Abstract
Background
Metastatic melanoma is a severe disease with one of the highest mortality rate in skin diseases. Overall survival has significantly improved with immunotherapy and targeted therapies. Kinase inhibitors targeting BRAF V600 showed promising results. BRAF genotyping is mandatory for the prescription of anti-BRAF therapies.
Methods
Fifty-nine formalin-fixed paraffin-embedded melanoma samples were assessed using High-Resolution-Melting (HRM) PCR, Real-time allele-specific amplification (RT-ASA) PCR, Next generation sequencing (NGS), immunohistochemistry (IHC) and the fully-automated molecular diagnostics platform IdyllaTM. Sensitivity, specificity, positive predictive value and negative predictive value were calculated using NGS as the reference standard to compare the different assays.
Results
BRAF mutations were found in 28(47.5%), 29(49.2%), 31(52.5%), 29(49.2%) and 27(45.8%) samples with HRM, RT-ASA, NGS, IdyllaTM and IHC respectively. Twenty-six (81.2%) samples were found bearing a c.1799T>A (p.Val600Glu) mutation, three (9.4%) with a c.1798_1799delinsAA (p.Val600Lys) mutation and one with c.1789_1790delinsTC (p.Leu597Ser) mutation. Two samples were found bearing complex mutations.
Conclusions
HRM appears the less sensitive assay for the detection of BRAF V600 mutations. The RT-ASA, IdyllaTM and IHC assays are suitable for routine molecular diagnostics aiming at the prescription of anti-BRAF therapies. IdyllaTM assay is fully-automated and requires less than 2 minutes for samples preparation and is the fastest of the tested assays.
Introduction
Metastatic melanoma is a severe disease with one of the highest mortality rate in skin diseases [1]. Response rate, progression-free survival and overall survival have significantly improved these last years with the development of immunotherapy and targeted therapies. Kinase inhibitors specifically targeting BRAF with V600 mutated phenotype, like vemurafenib or dabrafenib showed promising results compared to dacarbazine chemotherapy [2, 3]. The association of dabrafenib with an anti-MEK therapy i.e. trametinib, also showed a significantly higher overall survival than vemurafenib alone in BRAF mutated patients with metastatic melanoma [4]. Since these anti-BRAF targeted therapies are only effective on BRAF V600 mutated melanomas, the detection of BRAF mutations on exon 15, especially on the V600 hotspot is mandatory for prescription. BRAF mutations are found in approximately 50% of metastatic melanomas [5] with V600E (c.1799T>A; p.Val600Glu), V600K (c.1798_1799delinsAA; p.Val600Lys), V600R (c.1798_1799delinsAG; p.Val600Arg) and V600M (c.1798G>A; p.Val600Met) in 79%, 12%, 5% and 4% frequencies respectively.
Most of the assays for the routine detection of BRAF mutations in metastatic melanomas are PCR-based, but sequencing or immunohistochemistry assays are also widely used [6, 7]. The ideal assay should be easy, accurate and highly sensitive to ensure the detection of low BRAF mutant allele frequency.
In this study, we compared the new IdyllaTM fully-automated CE-IVD platform with four routine assays in our lab. Fifty-nine samples from patients with metastatic melanoma were assessed for BRAF mutations using IdyllaTM, high resolution amplicon melting (HRM), real-time allele specific amplification (RT-ASA), next generation sequencing (NGS) and immunohistochemistry (IHC).
Materials and Methods
Patients and samples
Fifty-nine formalin-fixed paraffin-embedded tumor samples from patients treated for a metastatic melanoma at Institut de Cancérologie de Lorraine between 2012 to 2015 have been retrospectively collected for this study. All patients gave their consent for the research of BRAF mutations and the use of their samples. Study has been approved by Institut de Cancérologie de Lorraine scientific board. All patients’ data have been anonymized and de-identified prior to analysis. Tumor specimens were macrodissected after hematoxylin-eosin slide examination by a qualified senior pathologist to evaluate the percent tumor nuclei in the sample selected for DNA extraction [8]. For HRM, RT-ASA and NGS assays, one macrodissected section of ten micrometer was used for DNA extraction with no restriction of tumor cell content. For IdyllaTM assay, one ten micrometer section from FFPE samples was used. According to Idylla manufacturer’s recommendations, the sample introduced in the cartridge must contain at least 50% of tumoral cells with an area included in the 25-300mm² range. Multiple sections of 10μm and macrodissection has been used for samples that do not meet these criteria to ensure a total content of 50% of tumor cells. Samples characteristics are detailed in Table 1.
Table 1. Samples characteristics.
| Sample # | Localization | Percent tumor nuclei | DNA concentration (ng/μL) |
|---|---|---|---|
| 1 | right side | 90% | 149.3 |
| 2 | axillary node | 90% | 54.5 |
| 3 | axillary node | 90% | 23.3 |
| 4 | elbow nodule | 90% | 29.5 |
| 5 | left foot | 95% | 27.3 |
| 6 | axillary node | 90% | 17.0 |
| 7 | left thigh | 90% | 52.0 |
| 8 | left arm | 90% | 99.5 |
| 9 | inguinal lymph node | 80% | 17.5 |
| 10 | sentinel node | 20% | 118.2 |
| 11 | node | 90% | 73.4 |
| 12 | cervix | 90% | 20.8 |
| 13 | node | 90% | 61.3 |
| 14 | right buttock | 90% | 22.5 |
| 15 | axillary node | 60% | 22.0 |
| 16 | mastoid | 80% | 51.5 |
| 17 | breast | 90% | 107.3 |
| 18 | heel | 40% | 33.5 |
| 19 | scapular bone | 80% | 130.8 |
| 20 | heel | 30% | 10.4 |
| 21 | right arm | 20% | 33.4 |
| 22 | node | 90% | 16.3 |
| 23 | iliac node | 90% | 55.8 |
| 24 | cheek | 70% | 10.6 |
| 25 | inguinal lymph node | 90% | 31.0 |
| 26 | arm node | 95% | 20.5 |
| 27 | axillary node | 30% | 115.8 |
| 28 | lumbar vertebrae | 10% | 2.1 |
| 29 | pubic symphysis | 90% | 145.5 |
| 30 | inguinal lymph node | 60% | 155.0 |
| 31 | inguinal lymph node | 90% | 52.0 |
| 32 | scapular bone | 75% | 103.5 |
| 33 | inguinal lymph node | 90% | 89.0 |
| 34 | inguinal lymph node | 30% | 114.5 |
| 35 | foot nodule | 90% | 6.8 |
| 36 | gluteal crease | 80% | 30.5 |
| 37 | pectoral muscle | 90% | 12.9 |
| 38 | right calf | 90% | 169.8 |
| 39 | inguinal lymph node | 20% | 64.8 |
| 40 | left thigh | 10% | 5.4 |
| 41 | axillary node | 95% | 109.3 |
| 42 | maxillofacial | 80% | 54.2 |
| 43 | right thigh | 90% | 40.5 |
| 44 | right arm | 75% | 65.8 |
| 45 | axillary node | 40% | 121.7 |
| 46 | axillary node | 50% | 56.3 |
| 47 | scapular bone | 90% | 51.0 |
| 48 | abdominal lesion | 85% | 16.5 |
| 49 | sentinel node | 60% | 39.8 |
| 50 | scalp | 60% | 20.2 |
| 51 | axillary node | 20% | 62.5 |
| 52 | vagina | 50% | 76.5 |
| 53 | axillary node | 50% | 127.5 |
| 54 | back | 25% | 36.2 |
| 55 | iliac node | 90% | 82.3 |
| 56 | right arm | 70% | 66.5 |
| 57 | axillary node | 90% | 23.8 |
| 58 | anal margin | 70% | 17.8 |
| 59 | right arm | 50% | 30.3 |
DNA isolation
For HRM, RT-ASA and NGS, DNA isolation was performed using COBAS® DNA Sample preparation kit (Roche Diagnostics, Meylan, France), as described in the manufacturers protocol. DNA was finally eluted in a volume of 100μL of buffer. DNA concentrations were assessed in duplicate using NanoVue spectrophotometer (GE Healthcare, Buc, France). For IdyllaTM, DNA was automatically extracted in the cartridge. No DNA extraction was needed for IHC.
BRAF mutation detection
All workflows are described in Fig 1.
Fig 1. Workflows for the different assays used for BRAF mutation analysis.
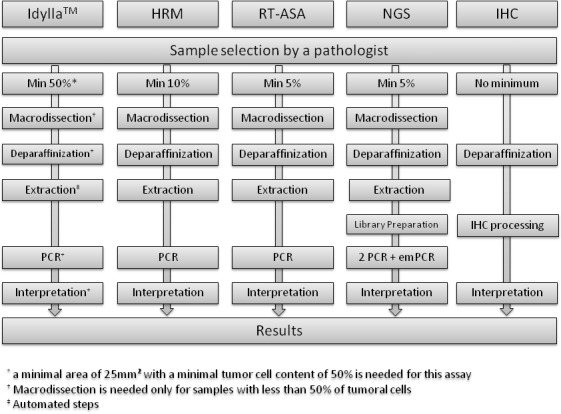
IdyllaTM platform
IdyllaTM platform (Biocartis, Mechelen, Belgium) is a fully catridge-based automated platform and uses microfluidics processing with all reagents on-board. The platform comprises a console and up to eight independent processing units allowing the detection of 8 samples at the same time (e.g. BRAF, KRAS, NRAS or EGFR mutations in different samples). All 59 melanoma samples were assessed for the detection of BRAF p.V600E (c.1799T>A; p.Val600Glu), p.V600E2 (c.1799_1800delinsAA; p.Val600Glu), p.V600D (c.1799_1800delinsAT and c.1799_1800delinsAC; p.Val600Asp), p.V600K (c.1798_1799delinsAA; p.Val600Lys), p.V600R (c.1798_1799delinsAG; p.Val600Arg) and p.V600M (c.1798T>A; p.Val600Met) mutations. Macrodissected FFPE samples sections were transferred to wetted (nuclease-free water) filter papers. A second wetted filter paper was then added on top of the FFPE material. Sample with the two wetted filter papers was finally placed on the lysis pad in the IdyllaTM BRAF mutation test cartridge (Biocartis) and inserted in the instrument. Inside the cartridge, the sample is homogenized and cells lysed using a combination of high intensity focused ultrasound, enzymatic/chemical digestion and heat. The nucleic acids are liberated and ready for subsequent PCR amplification. The PCR is real-time and uses a fluorophore-based detection system.
After a 90 minutes run, all steps were automatically performed inside the cartridge and final reports were directly available on the system after an automatic on-board post-PCR curve analysis. BRAF V600E, V600E2 and V600D were detected as “V600E/E2/D Mutation” and V600K, V600R and V600M as “V600K/R/M Mutation” by the system. The system had been previously calibrated using BRAF V600E, V600K and wild-type standards (Horizon Diagnostics).
High Resolution Melting
HRM analysis was performed using the LC 480 HRM Master kit (Roche Diagnostics, Meylan, France) and 96 well plates (Roche Diagnostics) using LC480 thermocycler (Roche Diagnostics) as previously described [9, 10]. Melting curves were analyzed using LightCycler 480 software v.1.5. (Roche Diagnostics).
DNA extracted from WidR cell line (ATCC® CCL-218TM) was used as BRAF V600E (c.1799T>A; p.Val600Glu) mutated positive control and DNA extracted from LoVo cell line (ATCC® CCL-229TM) was used as BRAF wild-type control.
Real-time allele specific amplification
Real-time allele specific amplification analysis was performed using the LC 480 SybrGreen Master kit (Roche Diagnostics) and 384 well plates (Roche Diagnostics) using LC480 thermocycler (Roche Diagnostics) as previously described [9, 11]. Specific primers for the detection of BRAF p.V600E (c.1799T>A; p.Val600Glu), p.V600K (c.1798_1799delinsAA; p.Val600Lys), p.V600R (c.1798_1799delinsAG; p.Val600Arg) and p.V600D (c. 1799_1800delinsAT; p.Val600Asp) were used for this study.
DNA extracted from WidR cell line was used as BRAF V600E (c.1799T>A; p.Val600Glu) mutated positive control and DNA extracted from LoVo cell line was used as BRAF wild-type control. DNA extracted from standard FFPE samples (Horizon Diagnostics, Cambridge, UK) were used for V600K (Horizon HD268) and V600R (Horizon HD275) mutated positive controls. No positive control was used for V600D mutation because of the rarity of this mutation.
Next Generation Sequencing
Ultra-deep pyrosequencing (Roche Diagnostics) was used to detect mutations on exon 15 of BRAF. Fifty nanograms of DNA were used for PCR amplification (High Fidelity PCR System, Roche Diagnostics, Meylan, France) with specific primers (Forward 5’- ACCTAAACTCTTCATAATGCTTGCT-3’; Reverse 5’-AACTCAGCAGCATCTCAGGG-3’) designed using Primer3Plus online software v.2.3.6 [12]. Universal M13 tails (Forward and Reverse) were added to the specific designed primers for the amplification of the target regions (exon 15 of BRAF). A second PCR was then assessed including multiplex identifiers (MIDs), adaptators and complementary sequence of the universal tail used for the first PCR. Quality of synthesized amplicons was assessed using 1% agarose gel for proper amplification. Amplicon processing was done as described by the Amplicon Library Preparation and emulsion PCR (emPCR; Lib-A) Method GS Junior Titanium Series manual from Roche Diagnostics. Amplicons were purified using Agencourt AMPure XP beads (Beckman Coulter, SA, Nyon, Switzerland) and High Pure PCR Product purification kit (Roche Diagnostics) and finally quantified with the Quant-it TM PicoGreen dsDNA Assay Kit (Life Technologies, Oregon, USA). An emulsion PCR (emPCR) was finally assessed with 1.106 molecules, and 5.106 enriched beads were loaded on a picotiter plate for GS Junior Sequencer (Roche Diagnostics). Data were treated with Amplicon Variant Analyzer software (454 Life Sciences Corp. Roche, Branford, Connecticut, USA). Sequences were aligned with NM_004333 for reference sequence and variant calling was processed. At X1000 depth, NGS sensitivity was 1%. A second data analysis using SeqPilot (SeqNext, JSI medical systems, Ettenheim, Germany) was performed. A final analysis using BWA 0.7.12 (mem algorithm, default parameters) for mapping and sorting and indexing using SAMtools was performed. VarScan2 (mpileup2snp algorithm, with filters—min-coverage 100—minreads 20—min-var-freq 0.01—p-value 0.05) was used for variant calling [13].
Immunohistochemistry
IHC was assessed on 5μm tissue sections from the same tissue block used for mutation testing and VE1 V600E specific antigen (Spring Bioscience, Pleasanton, CA, USA) diluted at 1/50 was used as previously described [14]. All process was automated using BenchMark Ultra (Ventana, Meylan, France). Briefly, staining procedures included sample deparaffinization using EasyPrep (Roche Diganostics), drying at 72°C for 15 minutes, pretreatment with cell conditioning 1 pH 8 (Roche Diagnostics) for 52 minutes and incubation with the primary antibody at 42°C for 44 minutes. Antibody incubation was followed by counterstaining with one drop of hematoxylin for 12 minutes and one drop of bluing reagent for 4 minutes. For chromogenic detection, OptiView DAB IHC Detection Kit (Roche Molecular Diagnostics) was used as previously described [15, 16]. Slides were finally washed using detergent and mounted for observation. Two BRAF V600E-positive and one BRAF V600E-negative malignant melanoma confirmed samples were used as controls. Each run also contained buffer with no primary antibody as negative control. Staining scoring was finally assessed by a pathologist considering granulocytoplasmic intensity (Fig 2). Staining was defined as 0+ staining intensity when comparable to negative BRAF V600E-negative control sample. Low, moderate and strong cytoplasmic staining intensities were defined as 1+, 2+ and 3+, respectively. All scoring were assessed blinded to other outcomes.
Fig 2. Immunohistochemistry staining according to BRAF mutational status.
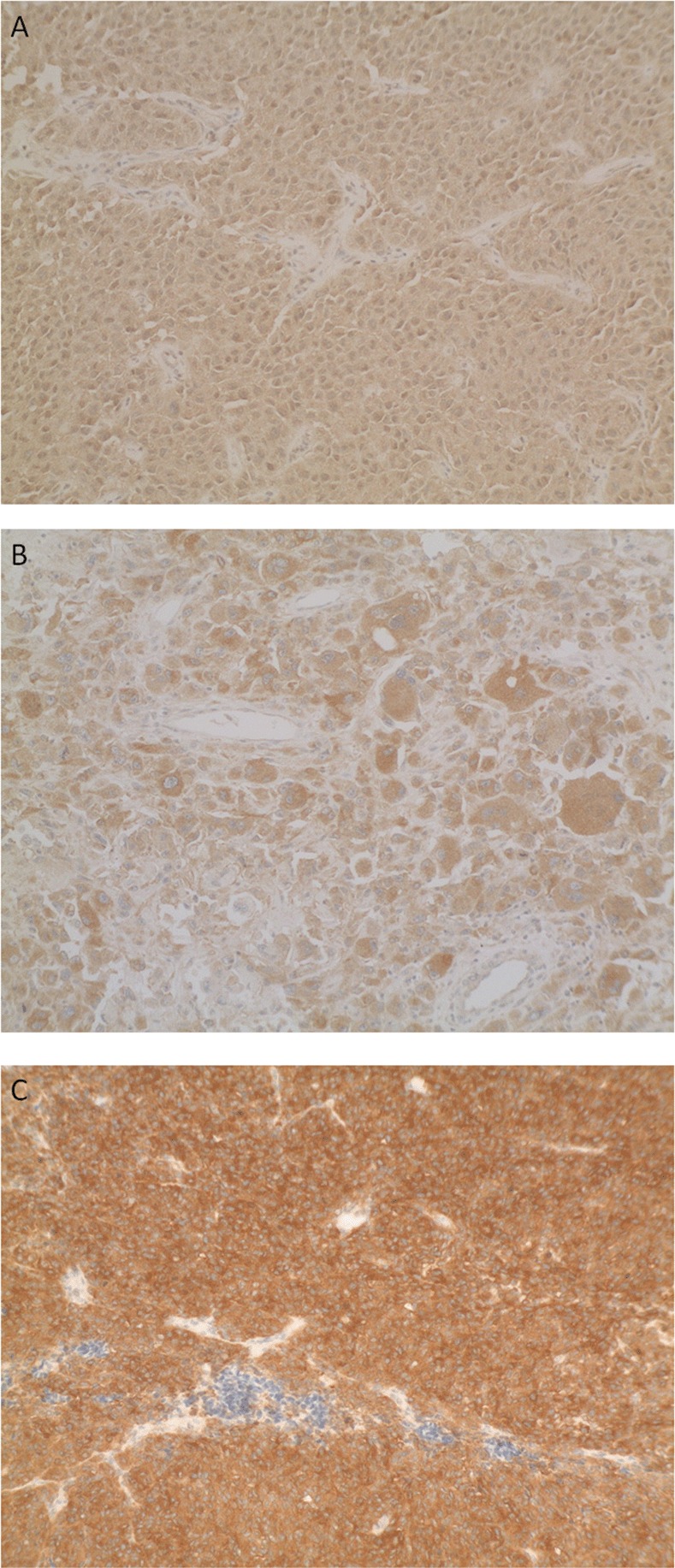
No signal can be observed in BRAF wild-type sample (A), but medium (B) to strong signal (C) is observed in samples presenting a BRAF V600E mutation.
Statistical analysis
Qualitative parameters were described by frequency and percentage. Specificity (Sp), sensitivity (Se), positive predictive value (PPV) and negative predictive value (NPV) were calculated using NGS as the reference standard.
Statistical analysis was performed using SAS software (SAS Institute, Cary, NC 25513; version 9.2).
Results
All samples were processed using the 5 assays and provided interpretable results in all cases. Mutations on BRAF were found in 28 (47.5%), 29 (49.2%), 31 (52.5%), 29 (49.2%) and 27 (45.8%) samples with HRM, RT-ASA, NGS, IdyllaTM and IHC respectively (Table 2). Twenty-six (81.2%) samples were found bearing a c.1799T>A (p.Val600Glu) mutation, three (9.4%) with a c. 1798_1799delinsAA (p.Val600Lys) mutation and one with c.1789_1790delinsTC (p.Leu597Ser) mutation. Two samples were found bearing complex mutations. NGS data suggest that sample #40 contains 3 different tumoral clones: one clone bearing c.1796C>T (p.Thr599Ile) mutation, one clone bearing c.1799T>A (p.Val600Glu) mutation and one clone bearing both mutations. Sample #49 also bears a complex mutation c.1797delinsTACT (p.Tthr599delinsThrThr). Mutations details are presented in Table 3.
Table 2. Mutation results per assay.
| Wild-type | (%) | BRAF mutation* | (%) | |
|---|---|---|---|---|
| High Resolution Melting | 31 | (52,5) | 28 | (47,5) |
| Real-time allele specific amplification | 30 | (50,8) | 29 | (49,2) |
| Next Generation Seqencing | 28 | (47,5) | 31 | (52,5) |
| IdyllaTM | 30 | (50,8) | 29 | (49,2) |
| Immunohistochemistry | 32 | (54,2) | 27 | (45,8) |
*BRAF mutations are detailed in Table 3
Table 3. Mutations details.
| Mutations | Number of samples with mutation | |||||||
|---|---|---|---|---|---|---|---|---|
| CDS mutation | AA variation | Cosmic ID | Frequency* | HRM† | RT-ASA | NGS | IdyllaTM | IHC |
| c.1799T>A | p.Val600Glu | COSM476 | 80~90% | 21 | 25 | 25 | 25 | 24 |
| c.1798_1799delinsAA | p.Val600Lys | COSM473 | 8% | 3 | 3 | 3 | 3 | 2 |
| c.1789_1790delinsTC | p.Leu597Ser | COSM1126 | < 1% | 1 | 0 | 1 | 0 | 0 |
| c.1796_1799delinsTAGA | p.Thr599_Val600delinsIleGlu | n/a | n/a | 1 | 1 | 1 | 1 | 1 |
| c.1797delinsTACT | p.Thr599delinsThrThr | n/a | n/a | 1 | 0 | 1 | 0 | 0 |
*According to COSMIC database
† One sample is considered as false positive with HRM and is not mentioned in this table Sp, Se, PPV and NPV for each of the four validated assays are summarized in Table 4. Lower specificity and PPV were seen when using HRM. IdyllaTM and RT-ASA yielded the highest sensitivity and NPV. Based on V600 cases, calculated Se, Sp, PPV and NPV were 100% for IdyllaTM and RT-ASA and 93.1%, 100%, 100% and 93.8% for IHC for Se, Sp, PPV and NPV respectively.
Table 4. Performance statistics of the BRAF detection with the four validated assays compared to NGS.
| HRM | RT-ASA | IdyllaTM | IHC | |
|---|---|---|---|---|
| Sensitivity | 87.1% | 93.5% | 93.5% | 81.7% |
| Specificity | 96.4% | 100.0% | 100.0% | 100.0% |
| PPV | 96.4% | 100.0% | 100.0% | 100.0% |
| NPV | 87.1% | 93.3% | 93.3% | 87.5% |
Abbreviations: IHC, immunohistochemistry; NGS, next-generation sequencing; HRM: High Resolution Melting; RT-ASA: Real-Time Allele Specific Amplification; NPV, negative predictive value; PPV, positive predictive value.
Ten samples were found with discrepancies between the 5 assays (Table 5). Samples #1, #10 and #39 were identified as wild-type using HRM assay and as mutated with all the other assays. Sample #36 was found wild-type with HRM and IHC assays and bearing a mutation with the other assays. Sample #21 was found bearing a mutation with HRM assay and wild-type with all other assays. Samples #40 and #47 were found bearing a mutation with HRM and NGS assays and wild-type with the other assays. Sample #28 was found wild-type with IHC and mutated with the other assays. Finally, samples #16 and #50 have been found bearing a V600E with IHC and a V600K mutation with all the other assays.
Table 5. Samples with discrepancies.
| Sample # | Tumoral cells content (%) | Mutant allele frequency (%) | Mutation* | Results | |||
|---|---|---|---|---|---|---|---|
| HRM | SybrGreen | Idylla | IHC | ||||
| 1 | 90 | 84.22 | c.1799T>A p.Val600Glu | wild-type | V600E | V600E-E2-D | V600E |
| 10 | 20 | 1.86 | c.1799T>A p.Val600Glu | wild-type | V600E | V600E-E2-D | V600E |
| 16 | 80 | 58.23 | c.1798_1799delinsAA p.Val600Lys | Mutation on exon 15 | V600K | V600K-R-M | V600E |
| 21 | 20 | - | wild-type | Mutation on exon 15 | wild-type | wild-type | wild-type |
| 28 | 10 | 7.59 | c.1798_1799delinsAA p.Val600Lys | Mutation on exon 15 | V600K | V600K-R-M | wild-type |
| 36 | 80 | 39.44 | c.1799T>A p.Val600Glu | wild-type | V600E | V600E-E2-D | wild-type |
| 39 | 20 | 5.38 | c.1799T>A p.Val600Glu | wild-type | V600E | V600E-E2-D | V600E |
| 40 | 10 | 4.87 | c.1796_1799delInsTAGA p.Thr599_Val600delinsIleGlu | Mutation on exon 15 | wild-type | wild-type | wild-type |
| 47 | 90 | 74.25 | c.1789_1790delinsTC p.Leu597Ser | Mutation on exon 15 | wild-type | wild-type | wild-type |
| 50 | 60 | 37.11 | c.1798_1799GT>AA p.Val600Lys | Mutation on exon 15 | V600K | V600K-R-M | V600E |
*Mutations characterized and confirmed by NGS
Discussion
BRAF genotyping is important for the treatment of patients with metastatic melanoma. The use of anti-BRAF drugs (vemurafenib or dabrafenib) associated or not with anti-MEK therapy (trametinib or cobimetinib), has considerably improved progression-free and overall survival for patients bearing mutated V600 metastatic melanoma [4, 17]. In this study, we compared the new IdyllaTM BRAF Mutation test with standard methods already compared in some studies for the detection of BRAF somatic mutations [6, 18].
Numerous pre-analytical parameters may influence molecular mutations detection. The theoretical analytical sensitivities used in this study were 10% for HRM and 1% for IdyllaTM, RT-ASA, IHC and NGS. IdyllaTM and RT-ASA yielded the highest sensitivity (both 93.5% and 100% when considering only V600 mutated samples). IHC yielded a 81.7% sensitivity when calculated with all the samples, but yielded 93.1% for V600 mutated samples which is conform with previously published data [19, 20]. Calculated specificity for IdyllaTM, RT-ASA and IHC where 100%. Se and Sp for HRM were 87.1% and 96.4% respectively and was the less accurate assay for the detection of BRAF mutation on exon 15. The interpretation of HRM curves requires standardization and interpretation is sometimes difficult for samples with low quality DNA. Some false positive can be observed with IHC because of melanin pigmentation but a simple treatment with alkaline phosphatase can prevent this issue. No false positive sample was found with IHC in this study. No false positive sample was found in this study with PCR assays except for sample #21 which has been identified as mutated with HRM and as wild-type with all the other assays. We confirmed that no mutation was present in this sample by retesting it with all assays and NGS depth was set above 9500x. Samples #1, #10 and #39 were found as wild-type with HRM and are considered as false negative. Mutant allele/wild-type allele frequencies were 84.22%, 1.86% and 5.38% for these samples respectively and tumor cells proportion were 90%, 20% and 20% respectively. The lack of sensitivity of HRM assay can probably explain the false-negative results for samples #10 and #39, but cannot explain the results obtained for sample #1. We think that the lack of DNA quality of this sample did not allow the detection of the mutation.
Ten samples were found with discrepancies. Samples #40, #47 and #49 were found with c.1797delinsTACT (p.Thr599delinsThrThr), c.1789_1790delinsTC (p.Leu597Ser) and c.1796_1799delinsTAGA (p.Thr599_Val600delinsIleGlu) respectively. The c.1797delinsTACT (p.Thr599delinsThrThr) and c.1789_1790delinsTC (p.Leu597Sser) mutations were only detected with HRM and NGS which is obvious according to other assays technology based on the specific detection of V600 mutations. Samples #16, #28 and #50 were found bearing a c.1798_1799delinsAA p.Val600Lys mutations and surprisingly, samples #16 and #50 were found as V600E positive samples using IHC whereas the specificity of the VE1 BRAF V600E specific antigen should not allow the detection of these two cases as it did not for sample #28. Cross-reactivity has already been described with detection of BRAF V600E and V600R mutation [19]. We also hypothesize that the high percentage of tumoral cells in these two samples, 80% and 60% for samples #16 and #50 respectively, may allow the detection of low-V600E subclones in these samples. The low percent of tumor nuclei in sample #28 (10%) may have an impact on the no detection of potential of low-V600E subclones, or may simply prevent cross-reactivity. Finally, sample #36 was found wild-type with IHC and bearing a BRAF Val600Glu mutation with all other assays. Whereas the total proportion of tumor nuclei in this sample was 80%, the tissue necrosis present in this sample may explain this result. Moreover, interpretation of IHC may be complicated for some cases. In our data, 30% of IHC scores were 0+/1+ scoring because of the difficulty to distinguish control nonspecific (0+) staining from cytoplasmic staining of tumoral cells (+1). Moreover, in our study, scoring has been made by only one pathologist whereas some studies showed that a highly reproducible and accurate IHC scoring can be achieved in challenging specimens using stringent IHC criteria and consensus review [20].
Finally, NGS bioinformatics may also been a parameter that may influence the final result. In our study, we decided to analyze the data using 3 different bioinformatics pipelines as described in the material and methods section. No difference has been found using the 3 different pipelines.
The complex mutation c.1796_1799delinsTAGA can be considered as two mutations c.1796C>T (p.Thr599Ile) and c.1799T>A (p.Val600Glu) and was found with all the assays because of the presence of the p.Val600Glu mutation. Response to anti-BRAF therapies in patients with metastatic melanoma bearing complex mutations or single mutation other than V600 have already been described in the literature [21–24] but only p.V600 mutations are officially recognized as response biomarkers for the use of vemurafenib and dabrafenib. NGS and HRM assays allow the extensive detection of all BRAF exon 15 including off-hot-spot mutations whose anti-BRAF response prediction value has not been validated. On the other hand, specific BRAF V600 assays (RT-ASA, IdyllaTM and IHC) are more restrictive but are designed to match with drugs registration.
Finally, time from sample qualification to BRAF mutation analysis result including running-time was 95, 290, 320, 940 and 2,200 minutes for IdyllaTM, HRM, RT-ASA, IHC and NGS respectively (data not shown). Total hands-on time was 2, 210, 210, 10 and 1,780 minutes for IdyllaTM, HRM, RT-ASA, IHC and NGS respectively. IdyllaTM assay is the fastest assay with a total time of about 95 minutes per sample, but one sample must be assessed in one cartridge inserted in one instrument at time. Up to 8 instruments can be connected to one console allowing the simultaneous detection of 8 samples. In our study, we used two instruments connected to one console, thus this configuration allows the assessment of BRAF mutations in a routine configuration but is not the most appropriate assay for a large batch assessment.
In conclusion, in the present study HRM appears the less sensitive assay for the detection of BRAF V600 mutations. The RT-ASA and IdyllaTM assays are fully suitable for routine PCR-based molecular diagnostics aiming at the prescription of anti-BRAF therapies. The NGS allows the extensive detection and characterization of all exon 15 BRAF mutations, but lacks usefulness until now since no off-hotspots mutations have been described as predictive markers for anti-BRAF therapies. Finally, the IdyllaTM assay is easier and faster because it requires less than 2 minutes for sample preparation and all the other steps are fully-automated and assessed in about 90 minutes.
Data Availability
All relevant data are within the paper and BioProject accession Number: PRJNA316379.
Funding Statement
The authors have no support or funding to report.
References
- 1.Boyers LN, Karimkhani C, Naghavi M, Sherwood D, Margolis DJ, Hay RJ, et al. Global mortality from conditions with skin manifestations. J Am Acad Dermatol. 2014;71(6):1137–43. 10.1016/j.jaad.2014.08.022 [DOI] [PubMed] [Google Scholar]
- 2.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–16. 10.1056/NEJMoa1103782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380(9839):358–65. 10.1016/S0140-6736(12)60868-X [DOI] [PubMed] [Google Scholar]
- 4.Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372(1):30–9. 10.1056/NEJMoa1412690 [DOI] [PubMed] [Google Scholar]
- 5.Lovly CM, Dahlman KB, Fohn LE, Su Z, Dias-Santagata D, Hicks DJ, et al. Routine multiplex mutational profiling of melanomas enables enrollment in genotype-driven therapeutic trials. PLoS One. 2012;7(4):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mourah S, Denis MG, Narducci FE, Solassol J, Merlin JL, Sabourin JC, et al. Detection of BRAF V600 Mutations in Melanoma: Evaluation of Concordance between the Cobas(R) 4800 BRAF V600 Mutation Test and the Methods Used in French National Cancer Institute (INCa) Platforms in a Real-Life Setting. PLoS One. 2015;10(3):2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Na JI, Kim JH, Kim HJ, Kim HK, Moon KS, Lee JS, et al. VE1 immunohistochemical detection of the BRAF V600E mutation in thyroid carcinoma: a review of its usefulness and limitations. Virchows Arch. 2015;2015:19. [DOI] [PubMed] [Google Scholar]
- 8.Harle A, Busser B, Rouyer M, Harter V, Genin P, Leroux A, et al. Comparison of COBAS 4800 KRAS, TaqMan PCR and high resolution melting PCR assays for the detection of KRAS somatic mutations in formalin-fixed paraffin embedded colorectal carcinomas. Virchows Arch. 2013;462(3):329–35. 10.1007/s00428-013-1380-x [DOI] [PubMed] [Google Scholar]
- 9.Pichler M, Balic M, Stadelmeyer E, Ausch C, Wild M, Guelly C, et al. Evaluation of high-resolution melting analysis as a diagnostic tool to detect the BRAF V600E mutation in colorectal tumors. J Mol Diagn. 2009;11(2):140–7. 10.2353/jmoldx.2009.080100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willmore-Payne C, Holden JA, Tripp S, Layfield LJ. Human malignant melanoma: detection of BRAF- and c-kit-activating mutations by high-resolution amplicon melting analysis. Hum Pathol. 2005;36(5):486–93. [DOI] [PubMed] [Google Scholar]
- 11.Jarry A, Masson D, Cassagnau E, Parois S, Laboisse C, Denis MG. Real-time allele-specific amplification for sensitive detection of the BRAF mutation V600E. Mol Cell Probes. 2004;18(5):349–52. [DOI] [PubMed] [Google Scholar]
- 12.Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35(Web Server issue):W71–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harle A, Filhine-Tresarrieu P, Husson M, Boidot R, Rouyer M, Dubois C, et al. Rare RAS Mutations in Metastatic Colorectal Cancer Detected During Routine RAS Genotyping Using Next Generation Sequencing. Targeted oncology. 2015. Epub 2015/12/15. 10.1007/s11523-015-0404-7 . [DOI] [PubMed] [Google Scholar]
- 14.Long GV, Wilmott JS, Capper D, Preusser M, Zhang YE, Thompson JF, et al. Immunohistochemistry is highly sensitive and specific for the detection of V600E BRAF mutation in melanoma. The American journal of surgical pathology. 2013;37(1):61–5. 10.1097/PAS.0b013e31826485c0 [DOI] [PubMed] [Google Scholar]
- 15.Capper D, Preusser M, Habel A, Sahm F, Ackermann U, Schindler G, et al. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta neuropathologica. 2011;122(1):11–9. 10.1007/s00401-011-0841-z [DOI] [PubMed] [Google Scholar]
- 16.Uguen A, Talagas M, Costa S, Samaison L, Paule L, Alavi Z, et al. NRAS (Q61R), BRAF (V600E) immunohistochemistry: a concomitant tool for mutation screening in melanomas. Diagnostic pathology. 2015;10:121 10.1186/s13000-015-0359-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahman A. Vemurafenib and cobimetinib in BRAF-mutated melanoma. The Lancet Oncology. 2014;15(12):e535. [DOI] [PubMed] [Google Scholar]
- 18.Lopez-Rios F, Angulo B, Gomez B, Mair D, Martinez R, Conde E, et al. Comparison of testing methods for the detection of BRAF V600E mutations in malignant melanoma: pre-approval validation study of the companion diagnostic test for vemurafenib. PLoS One. 2013;8(1):e53733 10.1371/journal.pone.0053733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ihle MA, Fassunke J, Konig K, Grunewald I, Schlaak M, Kreuzberg N, et al. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p.V600E and non-p.V600E BRAF mutations. BMC cancer. 2014;14:13 10.1186/1471-2407-14-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher KE, Cohen C, Siddiqui MT, Palma JF, Lipford EH 3rd, Longshore JW. Accurate detection of BRAF p.V600E mutations in challenging melanoma specimens requires stringent immunohistochemistry scoring criteria or sensitive molecular assays. Human pathology. 2014;45(11):2281–93. 10.1016/j.humpath.2014.07.014 [DOI] [PubMed] [Google Scholar]
- 21.Busser B, Leccia MT, Gras-Combe G, Bricault I, Templier I, Claeys A, et al. Identification of a novel complex BRAF mutation associated with major clinical response to vemurafenib in a patient with metastatic melanoma. JAMA dermatology. 2013;149(12):1403–6. 10.1001/jamadermatol.2013.8198 [DOI] [PubMed] [Google Scholar]
- 22.Dahlman KB, Xia J, Hutchinson K, Ng C, Hucks D, Jia P, et al. BRAF(L597) mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer discovery. 2012;2(9):791–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trudel S, Odolczyk N, Dremaux J, Toffin J, Regnier A, Sevestre H, et al. The clinical response to vemurafenib in a patient with a rare BRAFV600DK601del mutation-positive melanoma. BMC cancer. 2014;14:727 10.1186/1471-2407-14-727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson MA, Morrissette JJ, McGettigan S, Roth D, Elder D, Schuchter LM, et al. What you are missing could matter: a rare, complex BRAF mutation affecting codons 599, 600, and 601 uncovered by next generation sequencing. Cancer genetics. 2014;207(6):272–5. 10.1016/j.cancergen.2014.06.022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper and BioProject accession Number: PRJNA316379.


