Abstract
In June 2015, the fourth European Workshop on Plant Chromatin took place in Uppsala, Sweden, bringing together 80 researchers studying various aspects of plant chromatin and epigenetics. The intricate relationships between plant chromatin dynamics and gene expression change, chromatin organization within the plant cell nucleus, and the impact of chromatin structure on plant development were discussed. Among the main highlights of the meeting were an ever-growing list of newly identified players in chromatin structure establishment and the development of novel tools and approaches to foster our understanding of chromatin-mediated gene regulation, taking into account the context of the plant cell nucleus and its architecture. In this report, we summarize some of the main advances and prospects of plant chromatin research presented at this meeting.
Keywords: chromatin, development, epigenetics, histone variant, histone modification, nucleus, plant
Abbreviations
- PTM
Posttranslational modification
- PRC
Polycomb repressive complex
- PcG
Polycomb group
- CLF
curly leaf
- TRB
telomere repeat binding protein
- LHP1
like heterochromatin 1
- HSF
Heat-shock factor
- RNAP
RNA polymerase
- MBD
Methyl-CpG-binding domain
- TE
Transposable element
- CAF-1
chromatin assembly factor 1
- SWR1-C
SWR1 complex
Introduction
In June 2015, the fourth European Workshop on Plant Chromatin (EWPC) took place in Uppsala, Sweden, as part of the biannual plant chromatin meeting series (https://colloque6.inra.fr/ewpc_series_on_plant_chromatin).1-3 The 2-day workshop provided an excellent opportunity to present and discuss recent advances, current focus, and challenges in the field of plant chromatin research.
Much progress has been made in recent years to unravel the complexity of mutual interplay between plant chromatin structure and gene expression, and how this translates into developmental phenotypes. In spite of all the advances, the dynamics of plant chromatin structure, its organization within the plant nucleus, and its contribution to gene expression regulation and phenotype establishment are far from understood.
These and other topics were covered during the 4th EWPC, and the results presented demonstrated the fast pace of advance in plant chromatin research. Novel components of chromatin-modifying and chromatin-associated complexes have been identified, increasing the spectra of players and mechanisms of chromatin structure establishment. The impact of histone posttranslational modifications (PTMs) and chromatin structure on gene expression and the dynamics of chromatin in response to transcriptional change were topics addressed by multiple reports, pinpointing a major focus in the field. Novel plant histone modifications and novel chromatin readers have been identified, extending the number of factors that determine chromatin states. Mechanisms of establishment and function of higher-order chromatin structure and chromatin organization within the plant cell nucleus were discussed, highlighting the dynamic nature of chromatin in response to developmental and environmental cues. Finally, the importance of chromatin dynamics for correct execution of developmental programs in Arabidopsis as well as in trees and other non-model species demonstrated the increasing focus of plant epigenetics and epigenomics on species with high environmental and agronomical relevance. Here, we report some findings presented during the 4th EWPC, which highlight future challenges in this field of research.
Family or friends? The search for interactors of Polycomb repressive complexes continues
Polycomb repressive complexes (PRCs) are histone modifying complexes that deposit the histone 3 lysine 27 trimethylation (H3K27me3, PRC2) and histone 2A ubiquitination (H2Aub, PRC1), which mark repressive chromatin. Although the core subunits of the PRC2 and the catalytic subunits of PRC1 are known, the exact composition of the complexes and how they are integrated into larger functional protein complexes remains unresolved.4-6
Sara Farrona (Galway, Ireland) presented recent advances in the research of the PWO-proteins, a plant-specific clade of Pro-Trp-Trp-Pro (PWWP) motif domain proteins that were identified in the group of Daniel Schubert in Düsseldorf as genetic interactors of CURLY LEAF (CLF), which encodes a catalytic as subunit of PRC2, and as physical interactors of the PRC2 components CLF, SWINGER (SWN), MEDEA (MEA) and MULTICOPY SUPRESSOR OF IRA 1 (MSI1). PWO1–3 are proteins evolutionarily conserved from lycophytes to angiosperms indicating that the acquisition of PWO proteins coincided with the origin of the vascular plants. PWO proteins are characterized by the N-terminal PWWP domain responsible for nuclear localization and histone H3 and H3K27me3 binding, and an unknown conserved domain in the C-terminal part of the protein that is required for the interaction with Polycomb group (PcG) proteins. Structural conservation of the proteins among vascular plant groups suggests that the role of the PWO protein interaction with PRC2 may be evolutionarily conserved.
Recently, a number of studies in plants have revealed the contribution of transcription factors to sequence-specific targeting of the PRCs to DNA.4,6 An additional role of transcription factors as mediators of repression in cooperation with the PRCs was proposed by Franziska Turck (Cologne, Germany). Mutants in 2 genes encoding Myb-domain-containing transcription factors, TELOMERE REPEAT BINDING PROTEIN (TRB) 1 and TRB3, were identified as enhancers of the Arabidopsis lhp1 mutant, deficient in the PRC protein LIKE HETEROCHROMATIN 1 (LHP1). Genome-wide profiling revealed that TRBs bind sequences enriched for the telobox or related cis-elements without preference for H3K27me3-targets. In fact, H3K27me3 marked regions are less likely to be bound by TRBs despite their relative enrichment for the cognate cis-elements. In lhp1, there is a genome-wide increase in the number of TRB-bound sites, supporting a model in which TRBs may act as transcriptional repressors at PcG target genes under conditions of LHP1—or H3K27me3—depletion when the accessibility of telobox-enriched TRB-binding sites is increased (Fig. 1).
Figure 1.
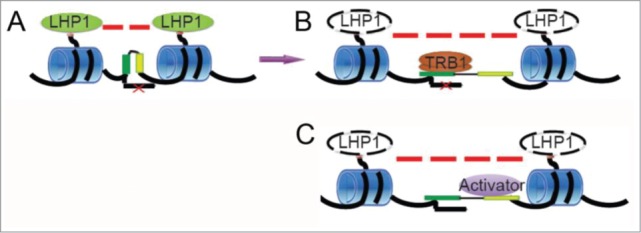
Possible scenario for the role of TRBs at PcG target genes. In the default state (A), PcG-mediated chromatin compaction prohibits access to transcription factor binding sites. In case of compromised repression (B), TRBs can access and bind their cognate sites thereby interfering with the binding of other factors acting as transcriptional activators (C). (Courtesy of Franziska Turck, Cologne, Germany).
Narrowing the gap between repression and activation
The traditional view of chromatin structure as being either “repressed” or “active” for transcription has in recent years been challenged by the identification of multiple chromatin states associated with different combinations of histone variants and histone PTMs,7-9 and especially by the identification of bivalent chromatin states where active and repressive chromatin marks co-localize.9
One of the histone variants that confer specific properties to nucleosomes is the non-canonical histone 2A variant H2A.Z. H2A.Z plays important roles in plant developmental, cell-cycle-related, metabolic, and stress-response-related processes,10 marking especially genes responsive to environmental or developmental stimuli.11 How H2A.Z deposition is targeted to specific genes and how its presence influences transcriptional activation remains unclear. Pedro Crevillén from the group of José Jarillo (Madrid, Spain) presented a targeting mechanism for H2A.Z deposition by SWR1 complex (SWR1-C), a Snf2 ATP-dependent chromatin remodeling complex. The interaction of SWR1-C with A/T-rich regions in the promoters of target genes is mediated by SWC4, a newly identified binding partner of the SWR1-C core subunit SWC6 that was found to be essential for Arabidopsis development.
In plants, H2A.Z is evicted in the vicinity of the transcription start sites (TSSs) of temperature-responsive genes upon elevation of ambient temperature.12 but the mechanism of the eviction and its contribution to transcriptional activation remain elusive. Using a genome-wide H2A.Z and transcriptome profiling, Sandra Cortijo from the group of Philip Wigge (Cambridge, UK) showed that elevation of ambient temperature from 17°C to 27°C initially results in a fast and transient site-specific eviction of H2A.Z associated with transcriptional change. The presence of strong heat shock elements and heat-shock factor (HSF) binding in proximity of the fast-responding genes suggests an interplay between HSFs, H2A.Z eviction, and rapid chromatin remodeling at these sites (Fig. 2), which is being further explored in the Wigge laboratory.
Figure 2.
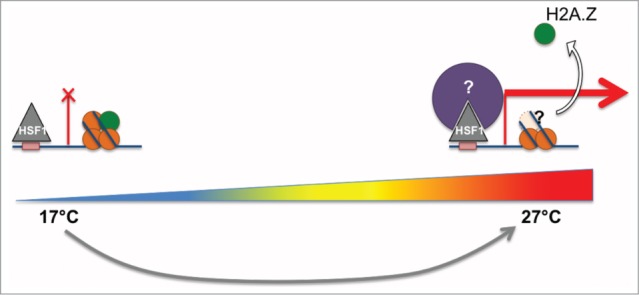
Hypothetical model depicting the eviction of H2A.Z from nucleosomes and transcriptional activation of target genes after ambient temperature increase. HSF - Heat-shock factor. (Courtesy of Sandra Cortijo and Philip Wigge, Cambridge, UK).
While transcriptional activation of temperature-sensitive genes is associated with the eviction of H2A.Z, its presence marks active metabolic gene clusters. Metabolic clusters contain functionally related but non-homologous genes that are required in a common metabolic pathway and that are often expressed in a coordinate fashion. Although traditionally associated with bacterial and fungal genomes, metabolic gene clusters have recently been identified in plant genomes.13 Hans-Wilhelm Nützmann from the group of Anne Osbourn (Norwich, UK) presented evidence that the active state of genes within metabolic clusters in Arabidopsis is marked by the presence of H2A.Z,14 while repression is associated with increased abundance of H3K27me3. The presence of H3K27me3 in metabolic gene clusters presents another example demonstrating that the role of PRC2 extends beyond repressing developmentally-regulated genes, adding to the growing list of responsive genes that are subject to PcG repression.15,16
An example of H3K27me3 involvement in establishing optimal expression levels of environmentally responsive genes was presented by Antoine Martin from the group of Alain Gojon (Montpellier, France). Transcriptional repression of the gene encoding the nitrate transporter NRT2.1 in nitrogen-rich environment is associated with deposition of H3K27me3.17 Surprisingly, NRT2.1 is marked by H3K27me3 even under inductive (low nitrogen) conditions, its transcription is further increased in PRC2 mutants but its repression under non-inductive (high nitrogen) conditions is independent of PRC2 function. Therefore, apart from “locking” a repressed state of a gene, the relative abundance of H3K27me3 may also be modulating the final (active) transcriptional output.
The presence of H3K27me3 being compatible with gene activation was also suggested by Julia Engelhorn from the group of Cristel Carles (Grenoble, France), who studied the sequence of chromatin events accompanying transcriptional activation of PcG-repressed genes. Upon synchronized induction of flower development,18 deposition of H3K4me3 on several PcG-target genes preceded reduction of H3K27me3, indicating that presence of H3K27me3 does not necessarily prevent transcription and suggesting that the relative abundance of opposing chromatin marks may be the final determinant of transcription level.
The primary chromatin structure is a complex template for the transcription machinery that needs to overcome the barrier imposed by the nucleosomes and other DNA-associated proteins. Klaus Grasser (Regensburg, Germany) presented recent advances in the identification of transcript elongation factors that physically interact with the subunits of the RNA polymerase II (RNAP II) complex and facilitate the progression of the transcription machinery through the chromatin template. The complex chromatin remodeling that accompanies transcript elongation was highlighted by the identification of the H3/H4 and H2A/H2B histone chaperones SPT6 and FACT, histone acetylation, methylation, or ubiquitination proteins, as well as modulators of RNAP II activity.19
A flexible transition between higher-order repressive and accessible chromatin structure in response to stress was suggested by Sascha Waidmann from the group of Claudia Jonak (Vienna, Austria) who discussed the role of the DEK-domain containing protein DEK3 in negative regulation of salt-stress responses.20 Waidmann showed that upon salt treatment, the stress-activated protein kinase ASK7 phosphorylates DEK3, disrupting its interaction with the SCC3-subunit of the cohesin complex. Based on the presented hypothetical model, DEK3 interaction with the cohesin complex under non-inductive conditions establishes a repressive higher-order chromatin structure. During salt stress, ASK7-mediated phosphorylation of DEK3 may prevent the DEK3-cohesin interaction and facilitate the expression of salt-responsive genes. This could suggest a potential mechanism for rapid transition between chromatin states in response to environmental cues.
Novel chromatin states need novel writers and readers
Despite the crucial role of histone PTMs in specifying the chromatin structure, the repertoire of plant histone modifications and their combinations remains largely unexplored. Minerva Trejo from the group of Lars Hennig (Uppsala, Sweden) presented the identification of a novel heterochromatic histone mark that co-localizes with H3K9me2 in transposable elements in a manner depending on the H3K9me2 DNA methyltransferase KRYPTONITE (KYP, SUVH4).
Histone arginine methylation and its role during gametogenesis and embryogenesis in Arabidopsis were addressed by Daphné Autran and colleagues (Montpellier, France). While histone H3 arginine methylation has been shown to regulate the pluripotency of mouse embryonic cells,21 its possible function in plants is unknown. Autran reported the presence of H3R26me2 in the female gametophyte and zygote, and identified mutants in histone arginine methyltransferases that cause fertility defects, which together support a role of histone arginine methylation in Arabidopsis reproductive development.
Co-occurrence of different epigenetic modifications may be important for a specific functional output, as is suggested by the existence of proteins with combinations of chromatin-mark-recognition domains, such as the members of the Methyl-CpG-binding domain (MBD) protein family that, in addition to the MBD module, contain the H3K4me-binding CW domain.22 Hatice Zeynep Nenseth from the group of Reidunn Aalen (Oslo, Norway) focused on MBD1, MBD2, and MBD4 and presented evidence for the requirement of both the MBD and CW domains for chromatin binding. The studied MBDs localized to euchromatin in vivo and predominantly affected light-related stress responses. The authors suggested that MBD proteins containing both MBD and the CW domains may function within the chromatin of moderately-expressed genes, where both DNA methylation and H3K4me coexist.
Organized genome in an organized nucleus
Chromatin within the plant nucleus is well ordered but, at the same time, dynamic to organize the genome and modulate its expression. Several approaches have recently been developed to decipher genome organization and the dynamics of various nuclear compartments to better understand the relationship between nuclear structure and genome functions, such as transcription, in the 3-dimensional nuclear space.23
An example of chromatin-context-dependent genome expression is the transcriptional silencing of transposable elements (TEs). Transposition is a naturally occurring event that may contribute to inter- and intra-species variation but that is tightly controlled. Isabel Bäurle (Potsdam, Germany) investigated the silencing of a TE in natural accessions and recombinant inbred lines of Arabidopsis at high resolution. While intergenic localization of the TE AtMu1c is more likely to be subject to epigenetic silencing, its transposition into the 3´ region of a protein-coding gene prevented the TE repression. This in general suggested that specific genomic locations and chromatin environments may favor escape of TEs from epigenetic silencing.24
Within the nucleus, silent heterochromatin regions are visible as densely stained chromocenters that in Arabidopsis mainly comprise centromeric and pericentromeric heterochromatin. Chromocenters de-condense during imbibition and seed germination and progressively reform during the first days of post-germination seedling growth, following extensive chromatin rearrangements.25 Aline Probst (Clermont-Ferrand, France) reported that chromocenter formation correlates with enrichment of the canonical histone H3.1 and the repressive chromatin mark H3K9me2 at heterochromatic sequences. This process depends on the H3.1 chaperone CHROMATIN ASSEMBLY FACTOR 1 (CAF-1), while HISTONE REGULATOR A (HIRA), the Arabidopsis homolog of the animal H3.3 histone chaperone complex,26,27 is dispensable, suggesting a key role of H3.1 deposition in post-germination chromocenter establishment.
The developmental importance of chromocenter decondenzation, on the other hand, was demonstrated by Zsuzsanna Mérai from the group of Hisashi Tamaru (Vienna, Austria). During male gametogenesis, the pollen vegetative cell nucleus undergoes heterochromatin decondenzation coupled with active removal of CenH3.28 Mérai and colleagues showed that the AAA-ATPase chaperone CDC48A is required for the unloading of sumoylated CenH3, chromatin decondenzation and global activation of silent rRNA genes. As the cdc48a allele is rarely paternally transmitted due to defective pollen tube formation, the authors suggested that the bulk rRNA gene activation may be essential to meet the increased metabolic demand during pollen tube growth.29
The expression of 45S rRNA genes depends on their epigenetic state and position within the nucleus—while nucleolar-localized rRNA genes are transcribed by RNAP I, extranucleolar rRNA genes clusters in heterochromatic knobs remain repressed. Distinct sequence variants of the 45S rRNA genes exist, which have either extra- or intra-nucleolar localization.30 This homeostasis is compromised in the CAF-1 mutants fasciata, where rDNA copies are progressively lost and the originally inactive rRNA gene copies translocate into the nucleolus and are expressed.30,31 Jiri Fajkus (Brno, Czech Republic) showed that the reintroduction of a wild-type FAS allele into fas mutant plants that have been inbred for several generations can lead to reversion of the rRNA gene copy-number but causes major changes in the bulk variant composition, their transcriptional state and nuclear localization, suggesting reprogramming and establishment of a novel epigenetic equilibrium at the nucleolus organizer regions in these plants.
The nucleolus is the site of RNAP I-mediated transcription while RNAP II is excluded.32 A potential role of the nucleolus as a compartment sequestering transcriptionally repressed parts of genome away from the RNAP II territory was presented by Frédéric Pontvianne (Perpignan, France). Fluorescence-assisted sorting of nucleoli30 followed by DNA sequencing revealed that, in addition to rRNA genes, also RNAP II-transcribed genomic DNA sequences can localize within the nucleolus, defining nucleolus-associated domains (NADs) in plants (Fig. 3). Spatial sequestration of these sequences into the nucleolus may contribute to their silencing, highlighting a novel role as a silencing center for this nuclear compartment.
Figure 3.
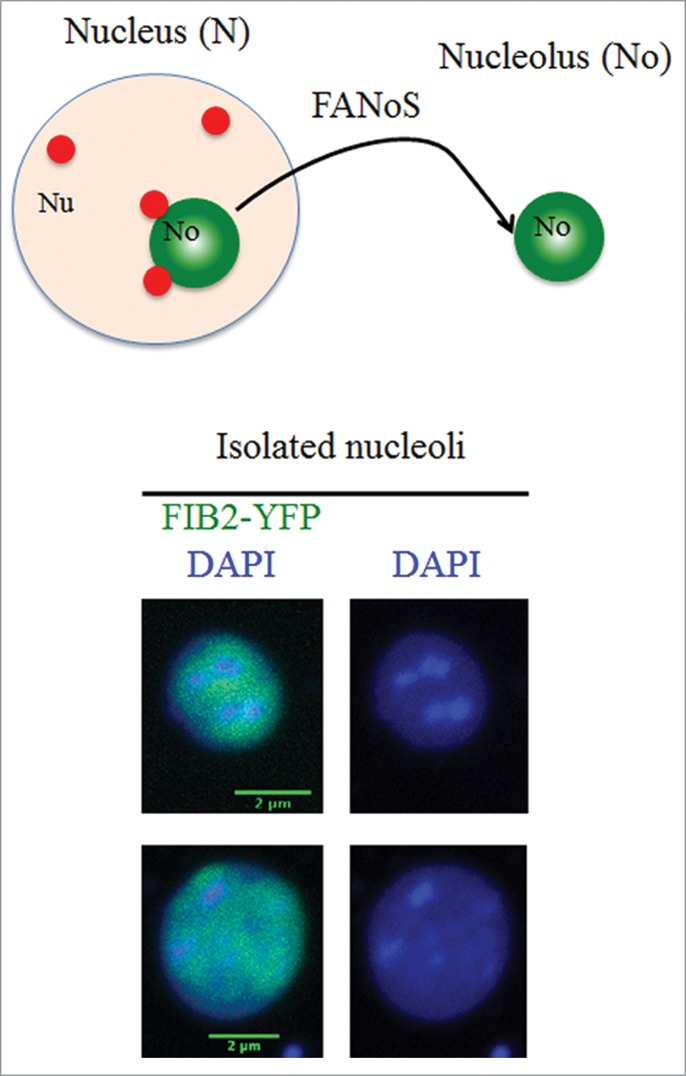
Schematic representation of nucleoli purification using fluorescence-activated nucleolar sorting (FANoS). Lower part: Examples of DAPI-stained nucleoli marked by the nucleolar protein FIBRILLARIN 2 fused to YFP (FIB2-YFP). (Courtesy of Frédéric Pontvianne, Perpignan, France).
To follow the in vivo localization and dynamics of epigenetic marks such as DNA methylation, Mathieu Ingouff (Montpellier, France) developed a powerful approach based on fluorescent biosensors (“DYNAMETs”) and demonstrated their use on the changes of DNA methylation that occur during female sporogenesis and gametogenesis (Fig. 4).
Figure 4.
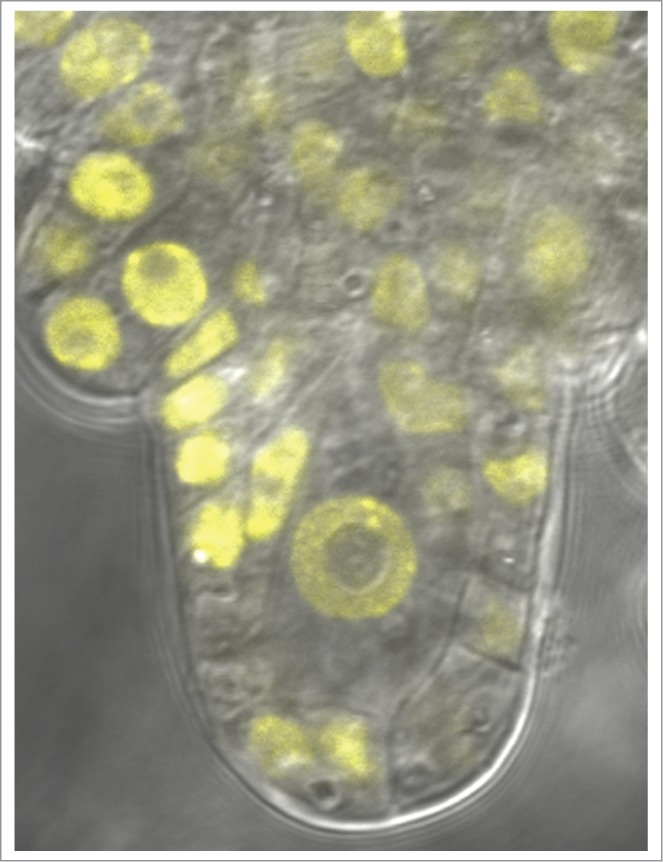
CHH methylation pattern during megaspore mother cell (MMC) development visualised by DYNAMET. (Courtesy of Mathieu Ingouff, Montpellier, France).
In addition to the chromocenters, interspersed heterochromatin foci were previously observed in the endosperm nuclei, and it was suggested that excessive maternal genome sequences may be heterochromatinised.33 Carrying on the characterization of endosperm nuclei, Célia Baroux (Zürich, Switzerland) presented evidence for frequent associations of the centromeres and chromosomal segments in endosperm nuclei fitting a non-random, pair-wise association model. The analysis of nuclei with imbalanced parental genome contributions led the authors to propose a hypothetical model whereby association of parental homologs together with the formerly reported heterochromatinization of excessive maternal genomic regions may play a role in regulating dosage compensation in the endosperm.
Valérie Gaudin and colleagues (Versailles, France) investigated the rules of the 3 -dimensional organization of interphase nuclei, taking into account the variability of nuclear morphology and of nuclear compartments. Previous results demonstrated a non-random, regular spatial distribution of chromocenters in Arabidopsis leaf cell nuclei with an apparent repulsion.34 New spatial statistical models (sets of mathematical and/or computational rules that generate spatial patterns) incorporating novel constraints between compartments and nuclear envelope were presented, demonstrating a preferential peripheral localization of chromocenters.
Chromatin in development
Developmental phenotypes form the top of the iceberg of chromatin structure-modulated gene expression, and assigning a developmental phenotype to a specific chromatin-based mechanism is often challenging.
The effects of DNA hypomethylation are often studied using non-methylable cytidine analogs such as 5-azacytidine or zebularine.35 that interfere with transcriptional gene silencing. A previously unknown effect associated with the use of zebularine in plants was reported by Andreas Finke from the group of Ales Pecinka (Cologne, Germany). Independently of reducing DNA methylation, zebularine induced DNA-replication-coupled DNA damage, resulting in cell cycle retardation, onset of endoreduplication, DNA damage response and homologous-recombination-based repair.36 It will be therefore necessary to carefully dissect phenotypic defects resulting from DNA hypomethylation by zebularine from those related to DNA damage.
The juvenile-to-adult phase transition in Arabidopsis is governed by the opposing activities of the 2 microRNAs, miR156 and miR172, that respectively target promoters or repressors of adult phase transition and flowering. Myriam Calonje (Seville, Spain) presented data supporting roles of the PRC1 components BMI1A/B and EMF1 in the transcriptional repression of the 2 opposing miRNA genes. While the BMI1A/B-containing PRC1 complex is required for the direct repression of the pre-MIR156 genes and the downstream pathway, EMF1 is involved in repressing the MIR172-related pathway,37 explaining the difference in flowering time of the respective PRC1 mutants. These data highlight the involvement of the PcG repression in ways that contribute to both repression as well as activation of developmental pathways.
The developmental importance of epigenetic regulation of gene expression in response to temperature and environmental cues was highlighted in several talks. The digital nature of the cold-induced epigenetic silencing of the FLOWERING LOCUS C (FLC) during vernalization.38-40 was discussed by Rea Antoniou-Kourounioti from the group of Caroline Dean and Martin Howard (Norwich, UK), who highlighted the potential of combining experimental with theoretical modeling approaches in understanding biological systems. A mechanistic model was presented that captures the vernalization behavior of Arabidopsis under lab conditions and makes predictions about this response in the wild.
Trees as experimental model systems with specific developmental transitions were a new addition to the meeting. Daniel Conde from the group of Isabel Allona (Madrid, Spain) reported the identification and possible role of 2 poplar homologs of the Arabidopsis DNA demethylase DEMETER (DME), PtaDML8 and PtaDML10, in the reduction of DNA methylation that is required for the reactivation of bud growth in the spring. Pal Miskolczi from the group of Rishikesh Bhalerao (Umeå, Sweden) showed that global H3K27me3 reprogramming accompanies and is required for the transition from active growth to dormancy and back in the apical buds of hybrid aspen. Together, the studies highlight the complexity of global chromatin reprogramming involved in dormancy establishment and release, and the emergence of translational research in plant epigenomics to species with more complex genomes but with high agronomical and environmental impacts.
More faces of plant chromatin to come?
Increased knowledge of plant chromatin structure and its dynamics are challenging traditional views of chromatin organization. It is becoming evident that multiple functionally distinct chromatin states marked by various combinations of DNA methylation and histone PTMs comprise the traditional categories of constitutive and facultative heterochromatin or euchromatin. Which combinations of chromatin marks specify the chromatin states, how they are established and how they influence higher order chromatin structure and gene expression remains to be explored. The emerging use of sequential chromatin immunoprecipitation (ChIP)9 and chromosome conformation capture techniques.41,42 in plants will allow these questions to be addressed in the future. Although chromatin structure is highly dynamic, methods for its visualization have so far been limited to performing ex vivo experiments on arrested cells. Chromatin mark biosensors are emerging powerful tools to follow the dynamics of chromatin marks in single cells in vivo. Development of biosensors for different chromatin marks combined with live-cell imaging techniques will be a great asset to chromatin dynamics research. Novel tools for studying the organization of chromatin within the nuclear space were presented,30,34 which can help to address the molecular mechanisms governing the 3-dimensional nuclear organization, its dynamics and role in plant development.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank all the participants for sharing their published and unpublished results, for fruitful discussions and comments on this manuscript, and apologize to all our colleagues whose contributions could not be mentioned due to space limitation.
References
- 1.Houba-Herin N, Hennig L, Kohler C, Gaudin V. A fruitful chromatin harvest: meeting summary of the Second European Workshop on Plant Chromatin 2011 in Versailles, France. Epigenetics 2012; 7:307-11; PMID:22430807; http://dx.doi.org/ 10.4161/epi.7.3.19104 [DOI] [PubMed] [Google Scholar]
- 2.Jarillo JA, Gaudin V, Hennig L, Kohler C, Pineiro M. Plant chromatin warms up in Madrid: meeting summary of the 3rd European Workshop on Plant Chromatin 2013, Madrid, Spain. Epigenetics 2014; 9:644-52; PMID:24504145; http://dx.doi.org/ 10.4161/epi.28094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohler C, Gaudin V, Hennig L. Green chromatin dynamics in Zurich: meeting summary based on the European Workshop on Plant Chromatin 2009 in Zurich, Switzerland. Epigenetics 2010; 5:80-3; PMID:20009573; http://dx.doi.org/ 10.4161/epi.5.1.10376 [DOI] [PubMed] [Google Scholar]
- 4.Del Prete S, Mikulski P, Schubert D, Gaudin V. One, Two, Three: Polycomb Proteins Hit All Dimensions of Gene Regulation. Genes (Basel) 2015; 6:520-42; PMID:26184319; http://dx.doi.org/ 10.3390/genes6030520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merini W, Calonje M. PRC1 is taking the lead in PcG repression. Plant J 2015; 83:110-20; PMID:25754661; http://dx.doi.org/ 10.1111/tpj.12818 [DOI] [PubMed] [Google Scholar]
- 6.Mozgova I, Hennig L. The polycomb group protein regulatory network. Annu Rev Plant Biol 2015; 66:269-96; PMID:25621513; http://dx.doi.org/ 10.1146/annurev-arplant-043014-115627 [DOI] [PubMed] [Google Scholar]
- 7.Engelhorn J, Blanvillain R, Carles CC. Gene activation and cell fate control in plants: a chromatin perspective. Cell Mol Life Sci 2014; 71:3119-37; PMID:24714879; http://dx.doi.org/ 10.1007/s00018-014-1609-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roudier F, Ahmed I, Berard C, Sarazin A, Mary-Huard T, Cortijo S, Bouyer D, Caillieux E, Duvernois-Berthet E, Al-Shikhley L, et al.. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J 2011; 30:1928-38; PMID:21487388; http://dx.doi.org/ 10.1038/emboj.2011.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sequeira-Mendes J, Araguez I, Peiro R, Mendez-Giraldez R, Zhang X, Jacobsen SE, Bastolla U, Gutierrez C. The Functional Topography of the Arabidopsis Genome Is Organized in a Reduced Number of Linear Motifs of Chromatin States. Plant Cell 2014; 26:2351-66; PMID:24934173; http://dx.doi.org/ 10.1105/tpc.114.124578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarillo JA, Pineiro M. H2A.Z mediates different aspects of chromatin function and modulates flowering responses in Arabidopsis. Plant J 2015; 83:96-109; PMID:25943140; http://dx.doi.org/ 10.1111/tpj.12873 [DOI] [PubMed] [Google Scholar]
- 11.Coleman-Derr D, Zilberman D. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet 2012; 8:e1002988; PMID:23071449; http://dx.doi.org/ 10.1371/journal.pgen.1002988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 2010; 140:136-47; PMID:20079334; http://dx.doi.org/ 10.1016/j.cell.2009.11.006 [DOI] [PubMed] [Google Scholar]
- 13.Nutzmann HW, Osbourn A. Gene clustering in plant specialized metabolism. Curr Opin Biotechnol 2014; 26:91-9; PMID:24679264; http://dx.doi.org/ 10.1016/j.copbio.2013.10.009 [DOI] [PubMed] [Google Scholar]
- 14.Nutzmann HW, Osbourn A. Regulation of metabolic gene clusters in Arabidopsis thaliana. New Phytol 2015; 205:503-10; PMID:25417931; http://dx.doi.org/ 10.1111/nph.13189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Lucia F, Gaudin V. Epigenetic control by plant polycomb proteins: New perspectives and emerging roles in stress response In: Palmiro Poltronieri NB, Fogher C, eds. From plant genomics to plant biotechnology. Cambridge UK: Woodhead Publishing, Elsevier, 2013:31-48 [Google Scholar]
- 16.Kleinmanns JA, Schubert D. Polycomb and Trithorax group protein-mediated control of stress responses in plants. Biol Chem 2014; 395:1291-300; PMID:25153238; http://dx.doi.org/ 10.1515/hsz-2014-0197 [DOI] [PubMed] [Google Scholar]
- 17.Widiez T, El Kafafi El S, Girin T, Berr A, Ruffel S, Krouk G, Vayssieres A, Shen WH, Coruzzi GM, Gojon A, et al.. High nitrogen insensitive 9 (HNI9)-mediated systemic repression of root NO3- uptake is associated with changes in histone methylation. Proc Natl Acad Sci U S A 2011; 108:13329-34; PMID:21788519; http://dx.doi.org/ 10.1073/pnas.1017863108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wellmer F, Alves-Ferreira M, Dubois A, Riechmann JL, Meyerowitz EM. Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet 2006; 2:e117; PMID:16789830; http://dx.doi.org/ 10.1371/journal.pgen.0020117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Lijsebettens M, Grasser KD. Transcript elongation factors: shaping transcriptomes after transcript initiation. Trends Plant Sci 2014; 19:717-26; PMID:25131948; http://dx.doi.org/ 10.1016/j.tplants.2014.07.002 [DOI] [PubMed] [Google Scholar]
- 20.Waidmann S, Kusenda B, Mayerhofer J, Mechtler K, Jonak C. A DEK domain-containing protein modulates chromatin structure and function in Arabidopsis. Plant Cell 2014; 26:4328-44; PMID:25387881; http://dx.doi.org/ 10.1105/tpc.114.129254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 2007; 445:214-8; PMID:17215844; http://dx.doi.org/ 10.1038/nature05458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoppmann V, Thorstensen T, Kristiansen PE, Veiseth SV, Rahman MA, Finne K, Aalen RB, Aasland R. The CW domain, a new histone recognition module in chromatin proteins. EMBO J 2011; 30:1939-52; PMID:21522130; http://dx.doi.org/ 10.1038/emboj.2011.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Del Prete S, Arpon J, Sakai K, Andrey P, Gaudin V. Nuclear architecture and chromatin dynamics in interphase nuclei of Arabidopsis thaliana. Cytogenet Genome Res 2014; 143:28-50; PMID:24992956; http://dx.doi.org/ 10.1159/000363724 [DOI] [PubMed] [Google Scholar]
- 24.Kabelitz T, Kappel C, Henneberger K, Benke E, Noh C, Baurle I. eQTL mapping of transposon silencing reveals a position-dependent stable escape from epigenetic silencing and transposition of AtMu1 in the Arabidopsis lineage. Plant Cell 2014; 26:3261-71; PMID:25096782; http://dx.doi.org/ 10.1105/tpc.114.128512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benoit M, Layat E, Tourmente S, Probst AV. Heterochromatin dynamics during developmental transitions in Arabidopsis - a focus on ribosomal DNA loci. Gene 2013; 526:39-45; PMID:23410919; http://dx.doi.org/ 10.1016/j.gene.2013.01.060 [DOI] [PubMed] [Google Scholar]
- 26.Duc C, Benoit M, Le Goff S, Simon L, Poulet A, Cotterell S, Tatout C, Probst AV. The histone chaperone complex HIR maintains nucleosome occupancy and counterbalances impaired histone deposition in CAF-1 complex mutants. Plant J 2015; 81:707-22; PMID:25600486; http://dx.doi.org/ 10.1111/tpj.12758 [DOI] [PubMed] [Google Scholar]
- 27.Nie X, Wang H, Li J, Holec S, Berger F. The HIRA complex that deposits the histone H3.3 is conserved in Arabidopsis and facilitates transcriptional dynamics. Biol Open 2014; 3:794-802; PMID:25086063; http://dx.doi.org/ 10.1242/bio.20148680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schoft VK, Chumak N, Mosiolek M, Slusarz L, Komnenovic V, Brownfield L, Twell D, Kakutani T, Tamaru H. Induction of RNA-directed DNA methylation upon decondensation of constitutive heterochromatin. EMBO Rep 2009; 10:1015-21; PMID:19680290; http://dx.doi.org/ 10.1038/embor.2009.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merai Z, Chumak N, Garcia-Aguilar M, Hsieh TF, Nishimura T, Schoft VK, Bindics J, Slusarz L, Arnoux S, Opravil S, et al.. The AAA-ATPase molecular chaperone Cdc48/p97 disassembles sumoylated centromeres, decondenses heterochromatin, and activates ribosomal RNA genes. Proc Natl Acad Sci U S A 2014; 111:16166-71; PMID:25344531; http://dx.doi.org/ 10.1073/pnas.1418564111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pontvianne F, Blevins T, Chandrasekhara C, Mozgova I, Hassel C, Pontes OM, Tucker S, Mokros P, Muchova V, Fajkus J, et al.. Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev 2013; 27:1545-50; PMID:23873938; http://dx.doi.org/ 10.1101/gad.221648.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mozgova I, Mokros P, Fajkus J. Dysfunction of chromatin assembly factor 1 induces shortening of telomeres and loss of 45S rDNA in Arabidopsis thaliana. Plant Cell 2010; 22:2768-80; PMID:20699390; http://dx.doi.org/ 10.1105/tpc.110.076182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schubert V, Weisshart K. Abundance and distribution of RNA polymerase II in Arabidopsis interphase nuclei. J Exp Bot 2015; 66:1687-98; PMID:25740920; http://dx.doi.org/ 10.1093/jxb/erv091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baroux C, Pecinka A, Fuchs J, Schubert I, Grossniklaus U. The triploid endosperm genome of Arabidopsis adopts a peculiar, parental-dosage-dependent chromatin organization. Plant Cell 2007; 19:1782-94; PMID:17557811; http://dx.doi.org/ 10.1105/tpc.106.046235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andrey P, Kieu K, Kress C, Lehmann G, Tirichine L, Liu Z, Biot E, Adenot PG, Hue-Beauvais C, Houba-Herin N, et al.. Statistical analysis of 3D images detects regular spatial distributions of centromeres and chromocenters in animal and plant nuclei. PLoS Comput Biol 2010; 6:e1000853; PMID:20628576; http://dx.doi.org/ 10.1371/journal.pcbi.1000853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baubec T, Pecinka A, Rozhon W, Mittelsten Scheid O. Effective, homogeneous and transient interference with cytosine methylation in plant genomic DNA by zebularine. Plant J 2009; 57:542-54; PMID:18826433; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03699.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu CH, Finke A, Diaz M, Rozhon W, Poppenberger B, Baubec T, Pecinka A. Repair of DNA Damage Induced by the Cytidine Analog Zebularine Requires ATR and ATM in Arabidopsis. Plant Cell 2015; 27:1788-800; PMID:26023162; http://dx.doi.org/ 10.1105/tpc.114.135467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pico S, Ortiz-Marchena MI, Merini W, Calonje M. Deciphering the role of Polycomb Repressive Complex 1 (PRC1) variants in regulating the acquisition of flowering competence in Arabidopsis. Plant Physiol 2015; PMID:25897002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angel A, Song J, Dean C, Howard M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature 2011; 476:105-8; PMID:21785438; http://dx.doi.org/ 10.1038/nature10241 [DOI] [PubMed] [Google Scholar]
- 39.Angel A, Song J, Yang H, Questa JI, Dean C, Howard M. Vernalizing cold is registered digitally at FLC. Proc Natl Acad Sci U S A 2015; 112:4146-51; PMID:25775579; http://dx.doi.org/ 10.1073/pnas.1503100112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berry S, Hartley M, Olsson TS, Dean C, Howard M. Local chromatin environment of a Polycomb target gene instructs its own epigenetic inheritance. Elife 2015; 4; PMID:25955967; http://dx.doi.org/ 10.7554/eLife.07205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grob S, Schmid MW, Luedtke NW, Wicker T, Grossniklaus U. Characterization of chromosomal architecture in Arabidopsis by chromosome conformation capture. Genome Biol 2013; 14:R129; PMID:24267747; http://dx.doi.org/ 10.1186/gb-2013-14-11-r129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang C, Liu C, Roqueiro D, Grimm D, Schwab R, Becker C, Lanz C, Weigel D. Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res 2015; 25:246-56; PMID:25367294; http://dx.doi.org/ 10.1101/gr.170332.113 [DOI] [PMC free article] [PubMed] [Google Scholar]


