Abstract
Despite the fundamental importance of proteasomal degradation in cells, little is known about whether and how the 26S proteasome itself is regulated in coordination with various physiological processes. Here we show that the proteasome is dynamically phosphorylated during cell cycle at Thr25 of the 19S subunit Rpt3. CRISPR/Cas9-mediated genome editing, RNA interference and biochemical studies demonstrate that blocking Rpt3-Thr25 phosphorylation markedly impairs proteasome activity and impedes cell proliferation. Through a kinome-wide screen, we have identified dual-specificity tyrosine-regulated kinase 2 (DYRK2) as the primary kinase that phosphorylates Rpt3-Thr25, leading to enhanced substrate translocation and degradation. Importantly, loss of the single phosphorylation of Rpt3-Thr25 or knockout of DYRK2 significantly inhibits tumor formation by proteasome-addicted human breast cancer cells in mice. These findings define an important mechanism for proteasome regulation and demonstrate the biological significance of proteasome phosphorylation in regulating cell proliferation and tumorigenesis.
Introduction
The 26S proteasome is an essential protein complex responsible for degrading the majority of cellular proteins in eukaryotes1. An impaired proteasome system often underlies neurodegenerative diseases and the aging process2, 3. On the other hand, the rapid growth of cancer cells is often dependent on elevated proteasome activity, and proteasome inhibitors such as Bortezomib (Velcade™) have proven to be effective against multiple myeloma and certain solid cancers4, 5. Further understanding of proteasome regulation is of enormous biological and clinical importance.
The mature 26S proteasome consists of at least 33 distinct subunits. Fourteen of them (α1-7 and β1-7) form the 20S core particle (CP), a barrel-shaped structure that encloses three types of peptidase activities (trypsin-like, caspase-like and chymotrypsin-like). The remaining 19 subunits (Rpt1-6, Rpn1-3, 5-13 and 15) constitute the 19S regulatory particle (RP) that caps the CP on one or both ends. Protein substrates destined for proteasomal degradation are captured and processed by the 19S RP before they are threaded into the 20S CP for proteolysis. During this process, the ATPase subunits (Rpt1-6) play key roles in substrate engagement, unfolding, translocation and CP gate opening6-8.
Given its biological importance and biochemical complexity, the 26S proteasome is regulated at several levels by multiple mechanisms, ranging from transcriptional control to post-translational modifications (e.g. phosphorylation) of proteasome subunits9-14. Notably, the human 26S proteasome contains over 300 phosphorylation sites, over 99% of which have not been studied (http://www.phosphosite.org). It remains poorly understood how these regulations are achieved biochemically and how they influence the vast biological processes that require proteasome function.
Cell cycle regulation is one of the best appreciated functions of the 26S proteasome15, 16. Impaired degradation of key proteins caused by proteasome inhibitors or protein aggregation impedes cell proliferation, which underpins the pathogenesis and treatment of certain diseases4, 5, 17, 18. Recent phospho-proteomic studies have revealed a number of proteasome phosphorylation events at different cell cycle stages19-22, raising an important and intriguing question whether and how the proteasome itself is regulated during cell cycle to accommodate this process where protein degradation must be finely regulated.
Here we show that the 26S proteasome is dynamically phosphorylated at Thr25 of the 19S subunit Rpt3 in a cell cycle-regulated manner. Cells deficient of Rpt3-T25 phosphorylation exhibit reduced proliferation and decreased proteasome activity. We identify dual-specificity tyrosine-regulated kinase 2 (DYRK2) as the major kinase that phosphorylates Rpt3-T25. Loss of this single phosphorylation significantly inhibits tumor growth in vivo. Our study for the first time demonstrates the biological importance of proteasome phosphorylation in cell cycle and tumorigenesis, and suggests a possible approach of proteasome-oriented therapy by targeting proteasome kinases.
RESULTS
Cell cycle-dependent Rpt3-Thr25 phosphorylation
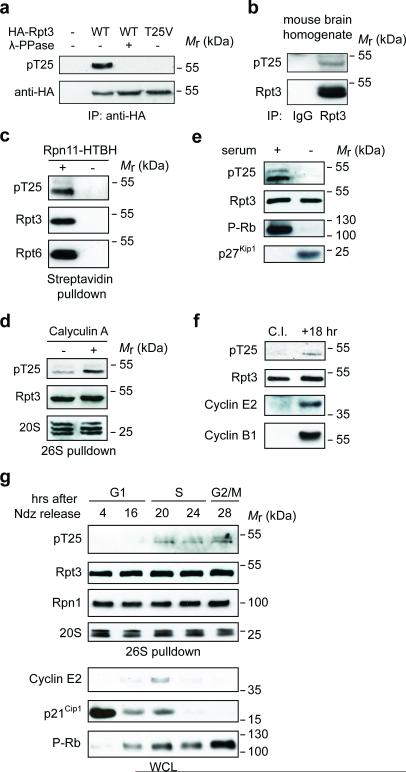
Rpt3-T25 phosphorylation has been documented in several proteomic studies19, 23, 24, although its function and regulation remained unknown. To characterize this event, we generated a phospho-T25-specific antibody (Fig. 1a). T25 phosphorylation of endogenous Rpt3 was found both in vivo (Fig. 1b) and in 26S proteasomes isolated from multiple cell lines (Fig. 1c and Supplementary Fig. 1a), establishing Rpt3-T25 as a bona fide proteasome phosphorylation site. Several lines of evidence indicate that Rpt3-T25 phosphorylation undergoes reversible and dynamic regulation. First, the phosphorylation was increased by treating cells with Calyculin A, a potent inhibitor of the PP1 and PP2A phosphatases (Fig. 1d). Second, Rpt3-T25 phosphorylation appeared to be associated with actively proliferating cells, as it was downregulated following serum starvation (Fig. 1e) or contact inhibition (Fig. 1f), both of which arrest cells in the G0/G1 phase of cell cycle. Interestingly, Rpt3-T25 phosphorylation was first reported to be present in nocodazole-synchronized mitotic cells but not in cells at late G1 phase19. Indeed, we consistently found higher levels of Rpt3-T25 phosphorylation at G2/M phase than at the G1/S boundary in multiple cell types (Supplementary Fig. 1b, c). Further analysis using HaCaT cells (immortalized human keratinocytes) showed that phospho-T25 was low during most of the G1 phase, became upregulated as cells entered S phase and remained relatively constant through S and G2/M phases (Fig. 1g). We estimated that at least 15% of total Rpt3 were T25-phosphorylated during G2/M in HaCaT and 293A cells (Supplementary Fig. 1d). Together, these data indicate that the 26S proteasome is modified in a cell cycle-dependent manner, and suggest that Rpt3-T25 phosphorylation may be functionally implicated in regulating cell proliferation.
Figure 1. Rpt3-T25 is dynamically phosphorylated during cell cycle.
(a) Validation of anti-pT25 phospho-specific antibody. 293T cells transfected with a vector control (−) or HA-Rpt3 (WT or T25V) were subjected to anti-HA immunoprecipitation (IP). Samples were then treated with or without λ-phosphatase and analyzed by western blot.
(b) In vivo phosphorylation of Rpt3-T25. Whole brain from E12.5 mouse embryos was homogenized and subjected to immunoprecipitation (IP) with normal IgG or anti-Rpt3 antibody. T25 phosphorylation was determined by western blot.
(c) Phospho-T25 was detected from the purified 26S proteasome. Lysates from 293T cells stably expressing Rpn11-HTBH (for proteasome purification) or the HTBH tag only (“−”, negative control) were subjected to streptavidin pulldown. T25 phosphorylation was determined by western blot.
(d) Reversible phosphorylation of Rpt3-T25. 293T Rpn11-TBHA cells were either untreated (−) or pretreated with 25 nM Calyculin A for 10 min before harvest. The 26S proteasome was purified and pT25 was determined by western blot.
(e) HaCaT cells were either untreated (“+” serum) or serum-starved (“−”) for 48 hours. Cell lysates were probed with the indicated antibodies.
(f) Contact inhibition reduces Rpt3-T25 phosphorylation. WT MEFs expressing Rpn11-TBHA were grown to 100% confluence and contact-inhibited (C. I.) in the presence of serum for 48 hrs. Half of the cells were frozen immediately (in G1 phase) while the other half were allowed to resume growth at a lower density for 18 hrs. The proteasomes were purified from both samples at the same time and phospho-T25 was probed.
(g) Cell cycle-regulated Rpt3-T25 phosphorylation. HaCaT Rpn11-TBHA cells enriched in M phase with nocodazole (Ndz) were collected by mitotic shake-off and released. At the indicated time points. proteasomes from each sample were affinity-purified by streptavidin pulldown, and phospho-T25 and proteasome subunits were probed. Cell cycle proteins from whole cell lysate (WCL) were also probed to show progression along cell cycle.
Lack of Rpt3-T25 phosphorylation impedes cell proliferation
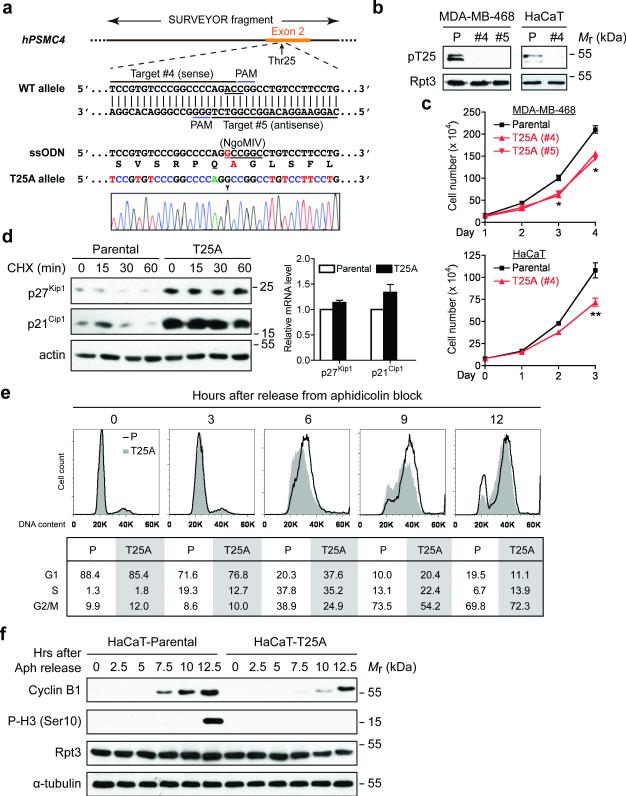
To understand the functional importance of Rpt3-T25 phosphorylation, we utilized the CRISPR/Cas9 system to create a T25A mutation in both copies of the PSMC4 gene (encoding Rpt3) in HaCaT cells and MDA-MB-468 human breast cancer cells (Fig. 2a). Homozygous T25A knock-in was confirmed by Sanger sequencing and it completely abrogated T25 phosphorylation as expected (Fig. 2a and b). The mutant Rpt3 was expressed at the same level as the wild-type protein and was properly assembled into the 26S proteasome (Fig. 2b and Supplementary Fig. 2a). Interestingly, all of the T25A knock-in cells proliferated more slowly than the corresponding control/parental cells (Fig. 2c). Consistent with this result, knockdown of endogenous Rpt3 by shRNA and simultaneous re-expression of a phospho-deficient mutant Rpt3-T25V also significantly reduced the proliferation of HaCaT and MCF10A cells (Supplementary Fig. 2b). These data demonstrate that Rpt3-T25 phosphorylation has a considerable contribution to cell proliferation.
Figure 2. Blockade of Rpt3-T25 phosphorylation impedes cell proliferation.
(a) Guide RNA design and sequencing verification of Rpt3-T25A knock-in using CRISPR/Cas9. The codon of Thr25 (ACC) is underlined in the WT allele. The single point mutation (A→G) is marked by an arrowhead in the sequencing result, and the resulting Ala25 is highlighted in red.
(b) T25A knock-in abolishes T25 phosphorylation. Parental (P) and T25A knock-in clones of MDA-MB-468 cells and HaCaT cells were engineered to stably express Rpn11-TBHA (not shown). After affinity purification, proteasome-associated T25 phosphorylation was probed.
(c) Growth curves of the indicated parental and T25A knock-in cells. Results are mean ± s.e.m. from n=3 independent experiments. **p<0.01, *p<0.05, Student's T-test (paired two-tailed test).
(d) Stabilization of p27Kip1 and p21Cip1 by T25A knock-in. HaCaT cells were enriched at early S phase with 0.4 mM hydroxyurea (HU) treatment then released into regular medium containing cycloheximide for the indicated lengths of time. Protein levels of p27Kip1 and p21Cip1 were determined from total cell extracts (left). No significant difference in their mRNA levels was seen between the parental and T25A cells at the end of HU treatment. Results are mean ± s.e.m. from n=3 independent experiments (right).
(e) Cell cycle analysis of HaCaT cells. After synchronization at G1/S boundary, cells were released and harvested at the indicated time points, stained with propidium iodide and analyzed by FACS. The percentage of cells in G1, S and G2/M phases are shown at the bottom.
(f) HaCaT cells were synchronized and released as in (e). Cell lysates collected at each time point were analyzed by western blot.
Source data for c and e can be found in Supplementary Table 3.
The proliferation defect of T25A knock-in cells suggests deregulated proteasomal degradation of cell cycle regulators. To test this idea, we examined a panel of cell cycle proteins and compared their expression levels and turnover rates between control and T25A cells. A remarkable accumulation and stabilization of the cell cycle inhibitors p21Cip1 and p27Kip1 was observed in T25A cells as compared with control cells, while their gene transcription was not affected (Fig. 2d and Supplementary Fig. 2c). This phenomenon was particularly evident at early S phase, which coincided with the upregulation of T25 phosphorylation in control cells (Fig. 1g). Consistently, after synchronization and release from G1/S boundary, the T25A cells clearly lagged behind the control cells and showed a 2.5-3 hr delay in the induction of G2/M markers such as cyclin B1 and phospho-Histone H3 (Ser10) (Fig. 2e, f and Supplementary Fig. 2d, e). No signs of DNA damage responses or ubiquitin depletion were detected under our assay conditions (Supplementary Fig. 2f, g). These data indicate that timely progression through S phase requires concomitant Rpt3-T25 phosphorylation, which likely promotes proteasomal degradation of cell cycle regulators such as p21Cip1/p27Kip1.
Blocking Rpt3-T25 phosphorylation inhibits 26S proteasome activity
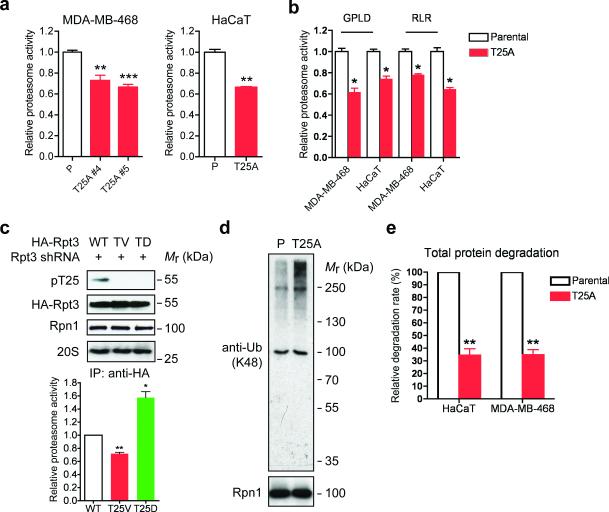
We next measured proteasome activity in control and T25A knock-in cells. In both HaCaT and MDA-MB-468 cells, loss of Rpt3-T25 phosphorylation reduced endogenous proteasome activity towards a fluorogenic peptide substrate Suc-LLVY-AMC25 by approximately 30% (Fig. 3a). In fact, all three types of peptidase activity of the proteasome were similarly downregulated (Fig. 3a, b), suggesting an overall weakening of proteasome function in the absence of T25 phosphorylation. We also confirmed these findings in additional cell lines by replacing endogenous Rpt3 with HA-tagged WT or mutant Rpt3 using the aforementioned shRNA/add-back approach (Supplementary Fig. 3a). Similar to T25A knock-in, substitution of endogenous Rpt3 with the phospho-deficient T25V mutant significantly reduced proteasome activity against Suc-LLVY-AMC, while the phospho-mimetic T25D mutation had the opposite effect (Fig. 3c and Supplementary Fig. 3b). Moreover, GFPu, a short-lived reporter protein that undergoes rapid ubiquitination and proteasome degradation17, was cleared at a significantly slower rate in Rpt3-T25V-expressing cells than in control cells (Supplementary Fig. 5c). Together, these data demonstrate that Rpt3-T25 phoshorylation is required for efficient proteasomal degradation.
Figure 3. Loss of Rpt3-T25 phosphorylation downregulates 26S proteasome activity.
(a) Proteasome activity in total cell lysates from the indicated MDA-MB-468 and HaCaT cells was measured with Suc-LLVY-AMC. **p<0.01, ***p<0.001 (compared to parental line, two-tailed paired Student's t-test, mean ± s.e.m from n=3 independent experiments).
(b) Proteasome activity was measured as in (a) with Ac-GPLD-AMC (caspase-like activity) or Ac-RLR-AMC (trypsin-like activity) as substrate. *p<0.05, two-tailed paired Student's t-test, mean ± s.e.m from n=3 independent experiments.
(c) 26S proteasomes were isolated by anti-HA IP from the indicated HA-Rpt3-expressing 293T cells. Proteasome components were shown by western blot (top) and proteasome activity from the immunoprecipitates was determined by Suc-LLVY-AMC cleavage (bottom). **p<0.01, *p<0.05 (compared to WT, two-tailed paired Student's t-test, mean ± s.e.m from n=3 independent experiments).
(d) Accumulation of ubiquitinated proteins in T25A cells. Parental and T25A HaCaT cells were enriched at early S phase with 0.4 mM HU treatment, and whole cell extracts were probed for K48-linked ubiquitination. Rpn1 is shown as a loading control.
(e) Total protein degradation rate in parental and T25A knock-in cells was determined by 3H-Phe pulse-chase assay. The protein degradation rates were calculated from n=3 independent experiments and presented as the percentage of that observed in each parental line. **p<0.01 (two-tailed paired Student's t-test, mean ± s.e.m from n=3 independent experiments).
Source data for a, b and c can be found in Supplementary Table 3.
We further assessed the role of Rpt3-T25 phosphorylation in cellular protein turnover. Consistent with the overall decrease of proteasomal function, the Rpt3-T25A knock-in cells accumulated higher levels of K48-linked polyubiquitinated proteins than control cells (Fig. 3d). A more pronounced effect was seen when cellular protein degradation was determined using 3H-Phe pulse-chase assay26. In both HaCaT and MDA-MB-468 cells, the single T25A substitution reduced the rate of total protein degradation by approximately 66% (Fig. 3e), indicating an important role of T25 phosphorylation in regulating proteasome function. Loss of this phosphorylation is therefore expected to impact a variety of biological processes.
DYRK2 is the primary kinase that phosphorylates Rpt3-Thr25
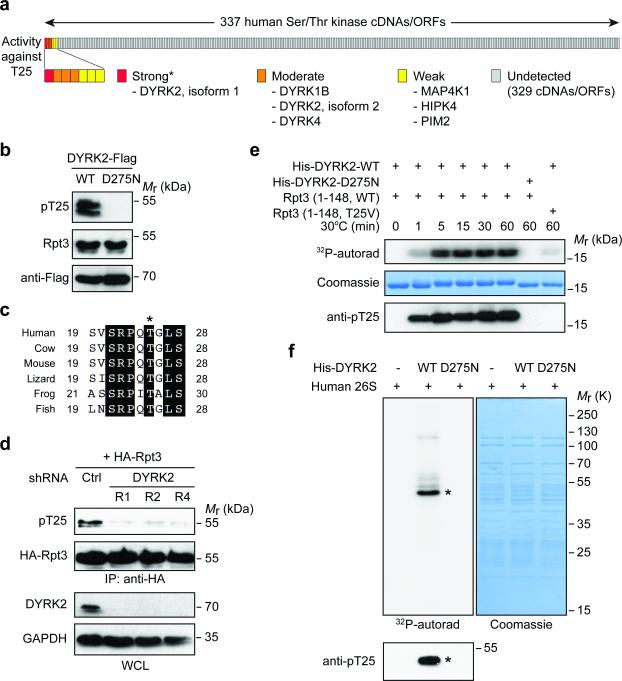
The biological and biochemical importance of Rpt3-T25 phosphorylation prompted us to identify the responsible kinase(s). To this end, we performed a kinome-wide screen of more than 300 human Ser/Thr kinases27 by individually overexpressing each cDNA in 293T cells (Fig. 4a). Strikingly, only the DYRK subfamily of kinases were capable of phosphorylating endogenous Rpt3-T25 to a considerable level (Fig. 4a, b and Supplementary Fig. 4a). Indeed, the sequence surrounding Rpt3-T25 (SRPQTGLS) conforms to a DYRK consensus motif (RPXS/T, where X is any amino acid) and is evolutionarily conserved among vertebrate Rpt3 proteins (Fig. 4c). Five DYRK family members exist in vertebrates (DYRK1A, 1B, 2, 3 and 4)28, among which only DYRK2 knockdown greatly reduced T25 phosphorylation in cells (Fig. 4d and Supplementary Fig. 4b). In addition, DYRK2 protein and mRNA levels were both regulated in response to serum deprivation and during cell cycle in a manner very similar to Rpt3-T25 phosphorylation (Supplementary Fig. 4c). These data indicate that DYRK2 is the primary kinase responsible for T25 phosphorylation.
Figure 4. DYRK2 is the primary Rpt3-T25 kinase.
(a) Summary of the Rpt3-T25 kinase screen. Each vertical bar represents an individual kinase cDNA/open reading frame (ORF). The activity of each kinase towards endogenous Rpt3-T25 in 293T cells is marked as strong (red), moderate (orange), weak (yellow) or undetected (gray). *DYRK3 cDNA from the kinase library failed to express properly, although the kinase could strongly phosphorylate T25 when overexpressed (see Supplementary Fig. 4a).
(b) 293T cells were transfected with WT DYRK2 or the catalytically inactive mutant, D275N. T25 phosphorylation of endogenous Rpt3 is blotted from whole cell lysates.
(c) An alignment of vertebrate Rpt3 protein sequences surrounding Thr25 (asterisk). The sequences shown are NP_006494 (human), NP_001030255 (cow), NP_036004 (mouse), XP_008119139 (lizard), NP_001008010 (frog) and NP_956044 (fish).
(d) DYRK2 knockdown decreases T25 phosphorylation. HA-Rpt3 (WT) was transfected into 293T cells stably expressing control or three independent DYRK2 shRNAs. Following anti-HA IP, T25 phosphorylation was determined by western blot.
(e) Time-dependent phosphorylation of recombinant human Rpt3 (aa 1-148) by bacterially expressed DYRK2 (aa 74-479), indicated by 32P-phosphate incorporation (top), gel shift (middle) and anti-pT25 western blot (bottom).
(f) In vitro kinase assay with DYRK2 and purified human 26S proteasome. Asterisk indicates the most strongly phosphorylated band, which matched with the anti-pT25 blot (bottom).
In vitro, bacterially expressed recombinant DYRK2-WT, but not the catalytically inactive mutant DYRK2-D275N, efficiently phosphorylated a bacterially purified N-terminal fragment of hRpt3 (amino acids 1-148, ref.29), while this phosphorylation was essentially abolished by the T25V mutation (Fig. 4e and Supplementary Fig. 4d). Furthermore, DYRK2 readily phosphorylated Rpt3-T25 within purified human 26S proteasomes as confirmed by tandem mass spectrometry and western blot (Fig. 4f and Supplementary Fig. 4e). Therefore, DYRK2 is a proteasome kinase that directly phosphorylates Rpt3-T25.
DYRK2 activates the 26S proteasome
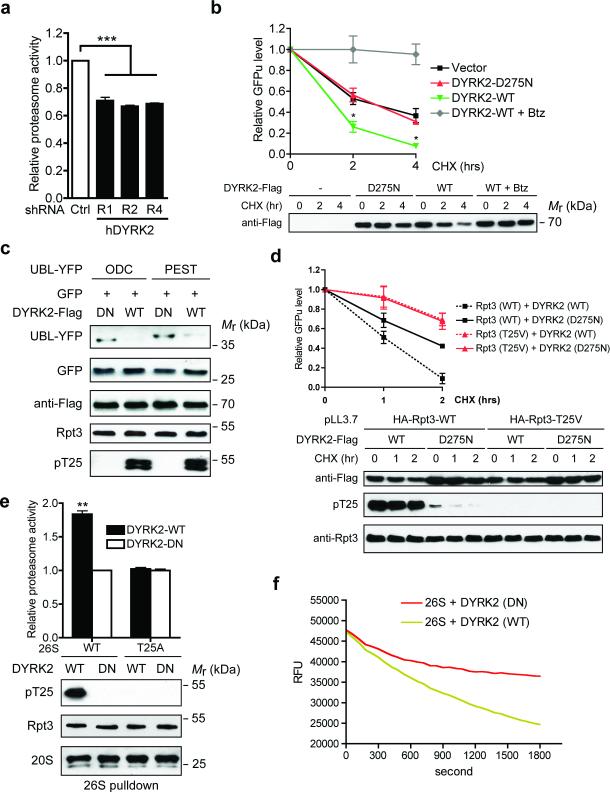
We next evaluated the role of DYRK2 in proteasome regulation. In 293T cells, knockdown of DYRK2 by three independent shRNAs (Fig. 4d) consistently reduced proteasome activity against Suc-LLVY-AMC by approximately 30% (Fig. 5a). Similar results were seen with CRISPR/Cas9-mediated DYRK2 knockout (KO) in MDA-MB-468 cells (Supplementary Fig. 5a, b), and the degree of proteasome inhibition was comparable to that caused by Rpt3-T25A knock-in (Fig. 3a). On the other hand, overexpression of wild-type DYRK2, but not DYRK2-D275N, markedly increased proteasomal degradation of GFPu (Fig. 5b, d) as well as of two additional model substrates, UBL-YFP-ODC and UBL-YFP-PEST, which are degraded by the proteasome in a ubiquitination-independent manner (Fig. 5c). Importantly, DYRK2 had no effect on GFPu degradation in Rpt3-T25V-expressing cells, nor did it affect the level of EGFP that is resistant to proteasomal degradation (Fig. 5c, d). These data demonstrate that DYRK2 enhances proteasome activity via Rpt3-T25 phosphorylation.
Figure 5. DYRK2 is a positive regulator of proteasome activity.
(a) Proteasome activity in total cell lysates from 293T cells stably expressing control or DYRK2 shRNAs was measured using Suc-LLVY-AMC as substrate. ***p<0.001 (One-way ANOVA, mean ± s.e.m from n=3 independent experiments)..
(b) DYRK2 promotes GFPu degradation. 293T cells were co-transfected with vector control, DYRK2-WT or DYRK2-D275N and GFPu. After 1-hour pre-treatment of DMSO or 1 μM Bortezomib (Btz), cycloheximide (CHX, 50 μg/ml) was added for 0, 2 or 4 hours. GFP fluorescence in cell lysates was determined at each time point, background-subtracted and normalized to the starting level at time 0. *p<0.05 (DYRK2-WT vs. Vector or DYRK2-DN, One-way ANOVA, mean ± s.e.m from n=3 independent experiments). Expression of the DYRK2-3xFlag-V5 constructs was determined by anti-Flag western blot.
(c) DYRK2 promotes the degradation of additional proteasome reporters. 293T cells were co-transfected with the indicated constructs as in (b) and cell lysates were analyzed by western blot.
(d) DYRK2 promotes GFPu degradation in a T25-dependent manner. 293T cells stably expressing HA-Rpt3-WT or T25V was transfected and treated with CHX as in (b). GFPu levels are shown as mean ± s.e.m from n=3 independent experiments (top). DYKR2 expression and T25 phosphorylation were confirmed by western blot (bottom). Note that no T25 phosphorylation was detected in 293T pLL3.7-HA-Rpt3-T25V cells even in the presence of overexpressed DYRK2-WT, indicating a complete switch from endogenous WT Rpt3 to HA-Rpt3-T25V.
(e) DYRK2 activates wild-type proteasome in vitro via T25 phosphorylation. 26S proteasomes were purified from parental and T25A MDA-MB-468 cells in the absence of phosphatase inhibitors, followed by in vitro phosphorylation with DYRK2. After removal of DYRK2, proteasome activity was measured with Suc-LLVY-AMC (top, **p<0.01, two-tailed paired Student's T-test, mean ± s.e.m from n=3 independent experiments). Total Rpt3, T25 phosphorylation and 20S subunits from the pulldown are shown by western blot (bottom).
(f) In vitro degradation of polyubiquitinated GFP-titinV15P-cyclin-PY fusion protein by proteasomes treated with DYRK2-WT or D275N (in triplicates). RFU, relative fluorescence units.
Source data for a, b, d and e can be found in Supplementary Table 3.
We further characterized proteasome activation by DYRK2 using in vitro assays. Affinity-purified 26S proteasome treated with wild-type DYRK2 showed strong Rpt3-T25 phosphorylation and markedly increased peptidase activity towards Suc-LLVY-AMC when compared with that treated with inactive DYRK2 (Fig. 5e). This stimulatory effect of DYRK2 was completely lost on mutant 26S proteasomes purified from the Rpt3-T25A knock-in cells (Fig. 5e), again highlighting the importance of this site in DYRK2-mediated proteasome regulation. T25-phosphorylated 26S proteasome also more efficiently degraded the unstructured protein casein (Supplementary Fig. 5c) and a polyubiquitinated GFP fusion protein substrate30 (Fig. 5f). These data fully recapitulate our observations in cells and indicate that DYRK2 can directly activate the 26S proteasome.
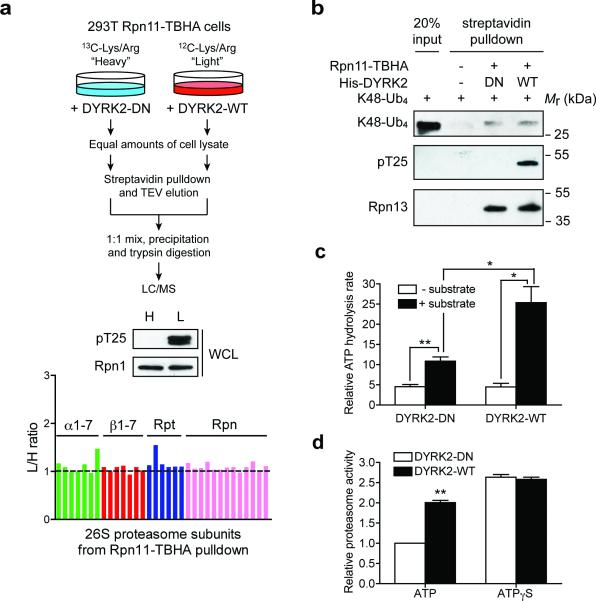
To gain insights into the possible mechanism of such activation, we first confirmed using quantitative mass spectrometry that DYRK2 overexpression does not alter proteasome abundance or assembly in cells (Fig. 6a, see also Fig. 4b, 5c and Supplementary Table 1). Nor does T25 phosphorylation affect proteasome binding with K48-linked ubiquitin chain (Fig. 6b). Next, we examined proteasome ATPase activity. Although Rpt3-T25 phosphorylation did not change the basal ATPase activity, it did increase substrate-stimulated ATP hydrolysis (Fig. 6c), which is generally believed to correlate with the rate of substrate unfolding and translocation31-33. In addition, in the presence of ATPγS, a weakly hydrolysable ATP analog that keeps the proteasome in a configuration competent for substrate translocation34, wild-type DYRK2 could not further increase proteasome peptidase activity (Fig. 6d). Therefore, a likely consequence of Rpt3-T25 phosphorylation by DYRK2 is enhanced substrate translocation, although other mechanisms may exist.
Figure 6. Mechanisms by which DYRK2 regulates the proteasome.
(a) DYRK2 does not affect 26S proteasome assembly. 293T Rpn11-TBHA cells labeled with “heavy (H)” or “light (L)” isotopes were transfected with inactive or WT DYRK2, respectively. Proteasomes were isolated from these cells and analyzed by quantitative mass spectrometry (top). DYRK2-WT strongly phosphorylated Rpt3-T25 (middle), without affecting the relative abundance of each subunit in the purified 26S proteasomes (bottom).
(b) In vitro binding of K48-linked tetraubiquitin (K48-Ub4) to the proteasome. Control 293T cells and 293T Rpn11-TBHA cells were subjected to streptavidin pulldown. The purified proteasome was treated with DYRK2-WT or DN in vitro and then incubated with K48-Ub4. Proteasome-bound K48-Ub4 was probed with anti-ubiquitin antibody. Western blot of the ubiquitin receptor Rpn13/ADRM1 shows equal amounts of proteasome in the DYRK2-treated samples.
(c) Affinity-purified 26S proteasomes (~ 1 μg) was treated with DYRK2-WT or D275N in vitro. ATPase activity was monitored at 37°C using the malachite green method in the absence or presence of a proteasome substrate, UBL-YFP-ODC (1 μg). **p<0.01, *p<0.05 (two-tailed paired Student's t-test, mean ± s.e.m from n=3 independent experiments).
(d) 26S proteasomes from 293T cells overexpressing WT or DN DYRK2 were immobilized on streptavidin beads, washed extensively with reaction buffer containing 1 mM ATP or 1 mM ATPγS, and assayed for Suc-LLVY-AMC cleavage in the same buffers. Reaction was carried out for 10 min at 37°C. **p<0.01 (two-tailed paired Student's t-test, mean ± s.e.m from n=3 independent experiments).
Source data for c and d can be found in Supplementary Table 3.
DYRK2 positively regulates cell growth
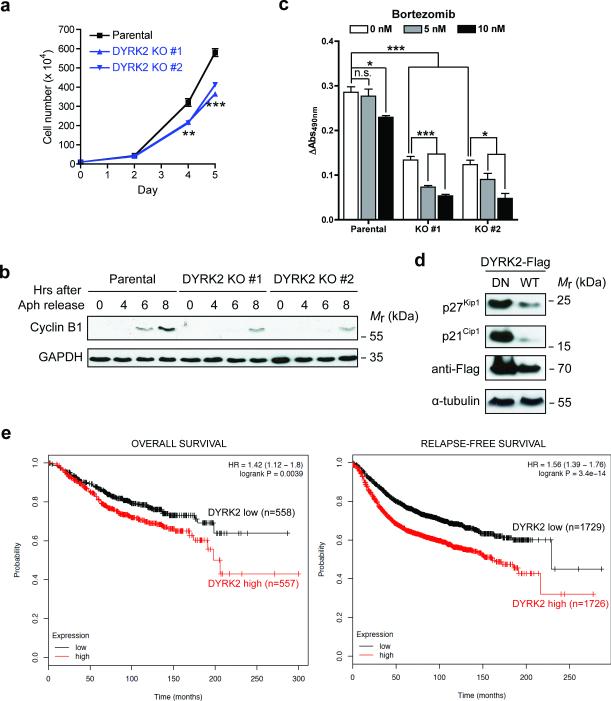
Based on these characterizations, we hypothesized that loss of DYRK2 would phenocopy the anti-proliferative effect of Rpt3-T25A knock-in. Indeed, CRISPR/Cas9-mediated knockout of DYRK2 in MDA-MB-468 cells reduced cell proliferation and caused a cell cycle delay comparable to that seen in Rpt3-T25A cells (Fig. 7a, b). Intriguingly, loss of DYRK2 not only attenuated the growth of MDA-MB-468 cells but also sensitized them to the proteasome inhibitor Bortezomib (Fig. 7c). A similar effect was also noted in HaCaT cells (Supplementary Fig. 6). Therefore, direct inhibition of proteasome activity and concurrent blocking of a proteasome-activating kinase can synergize in suppressing cell growth.
Figure 7. DYRK2 positively regulates cell growth.
(a) Growth curves of MDA-MB-468 parental and DYRK2 KO cells. **p<0.01, ***p<0.001 (One-way ANOVA, mean ± s.e.m from n=3 independent experiments)
(b) MDA-MB-468 parental and DYRK2 KO cells were synchronized by aphidicolin and released. Cell lysates collected at each time point were analyzed by western blot.
(c) MTS assay of MDA-MB-468 parental and DYRK2 KO cells. Equal number of cells (2.0 × 104/well) were plated in triplicates in a 96-well plate. MTS activity was measured before and after DMSO or low-dose Bortezomib treatment for 24 hours. Data are presented as the increase of MTS activity from Day 1 to Day 2. *p<0.05, ***p<0.001 (two-tailed non-paired Student's T-test, mean ± s.e.m from n=3 independent experiments). n.s., non-significant.
(d) DYRK2 downregulates p27Kip1 and p21Cip1. Cells were transiently transfected with DYRK2 (WT or D275N). Whole cell lysates were probed with the indicated antibodies.
(e) Kaplan-Meier curves of overall survival (left) and relapse-free survival (right) of breast cancer patients with differential levels of DYRK2 mRNA.
Source data for b and c can be found in Supplementary Table 3
On the other hand, overexpression of WT DYRK2 markedly downregulated the cell cycle inhibitors p21Cip1 and p27Kip1 (Fig. 7d). In fact, the most prevalent changes of DYRK2 in the majority of cancer types is gene amplification (Supplementary Fig. 6 and ref.35). In breast cancer patients, high levels of DYRK2 mRNA correlate with significantly poorer prognosis (Fig. 7e and ref.36). These result further support the notion that DYRK2 is a positive regulator of cancer cell growth.
Rpt3-T25 phosphorylation is required for tumor growth in vivo
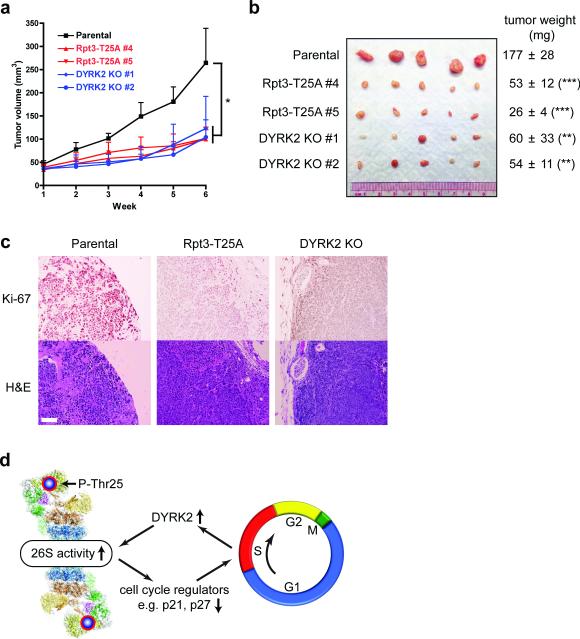
MDA-MB-468 is one of the basal-like triple-negative breast cancer cell lines recently shown to be “addicted” to proteasome activity and particularly vulnerable to proteasome inhibition37. Our in vitro results made us wonder whether preventing Rpt3-T25 phosphorylation or inactivation of DYRK2 could inhibit the tumorigenic growth of these cells in vivo. As a proof of principle, control and genome-edited MDA-MB-468 cells were injected subcutaneously into nude mice to form tumors. Indeed, tumors derived from the T25A knock-in cells and DYRK2 KO cells all grew at a significantly lower rate than those formed by the parental cells (Fig. 8a, b). Histological examination of the xenograft tumors also showed greatly attenuated Ki-67 staining (a cellular marker for proliferation) in cancer cells lacking T25 phosphorylation (Fig. 8c). These data strongly support the biological importance of Rpt3-T25 phosphorylation in regulating cell proliferation in vivo, and suggest that targeting proteasome regulators (such as DYRK2) in combination with proteasome inhibitors may be a promising approach of anti-cancer therapy.
Figure 8. Rpt3-T25 phosphorylation is required for tumor growth in vivo.
(a) Tumor xenograft studies with parental and genome-edited MDA-MB-468 cells injected subcutaneously into nude mice. Tumor volumes at each time point after injection are shown (mean ± s.e.m). *p<0.05 (n = 5 mice, One-way ANOVA).
(b) Xenograft tumors from (a) were resected at 6 weeks post-injection and imaged. Tumor weights are shown on the right as mean ± SEM. **p<0.01, ***p<0.001 (n =5 mice, compared to parental line, two-tailed non-paired Student's T-test).
(c) Histological examination of consecutive sections of the tumors with Ki-67 and hematoxylin/eosin staining. Scale bar = 100 μm.
(d) A model of reversible phospho-regulation of the 26S proteasome (adapted from ref.53). The approximate position of Rpt3-T25 in the 26S proteasome complex is highlighted. Cell cycle-dependent Rpt3-T25 phosphorylation regulated by DYRK2 facilitates the degradation of key proteins such as p21 and p27, which in turn promotes cell cycle transition. Pharmacological intervention of this process by targeting proteasome kinases can have therapeutic potentials.
Discussion
In this study we have demonstrated that the 26S proteasome undergoes dynamic phosphorylation during cell cycle that in turn contributes to cell proliferation by regulating proteasome activity (Fig. 8d). Despite its quintessential role in almost every cellular activity, the 26S proteasome has long been viewed as a housekeeping machinery that does not require significant regulation. This perception has drastically changed with recent studies showing that proteasome activity and abundance are dynamically regulated under physiological and pathological conditions2-4, 38. Our present work has provided a first glimpse at the phospho-regulation of the proteasome during cell cycle. Rpt3-T25 phosphorylation is associated with actively proliferating cells and upregulated as cells transit from G1 to S phase. This pattern very likely results from the parallel change of DYRK2 expression, although a role for phosphatases such as PP1 and PP2A cannot be excluded. Remarkably, blocking this single phosphorylation is sufficient to downregulate proteasome activity and slow cell proliferation. Therefore, proteasome function and cell cycle progression are coordinated by Rpt3-T25 phosphorylation. It should be noted though that cell cycle of the T25A knock-in cells is delayed but not blocked, consistent with that only a fraction of Rpt3 is phosphorylated at T25, and that the proteasome activity is only partly inhibited in the knock-in cells. We do not anticipate that loss of one phosphorylation site would cause a complete inhibition of the proteasome, which is required for an immediate full block of cell cycle39. Nevertheless, the fine-tuning role of Rpt3-T25 phosphorylation in proteasome activity is still biologically important, and the cumulative outcome of cell cycle slowing is demonstrated by significant tumor inhibition in vivo (Fig. 8). We also argue that the Rpt3-T25A knock-in not only impairs p21Cip1/p27Kip1 degradation but also causes many other proteins to accumulate in hyper-ubiquitinated forms (Fig. 3d). Therefore, T25 phosphorylation may affect a variety of other cellular functions that require the 26S proteasome. Besides Rpt3-T25, mass spectrometry studies suggest that several other proteasome phosphorylation sites may also be implicated with cell cycle regulation19-22. Taken together, these modifications may have a profound impact on cell proliferation and therefore warrant further investigation.
Over 300 proteasome phosphorylation sites have been documented13, 19, 40-49, although the biological meanings of the vast majority of them remain unknown. To date only a handful of kinases have been implicated in proteasome regulation. The best characterized example is CaMKIIα–mediated Rpt6-Ser120 phosphorylation in neuronal synapses45-48. In addition, proteasome phosphorylation by protein kinase G (PKG) protects cardiomyocytes from toxic misfolded proteins49. A theme emerging from these and other studies is that proteasome activity is often increased by phosphorylation, although the molecular mechanisms remain a mystery and probably vary among different phosphorylation events.
Here we have identified DYRK2 as the Rpt3-T25 kinase. DYRK2 is known to trigger ubiquitination and degradation of several proteins50, 51. Our findings now suggest that DYRK2-stimulated protein degradation can be partly attributed to proteasome activation. DYRK2-mediated Rpt3-T25 phosphorylation does not appear to affect proteasome abundance, assembly, ubiquitin capture or its basal ATPase activity (Figs. 2, 5, 6, Supplementary Figs. 2 and 5). On the other hand, T25 phosphorylation increases substrate-induced ATP hydrolysis but fails to enhance the peptidase activity of the proteasome that is already in an ATPγS-induced, translocation-competent conformation. These results support a role of T25 phosphorylation in promoting substrate translocation. How this happens remains unclear. Thr25 is located in a flexible region N-terminal to the coiled coil domain of Rpt3, which is almost at the apex of the 26S proteasome complex hence far away from the ATPase domains and RP-CP interface52, 53 (Fig. 8d). This region has not been resolved in any of the available crystal or electron microscopy structures of the proteasome, making it difficult to predict the structural consequences of Thr25 phosphorylation. However, this N-terminal region extends from the Rpt3-Rpt6 coiled coil and potentially makes direct contact with the Rpn2 subunit52, 53. Upon substrate engagement, Rpn2 and several other 19S subunits (known as the “lid”) rotate around the Rpt3-Rpt6 coiled coil, which realigns the ATPases and the 20S CP to form a continuous central channel for substrate entry54---a similar configuration induced by ATPγS binding34. Therefore, one possible scenario is that Thr25 phosphorylation may facilitate or stabilize the rearrangements of Rpn2 and the lid, leading to enhanced substrate translocation and degradation. Further studies are required to reveal the molecular details of this regulation.
That said, other mechanisms are also possible underlying DYRK2-mediated 26S proteasome activation, especially inside the cells. For example, T25 phosphorylation may alter the binding of a yet-to-be-identified proteasome interacting protein, thereby changing the substrate degradation rate. The phosphorylation may also affect substrate selectivity of the proteasome. Alternatively, T25 phosphorylation may lead to and/or function together with additional modifications of the proteasome. In fact, our anti-pT25 antibody often detects two species of T25-phosphorylated Rpt3 seen as a doublet band. The upper band likely results from dual phosphorylation of Rpt3 at T25 and another residue that we are currently investigating. Given the complexity of proteasomal structure and function, we believe that much more work is needed to pinpoint the mechanism(s) of proteasome regulation by phosphorylation of Rpt3-T25 (or any functionally relevant phospho-site).
Hyperactivity of the proteasome is a known feature of many cancers, and proteasome-oriented anti-cancer therapies have been relying on the development and delivery of various proteasome inhibitors including Bortezomib (Velcade™). Our findings suggest that the proteasome system can be perturbed by means other than proteasome inhibitors, i.e., preventing proteasome phosphorylation. DYRK2 inactivation reduces proteasome activity, interferes with cell proliferation and potentiates the anti-growth effect of Bortezomib. Although DYRK2 has been suggested to have both pro- and anti-tumor potentials55-58, our data clearly favor a pro-proliferative function of this kinase in both cancer and non-cancer cells. The requirement of DYRK2-meditated proteasome phosphorylation for in vivo tumor formation (Fig. 8) and the correlation between DYRK2 expression and breast cancer prognosis (Fig. 7) further indicate that this kinase may be a worthy drug target for cancer treatment. Combinatory therapies using inhibitors of the proteasome and of its activating kinase(s) such as DYRK2 are expected to have enhanced efficacy against cancer cells, especially those with deregulated proteasome phosphorylation and/or strong dependence on proteasome function.
Methods
Cell culture, transfection and infection
All cell lines were originally obtained from American Type Culture Collection (ATCC) and maintained in DMEM with 10% fetal bovine serum (FBS) and Penicillin/Streptomycin (Life Technologies), except that MCF10A cells were cultured in DMEM/F-12 medium supplemented with 5% horse serum, 20 ng/ml EGF, 0.5 μg/ml hydrocortisone, 100 ng/ml cholera toxin, 10 μg/ml insulin and Penicillin/Streptomycin (Life Technologies). Cells were intermittently treated with Ciprofloxacin (GenHunter) to prevent mycoplasma contamination. Transient transfection of 293T cells was done with the X-tremeGENE 9 reagent (Roche). Retroviruses were produced from 293T cells co-transfected with the retroviral backbone and the pCL10A1 helper vector59, and lentiviruses were produced using the psPAX2 and pMD2.G packaging vectors. Viral media were passed through a pre-wetted 0.45 μm filter and mixed with 10 μg/ml polybrene (Sigma) before being added to recipient cells. Infected cells were selected with puromycin (1–2 μg/mL, Life Technologies) to generate stable populations.
Plasmids and shRNAs
Human Rpt3 cDNA was kindly provided by Dr. Shigeo Murata (the University of Tokyo). Full-length Rpt3 was subcloned into the pQCXIP vector (Clontech) with an engineered N-terminal HA tag. GFPu was reported (ref. 18) and provided by Dr. Gentry Patrick (University of California, San Diego). We are deeply grateful for Dr. Andreas Matouschek (University of Texas, Austin) for providing the unpublished UBL-YFP proteasome activity reporters. The human kinase expression library28 was generously provided by Dr. Susan Lindquist at Massachusetts Institute of Technology. GFP-tagged Human DYRK1B, 2, 3, 4 and rat DYRK1A were gifts from Dr. Walter Becker (RWTH Aachen University, Germany). All point mutations were introduced following the QuikChange protocol (Agilent). To replace endogenous Rpt3 with exogenous WT or mutant forms, a shRNA sequence (designated as R2, targeting Rpt3 coding sequence) was inserted into the pLL3.7 vector as suggested (Dr. Tyler Jacks’ laboratory, Massachusetts Institute of Technology). The GFP coding sequence in the original pLL3.7 vector was then removed and replaced by the HA-Rpt3 (WT or mutant)-IRES-Puromycin fragment amplified from the above pQCXIP-HA-Rpt3 constructs using PCR. RNAi-resistant silent mutations was introduced, and the final pLL3.7-R2-HA-Rpt3-IRES-Puro constructs were built using the Gibson Assembly kit (New England BioLabs).
The targeting sequences of human DYRK2, 1A, 1B and 4 were also expressed from the pLL3.7 vector, while shRNAs against human DYRK3 were cloned into the pLKO.1 vector (Addgene).
All shRNA targeting sequences are listed in Supplementary Table 2. All plasmids were verified by DNA sequencing.
Antibodies and reagents
Antibodies used in this study were: Rpt3 (clone TBP7-27, BML- PW-8765), Rpt6 (clone p45-110, BML-PW9625), Rpn13/ADRM1 (BML-PW9910), 20S α subunits (clone MCP231, BML-PW8195) from Enzo Life Sciences; PSMC4 (clone H-2, sc-166003, 1:200), Rpn1 (clone A-11, sc-271775, 1:500), Rpn2 (clone C-7, sc-166038) from Santa Cruz Biotechnology; Rpt3 (PSMC4-A303-849A) from Bethyl Laboratories; anti-Flag M2 (F3165) and actin (clone AC-74, A2228) from Sigma; anti-HA (clone HA.11, MM5101R), Ubiquitin (P4G7) from Covance; DYRK2 (#8143), poly-Ub (K48 linkage, #4289), p21 (#2947), p27 (#3686), Phospho-Rb (#8516), cyclin B1 (#12231), cyclin E2 (#4132), P-H3 (Ser10, #3377), H2AX (#2595), γ-H2AX (#9718) from Cell Signaling; α-tubulin (clone DM1A, CP06) from Oncogene; GFP (clone JL-8, 632381) from BD Clontech; and GAPDH (clone 6C5, CB1001, 1:5,000), Ub-H2B (clone 56, 05-1312) from Millipore. All antibodies were diluted 1:1000 for western blot unless otherwise noted. Rabbit anti-pT25 polyclonal antibody was generated using the following phospho-peptide as immunogen: LSVSRPQ(pT)GLSFLGP. After multiple rounds of negative depletion with the non-phosphorylated peptide, the antisera were affinity-purified and concentrated. Cycloheximide (CHX), hydroxyurea (HU) and L-Phenylalanine were purchased from Sigma, Bortezomib from Selleck Chemicals, λ-phosphatase from New England Biolabs, aphidicolin from Millipore and nocodazole from Tocris. ATP [γ-32P] and 3H-Phe were obtained from PerkinElmer.
In vitro kinase assay
Human DYRK2 (aa 74-479, WT or D275N) with a N-terminal 8×His tag was expressed in BL21 (DE3)-RILP cells and purified using Ni-NTA resin (Thermo) as described60. Human Rpt3 (1-148, WT or T25V) with a C-terminal 6×His tag was generated based on ref. 29 and purified in a similar manner. For in vitro phosphorylation, Rpt3 (1-148) proteins were incubated with DYRK2 kinase at 30°C in 1X kinase buffer (50 mM Tris, pH7.5, 10 mM MgCl2) supplemented with 100 μM ATP, 5,000 cpm/pmol ATP [γ-32P] and 1 mM Na3VO4. At the indicated time points an fixed amount of reaction containing 4 μg of Rpt3 (1-148) and 50 ng of DYRK2 was withdrawn and immediately boiled in Laemmli sample buffer supplemented with 20 mM EDTA. Samples were then separated on a 12% SDS-PAGE gel followed by autoradiography on a Typhoon storage phosphorimager (GE Healthcare). The final stoichiometry of phosphorylation was calculated to be 0.5-1.0 mole of phosphate/mole of Rpt3 (1-148) based on scintillation counting. Smaller aliquots each containing 150 ng of Rpt3 (1-148) were used for anti-pT25 western blot. For whole proteasome phosphorylation, 2 μg of affinity-purified 26S proteasome from 293T cells was incubated with 50 ng of DYRK2 and radio-labeled ATP at 30°C for 1 hr and imaged as above.
Liquid Chromatography Tandem Mass Spectrometry (LC MS/MS) and SILAC
DYRK2-treated purified proteasome complexes were digested in solution with trypsin and analyzed by LC-MS/MS using Easy-nanoLC 1000 (Thermo Fisher, San Jose, CA) coupled with a linear ion trap (LTQ) Orbitrap XL mass spectrometer (Thermo Electron Corp) as described61. LC MS/MS data were searched using the Batch-Tag within the developmental version (v 5.10.10) of Protein Prospector at University of California, San Francisco against a decoy database consisting of a normal Swissprot database concatenated with its randomized version (SwissProt.2013.06.17.random.concat with a total of 455294 protein entries). The mass accuracy for parent ions and fragment ions was set as ± 20 ppm and 0.6 Da, respectively. Trypsin was set as the enzyme, and a maximum of two missed cleavages were allowed. Protein N-terminal acetylation, methionine oxidation, N-terminal conversion of glutamine to pyroglutamic acid, and phosphorylation of serine or threonine were selected as variable modifications. The proteins were identified by at least two peptides with a false-positive rate of ≤0.5%.
For SILAC (stable isotope labeling by amino acids in cell culture) experiments, 293T Rpn11-TBHA cells were labeled with 12C-Lys/Arg (light, Sigma) or 13C-Lys/Arg (heavy, Cambridge Isotope Lab) as described62 for at least seven passages and seeded into 4 × 15 cm plates each. WT or inactive DYRK2 (20 μg/plate) was transfected into light- or heavy-labeled cells, respectively, using X-tremeGENE 9 with serum-free SILAC medium (Thermo). Cells were harvested 24 hrs later and proteasomes were purified separately from 26 mg (determined by Bradford protein assay) of each sample as described in the following section. Heavy and light proteasomes were mixed 1:1 and precipitated using the methanol:chloroform method. The sample pellet was then air-dried and stored at −80°C. LC-MS/MS was carried out by nanoflow reversed-phase liquid chromatography (Eksigent, Dublin, CA) coupled online to the LTQ-Orbitrap XL mass spectrometer as described above. The Monoisotopic masses of parent ions and corresponding fragment ions, parent ion charge states, and ion intensities from LC-MS/MS spectra were extracted using in-house software based on Raw_Extract script from Xcalibur v2.4. Following automated data extraction, the resultant peak lists for each LC-MS/MS experiment were submitted to the development version (5.13.1) of Protein Prospector (UCSF) for database searching using a concatenated Swissprot database (540546 sequence entries) composed of a SwissProt database (6/27/2013) and its randomized version. In addition to the modifications described above, 13C6-Arg and 13C6-Lys were also chosen as variable modifications. The Search Compare program in Protein Prospector was used for summarization, validation, comparison of results and calculation of the relative abundance ratios of Arg/Lys-containing peptides based on ion intensities of monoisotopic peaks observed in the LC MS spectra.
Proteasome purification and activity assays
Human 26S proteasomes were affinity-purified from cells stably expressing pQCXIP-Rpn11-HTBH (ref.13) or Rpn11-TBHA (TEV-Biotin-HA tag63). Briefly, cells were lysed with Tris buffer (50 mM, pH 7.5) plus 0.5% Nonidet P-40, 1 mM ATP, 10 mM MgCl2, 1 mM DTT and a phosphatase inhibitor cocktail (10 mM NaF, 20 mM β-glycerophosphate and 50 nM Calyculin A). The proteasomes were captured by incubating cleared cell lysates with high capacity streptavidin agarose resin (Thermo) for 30-60 min at 4°C. Beads were washed once with Tris buffer (+ 1 mM ATP, 250 mM NaCl) and three times with the same buffer without salt. When indicated, His-DYRK2-WT or D275N was added to the immobilized proteasome for in vitro kinase reactions at 30°C for 10 min. The kinases were then washed out with Tris buffer or 26S proteasome assay buffer (ref.25) containing 1 mM ATP. To elute proteasomes, the beads were further incubated with His-tagged Tobacco Etch Virus (TEV) protease prepared in our laboratory in the presence of 0.5 mM tris(2-carboxyethyl)phosphine (TCEP) at 4°C for 1.5-2.5 hours. The TEV protease was then removed by binding to Ni-NTA resin, and the flow-through containing purified proteasome was measured for protein concentration and then stored at −80°C. Peptidase activities of purified proteasome or proteasomes in whole cell lysates were assayed using fluorogenic peptide substrates (Enzo Life Sciences) according to ref.25 with phosphatase inhibitors present in the lysis and assay buffers. When cell lysate was used, the measured activity was normalized against total proteasome levels determined by parallel western blots. For casein degradation, 1 μg of self-quenching BODIPY-casein (EnzChek® Protease Assay Kit, green fluorescence, Life Technology) and 1 μg of purified, DYRK2-treated 26S proteasome were mixed in the 26S assay buffer with 5 mM ATP. Fluorescence (Ex 480/Em 530) was measured every minute for 30-40 min at 37°C in a Tecan Infinite® M200 Pro multi-well plate reader. Degradation of ubiquitinated substrate was performed according to the described method (ref.30). A mixture of 1 μM in vitro ubiquitinated GFP fusion substrate30 and 1X ATP-regenerating system (both generously provided by Dr. Andreas Martin, University of California, Berkeley) were incubated with 1 μg of DYRK2-treated 26S proteasome in HEPES reaction buffer30 containing 5 mM ATP. GFP fluorescence was measured at 37°C as described above. Measurements of protein degradation rate in live cells using 3H-Phe labeling were essentially performed as reported26. Briefly, equal numbers of control and T25A knock-in cells were seeded in 6-well plates and labeled with 5 μCi/ml of 3H-Phe (1.5 ml per well) in the presence of 0.4 mM HU (for S phase synchronization) for 24 hrs. Cells were then washed 3 times with PBS to remove HU and 3H-Phe, and chased in 2 ml/well of medium containing 2 mM cold Phe with DMSO or 1 μM Bortezomib. A 100 μl aliquot of medium was withdrawn at different time points and precipitated with 40 μl of trichloroacetic acid (TCA, Sigma) at 4°C overnight. At the last time point, cells were lysed with 0.1 M NaOH. Acid-soluble 3H-Phe from each sample and total 3H-Phe were measured by scintillation counting. After subtraction of non-proteasomal degradation (determined from Bortezomib-treated cells), the rate of 3H-Phe release into media (proportional to protein degradation) was calculated. Experiments were repeated three times for each cell type.
Proteasome ATPase activity was measured using the malachite green method64. Bacterially purified His8-UBL-YFP-ODC was included in the reaction as indicated. For ubiquitin chain binding assay, 26S proteasomes from 1 mg of 293T Rpn11-TBHA cell extracts were immobilized on streptavidin beads and in vitro phosphorylated by DYRK2 (WT or DN). Samples were briefly treated with 2.5 μM ubiquitin aldehyde (Ub-Al, UBPBio) on ice for 5-10 min. K48-linked tetraubiquitin (K48-Ub4, UBPBio) was then added at a final concentration of 500 nM65 and incubated with the proteasome for 30 min at 4°C. Excess K48-Ub4 was washed off with 1X kinase buffer (50 mM Tris, pH7.5, 10 mM MgCl2 with 1 mM ATP). Proteasome-bound ubiquitin chain was determined by western blotting.
CRISPR/Cas9-mediated genome editing
Guide RNA design, plasmid cloning and cell transfection were essentially done as described66. To create Rpt3-T25A knock-in alleles in human cells, two CRISPR guide RNA (gRNA) sequences near the T25 codon were chosen based on their specificity scores (http://crispr.mit.edu/) and targeting efficiency determined by the SURVEYOR assay. The gRNA sequences were cloned into the hSpCas9 plasmid (pX330, Addgene). The constructs were then independently electroporated into HaCaT or MDA-MB-468 cells together with pEGFP-C1 (Clontech) and a corresponding single-stranded DNA oligonucleotide (ssODN) containing the T25A mutation (ACC→GCC) and mutated PAM sequences. GFP-positive cells were isolated by FACS and single cell-seeded into each well of 96-well plates. After clonal expansion, genomic DNA was purified and amplified by PCR using SURVEYOR assay primers flanking the T25 site. The PCR products were digested with NgoMIV to identify cell clones with homozygous T25A mutation. The PCR fragments from the positive clones (without NgoMIV digestion) were then ligated into the pBlueScript II KS (+) vector for sequencing.
For DYRK2 knockout, a similar procedure was followed. The PCR products of genomic DNAs were directly cloned into pBlueScript II KS (+). Since both MDAMB-468 and HaCaT cells have three copies of DYRK2, at least 6 bacteria colonies were sequenced for each cell clone to ensure complete coverage of all alleles.
All guide RNA sequences are listed in Supplementary Table 2.
Cell proliferation assay, synchronization and cell cycle analysis
To measure cell proliferation, actively proliferating cells were seeded in 12-well and/or 6-well plates at a density of 0.8 or 1.0 × 105 cells/well. Every 24 hours afterwards, cells were trypsinized and quantified using the Countess® automated cell counter (Life Technologies). The time course experiments were repeated three times for each cell type. MTS assays were performed with the CellTiter 96® AQueous One Solution Cell Proliferation Assay kit (Promega) according to manufacturer's instruction. To synchronize HaCaT cells at G1/S boundary, cells were first starved in serum-free medium for 48 hours, released into full medium with 10% FBS for 12 hours, and treated with 10 μM aphidicolin for another 12 hours. When indicated, cells were also synchronized at late G1/early S with hydroxyurea. Nocodazole synchronization and mitotic shake-off were performed as described67. Bromodeoxyuridine (BrdU) and anti-BrdU antibody were purchased from BD Biosciences. BrdU labeling and propidium iodide (Sigma) staining were performed according to manufacturer's protocol. Cells were analyzed using a BD FACSJazz cell sorter and data analysis was done with Flowjo.
Quantitative RT-PCR
Total RNA from cells was extracted using the RNeasy Mini Kit (Qiagen) and reverse-transcribed with the iScript™ cDNA Synthesis Kit (Bio-Rad). Gene-specific primers and cDNAs were mixed with SYBR Premix Ex Taq II (Tli RNase H Plus, Takara) for PCR reactions in a 7500 Real-Time PCR System (Applied Biosystems). ACTB (actin) or GAPDH mRNA levels were used as internal control. Data were analyzed using the ddCt method. Results averaged from three independent experiments were shown. Primer sequences used for qRT-PCR are listed in Supplementary Table 2.
Tumor study
Female athymic Nude-Foxn1nu mice (Harlan) were housed and maintained at the University of California-San Diego (UCSD) in full compliance with policies of the Institutional Animal Core and Use Committee (IACUC). The mice (6-week old) were irradiated (300 Rads) at 24 hrs before injection. MDA-MB-468 cells were counted and suspended at 1.0 × 107/ml in PBS containing 50% growth factor-reduced Matrigel (BD Bioscience). One million cells (100 μl) were injected subcutaneously into the flank of each mouse, 5 mice per cell line. Tumor dimensions were measured twice per week using a digital caliper and tumor volume was calculated as (length × width2)/2. Mice were euthanized 42 days after injection and tumors were excised and weighed. For immunohistochemistry, tumor tissue was fixed in 10% neutral buffered formalin over night at room temperature, rinsed in 70% ethanol and placed in histology cassettes for paraffin embedment. Hematoxylin and eosin (H & E) staining and anti-Ki-67 staining were performed at the Histology Core Facility at University of California, San Diego.
Kaplan-Meier curves of DYRK2 were generated at http://www.kmplot.com using the 202971_s_at probe set, and the conclusion holds true for all four DYRK2 probe sets. Gene expression data and patient information used by this online tool were all downloaded from public databases including GEO, EGA and TCGA (http://www.kmplot.com/analysis/index.php?p=background and ref. 36).
Statistics and data presentation
Most experiments were repeated at least 3 times to be eligible for the indicated statistical analyses, and the data exhibited normal distribution. There was no estimation of group variation prior to experiments. All results are presented as mean ± s.e.m. unless otherwise noted. For animal studies, five mice per group is the standard sample size for tumor xenograft experiments, and no statistical method was used to predetermine sample size. None of samples/animals were excluded from the experiment, and the animals were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Among the following representative images:
Fig.1a,c,e, 2b, 4b,e, 5e, S2c,d, S4a,c have been repeated more than 4 times.
Fig.1d-g, 2d, f, 3c,d, 4d,f, 5b,c,d, 6b, 7b,d, S1a, S2a,g, S3b, S4b, S5b have been reproduced 3 times.
The remaining figures have been repeated twice each.
Supplementary Material
Acknowledgements
The authors would like to thank members of the Dixon lab and Drs. Kun-Liang Guan, Eric Bennett, Bing Yang, Manuel Kaulich and Gabriel Lander for insightful suggestions and discussions of the manuscript. We acknowledge Eric Durrant and Andrea Ly for excellent technical assistance. We thank Drs. Shigeo Murata, Andreas Matouschek, Susan Lindquist and Walter Becker for providing crucial cDNAs. We owe a debt of gratitude to Charlene Bashore and Dr. Andreas Martin for the polyubiquitinated GFP substrate. This work was supported by the NIH grants RO1DK018849 to JED, RO1GM074830 to L.H., and 1RO1CA168689 and 1R01CA174869 to J.Y. XG was partly supported by the Susan G. Komen postdoctoral fellowship for breast cancer research (KG111280).
Footnotes
Author contributions
X.G. and J.E.D. conceived the study. X.G., X.W. and L.H. made the original observation of Rpt3-T25 phosphorylation. X.G. designed the the majority of experiments. X.G., X.W., Z.W., S.B. executed the experiments. X.G., X.W., S.B. and L.H. performed data analyses. X.W. and L.H. generated and analyzed all mass spectrometry data. Tumor xenograft study was performed by X.G. with J.Y.'s support and guidance. X.G., Z.W. and J.E.D. wrote the paper.
Reference
- 1.Coux O, Tanaka K, Goldberg AL. Structure and Functions of the 20S and 26S Proteasomes. Annual Review of Biochemistry. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 2.Tai HC, Schuman EM. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nature reviews. Neuroscience. 2008;9:826–838. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- 3.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The Hallmarks of Aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 5.Orlowski RZ, Kuhn DJ. Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin Cancer Res. 2008;14:1649–1657. doi: 10.1158/1078-0432.CCR-07-2218. [DOI] [PubMed] [Google Scholar]
- 6.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharyya S, Yu H, Mim C, Matouschek A. Regulated protein turnover: snapshots of the proteasome in action. Nat Rev Mol Cell Biol. 2014;15:122–133. doi: 10.1038/nrm3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehlinger A, Walters KJ. Structural insights into proteasome activation by the 19S regulatory particle. Biochemistry. 2013;52:3618–3628. doi: 10.1021/bi400417a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt M, Finley D. Regulation of proteasome activity in health and disease. Biochimica et biophysica acta. 2014;1843:13–25. doi: 10.1016/j.bbamcr.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Radhakrishnan SK, et al. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol Cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomko RJ, Hochstrasser M. Molecular Architecture and Assembly of the Eukaryotic Proteasome. Annual Review of Biochemistry. 2013;82:415–445. doi: 10.1146/annurev-biochem-060410-150257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, et al. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46:3553–3565. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Huang L. Identifying dynamic interactors of protein complexes by quantitative mass spectrometry. Mol Cell Proteomics. 2008;7:46–57. doi: 10.1074/mcp.M700261-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Hershko A. Roles of ubiquitin-mediated proteolysis in cell cycle control. Current Opinion in Cell Biology. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- 16.Teixeira LK, Reed SI. Ubiquitin ligases and cell cycle control. Annu Rev Biochem. 2013;82:387–414. doi: 10.1146/annurev-biochem-060410-105307. [DOI] [PubMed] [Google Scholar]
- 17.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 18.Min M, Lindon C. Substrate targeting by the ubiquitin–proteasome system in mitosis. Seminars in Cell & Developmental Biology. 2012;23:482–491. doi: 10.1016/j.semcdb.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 19.Dephoure N, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsen JV, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Science signaling. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 21.Nagano K, et al. Phosphoproteomic analysis of distinct tumor cell lines in response to nocodazole treatment. PROTEOMICS. 2009;9:2861–2874. doi: 10.1002/pmic.200800667. [DOI] [PubMed] [Google Scholar]
- 22.Kettenbach AN, et al. Quantitative Phosphoproteomics Identifies Substrates and Functional Modules of Aurora and Polo-Like Kinase Activities in Mitotic Cells. Sci. Signal. 2011;4:rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bian Y, et al. An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. Journal of Proteomics. 2014;96:253–262. doi: 10.1016/j.jprot.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 24.Mayya V, et al. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Science signaling. 2009;2:ra46. doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- 25.Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol. 2005;398:364–378. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- 26.Besche HC, et al. Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J. 2014;33:1159–1176. doi: 10.1002/embj.201386906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taipale M, et al. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 29.Djuranovic S, et al. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell. 2009;34:580–590. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- 30.Beckwith R, Estrin E, Worden EJ, Martin A. Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase. Nature structural & molecular biology. 2013;20:1164–1172. doi: 10.1038/nsmb.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu CW, et al. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol Cell. 2006;24:39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith DM, et al. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 33.Peth A, Kukushkin N, Bosse M, Goldberg AL. Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs. J Biol Chem. 2013;288:7781–7790. doi: 10.1074/jbc.M112.441907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Śledź P, et al. Structure of the 26S proteasome with ATP-γS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proceedings of the National Academy of Sciences. 2013;110:7264–7269. doi: 10.1073/pnas.1305782110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer. 2010;10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- 36.Györffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Research and Treatment. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 37.Petrocca F, et al. A genome-wide siRNA screen identifies proteasome addiction as a vulnerability of basal-like triple-negative breast cancer cells. Cancer Cell. 2013;24:182–196. doi: 10.1016/j.ccr.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vilchez D, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shabaneh TB, et al. Molecular basis of differential sensitivity of myeloma cells to clinically relevant bolus treatment with bortezomib. PloS one. 2013;8:e56132. doi: 10.1371/journal.pone.0056132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mason GG, Murray RZ, Pappin D, Rivett AJ. Phosphorylation of ATPase subunits of the 26S proteasome. FEBS Lett. 1998;430:269–274. doi: 10.1016/s0014-5793(98)00676-0. [DOI] [PubMed] [Google Scholar]
- 41.Bose S, Stratford FL, Broadfoot KI, Mason GG, Rivett AJ. Phosphorylation of 20S proteasome alpha subunit C8 (alpha7) stabilizes the 26S proteasome and plays a role in the regulation of proteasome complexes by gamma-interferon. Biochem J. 2004;378:177–184. doi: 10.1042/BJ20031122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satoh K, Sasajima H, Nyoumura K.-i., Yokosawa H, Sawada H. Assembly of the 26S Proteasome Is Regulated by Phosphorylation of the p45/Rpt6 ATPase Subunit. Biochemistry. 2000;40:314–319. doi: 10.1021/bi001815n. [DOI] [PubMed] [Google Scholar]
- 43.Feng Y, Longo DL, Ferris DK. Polo-like Kinase Interacts with Proteasomes and Regulates Their Activity. Cell Growth Differ. 2001;12:29–37. [PubMed] [Google Scholar]
- 44.Zhang F, et al. Proteasome function is regulated by cyclic AMP- dependent protein kinase through phosphorylation of Rpt6. J Biol Chem. 2007;282:22460–22471. doi: 10.1074/jbc.M702439200. [DOI] [PubMed] [Google Scholar]
- 45.Djakovic SN, Schwarz LA, Barylko B, DeMartino GN, Patrick GN. Regulation of the Proteasome by Neuronal Activity and Calcium/Calmodulin-dependent Protein Kinase II. Journal of Biological Chemistry. 2009;284:26655–26665. doi: 10.1074/jbc.M109.021956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bingol B, et al. Autophosphorylated CaMKIIα Acts as a Scaffold to Recruit Proteasomes to Dendritic Spines. Cell. 2010;140:567–578. doi: 10.1016/j.cell.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 47.Djakovic SN, et al. Phosphorylation of Rpt6 regulates synaptic strength in hippocampal neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:5126–5131. doi: 10.1523/JNEUROSCI.4427-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamilton AM, et al. Activity-dependent growth of new dendritic spines is regulated by the proteasome. Neuron. 2012;74:1023–1030. doi: 10.1016/j.neuron.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ranek MJ, Terpstra EJ, Li J, Kass DA, Wang X. Protein kinase g positively regulates proteasome-mediated degradation of misfolded proteins. Circulation. 2013;128:365–376. doi: 10.1161/CIRCULATIONAHA.113.001971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aranda S, Laguna A, de la Luna S. DYRK family of protein kinases: evolutionary relationships, biochemical properties, and functional roles. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2011;25:449–462. doi: 10.1096/fj.10-165837. [DOI] [PubMed] [Google Scholar]
- 51.Becker W. Emerging role of DYRK family protein kinases as regulators of protein stability in cell cycle control. Cell Cycle. 2012;11:3389–3394. doi: 10.4161/cc.21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.da Fonseca, Paula CA, He J, Morris Edward P. Molecular Model of the Human 26S Proteasome. Molecular Cell. 2012;46:54–66. doi: 10.1016/j.molcel.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 54.Matyskiela ME, Lander GC, Martin A. Conformational switching of the 26S proteasome enables substrate degradation. Nature structural & molecular biology. 2013;20:781–788. doi: 10.1038/nsmb.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller CT, et al. Amplification and overexpression of the dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 (DYRK2) gene in esophageal and lung adenocarcinomas. Cancer Res. 2003;63:4136–4143. [PubMed] [Google Scholar]
- 56.Bonifaci N, et al. Exploring the link between germline and somatic genetic alterations in breast carcinogenesis. PloS one. 2010;5:e14078. doi: 10.1371/journal.pone.0014078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gorringe KL, Boussioutas A, Bowtell DD, Melbourne Gastric Cancer Group P.M.M.A.F. Novel regions of chromosomal amplification at 6p21, 5p13, and 12q14 in gastric cancer identified by array comparative genomic hybridization. Genes, chromosomes & cancer. 2005;42:247–259. doi: 10.1002/gcc.20136. [DOI] [PubMed] [Google Scholar]
- 58.Taira N, et al. DYRK2 priming phosphorylation of c-Jun and c-Myc modulates cell cycle progression in human cancer cells. J Clin Invest. 2012;122:859–872. doi: 10.1172/JCI60818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo X, et al. Axin and GSK3-β control Smad3 protein stability and modulate TGF-β signaling. Genes Dev. 2008;22:106–120. doi: 10.1101/gad.1590908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soundararajan M, et al. Structures of Down syndrome kinases, DYRKs, reveal mechanisms of kinase activation and substrate recognition. Structure. 2013;21:986–996. doi: 10.1016/j.str.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaake RM, Milenković T, Pržulj N, Kaiser P, Huang L. Characterization of Cell Cycle Specific Protein Interaction Networks of the Yeast 26S Proteasome Complex by the QTAX Strategy. Journal of proteome research. 2010;9:2016–2029. doi: 10.1021/pr1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nature protocols. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 63.Guo X, et al. UBLCP1 is a 26S proteasome phosphatase that regulates nuclear proteasome activity. Proceedings of the National Academy of Sciences. 2011;108:18649–18654. doi: 10.1073/pnas.1113170108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011;144:526–538. doi: 10.1016/j.cell.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jacobson AD, MacFadden A, Wu Z, Peng J, Liu C-W. Autoregulation of the 26S proteasome by in situ ubiquitination. Molecular Biology of the Cell. 2014;25:1824–1835. doi: 10.1091/mbc.E13-10-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.