully maintained in optimised serum-free conditions for 30 days without changes in viability while maintaining high Cyp (cytochrome P450) enzyme activity. Toxicity and kinetics of valproic acid was successfully studied after repeated exposure over a two week period.
Another aspect of predictive in vitro models being considered in Tox21 Phase III is that the conventional tissue culture models could be improved to more accurately reflect the organisational complexity of three-dimensional (3D) organs like the liver to better model the pharmacologic and toxicological actions of chemical exposure. Compared to 2D cultures, 3D HepaRG cultures exhibit greater sensitivity to aflatoxin B1, amiodarone, valproic acid, and chlorpromazine (Mueller et al., 2014), that might be due to greater metabolic competency (e.g., higher CYP3A4 activity). The additional effort to create 3D cell culture models may limit their immediate application to screening large chemical libraries but as a secondary screen, the models will likely better mimic the in vivo conditions and improve cellular functionality. Many in the field hope that 3D systems for liver and other organs represent realistic cellular systems for rapid toxicity testing that may reduce animal testing and provide toxicological data for potential adverse effects to humans of environmental compounds.
Another area of increased emphasis in Tox21 Phase III will be an expanded utilisation of alternative lower organisms to screen chemical compounds for toxicity. Several species have served in chemical toxicity screening models including the fruitfly Drosophila melanogaster, the nematode Caenorhabditis elegans, and the zebrafish Danio rerio. For example, in order to being able to link HTS cellular toxicity data to molecular targets in multicellular organisms, investigators have measured behavioural responses and motor activity of D. melanogaster to airborne VOCs as a model system for discovering adverse outcome pathways and as a method of toxicity screening (Tatum-Gibbs et al., 2015). Other researchers have developed toxicity platforms using C. elegans to study environmental chemical effects on germline function, induction of aneuploidy and prediction of reproductive deficits (Allard et al., 2013). In related efforts, more high-throughput methods are being developed for assessing chemical toxicity in a C. elegans reproduction assay (Boyd et al., 2010).
There is particular interest in using zebrafish as an alternative testing species for toxicity screening. Zebrafish are small multicellular organisms with a short life cycle with transparent embryos during rapid organ and tissue development. Zebrafish larvae measure 1–4 mm in length, can live in a 96-well plate for several days, are inexpensively maintained, and organogenesis is completed by 72 hr after fertilisation (Pack et al., 1996). Automated systems are being developed for high content screening of zebrafish embryos to speed the process for evaluating potential teratogens and developmental toxicants (Lantz-McPeak et al., 2015). In a recent study (Truong et al., 2014), a total of 1,060 unique ToxCast Phase I and II compounds from the EPA were evaluated in embryonic zebrafish; 487 of these compounds induced significant adverse biological responses. The authors used 18 simultaneously measured endpoints in embryonic zebrafish development as a biological sensor for chemical hazard evaluation. Developmental endpoints included yolk sac and pericardial edema; bent body axis; malformations of the eye, snout, jaw, otic vesicle, brain, somite, pectoral fin, caudal fin, circulation, pigmentation, trunk length, swim bladder, and notochord; alterations in touch response; and mortality. The experimental approach involved plating dechorionated fish embryos via an automated embryo placement system at 4 hours post fertilisation (hpf), and then exposing 32 embryos per concentration at five concentrations at ten-fold serial dilutions ranging from 0.0064–64 μM until 5 days after fertilisation. Larval behaviour and morphological and behavioural endpoints were assessed. The authors described global patterns of variation across tested compounds, evaluated the concordance of the available in vitro and in vivo data with this study, and highlighted specific mechanisms and novel biology related to notochord development. These promising studies in the use of alternative species for toxicological evaluation demonstrate that the zebrafish exposure model can detect adverse responses that might not be found by less comprehensive testing strategies.
High dimensional transcript profiling is another area identified for growth in the Tox21 program. Phases I and II have primarily concentrated on reporter gene assays with single or dual fluorescent signal readouts. While thousands of chemicals can be evaluated in a single assay, the resulting data are limited in terms of biology coverage. As a result, hundreds of different assays might be needed to construct a complete portrait of adverse chemical effects. Another approach considers the possibility that large-scale transcript profiling might provide more readouts that can be used in constructing a more comprehensive toxicity profile for each chemical.
3.4 High throughput toxicogenomics
High dimensional toxicogenomics would involve measurement of multiple transcripts for each chemical at each concentration across exposure duration. The concept would eventually test the idea that transcriptional changes in specific cell types from various tissues (e.g., liver, kidney, heart, neurons) has predictive power for toxicity that can be extrapolated to human health. Technological solutions may be available in the near future to meet this need for a rapid and inexpensive method to determine, or ‘multiplex’, many transcriptional measures for thousands of chemicals at multiple concentrations and exposure times.
The practicality of performing whole transcriptome profiling in high throughput mode, such as the 40,000 probe microarrays currently available in conventional toxicogenomics experiments, is not yet technically feasible. Traditional qPCR measures one transcript per sample well and is not sufficiently high throughput for high dimensional gene expression. However, measuring many cellular transcripts from each well after in vitro chemical exposure is a more tractable goal that is being addressed by a variety of technology platforms including Luminex® beads, gene subarrays, Nanostring®, qNPA®, TaqMan® arrays, and others. Each of these platforms can simultaneously measure (e.g., multiplex) many transcripts per sample well. Depending on the platform, the multiplexing capabilities of these technologies may measure 10 to 50 transcripts per well or they may measure up to several hundred mRNA transcripts per well. The numbers of cells needed per well for multiplexing platforms are still relatively large (thousands of cells) but improvements in sensitivity continue to require fewer numbers of cells. The integration of NextGen sequencing that can generate hundreds of millions of short reads shows promise for generating sufficient numbers of reads to quantitatively measure > 1,000 transcripts with the very small sample sizes required by qHTS platforms. One approach – ‘RASL-seq’ that stands for ‘RNA-mediated oligonucleotide annealing, selection, and ligation with NextGen sequencing’ – can be adapted for large-scale quantitative analysis of gene expression to analyse key genes involved in dysregulated pathways that lead to toxicity (Li et al., 2012). The method of mRNA analysis levels in cell lysates is adaptable to full automation, making it ideal for large-scale analysis of multiple biological pathways or regulatory gene networks in the context of systematic genetic or chemical perturbations.
In order to implement a subgenomic approach to the transcriptome, relevant specific transcripts must be selected. Considerations for the criteria and objectives of choosing a subset of transcripts resulted in a decision at NIEHS for further input from the scientific community. As part of this effort, the NIEHS solicited public comment (NIEHS announcement in the Federal Register at http://www.gpo.gov/fdsys/pkg/FR-2013-07-29/html/2013-18058.htm) on this approach and sponsored a workshop on September 16–17, 2013 entitled, ‘High throughput transcriptomics workshop – gene prioritisation criteria’. Considerations involved genes that should be responsive to toxicity and disease, life-stage susceptibility, gene-environment interactions, and exposure-induced heritable changes. Some of the questions explored during this work shop involved:
those 1,000 genes (transcripts) that should be selected and was this number too few or too many
should data selection be a ‘data-driven’ approach – those genes known to respond to known toxicants, or a ‘knowledge-based’ approach – those genes representative of all known biological pathways
should genes be selected from a disease-driven, chemical-responsive, or toxicological basis, or all of these areas
which cells should be of primary interest.
On this last point, should the focus be on metabolic competency such as hepatocytes for liver, Clara cells for lung epithelium, proximal tubules for kidney; should there be a focus on stem cell such as iPSC’s (induced pluripotent stem cells) or ESC’s (embryonic stem cells); should cells include differentiated or dividing cells or primary versus immortalised (transformed) cells.
3.5 S1500
As an outgrowth of this workshop and deliberations at NIEHS, Dr. Richard S. Paules presented, for internal review to the NTP Board of Scientific Counselors, the progress in this area and discussed some of the guiding principles in constructing a sentinel 1,500 gene set or ‘S1500’ (http://ntp.niehs.nih.gov/ntp/about_ntp/bsc/2014/june/tox21phase3_1500genes_508.pdf). The highlights of the S1500 are that it should encompass several facets deemed critical to program objectives.
The S1500 will attempt to capture maximal variation in gene expression and dynamics to ensure that perturbagens (e.g., chemicals) will trigger changes in many of the S1500 genes in various cell types.
This gene subset will aim for extrapolability, meaning the ability to predict with some accuracy the expression changes in all genes from those observed in this reduced set of sentinel genes.
Maximal pathway coverage will be attempted based on inclusion of specifically selected genes to ensure maximal biological pathway coverage. While not complete, both pathway and signaling knowledge are sufficiently robust such that pathway genes should help toxicologists interpret S1500-generated data.
Toxicity and disease relevant genes will be selected for inclusion in the S1500 for recognised roles in toxicity-related and disease-related processes so that readouts from such annotated transcripts should be useful in toxicity and risk assessment evaluations.
The S1500 gene list is being built computationally against publically available human gene expression datasets. A manuscript describing the construction of the gene set and its initial validation is in preparation. Performance is being evaluated on large Affymetrix datasets of rat gene expression and then an independent external dataset of human gene expression data. By incorporating gene-gene coexpression relationships to represent known pathways, the aim is to balance data-driven and knowledge-based approaches to extrapolate S1500 data to whole transcriptome changes for any given sample. Future testing plans will involve characterisation across a diversity of cell types and eventually include tissue responses across exposure, time and different modes of toxicological action. The S1500 gene list has been released for public comment (https://federalregister.gov/a/2015-08529).
3.6 Validation and regulatory guidance
The development of high throughput in vitro assays has proceeded at a rapid pace over the last decade. However, there are accompanying challenges in interpreting in vitro results for their usefulness in predictive toxicology and their adoption by regulatory communities as detailed in recent reviews (Elmore et al., 2014; Persson et al., 2014; Piersma et al., 2014; Zhang et al., 2014). In applying in vitro screening data to in vivo toxicity, some approaches carefully target the use of specific in vitro assays toward predicting adverse effects such as liver injury or bile salt transporter inhibition-mediated cholestasis (Persson et al., 2014) or genotoxicity (Zhang et al., 2014). Others have taken a broader approach toward providing regulatory toxicology with alternative test systems that reduce animal use, testing time, and cost. In general, the ability for a direct replacement of one complex in vivo test by a relatively simple in vitro assay system is often not feasible. Yet, progress has been made using in vitro screens for very specific in vivo endpoints such as allergic contact dermatitis (Forreryd et al., 2014).
Some of the challenges and lessons learned in the Tox21 program have been recently reviewed (Tice et al., 2013). Briefly, formulating a reasonably large set of substances of general environmental and therapeutic importance in toxicology was a major effort for Tox21 so that qHTS data could be biologically diverse enough to be used in prioritisation schemes and toxicology prediction models. In Tox21, many thousands of compounds have been screened in a single experiment in an assay across a broad concentration range to make concentration-response curves. This qHTS approach was designed to identify compounds with a wide range of activities with a much lower false-positive or false-negative rate than conventional drug discovery using qHTS approaches. The result of qHTS screening in Tox21 is to identify active and inactive compounds according to specifications of a defined qHTS assay that measures a specific biological activity (e.g., mitochondria membrane potential) or a specific chemical-biological interaction (e.g., estrogen receptor activation assay). The aggregate of combining multiple individual qHTS assays can be used to provide insight into the toxicity potential of chemicals that have little or no toxicology information. In addition, predictive toxicology models also take into account structure-activity relationships, chemoinformatics, and pharmacokinetics of known chemicals and toxicants and apply them to individual chemicals or groups of chemical of interest (Dix et al., 2012; Kleinstreuer et al., 2013, 2014; Sipes et al., 2011).
4 Conclusions
The realisation that the tens of thousands of chemicals in commerce and in the environment cannot be tested in animal models in a timely manner has been a primary motivation of the Tox21 initiative to more effectively inform and guide public health scientists and regulators. Early phases of Tox21 were built on in vitro qHTS assays using gene or protein reporter constructs after short-term chemical exposure in order to robotically screen hundreds and now thousands of chemicals. These automated in vitro assays have focused on activation of nuclear receptors and assays measuring activation of specific cellular stress response pathways. While these assays in Tox21 Phases I and II continue to be performed and are of value, the next stage of Tox21 – Phase III – will still be capable of screening thousands of chemicals but will address some limitations of the existing systems, including metabolic activation, the use of different cell types with more complex cellular organisational features and a more comprehensive transcriptional profile. Interpretation of in vitro data in extrapolating high throughput results to human toxicity and disease remains a challenge; however, the intersection of toxicogenomics with high throughput transcriptional screening presents a great opportunity to bring more data to bear on the complex challenges in risk assessment in protecting public health from environmental chemical exposures.
Acknowledgments
Funding
This research was supported by the Intramural Research Programs of the Division of the National Toxicology Program (DNTP) at the National Institute of Environmental Health Sciences (NIEHS) of the National Institutes of Health (NIH).
Biographies
B. Alex Merrick is a group leader and molecular toxicologist in the Biomolecular Screening Branch at NTP. His interests involve gene expression and NextGen sequencing to help better understand toxicological effects of environmental chemicals. He is particularly interested in applying advanced Omics technologies to screening fixed archival tissues to extract molecular information that will provide further insights into understanding chemical exposure and disease.
Richard S. Paules is an experimental cancer biologist at NIEHS and recently assumed the position of Acting Branch Chief of the Biomolecular Screening Branch at NTP. His basic research interests have involved the role of ATM protein kinase and checkpoint control of the cell cycle. He was instrumental in creating the National Center for Toxicogenomics at NIEHS to develop a research program integrating whole transcriptome analysis with phenotypic anchoring in toxicology studies. Since joining the NTP, he has forwarded the S1500 concept into high throughput screening as a new direction for Tox21.
Raymond R. Tice recently retired from the NTP. Early in his career, he and others were first to introduce the alkaline comet assay as a tool for detecting DNA damage. He joined the NTP in 2005 as the first Deputy Director of the NTP Interagency Center for the Evaluation of Alternative Toxicological Methods and in 2008 became the first Chief of the Biomolecular Screening Branch. As Chief, he played a key role in establishing the U.S. Interagency Tox21 program as an innovative way to advance the field of toxicology testing.
Footnotes
disclaimer
The statements, opinions, or conclusions contained therein do not necessarily represent the statements, opinions, or conclusions of NIH or the United States Government. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
B. Alex Merrick, Email: merrick@niehs.nih.gov.
Richard S. Paules, Email: paules@niehs.nih.gov.
Raymond R. Tice, Email: tice@niehs.nih.gov.
References
- Allard P, Kleinstreuer NC, Knudsen TB, Colaiacovo MP. A C. elegans screening platform for the rapid assessment of chemical disruption of germline function. Environmental Health Perspectives. 2013;121(6):717–724. doi: 10.1289/ehp.1206301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attene-Ramos MS, Miller N, Huang R, Michael S, Itkin M, Kavlock RJ, Austin CP, Shinn P, Simeonov A, Tice RR, Xia M. The Tox21 robotic platform for the assessment of environmental chemicals--from vision to reality. Drug Discovery Today. 2013;18(15–16):716–723. doi: 10.1016/j.drudis.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach SS, Phadke DP, Mav D, Holmgren S, Gao Y, Xie B, Shin JH, Shah RR, Merrick BA, Tice RR. RNA-Seq-based toxicogenomic assessment of fresh frozen and formalin-fixed tissues yields similar mechanistic insights. Journal of Applied Toxicology. 2014 doi: 10.1002/jat.3068. in press. [DOI] [PubMed] [Google Scholar]
- Boyd WA, McBride SJ, Rice JR, Snyder DW, Freedman JH. A high-throughput method for assessing chemical toxicity using a Caenorhabditis elegans reproduction assay. Toxicol Appl Pharmacol. 2010;245(2):153–159. doi: 10.1016/j.taap.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Wendl MC, Koboldt DC, Mardis ER. Analysis of next-generation genomic data in cancer: accomplishments and challenges. Human Molecular Genetics. 2010;19(R2):R188–196. doi: 10.1093/hmg/ddq391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix DJ, Houck KA, Judson RS, Kleinstreuer NC, Knudsen TB, Martin MT, Reif DM, Richard AM, Shah I, Sipes NS, Kavlock RJ. Incorporating biological, chemical, and toxicological knowledge into predictive models of toxicity. Toxicological sciences: an official journal of the Society of Toxicology. 2012;130(2):440–441. doi: 10.1093/toxsci/kfs281. author reply pp.442–443. [DOI] [PubMed] [Google Scholar]
- Elmore SA, Ryan AM, Wood CE, Crabbs TA, Sills RC. FutureTox II: contemporary concepts in toxicology: ‘pathways to prediction: in vitro and in silico models for predictive toxicology’. Toxicol Pathol. 2014;42(5):940–942. doi: 10.1177/0192623314537135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forreryd A, Johansson H, Albrekt AS, Lindstedt M. Evaluation of high throughput gene expression platforms using a genomic biomarker signature for prediction of skin sensitization. BMC Genomics. 2014;15:379. doi: 10.1186/1471-2164-15-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Sakamuru S, Martin MT, Reif DM, Judson RS, Houck KA, Casey W, Hsieh JH, Shockley KR, Ceger P, Fostel J, Witt KL, Tong W, Rotroff DM, Zhao T, Shinn P, Simeonov A, Dix DJ, Austin CP, Kavlock RJ, Tice RR, Xia M. Profiling of the Tox21 10K compound library for agonists and antagonists of the estrogen receptor alpha signaling pathway. Scientific Reports. 2014;4:5664. doi: 10.1038/srep05664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Xia M, Cho MH, Sakamuru S, Shinn P, Houck KA, Dix DJ, Judson RS, Witt KL, Kavlock RJ, Tice RR, Austin CP. Chemical genomics profiling of environmental chemical modulation of human nuclear receptors. Environmental Health Perspectives. 2011;119(8):1142–1148. doi: 10.1289/ehp.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SR, Baghdasarian A, Fahlman RP, Michail K, Siraki AG. Current status and future prospects of toxicogenomics in drug discovery. Drug Discovery Today. 2014;19(5):562–578. doi: 10.1016/j.drudis.2013.11.001. [DOI] [PubMed] [Google Scholar]
- Klein S, Mueller D, Schevchenko V, Noor F. Long-term maintenance of HepaRG cells in serum-free conditions and application in a repeated dose study. Journal of Applied Toxicology: JAT. 2014;34(10):1078–1086. doi: 10.1002/jat.2929. [DOI] [PubMed] [Google Scholar]
- Kleinstreuer NC, Dix DJ, Houck KA, Kavlock RJ, Knudsen TB, Martin MT, Paul KB, Reif DM, Crofton KM, Hamilton K, Hunter R, Shah I, Judson RS. In vitro perturbations of targets in cancer hallmark processes predict rodent chemical carcinogenesis. Toxicological Sciences: An Official Journal of the Society of Toxicology. 2013;131(1):40–55. doi: 10.1093/toxsci/kfs285. [DOI] [PubMed] [Google Scholar]
- Kleinstreuer NC, Yang J, Berg EL, Knudsen TB, Richard AM, Martin MT, Reif DM, Judson RS, Polokoff M, Dix DJ, Kavlock RJ, Houck KA. Phenotypic screening of the ToxCast chemical library to classify toxic and therapeutic mechanisms. Nat Biotechnol. 2014;32(6):583–591. doi: 10.1038/nbt.2914. [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155(1):27–38. doi: 10.1016/j.cell.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz-McPeak S, Guo X, Cuevas E, Dumas M, Newport GD, Ali SF, Paule MG, Kanungo J. Developmental toxicity assay using high content screening of zebrafish embryos. Journal of Applied Toxicology: JAT. 2015;35(3):261–272. doi: 10.1002/jat.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Qiu J, Fu XD. RASL-seq for massively parallel and quantitative analysis of gene expression. In: Ausubel FM, et al., editors. Current Protocols in Molecular Biology. Unit 4. Chapter 4. 2012. pp. 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Chen E, Yang C, Zhu J, Jayaraman P, De Pons J, Kaczorowski CC, Jacob HJ, Greene AS, Hodges MR, Cowley AW, Jr, Liang M, Xu H, Liu P, Lu Y. Improved rat genome gene prediction by integration of ESTs with RNA-Seq information. Bioinformatics. 2015;31(1):25–32. doi: 10.1093/bioinformatics/btu608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick BA, Phadke DP, Auerbach SS, Mav D, Stiegelmeyer SM, Shah RR, Tice RR. RNA-Seq profiling reveals novel hepatic gene expression pattern in aflatoxin B1 treated rats. PloS One. 2013;8(4):e61768. doi: 10.1371/journal.pone.0061768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, Kramer L, Hoffmann E, Klein S, Noor F. 3D organotypic HepaRG cultures as in vitro model for acute and repeated dose toxicity studies. Toxicology In Vitro: An International Journal published in Association with BIBRA. 2014;28(1):104–112. doi: 10.1016/j.tiv.2013.06.024. [DOI] [PubMed] [Google Scholar]
- Nettles JH, Jenkins JL, Bender A, Deng Z, Davies JW, Glick M. Bridging chemical and biological space: ‘target fishing’ using 2D and 3D molecular descriptors. J Med Chem. 2006;49(23):6802–6810. doi: 10.1021/jm060902w. [DOI] [PubMed] [Google Scholar]
- Ning B, Su Z, Mei N, Hong H, Deng H, Shi L, Fuscoe JC, Tolleson WH. Toxicogenomics and cancer susceptibility: advances with next-generation sequencing. Journal of Environmental Science and Health Part C, Environmental Carcinogenesis & Ecotoxicology Reviews. 2014;32(2):121–158. doi: 10.1080/10590501.2014.907460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NRC. [accessed 3 October 2014];Toxicity Testing in the 21st Century: A Vision and a Strategy. 2007 [online] http://www.nap.edu/openbook.php?record_id=11970.
- NTP. [accessed 25 May 2015];NTP Vision and Roadmap. 2004 [online] http://ntp.niehs.nih.gov/ntp/about_ntp/ntpvision/ntproadmap_508.pdf.
- Nuwaysir EF, Bittner M, Trent J, Barrett JC, Afshari CA. Microarrays and toxicology: the advent of toxicogenomics. Mol Carcinog. 1999;24(3):153–159. doi: 10.1002/(sici)1098-2744(199903)24:3<153::aid-mc1>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Pack M, Solnica-Krezel L, Malicki J, Neuhauss SC, Schier AF, Stemple DL, Driever W, Fishman MC. Mutations affecting development of zebrafish digestive organs. Development. 1996;123(1):321–328. doi: 10.1242/dev.123.1.321. [DOI] [PubMed] [Google Scholar]
- Persson M, Loye AF, Jacquet M, Mow NS, Thougaard AV, Mow T, Hornberg JJ. High-content analysis/screening for predictive toxicology: application to hepatotoxicity and genotoxicity. Basic Clin Pharmacol Toxicol. 2014;115(1):18–23. doi: 10.1111/bcpt.12200. [DOI] [PubMed] [Google Scholar]
- Petit-Zeman S. [accessed 25 May 2015];Exploring Biological Space. 2004 [online] http://www.nature.com/horizon/chemicalspace/background/explore.html.
- Piersma AH, Ezendam J, Luijten M, Muller JJ, Rorije E, van der Ven LT, van Benthem J. A critical appraisal of the process of regulatory implementation of novel in vivo and in vitro methods for chemical hazard and risk assessment. Crit Rev, Toxicol. 2014;44(10):876–894. doi: 10.3109/10408444.2014.940445. [DOI] [PubMed] [Google Scholar]
- Shukla SJ, Huang R, Austin CP, Xia M. The future of toxicity testing: a focus on in vitro methods using a quantitative high-throughput screening platform. Drug Discovery Today. 2010;15(23–24):997–1007. doi: 10.1016/j.drudis.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB. Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chemical Research in Toxicology. 2013;26(6):878–895. doi: 10.1021/tx400021f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipes NS, Martin MT, Reif DM, Kleinstreuer NC, Judson RS, Singh AV, Chandler KJ, Dix DJ, Kavlock RJ, Knudsen TB. Predictive models of prenatal developmental toxicity from ToxCast high-throughput screening data. Toxicological Sciences: An Official Journal of the Society of Toxicology. 2011;124(1):109–127. doi: 10.1093/toxsci/kfr220. [DOI] [PubMed] [Google Scholar]
- Tatum-Gibbs KR, McKee JM, Higuchi M, Bushnell PJ. Effects of toluene, acrolein and vinyl chloride on motor activity of Drosophila melanogaster. Neurotoxicol Teratol. 2015 Jan-Feb;47:114–124. doi: 10.1016/j.ntt.2014.11.008. [DOI] [PubMed] [Google Scholar]
- Thomas CJ, Auld DS, Huang R, Huang W, Jadhav A, Johnson RL, Leister W, Maloney DJ, Marugan JJ, Michael S, Simeonov A, Southall N, Xia M, Zheng W, Inglese J, Austin CP. The pilot phase of the NIH chemical genomics center. Current Topics in Medicinal Chemistry. 2009;9(13):1181–1193. doi: 10.2174/156802609789753644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice RR, Austin CP, Kavlock RJ, Bucher JR. Improving the human hazard characterization of chemicals: a Tox21 update. Environmental Health Perspectives. 2013;121(7):756–765. doi: 10.1289/ehp.1205784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolosa L, Gomez-Lechon MJ, Donato MT. High-content screening technology for studying drug-induced hepatotoxicity in cell models. Arch Toxicol. 2015 doi: 10.1007/s00204-015-1503-z. in press. [DOI] [PubMed] [Google Scholar]
- Truong L, Reif DM, St Mary L, Geier MC, Truong HD, Tanguay RL. Multidimensional in vivo hazard assessment using zebrafish. Toxicological Sciences: An Official Journal of the Society of Toxicology. 2014;137(1):212–233. doi: 10.1093/toxsci/kft235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Gong B, Bushel PR, Thierry-Mieg J, Thierry-Mieg D, Xu J, Fang H, Hong H, Shen J, Su Z, Meehan J, Li X, Yang L, Li H, Labaj PP, Kreil DP, Megherbi D, Gaj S, Caiment F, van Delft J, Kleinjans J, Scherer A, Devanarayan V, Wang J, Yang Y, Qian HR, Lancashire LJ, Bessarabova M, Nikolsky Y, Furlanello C, Chierici M, Albanese D, Jurman G, Riccadonna S, Filosi M, Visintainer R, Zhang KK, Li J, Hsieh JH, Svoboda DL, Fuscoe JC, Deng Y, Shi L, Paules RS, Auerbach SS, Tong W. The concordance between RNA-Seq and microarray data depends on chemical treatment and transcript abundance. Nat Biotechnol. 2014;32(9):926–932. doi: 10.1038/nbt.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Fuscoe JC, Zhao C, Guo C, Jia M, Qing T, Bannon DI, Lancashire L, Bao W, Du T, Luo H, Su Z, Jones WD, Moland CL, Branham WS, Qian F, Ning B, Li Y, Hong H, Guo L, Mei N, Shi T, Wang KY, Wolfinger RD, Nikolsky Y, Walker SJ, Duerksen-Hughes P, Mason CE, Tong W, Thierry-Mieg J, Thierry-Mieg D, Shi L, Wang C. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun. 2014;5:3230. doi: 10.1038/ncomms4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiger E, Margolin BH. The proportions of mutagens among chemicals in commerce. Regulatory Toxicology and Pharmacology: RTP. 2000;32(2):219–225. doi: 10.1006/rtph.2000.1422. [DOI] [PubMed] [Google Scholar]
- Zhang L, McHale CM, Greene N, Snyder RD, Rich IN, Aardema MJ, Roy S, Pfuhler S, Venkatactahalam S. Emerging approaches in predictive toxicology. Environ Mol Mutagen. 2014;55(9):679–688. doi: 10.1002/em.21885. [DOI] [PMC free article] [PubMed] [Google Scholar]