Abstract
Exposure to arsenic early in life has been associated with increased risk of several chronic diseases and is believed to alter epigenetic programming in utero. In the present study, we evaluate the epigenome-wide association of arsenic exposure in utero and DNA methylation in placenta (n = 37), umbilical artery (n = 45) and human umbilical vein endothelial cells (HUVEC) (n = 52) in a birth cohort using the Infinium HumanMethylation450 BeadChip array. Unadjusted and cell mixture adjusted associations for each tissue were examined along with enrichment analyses relative to CpG island location and omnibus permutation tests of association among biological pathways. One CpG in artery (cg26587014) and 4 CpGs in placenta (cg12825509; cg20554753; cg23439277; cg21055948) reached a Bonferroni adjusted level of significance. Several CpGs were differentially methylated in artery and placenta when controlling the false discovery rate (q-value<0.05), but none in HUVEC. Enrichment of hypomethylated CpG islands was observed for artery while hypermethylation of open sea regions were present in placenta relative to prenatal arsenic exposure. The melanogenesis pathway was differentially methylated in artery (Max F P < 0.001), placenta (Max F P < 0.001), and HUVEC (Max F P = 0.02). Similarly, the insulin-signaling pathway was differentially methylated in artery (Max F P = 0.02), placenta (Max F P = 0.02), and HUVEC (Max F P = 0.02). Our results show that prenatal arsenic exposure can alter DNA methylation in artery and placenta but not in HUVEC. Further studies are needed to determine if these alterations in DNA methylation mediate the effect of prenatal arsenic exposure and health outcomes later in life.
Keywords: arsenic, DNA methylation, epigenetics, environmental epigenetics, fetal programming, Illumina 450K, in utero exposure
Introduction
Over 200 million individuals worldwide are exposed to elevated levels of inorganic arsenic. This is a public health concern because arsenic is a known human carcinogen and chronic exposure is associated with the development of skin, lung, bladder, kidney, liver, and, potentially, prostate cancer.1 Particularly, early life exposure to arsenic has been associated with the development of many latent health effects including carcinogenesis.2 Human ecological studies from the Antofagasta region of Chile have associated prenatal and early childhood exposure to arsenic from contaminated municipal water with increased risk of lung and bladder cancer later in life.3 Increased mortality from acute myocardial infarction and cancers of the bladder, kidney, lung, and liver have also been reported from this population decades after the exposure declined.4,5
Of public health interest is the ability of early life arsenic exposure, particularly exposures occurring in utero, to increase disease risk and susceptibility to adverse health conditions later in life. For example, animal models support the involvement of transplacental arsenic exposure in the development and progression of atherosclerosis, consistent with human studies linking early life exposure and cardiovascular disease.6,7 Emerging evidence also indicates that exposure to arsenic can disrupt normal immune function and in utero exposure can increase the susceptibility and severity of infections later in life.8-10 Furthermore, exposure to high levels of arsenic in utero is associated with higher risk of respiratory diseases, including bronchiolitis and lung cancer.11,12 Lastly, latent adverse neurological health outcomes have also been documented with maternal exposure to arsenic during pregnancy.13,14
The exact molecular mechanisms of the toxicological effects attributed to arsenic exposure remains elusive and no single mechanism has been identified in the development of arsenic associated diseases and the observed latency of health effects.15 However, the latency of health effects documented in epidemiological studies and animal models along with the observed susceptibility of prenatal exposures are suggestive of an epigenetic mode of action. Fetal programming events involving DNA methylation occur at critical windows of fetal development in a cell-specific manner shown to be sensitive to environmental exposures.16 Experimental evidence from animal models demonstrate that transplacental exposure to arsenic leads to epigenetic alterations, changes in gene expression, and increased incidence of tumors in the offspring.17,18 Therefore, it is postulated that epigenomic regulation including, but not limited to, DNA methylation is a potential mechanism of arsenic induced carcinogenesis and latent disease risk.2,19,20 Other likely interacting mechanisms of early life exposure to arsenic and latent disease risk include the development of cancer stem cells and perturbations of immune function.2
Several human studies have evaluated the impact of prenatal arsenic exposure on the cord blood and whole blood epigenome.1 Among these epidemiological studies evaluating cord blood or whole blood DNA methylation, no common loci have been identified as differentially methylated across studies.21-26 However, significant DNA methylation disruption of unique loci along with enrichment of key regulatory CpG regions has been documented across different study populations.22-28 Besides studies that examined cord and whole blood epigenome, only 2 studies to date have evaluated the association between arsenic exposure and CpG methylation of target tissue by evaluating DNA methylation in urothelial carcinoma samples and CpG methylation of exfoliated urothelial cells, respectively.29,30 These studies found differentially methylated loci associated with arsenic exposure in key regulatory genes potentially involved in the development of arsenic-induced urothelial carcinoma.
Epigenetic reprogramming during fetal development resulting from transplacental exposure is one of the main hypothesized mechanisms of arsenic's associated disease.2 To further our understanding of how prenatal arsenic exposure could alter epigenetic programming, it is important to evaluate its effect on different tissues with diverse cellular compositions. Evaluating if exposure to arsenic in utero alters DNA methylation of different tissues could yield insights into the etiology of toxicant-mediated disease and epigenetic modifications of relevant tissues with specific biological functions. Subsequently, we examined the association between maternal drinking water arsenic as a proxy of transplacental exposure during fetal development and the epigenome of placenta, umbilical artery and Human Umbilical Vein Endothelial Cells (HUVEC) from a birth cohort conducted in arsenic affected regions of Bangladesh.
Results
The sample size varied by tissue type with a maximum of 52 samples present for HUVEC followed by 45 samples in umbilical artery and 37 placenta samples. Arsenic concentration in maternal drinking water at study enrollment ranged from below the detection limit of <1 µg/L to 510 µg/L with a mean exposure concentration of 63.7 µg/L. Selected sample characteristics are shown in Table 1.
Table 1.
Sample characteristics for the 52 mother-infant pairs eligible for the analysis.
| Sample characteristics | Mean±SD | Range |
|---|---|---|
| Drinking water arsenic at recruitment (µg/L) | 63.7±116 .5 | 1 – 510 |
| Gestational age at recruitment (weeks) | 12.2±2 .5 | 6 – 16 |
| Gestational age at delivery (weeks) | 37.6±2 .1 | 33 – 41 |
| Birth weight (grams) | 2923±372 | 2080 – 4050 |
| Gender | N (%) | |
| Male | 33 (63.5 %) | |
| Female | 19 (36.5%) | |
| Number of samples available by tissue | n | CpG loci analyzed |
| HUVEC | 52 | 347,650 |
| Artery | 45 | 374,320 |
| Placenta | 37 | 365,994 |
Arterial tissue
Locus-by-Locus Analysis
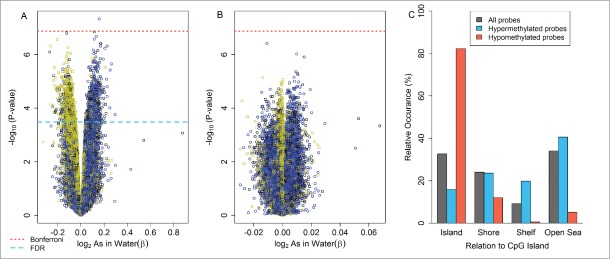
In the analysis that was unadjusted for cellular composition, one CpG loci (cg26587014) located in chromosome 19 and not annotated to any gene was differentially methylated in arterial tissue in relation to arsenic exposure using a Bonferroni threshold for statistical significance (P < 1.33 × 10−7). Controlling for the false discovery rate at 5% (q-value < 0.05) revealed 2,105 CpGs that were differentially methylated relative to log2-transformed maternal drinking water arsenic. However, after adjusting for cellular composition using the Houseman reference-free method, no loci reached a Bonferroni corrected level of significance or a q-value < 0 .05. Unadjusted and adjusted results are shown in Figure 1A–B, respectively. The top 100 differentially methylated loci ranked on lowest P-value are summarized in Table S1 and S2 for unadjusted and cell mixture adjusted analyses, respectively. In unadjusted analyses, differentially methylated loci with a q-value < 0.05 were disproportionately located in CpG islands (54%) compared to the distribution of CpG island probes in the rest of array (33%) (P < 1 × 10−4; Fig. S1A). The majority (83%) of unadjusted hypomethylated loci with a q-value < 0.05 were located in CpG islands (Fig. 1C). After adjusting for cellular heterogeneity, a similar enrichment of hypomethylated loci in CpG islands was observed among top loci having a nominal P-value < 1 x10−4, (Fig. S1B).
Figure 1.
Locus-by-locus epigenome-wide analysis for umbilical artery: volcano plots for the association between log2-transformed maternal drinking water arsenic: (A) unadjusted for cellular heterogeneity and (B) adjusting for cellular heterogeneity using the Houseman reference-free method. (C) Distribution of differentially methylated loci (q-value < 0.05) relative to CpG islands for the unadjusted cell mixture analysis. Volcano plot legend; Yellow: CpG Islands, Black: CpG shore, Blue: CpG shelfs and open sea. Square: Infinium I, Circle: Infinium II †Unadjusted analysis (A) performed on the logit transformed β methylation values (M-values) and adjusted analysis (B) performed on the β sclae methylation levels.
Biological Pathway Analysis
Omnibus permutation based tests revealed significant associations between in utero exposure to arsenic and epigenetic disruption of KEGG biological pathways in arterial tissue (Mean F-statistics P=0.009 and maximum F-statistic P=0.006). Pathways that were observed to have the strongest association based on the lowest mean F-static level of significance (P=0.004) were: maturity onset of diabetes of the young (hsa04950), primary immunodeficiency (hsa05340), ABC transporters (hsa02010), allograft rejection (hsa05330), and vibrio cholerae infection (hsa05110). Differentially methylated pathways observed to have a strong association using a maximum F-statistic level of significance (P < 0.001) included the Hedgehog signaling pathway (hsa04340), Melanogenesis (hsa04916), Wnt signaling pathway (hsa04310), Basal cell carcinoma (hsa05217), DNA replication (hsa03030), and the p53-signaling pathway (hsa04115). The summary for all associations between maternal drinking of arsenic water and epigenetic disruption of KEGG biological pathways are shown in Table S7.
Placenta tissue
Locus-by-Locus Analysis
In the analyses that were unadjusted for cellular composition, no single CpG loci reached Bonferroni adjusted significance in placenta (P < 1 .37×10−7). However, 2 CpG loci (cg26390526; cg03857453) annotated to the Epidermal Filaggrin gene (FLG) and the nuclear receptor subfamily 3, group C, member 1 glucocorticoid receptor gene (NR3C1) were hypermethylated relative to maternal drinking water arsenic after controlling for the false-discovery rate (q-value < 0.05). In analyses that adjusted for cell mixture in the placenta, 4 CpGs reached Bonferroni adjusted significance: cg12825509 (TRA2B gene), cg20554753, cg23439277 (PLCE1 gene) and cg21055948 (CD36 gene). Moreover, analyses adjusted for cellular heterogeneity revealed 518 CpG loci that were differentially methylated after controlling for the false discovery rate (q-value < 0.05). Unadjusted and cell mixture adjusted results are shown in Figure 2A–B, respectively. The top 100 differentially methylated loci ranked on lowest P-value are summarized in Tables S3 and S4 for unadjusted and cell mixture adjusted results, respectively. For the top unadjusted differentially methylated loci with a nominal P < 1 x10−4 a disproportionate amount of CpGs were located within open sea regions of CpG islands (76%) compared with the distribution of open sea loci in the rest of array (33%, P < 1 x10−4, Fig. S2A). Among these loci, the great majority of hypermethylated CpGs were located within open sea regions (89%) relative to CpG islands, Figure 2C. For the cell mixture adjusted analyses a similar enrichment of hypermethylated loci in open sea regions was observed for loci with a q-value < 0.05, Figure S2B.
Figure 2.
Locus-by-locus epigenome-wide analysis for placenta: volcano plots for the association between log2-transformed maternal drinking water arsenic: (A) unadjusted for cellular heterogeneity and (B) adjusting for cellular heterogeneity using the Houseman reference-free method. (C) Distribution of differentially methylated loci (nominal P<1 x10−4) relative to CpG islands for the unadjusted cell mixture analysis. Volcano plot legend; Yellow: CpG Islands, Black: CpG shore, Blue: CpG shelfs and open sea. Square: Infinium I, Circle: Infinium II †Unadjusted analysis (A) performed on the logit transformed β methylation values (M-values) and adjusted analysis (B) performed on the β sclae methylation levels.
Biological Pathway Analysis
Omnibus permutation based tests indicated that exposure to arsenic in utero disrupts methylation of a small number of CpGs within KEGG biological pathways in the placenta tissue (Omnibus maximum F-statistic P=0.004). However, a marginal association among KEGG biological pathways and arsenic exposure was observed using an omnibus Mean F-statistic test for association (P=0.108). KEGG biological pathways that were differentially methylated in relationship to arsenic exposure with a maximum F-statistic P < 1 x10−3 included: Melanogenesis (hsa04916), Neuroactive ligand-receptor interaction (hsa04080), Calcium signaling pathway (hsa04020), GnRH signaling pathway (hsa04912), Dilated cardiomyopathy (hsa05414), Gap junction (hsa04540), Vasopressin-regulated water reabsorption (hsa04962), Vascular smooth muscle contraction (hsa04270), Oocyte meiosis (hsa04114), Vibrio cholerae infection (hsa05110), Progesterone-mediated oocyte maturation (hsa04914), and the Peroxisome pathway (hsa04146). Several other pathways were significantly associated with arsenic exposure using a maximum F-statistic P < 0.05 and summarized in Table S7.
Umbilical vein endothelial cells (HUVEC)
Locus-by-Locus Analysis
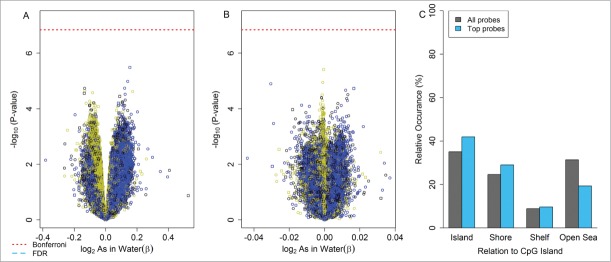
In both unadjusted and cell mixture adjusted analyses no single CpG loci was associated with arsenic exposure at a Bonferroni corrected level of significance (P < 1 .44×10−7) or a q-value < 0.05 after controlling for the false discovery rate, Figure 3A–B. Among the top 31 CpG loci with a nominal P < 1 x10−4 no significant differences were present for the occurrence of top loci relative to CpG island location compared to the rest of the array for unadjusted analyses, Figure 3C. The top 100 differentially methylated loci ranked by lowest P-value are summarized in Tables S5 and S6 for unadjusted and cell mixture adjusted results, respectively.
Figure 3.
Locus-by-locus epigenome-wide analysis for HUVEC: volcano plots for the association between log2-transformed maternal drinking water arsenic: (A) unadjusted for cellular heterogeneity and (B) adjusting for cellular heterogeneity using the Houseman reference-free method. (C) Distribution of differentially methylated loci (nominal P-value<1 x10−4) relative to CpG islands for the unadjusted cell mixture analysis.
Volcano plot legend; Yellow: CpG Islands, Black: CpG shore, Blue: CpG shelfs and open sea. Square: Infinium I, Circle: Infinium II
†Unadjusted analysis (A) performed on the logit transformed β methylation values (M-values) and adjusted analysis (B) performed on the β scale methylation levels.
Biological Pathway Analysis
Omnibus permutation tests for association among KEGG biological pathways indicated that arsenic exposure was not significantly associated with a large number of changes in DNA methylation across pathways in HUVEC (Mean F-statistic P =0.129) and the presence of a small number of strong associations was borderline significant (Max F-statistic P=0.06). Few individual biological pathways reached statistical significance using a maximum F-statistic level of significance. The top differentially methylated biological pathways (maximum F-statistic P=0.002) in HUVEC included: Melanogenesis (hsa04916), Wnt signaling pathway (hsa04310), Basal cell carcinoma (hsa05217) and the Insulin signaling pathway (hsa04910), all KEGG biological pathway based associations are summarized in Table S7.
The overlap among CpGs within each tissue for unadjusted and cell mixture adjusted analyses using the top 100 differentially methylated CpGs was 26 loci in artery, 21 loci in placenta and 33 loci for HUVEC, Figure S3. Among the top 100 differentially methylated loci, only one CpG (cg21002651) located within the body of the CASP1 gene was differentially methylated across 2 tissues in unadjusted analyses. This locus was hypomethylated in placenta (β=-0.20, P=5.73x10−6) but hypermethylated in HUVEC (β=0.20, P=1.29x10−4) in relationship to maternal drinking water arsenic. No other CpGs overlapped in unadjusted or adjusted analyses.
Discussion
Our study provides evidence that in utero exposure to arsenic can disrupt DNA methylation of artery and placenta tissues but the association with umbilical vein endothelial vein cells was marginal. However, the association of prenatal arsenic exposure on the epigenome on artery and placenta depended on the cell mixture adjustment. For instance, the association in artery was attenuated after controlling for cellular heterogeneity but strengthened in placenta. In utero exposure to arsenic was also associated with DNA methylation levels of key biological pathways across tissues providing new insights into the potential etiology of arsenic-mediated diseases with a plausible epigenetic reprogramming component.
In normal tissue, the majority of CpG islands remain unmethylated and methylation of CpG islands located within promoter regions of genes is usually restricted to genes at which there is long-term stabilization of repressed states such as in gene silencing of imprinted genes.31 However, CpG island methylation is not deterministic of gene expression and further studies are needed to determine if alterations in DNA methylation are associated with biological effects. We observed an enrichment of hypomethylated loci in CpG islands of arterial tissue relative to prenatal arsenic exposure. This is of particular interest because both animal and human studies have demonstrated that DNA hypomethylation occurs in atherosclerotic lesions and that hypomethylation of CpG islands is observed broadly in human atherosclerotic arteries and in arterial disease pathogenesis.32-34 Previous studies of arsenic exposure have characterized the hypermethylation of the promoter region of the p53 gene a mechanisms hypothesized to contribute to the carcinogenesis of arsenical compounds.35,36 Consistent with these reports, our gene set analysis shows that CpG methylation within the p53 signaling pathway is associated with arsenic exposure during pregnancy suggesting that artery might be a target tissue for the epigenetic toxicity of arsenic, and potentially involved in carcinogenesis. However, this hypothesis needs to be evaluated.
The placenta is an important regulator of fetal development and intrauterine growth that plays a crucial role mediating the maternal and fetal environment. Furthermore, the placenta is a unique epigenetic target organ as the majority of imprinted genes in animal models are both expressed and imprinted in the placenta and hypothesized to contribute to fetal neurodevelopment.37,38 In unadjusted analyses a CpG located in the body of the glucocorticoid receptor gene (NR3C1) was significantly hypermethylated in the placenta relative to prenatal arsenic exposure. Previous studies have shown that hypermethylation of the NR3C1 gene influences cortisol response, infant behavior and self-regulation.39,40 Interestingly a recent experimental study demonstrated that exposure to arsenic in utero lowers the activity of the glucocorticoid receptor pathway and these changes were maintained into adolescence of the mouse model.41 However, further studies are needed to confirm whether these biological findings influence behavioral outcomes. The placenta has also been characterized as one of the hypomethylated tissues as LINE-1 elements have lower levels of methylation when compared to other tissues. Furthermore, it has been shown that normal human placenta contains partially methylated domains (37%) with the ability to suppress genes and impact tissue-specific functions independent of the tissue of origin.42 The observed hypermethylation of open sea regions relative to CpG island location could have implications for normal methylation of LINE-1 elements and partially methylated domains, potentially affecting normal biological function and development of the placenta.
A few KEGG biological pathways were differentially methylated in relation to maternal drinking water arsenic in all 3 tissues. Namely, DNA methylation of the melanogenesis pathway was strongly associated with exposure to arsenic in artery, placenta, and HUVEC. An early clinical symptom of arsenicosis include the appearance of hyperpigmentation changes of the skin in the trunk, neck and chest regions of the body eventually progressing to the palmar and plantar regions and eventually leading to hyperkeratosis.43 Consistent with the differential methylation of this pathway in our data, arsenic-associated alterations in DNA methylation of leukocytes has been previously associated with increased risk of developing skin lesions.44 Lastly, the insulin-signaling pathway was observed to be differentially methylated across all 3 tissues with respect to arsenic exposure. Exposure to arsenic has been consistently associated with Type 2 diabetes and insulin resistance in both animal models and epidemiological studies.45 Previous studies have documented the epigenetic disruption of several genes involved in the development of diabetes and insulin resistance for individuals chronically exposed to arsenic.46 Although epigenetic perturbations were evaluated among these biological pathways relative to arsenic exposure, future studies need to test if these changes are associated with changes in gene expression, metabolism, and, ultimately, pathological phenotypes. It is also important to note that the permutation test used in this analysis evaluates the DNA methylation disruption of the biological pathways at a global level and not on a gene-by-gene basis. Therefore, it is not possible to determine if individual genes are differentially methylated with regards to arsenic exposure.
It is crucial to highlight that HUVEC is a homogenous tissue in terms of cellular composition and was not significantly disrupted in the locus-by-locus analysis and marginally associated among some biological pathways. However, artery and placenta, both representing a diverse mixture of cell types, were observed to be differentially methylated relative to prenatal arsenic exposure. DNA methylation is cell specific playing a key role in tissue differentiation and lineage commitment making this process particularly vulnerable to environmental stimuli and exposures during fetal development. The placenta represents the most diverse tissue composed of fetal vascular cells, mesenchymal cells, cytotrophoblast, and syncytiotrophoblast that originate from the trophoblast.47 Furthermore, it has also been observed that the human placenta contains both haematopoietic stem cells and mesenchymal stem cells.48 All tissues derived from the fetus are an extension of the mesoderm that differentiates during embryonic development to form the umbilical cord and placenta. Therefore, we further hypothesize that exposure to arsenic during fetal development could affect cellular differentiation and lineage commitment for placenta and artery but not HUVEC as this is a cellular homogenous tissue. Epidemiologic studies often rely on preserved samples and have limited fresh tissue availability making the sorting or isolation of target cell types not feasible. Therefore, future experimental studies should evaluate the development of cancer stem cells (CSCs) and alterations to the immune function as factors or intermediary mechanisms of the observed epigenetic perturbations, as others have also suggested.2 Moreover, the interaction between prenatal arsenic exposure and other transplacental contaminants should also be considered, as prenatal exposure to arsenic has been previously shown to interact with other prenatal exposures such as mercury.49
One of the major strengths for the present study is the epigenome-wide analysis of 3 different tissues collected from the same maternal-infant pairs yielding insights for the potential biological impact of arsenic exposure during fetal development. Also, the prospective design of this birth cohort along with the exposure assessment early during pregnancy are important qualities that strengthens the temporality of the epigenetic perturbations reported. Although the present study relies on a single water sample during early pregnancy and exposure misclassification cannot be ruled out, previous studies in rural Bangladesh have demonstrated that drinking water arsenic exposure is relatively constant and correlated with biomarkers of internal doses, such as urine and toenails.50,51 and that arsenic readily crosses the placenta.52 Additionally, the availability of umbilical samples at birth provides one of the few opportunities for examining epigenetic programming in cardiovascular target tissue in a non-invasive and feasible manner. There are a number of important limitations to our current study including the relatively small sample size and the lack of validation using a complimentary methylation method due to sample availability. The lack of reference methylomes for placenta, artery, and HUVEC also raise an important challenge when interpreting the observed epigenetic perturbations in tissues that might represent a mixture of cell types such as artery or placenta. However, we implemented a complementary bioinformatics method to adjust for cellular heterogeneity to identify potential perturbations in loci hypothesized to be associated with methylation levels independent of cellular heterogeneity. Furthermore, unique tissue samples were analyzed in separate plates raising the possibility that differences across tissue could be potentially attributed to technical plate effects. Finally, gene expression was not measured and the observed changes in DNA methylation need to be further confirmed and evaluated. Particularly significant association between DNA methylation among KEGG biological pathways might not result in functional gene expression or proteomic alterations within pathways.
In conclusion, we show that prenatal arsenic exposure is associated with altered DNA methylation of umbilical artery and placenta tissue but evidence of an association for HUVEC is limited. Furthermore, we present evidence of DNA methylation disruption of key biological pathways across different tissues holding the potential to mediate arsenic-associated diseases previously described from exposures in utero.
Materials & Methods
Study population
This pilot study was nested within an established birth cohort recruited in Bangladesh (2007–2011) and designed to characterize the potential epigenetic disruption associated with arsenic exposure during pregnancy in different tissues collected at birth. A more detailed explanation of the full birth cohort has been published previously.22 Briefly, pregnant women with < 16 weeks of gestation confirmed by ultrasound were enrolled in a prospective reproductive birth cohort in Bangladesh. Trained health care workers at community health clinics in Sirajdikhan and Birahimpur recruited pregnant women 18 y of age or older that used a tube-well as their primary drinking water source, planned to live at their current residency during the duration of the pregnancy and received prenatal health care at Dhaka Community Hospital (DCH) or affiliated community clinic. Study participants agreed to deliver at DCH or at home with a DCH trained midwife. Informed consent was obtained from all participants prior to enrollment. All participants were provided with prenatal care and prenatal vitamins offered by DCH. This study was approved by the Human Research Committees at the Harvard School of Public Health, Oregon State University and Dhaka Community Hospital Trust.
Three distinct tissues were collected at the time of delivery including: artery from the umbilical cord, placenta, and endothelial cells isolated from the umbilical vein. Since the goal of this pilot study was to examine the potential exposure-response relationship between arsenic and DNA methylation, specimens were selected based on maternal drinking water arsenic concentrations at study enrollment to cover a wide range of exposures (< 1 -510 µg/L). A total of 37 placenta samples, 45 artery samples, and 52 HUVEC samples were included in the final analysis.
Drinking water arsenic
Water samples were collected from the tube-well identified by participants as their main source of drinking water at the time of their enrollment into the study as previously described.22 Briefly, water samples were collected in a 50-mL polypropylene tubes (BD Falcon, BD Bioscience, Bedford, MA), preserved with Reagent Grade nitric acid (Merck, Germany) to a pH < 2 and stored at room temperature. Arsenic concentrations were measured by inductively coupled plasma-mass spectrometry (ICP-MS) using the US EPA method 200.8 to determined metals in water (Environmental Laboratory Services, North Syracuse, New York).53 Average percent recovery for Arsenic from plasmaCal multi-element QC standard #1 solution (SCP Science) was 102 ± 7%. The limit of detection (LOD) for arsenic in drinking water was 1 µg/L.
Tissue collection: umbilical artery, placenta & HUVEC
Trained medical workers present at delivery collected a sample of the umbilical cord and placenta immediately after the delivery was completed. Using sterile techniques, approximately 5–7 cm of umbilical vein was dissected out of fresh umbilical cord and rinsed with phosphate buffered saline solution to remove external contamination. The vein lumen was then bisected and the interior cavity was flushed with approximately 100 mL of phosphate buffered solution to remove blood. The interior lumen wall was gently rubbed using a sterile cytology brush to collected endothelial cells. The cytology brush was then vortexed in 1 mL of cell lysis solution (Qiagen) to transfer the cells. The cell lysis solution was then stored at 4°C. Samples were shipped to Harvard School of Public Health where the DNA was extracted using DNeasy Blood & Tissue Kit (Qiagen) following manufacturer's instructions.
Approximately 1 cm of umbilical cord artery was dissected out of fresh umbilical cord, the exterior of the artery was scraped to remove Wharton's Jelly, and rinsed with phosphate buffered saline solution to remove blood. The arterial cross section was placed in 2 mL of RNAse later and stored at −20°C. Samples were shipped to Harvard School of Public Health on dry ice. The artery sample was then minced using a sterile scalpel and added to Maxwell Cell DNA Purification kits (Promega) with an additional 20 µL of Proteinase K (Qiagen). Samples were allowed to sit for 30 mins before being extracted using the Maxwell 16 Research instrument following manufacturer's instructions.
For placenta samples, a one-centimeter tissue plug was excised from fresh placenta. The tissue plug was placed into a sterile vial and covered with Tissue-Tek O.C.T. gel (Electron Microscopy Sciences) and frozen at −20°C. Samples were then shipped to Harvard School of Public Health on dry ice. Next, approximately 10 g of placenta tissue was removed from the plug and minced using a sterile scalpel and added to Maxwell Cell DNA Purification kits (Promega) with an additional 20 µL of Proteinase K (Qiagen). Samples were allowed to sit for 30 min before being extracted using the Maxwell 16 Research instrument following manufacturer's instructions.
DNA methylation assessment and quality control
DNA was shipped to the University of Minnesota's Biomedical Genomic Center that quantified DNA methylation using the Illumina Infinium HumanMethylation450 BeadChip (Illumina, San Diego, CA) following standard manufacturer's protocols. The HumanMethylation450 BeadChip measures DNA methylation at > 485,000 CpG sites at single nucleotide resolution, covering 99% of the RefSeq genes.
Tissues were analyzed in separate plates and randomly allocated to different chips. Data were obtained and processed from raw methylation image files and normalized using internal control probes via the functional normalization method with 2 principal components to account for technical variation between samples using the minfi package of R.54 DNA methylation was estimated at each CpG as the fraction of DNA molecules whose target CpG loci is methylated and referred to as β-values. Measurements at CpG loci on X and Y chromosomes were excluded from the analysis to avoid gender-specific methylation bias. Previously identified non-specific and cross-reactive probes within the array along with polymorphic CpG loci ( > 5% of the minor allele frequency) were removed for the analysis.55 Furthermore, a detection P-value was computed for all CpGs and probes with non-significant detection (P>0 .01) in greater than 10% of the samples were removed from the analysis. After quality control, the total numbers of autosomal CpGs left in the analysis were 374,320 loci for artery, 365,994 loci for placenta, and 347,650 loci in HUVEC samples. Finally, a β-mixture quantile intra sample normalization procedure (BMIQ) was further applied to the data to reduce the potential bias that can arise from type 2 probes as previously described.56 Strip plots and signal intensities of control probes were visually examined for bisulfite conversion, probe hybridization, and single base extension. Density plots for the β-values were examined for all samples at each normalization step described above.
Statistical analysis
Unadjusted and Cell-adjusted Locus-by-Locus Analysis
We first aimed to identify differentially methylated CpG loci in relationship to prenatal arsenic exposure from maternal drinking water. Maternal arsenic concentration in water was right skewed and subsequently log2-tranformed. In order to evaluate linear associations between prenatal exposure to arsenic and differentially methylated CpG loci, β-values were logit-transformed to M-values previously described to be more appropriate for differential analysis of DNA methylation.57 In the locus-by-locus approach, 2 different but complementary methodologies were implemented. First, the linear association between individual CpG methylation on the M-value scale and log2-transformed arsenic was evaluated adjusting for infant sex using the limma function found in the minfi package of R. Second, due to the lack of reference methylomes of isolated cell types in placenta, artery or HUVEC, a novel reference-free method of adjusting for cellular heterogeneity was implemented using the RefFreeEWAS package of R. The reference-free method is an extension of the original Houseman method that utilizes a deconvolution approach similar to surrogate variable analysis (SVA) that is data driven to identify latent variables or dimensions as surrogates of cellular composition.58 Using this method, the sex-adjusted linear association between individual CpG methylation on the β-value scale and log2-transformed maternal drinking water arsenic was evaluated using 1000 bootstrap samples for estimating the standard errors of association in placenta, umbilical artery and HUVEC. Results from the unadjusted limma models and the reference-free cell mixture adjusted analyses were compared within tissues and across tissues. Enrichment analyses for the distribution of CpGs relative to CpG island location of the top differentially methylated loci based on a q-value < 0.05 or a nominal P < 1 x10−4, were compared to the distribution of probes on the rest of the array.
Biological Pathway Analysis
Omnibus permutation based tests and P-values were obtained by mapping subsets of CpGs to their associated genes in specific KEGG biological pathways. Gene sets were compiled from the Kyoto Encyclopedia of Genes and Genomes (KEGG) corresponding to specific biological pathways using the Entrez IDs matched to KEGG biological pathways using the Bioconductor library org.Hs.eg.db. The permutation distribution was obtained from unadjusted cell mixture models by permuting the exposure with respect to measured DNA methylation over subgroups of CpGs defined by biological pathways (1000 permutations). Unadjusted cell mixture methylation analyses were used for the KEGG pathways as the reference-free Houseman method is unable to accommodate for the permutation test of CpGs across individual biological pathways. Pathway based associations of DNA methylation with prenatal arsenic exposure as a continuous variable were summarized using a maximum nominal F-statistics P-value (akin to a minimum P-value) and an average nominal F-statistic P-value. The maximum and minimum F-statistic P-values are better suited for detecting a small number of strong associations and a large number of more variable associations, respectively, as previously described.22 This approach allowed us to test for significant DNA methylation disruption across single KEGG pathways and not over individual genes.
All statistical analyses were performed using the R statistical package version 3.2.0 (http://www.R-project.org).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the US National Institute of Environmental Health Sciences (NIEHS) grants R01 ES015533, K01 ES017800, R01 ES016454, P30 ES000210, and P30 ES000002.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- 1.Argos M. Arsenic Exposure and Epigenetic Alterations: Recent Findings Based on the Illumina 450K DNA Methylation Array. Curr Environ Health Rep 2015; 2:137-44; PMID:26231363; http://dx.doi.org/ 10.1007/s40572-015-0052-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey KA SA, Tokar EJ, Graziano JH, Kim KW, Navasumrit P, Ruchirawat M, Thiantanawat A, Suk WA, Fry RC. Mechanisms Underlying Latent Disease Risk Associated with Early-Life Arsenic Exposure: Current Research Trends and Scientific Gaps. Environ Health Perspect 2015; PMID:26115410; http://dx.doi.org/ 10.1289/ehp.1409360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinmaus C, Ferreccio C, Acevedo J, Yuan Y, Liaw J, Durán V, Cuevas S, García J, Meza R, Valdés R. Increased lung and bladder cancer incidence in adults after in utero and early-life arsenic exposure. Cancer Epidemiol Biomarkers Prevention 2014; 23:1529-38; http://dx.doi.org/ 10.1158/1055-9965.EPI-14-0059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan Y, Marshall G, Ferreccio C, Steinmaus C, Selvin S, Liaw J, Bates MN, Smith AH. Acute myocardial infarction mortality in comparison with lung and bladder cancer mortality in arsenic-exposed region II of Chile from 1950 to 2000. American J Epidemiol 2007; 166:1381-91; PMID:17875584; http://dx.doi.org/ 10.1093/aje/kwm238 [DOI] [PubMed] [Google Scholar]
- 5.Yuan Y, Marshall G, Ferreccio C, Steinmaus C, Liaw J, Bates M, Smith AH. Kidney cancer mortality: fifty-year latency patterns related to arsenic exposure. Epidemiology 2010; 21:103-8; PMID:20010213; http://dx.doi.org/ 10.1097/EDE.0b013e3181c21e46 [DOI] [PubMed] [Google Scholar]
- 6.Farzan SF, Karagas MR, Chen Y. In utero and early life arsenic exposure in relation to long-term health and disease. Toxicol Appl Pharmacol 2013; 272:384-90; PMID:23859881; http://dx.doi.org/ 10.1016/j.taap.2013.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srivastava S, D'Souza SE, Sen U, States JC. In utero arsenic exposure induces early onset of atherosclerosis in ApoE−/− mice. Reprod Toxicol 2007; 23:449-56; PMID:17317095; http://dx.doi.org/ 10.1016/j.reprotox.2007.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardenas A, Smit E, Houseman EA, Kerkvliet NI, Bethel JW, Kile ML. Arsenic Exposure and Prevalence of the Varicella Zoster Virus in the United States: NHANES (2003-2004 and 2009-2010). Environ Health Perspect 2015; 123:590-6; PMID:25636148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farzan SF, Korrick S, Li Z, Enelow R, Gandolfi AJ, Madan J, Nadeau K, Karagas MR. In utero arsenic exposure and infant infection in a United States cohort: A prospective study. Environ Res 2013; 126:24-30; PMID:23769261; http://dx.doi.org/ 10.1016/j.envres.2013.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahman A, Vahter M, Ekstrom E-C, Persson L-Å. Arsenic exposure in pregnancy increases the risk of lower respiratory tract infection and diarrhea during infancy in Bangladesh. Environ Health Perspectives 2010; 119:719-24; PMID:21147604; http://dx.doi.org/ 10.1289/ehp.1002265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dauphiné DC, Ferreccio C, Guntur S, Yuan Y, Hammond SK, Balmes J, Smith AH, Steinmaus C. Lung function in adults following in utero and childhood exposure to arsenic in drinking water: preliminary findings. Int Arch Occupational Environ Health 2011; 84:591-600; http://dx.doi.org/ 10.1007/s00420-010-0591-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith AH, Marshall G, Yuan Y, Ferreccio C, Liaw J, von Ehrenstein O, Steinmaus C, Bates MN, Selvin S. Increased mortality from lung cancer and bronchiectasis in young adults after exposure to arsenic in utero and in early childhood. Environ Health Perspectives 2006:1293-6; PMID:16882542; http://dx.doi.org/ 10.1289/ehp.8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamadani J, Tofail F, Nermell B, Gardner R, Shiraji S, Bottai M, Arifeen S, Huda SN, Vahter M. Critical windows of exposure for arsenic-associated impairment of cognitive function in pre-school girls and boys: a population-based cohort study. Int J Epidemiol 2011; 40:1593-604; PMID:22158669; http://dx.doi.org/ 10.1093/ije/dyr176 [DOI] [PubMed] [Google Scholar]
- 14.Tanaka H, Tsukuma H, Oshima A. Long-term prospective study of 6104 survivors of arsenic poisoning during infancy due to contaminated milk powder in 1955. J Epidemiol 2010; 20:439; http://dx.doi.org/ 10.2188/jea.JE20090131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailey KA, Fry RC. Arsenic-associated changes to the epigenome: what are the functional consequences? Curr Environ Health Rep 2014; 1:22-34; PMID:24860721; http://dx.doi.org/ 10.1007/s40572-013-0002-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsit CJ. Influence of environmental exposure on human epigenetic regulation. J Exp Biol 2015; 218:71-9; PMID:25568453; http://dx.doi.org/ 10.1242/jeb.106971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waalkes MP, Liu J, Diwan BA. Transplacental arsenic carcinogenesis in mice. Toxicol Appl Pharmacol 2007; 222:271-80; PMID:17306315; http://dx.doi.org/ 10.1016/j.taap.2006.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waalkes MP, Qu W, Tokar EJ, Kissling GE, Dixon D. Lung tumors in mice induced by “whole-life” inorganic arsenic exposure at human-relevant doses. Arch Toxicol 2014; 88:1619-29; PMID:25005685; http://dx.doi.org/ 10.1007/s00204-014-1305-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie Y, Liu J, Benbrahim-Tallaa L, Ward JM, Logsdon D, Diwan BA, Waalkes MP. Aberrant DNA methylation and gene expression in livers of newborn mice transplacentally exposed to a hepatocarcinogenic dose of inorganic arsenic. Toxicology 2007; 236:7-15; PMID:17451858; http://dx.doi.org/ 10.1016/j.tox.2007.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reichard JF, Puga A. Effects of arsenic exposure on DNA methylation and epigenetic gene regulation. Epigenomics 2010; 2:87-104; PMID:20514360; http://dx.doi.org/ 10.2217/epi.09.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koestler DC, Avissar-Whiting M, Houseman EA, Karagas MR, Marsit CJ. Differential DNA methylation in umbilical cord blood of infants exposed to low levels of arsenic in utero. Environ Health Perspectives 2013; 121:971-7; http://dx.doi.org/ 10.1289/ehp.1205925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kile ML, Houseman EA, Baccarelli AA, Quamruzzaman Q, Rahman M, Mostofa G, Cardenas A, Wright RO, Christiani DC. Effect of prenatal arsenic exposure on DNA methylation and leukocyte subpopulations in cord blood. Epigenetics 2014; 9:774-82; PMID:24525453; http://dx.doi.org/ 10.4161/epi.28153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rojas D, Rager JE, Smeester L, Bailey KA, Drobná Z, Rubio-Andrade M, Stýblo M, García-Vargas G, Fry RC. Prenatal arsenic exposure and the epigenome: identifying sites of 5-methylcytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes. Toxicol Sci 2015; 143:97-106; PMID:25304211; http://dx.doi.org/ 10.1093/toxsci/kfu210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broberg K, Ahmed S, Engström K, Hossain M, Jurkovic Mlakar S, Bottai M, Grandér M, Raqib R, Vahter M. Arsenic exposure in early pregnancy alters genome-wide DNA methylation in cord blood, particularly in boys. J Dev Origins Health Dis 2014; 5:288-98; PMID:24965135; http://dx.doi.org/ 10.1017/S2040174414000221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Zheng Y, Zhang W, Zhang X, LIoyd-Jones DM, Baccarelli AA, Ning H, Fornage M, He K, Liu K. Blood methylomics in response to arsenic exposure in a low-exposed US population. J Exposure Sci Environ Epidemiol 2014; 24:145-9; PMID:24368509; http://dx.doi.org/ 10.1038/jes.2013.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Argos M, Chen L, Jasmine F, Tong L, Pierce BL, Roy S, Paul-Brutus R, Gamble MV, Harper KN, Parvez F. Gene-specific differential DNA methylation and chronic arsenic exposure in an epigenome-wide association study of adults in Bangladesh. Environ Health Perspect 2015; 123:64-71; PMID:25325195; http://dx.doi.org/ 10.1289/ehp.123-A64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koestler DC, Avissar-Whiting M, Houseman EA, Karagas MR, Marsit CJ. Differential DNA Methylation in Umbilical Cord Blood of Infants Exposed to Low Levels of Arsenic in Utero. Environ Health Perspectives 2013; 121:971; PMID:23757598; http://dx.doi.org/ 10.1289/ehp.1205925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seow WJ, Kile ML, Baccarelli AA, Pan WC, Byun HM, Mostofa G, Quamruzzaman Q, Rahman M, Lin X, Christiani DC. Epigenome‐wide DNA methylation changes with development of arsenic‐induced skin lesions in Bangladesh: A case–control follow‐up study. Environ Mol Mutagenesis 2014; 55:449-56; PMID:24677489; http://dx.doi.org/ 10.1002/em.21860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang T-Y, Hsu L-I, Chiu AW, Pu Y-S, Wang S-H, Liao Y-T, Wu M-M, Wang Y-H, Chang C-H, Lee T-C. Comparison of genome-wide DNA methylation in urothelial carcinomas of patients with and without arsenic exposure. Environ Res 2014; 128:57-63; PMID:24268366; http://dx.doi.org/ 10.1016/j.envres.2013.10.006 [DOI] [PubMed] [Google Scholar]
- 30.Rager JE, Tilley SK, Tulenko SE, Smeester L, Ray PD, Yosim A, Currier JM, Ishida MC, González-Horta MdC, Sánchez-Ramírez B. Identification of Novel Gene Targets and Putative Regulators of Arsenic-Associated DNA Methylation in Human Urothelial Cells and Bladder Cancer. Chem Res Toxicol 2015; 28:1144-55; PMID:26039340; http://dx.doi.org/ 10.1021/tx500393y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012; 13:484-92; PMID:22641018; http://dx.doi.org/ 10.1038/nrg3230 [DOI] [PubMed] [Google Scholar]
- 32.Castillo-Díaz SA, Garay-Sevilla ME, Hernández-González MA, Solís-Martínez MO, Zaina S. Extensive demethylation of normally hypermethylated CpG islands occurs in human atherosclerotic arteries. Int J Mol Med 2010; 26:691-700 [DOI] [PubMed] [Google Scholar]
- 33.Hiltunen MO, Ylä-Herttuala S. DNA methylation, smooth muscle cells, and atherogenesis. Arteriosclerosis Thrombosis Vascular Biol 2003; 23:1750-3; PMID:12947012; http://dx.doi.org/ 10.1161/01.ATV.0000092871.30563.41 [DOI] [PubMed] [Google Scholar]
- 34.Coit P, De Lott LB, Nan B, Elner VM, Sawalha AH. DNA methylation analysis of the temporal artery microenvironment in giant cell arteritis. Ann Rheumatic Dis 2015. PMID:26038090; http://dx.doi.org/ 10.1136/annrheumdis-2014-207116 [DOI] [PubMed] [Google Scholar]
- 35.Mass MJ, Wang L. Arsenic alters cytosine methylation patterns of the promoter of the tumor suppressor gene p53 in human lung cells: a model for a mechanism of carcinogenesis. Mutation Res/Rev Mutation Res 1997; 386:263-77; http://dx.doi.org/ 10.1016/S1383-5742(97)00008-2 [DOI] [PubMed] [Google Scholar]
- 36.Intarasunanont P, Navasumrit P, Waraprasit S, Chaisatra K, Suk WA, Mahidol C, Ruchirawat M. Effects of arsenic exposure on DNA methylation in cord blood samples from newborn babies and in a human lymphoblast cell line. Environ Health 2012; 11:31; PMID:22551203; http://dx.doi.org/ 10.1186/1476-069X-11-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tunster SJ, Jensen AB, John RM. Imprinted genes in mouse placental development and the regulation of fetal energy stores. Reproduction 2013; 145:R117-R37; PMID:23445556; http://dx.doi.org/ 10.1530/REP-12-0511 [DOI] [PubMed] [Google Scholar]
- 38.Lesseur C, Paquette AG, Marsit CJ. Epigenetic regulation of infant neurobehavioral outcomes. Med Epigenetics 2014; 2:71-9; PMID:25089125; http://dx.doi.org/ 10.1159/000361026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics 2008; 3:97-106; PMID:18536531; http://dx.doi.org/ 10.4161/epi.3.2.6034 [DOI] [PubMed] [Google Scholar]
- 40.Conradt E, Fei M, LaGasse L, Tronick EZ, Guerin D, Gorman D, Marsit CJ, Lester B. Prenatal predictors of infant self-regulation: The contributions of placental DNA methylation of NR3C1 and neuroendocrine activity. Name: Front Behav Neurosci 2015; 9:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caldwell KE, Labrecque MT, Solomon BR, Ali A, Allan AM. Prenatal arsenic exposure alters the programming of the glucocorticoid signaling system during embryonic development. Neurotoxicol Teratol 2015; 47:66-79; PMID:25459689; http://dx.doi.org/ 10.1016/j.ntt.2014.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schroeder DI, Blair JD, Lott P, Yu HOK, Hong D, Crary F, Ashwood P, Walker C, Korf I, Robinson WP. The human placenta methylome. Proc Natl Acad Sci 2013; 110:6037-42; http://dx.doi.org/ 10.1073/pnas.1215145110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wahed M, Rahman M, Vahter M. Prevalence of arsenic exposure and skin lesions. A population based survey in Matlab, Bangladesh. J Epidemiol Community Health 2006; 60(3):242-8; PMID:16476755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pilsner RJ, Liu X, Ahsan H, Ilievski V, Slavkovich V, Levy D, Factor-Litvak P, Graziano JH, Gamble MV. Folate deficiency, hyperhomocysteinemia, low urinary creatinine, and hypomethylation of leukocyte DNA are risk factors for arsenic-induced skin lesions. Environ Health Perspectives 2009; 117:254-60; PMID:19270796; http://dx.doi.org/ 10.1289/ehp.11872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan WC, Seow WJ, Kile ML, Hoffman EB, Quamruzzaman Q, Rahman M, Mahiuddin G, Mostofa G, Lu Q, Christiani DC. Association of low to moderate levels of arsenic exposure with risk of type 2 diabetes in Bangladesh. Am J Epidemiol 2013; 178:1563-70; PMID:24049161; http://dx.doi.org/ 10.1093/aje/kwt195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailey KA, Wu MC, Ward WO, Smeester L, Rager JE, García‐Vargas G, Del Razo LM, Drobná Z, Stýblo M, Fry RC. Arsenic and the epigenome: interindividual differences in arsenic metabolism related to distinct patterns of DNA methylation. J Biochem Mol Toxicol 2013; 27:106-15; PMID:23315758; http://dx.doi.org/ 10.1002/jbt.21462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y. Vascular biology of the placenta. Colloquium Series Integrated Sys Physiology: from Mol Function: Morgan Claypool Life Sci 2010:1-98; http://dx.doi.org/ 10.4199/C00016ED1V01Y201008ISP009 [DOI] [PubMed] [Google Scholar]
- 48.Fukuchi Y, Nakajima H, Sugiyama D, Hirose I, Kitamura T, Tsuji K. Human placenta‐derived cells have mesenchymal stem/progenitor cell potential. Stem Cells 2004; 22:649-58; PMID:15342929; http://dx.doi.org/ 10.1634/stemcells.22-5-649 [DOI] [PubMed] [Google Scholar]
- 49.Cardenas A, Koestler DC, Houseman EA, Jackson BP, Kile ML, Karagas MR, Marsit CJ. Differential DNA Methylation in Umbilical Cord Blood of Infants Exposed to Mercury and Arsenic in utero. Epigenetics 2015; (10)6:508-15; PMID:25923418; http://dx.doi.org/ 10.1080/15592294.2015.1046026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kile ML, Hoffman E, Hsueh Y-M, Afroz S, Quamruzzaman Q, Rahman M, Mahiuddin G, Ryan L, Christiani DC. Variability in biomarkers of arsenic exposure and metabolism in adults over time. Environ Health Perspect 2009; 117:455-60; PMID:19337522; http://dx.doi.org/ 10.1289/ehp.11251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kile ML, Houseman EA, Rodrigues E, Smith TJ, Quamruzzaman Q, Rahman M, Mahiuddin G, Su L, Christiani DC. Toenail arsenic concentrations, GSTT1 gene polymorphisms, and arsenic exposure from drinking water. Cancer Epidemiol Biomarkers Prev 2005; 14:2419-26; http://dx.doi.org/ 10.1158/1055-9965.EPI-05-0306 [DOI] [PubMed] [Google Scholar]
- 52.Concha G, Vogler G, Lezcano D, Nermell B, Vahter M. Exposure to inorganic arsenic metabolites during early human development. Toxicol Sci 1998; 44:185-90; PMID:9742656; http://dx.doi.org/ 10.1093/toxsci/44.2.185 [DOI] [PubMed] [Google Scholar]
- 53.Creed J, Brockhoff C, Martin T. Method 200.8: Determination of trace elements in waters and wastes by inductively-coupled plasma-mass spectrometry. Environmental Monitoring Sys Lab Office Res Dev US Environmental Protection Agency Cincinnati OH Rev 1994; 5 [Google Scholar]
- 54.Fortin JP, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, Greenwood CM, Hansen KD. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol 2014; 15:503; PMID:25599564; http://dx.doi.org/ 10.1186/s13059-014-0503-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R. Discovery of cross-reactive probes and polymorphic cpgs in the Illumina Infinium humanmethylation450 microarray. Epigenetics 2013; 8:203-9; PMID:23314698; http://dx.doi.org/ 10.4161/epi.23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S. A β-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 2013; 29:189-96; PMID:23175756; http://dx.doi.org/ 10.1093/bioinformatics/bts680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du P, Zhang X, Huang C-C, Jafari N, Kibbe WA, Hou L, Lin SM. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 2010; 11:587; PMID:21118553; http://dx.doi.org/ 10.1186/1471-2105-11-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Houseman EA, Molitor J, Marsit CJ. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics 2014; 30:1431-9; PMID:24451622; http://dx.doi.org/ 10.1093/bioinformatics/btu029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.