Abstract
Prenatal smoke exposure, maternal obesity, aberrant fetal growth, and preterm birth are all risk factors for offspring metabolic syndrome. Cord blood aryl-hydrocarbon receptor repressor (AHRR) DNA methylation is responsive to maternal smoking during pregnancy. AHRR serves not only to inhibit aryl-hydrocarbon receptor (AHR) transcription, which is involved in mediating xenobiotic metabolism, but it is also involved in cell growth and differentiation. Other than maternal smoking, other predictors of offspring AHRR DNA methylation status remain unknown; we sought to identify them among newborns. We enrolled pregnant women in the PROGRESS birth cohort in Mexico City. Using pyrosequencing, we analyzed DNA methylation of 3 CpG sites within the AHRR gene promoter from the umbilical cord blood of 531 infants. We used generalized estimating equations to account for the correlation of DNA methylation between CpG sites. Multivariable models were used to adjust for maternal age, BMI, education, parity, smoke-exposure, infant sex, gestational age, and birth weight-for-gestational age. AHRR DNA methylation was positively associated with maternal BMI (P = 0.0009) and negatively associated with the length of gestation (P < 0.0001) and birth weight-for-gestational age (P < 0.0001). AHRR DNA methylation was 2.1% higher in offspring of obese vs. normal weight mothers and 3.1% higher in preterm vs. term infants, representing a third and a half standard deviation differences in methylation, respectively. In conclusion, offspring AHRR DNA methylation was associated with maternal obesity during pregnancy as well as infant gestational age and birth weight-for-gestational age. Further work to discover the health impacts of altered AHRR DNA methylation is warranted.
Keywords: AHRR, aryl-hydrocarbon receptor repressor, BMI, DNA methylation, epigenetics, preterm birth, pregnancy
Abbreviations
- 5mC
5-methylcytosine
- AGA
appropriate [birth weight]-for-gestational age
- AHR
aryl-hydrocarbon receptor
- AHRR
aryl-hydrocarbon receptor repressor
- ARNT
AHR nuclear translocator
- BMI
body mass index
- CpG
cytosine-phosphate-guanine
- IUGR
intrauterine growth restriction
- LGA
large [birth weight]-for-gestational age
- NFKB
nuclear factor kappa B
- SGA
small [birth weight]-for-gestational age
- XRE
xenobiotic response element.
Introduction
Over 1.4 billion adults are overweight or obese worldwide, and 65% of the world population live in countries where deaths from excess adiposity exceeds death from undernutrition.1 Risk factors for obesity start to accrue even in fetal life and include maternal obesity2 and slow or fast fetal growth.3 Preterm birth itself has been variably associated with later onset of metabolic disorders related to obesity such as insulin resistance and hypertension.4 How perinatal risk factors translate into the pathophysiology of obesity and metabolic syndrome remains unclear.
Epigenetic mechanisms may provide an explanation of how intrauterine programming of later obesity and metabolic syndrome occurs.5 Epigenetics refers to changes in gene expression and phenotype in the absence of DNA sequence variation. DNA methylation, one epigenetic mechanism, may be particularly sensitive to fetal exposures given the dynamic process of demethylation and remethylation after fertilization.6 Additionally, DNA methylation plays a critical role in cell differentiation, much of which occurs in utero.7
Maternal smoking during pregnancy affects fetal development leading to higher risks of intrauterine growth restriction (IUGR)8 and preterm delivery.9 Prenatal smoke exposure may also be associated with a higher risk of later obesity and cardiometabolic disorders.10 Recent studies demonstrate that smoke exposure in adults11 and among fetuses exposed to maternal smoking12 is associated with altered leukocyte DNA methylation of the aryl-hydrocarbon receptor repressor (AHRR) gene, located on chromosome 5.12,13 AHRR methylation is particularly interesting to study during pregnancy because the aryl-hydrocarbon receptor (AHR) is involved in metabolizing xenobiotics14 that might affect fetal development. While a growing body of evidence suggests that infant leukocyte DNA methylation of several genes varies according to maternal BMI,15 fetal growth,16-20 and preterm birth,21 no study has specifically assessed whether AHRR DNA methylation is independently associated with these perinatal predictors of longer-term infant metabolic dysregulation. In this study, we analyzed data from umbilical cord blood DNA to test the hypothesis that maternal and infant covariates would be associated with offspring AHRR DNA methylation.
Results
Descriptive characteristics of the cohort participants included in this study are provided in Table 1. Most women had a BMI <27 during the second trimester, but 160 (31%) were overweight and an additional 65 (13%) were obese. Fifty-two (10%) infants were born preterm. Infants in this study had lower birth weights-for-gestational age than the commonly used international reference population,22 with a mean Z score of −0.46; 92 (18%) infants were small-for-gestational age (SGA). Just 12 (2.3%) infants were large-for-gestational age (LGA).
Table 1.
Characteristics of PROGRESS birth cohort participants with umbilical cord DNA methylation from umbilical cord blood samples, n = 512, Mexico City, 2007–2010
| Mean | SD | Range | |
|---|---|---|---|
| Maternal age (years) | 27.8 | 5.5 | 18, 44 |
| Second trimester measured BMI (kg/m2) | 26.9 | 4.2 | 17.4, 44.7 |
| Pre-pregnancy self-reported BMI (kg/m2) | 25.2 | 4.2 | 16.0, 44.9 |
| Gestational age (weeks) | 38.8 | 1.8 | 24.4, 43.9 |
| Birth weight (kg) | 3.067 | 0.489 | 0.625, 4.625 |
| Fenton BWT/GA Z score | −0.46 | 0.95 | −5.72, 3.15 |
| n | % | ||
| Maternal education | |||
| Less than 12 years | 203 | 39.7 | |
| 12 years | 169 | 33.0 | |
| More than 12 years | 140 | 27.3 | |
| Multiparous | 279 | 54.7 | |
| Household smoke exposure | 239 | 46.8 | |
| Male infant | 282 | 55.2 |
Pearson correlation = 0.89 between pre-pregnancy BMI and measured BMI.
Missing values (n) for the following variables: birth weight (n = 1), infant sex (n = 1), parity (n = 2), household smoke defined as household member in home who smokes (n = 1).
BMI, body mass index; BWT, birth weight; GA, gestational age.
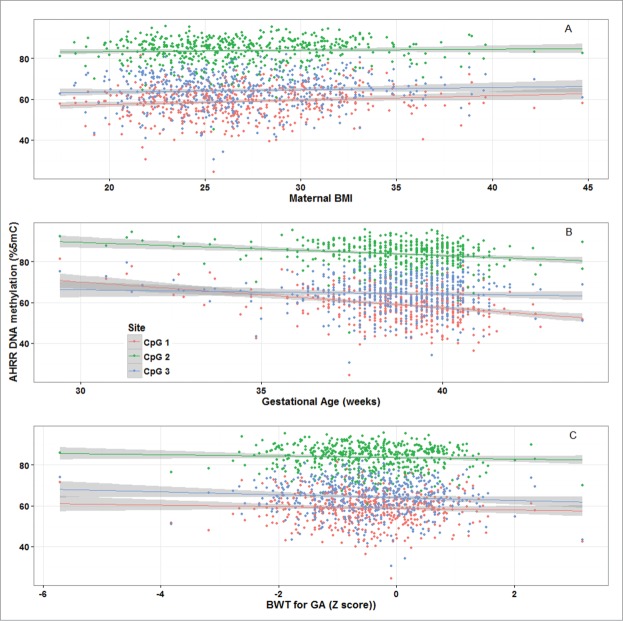
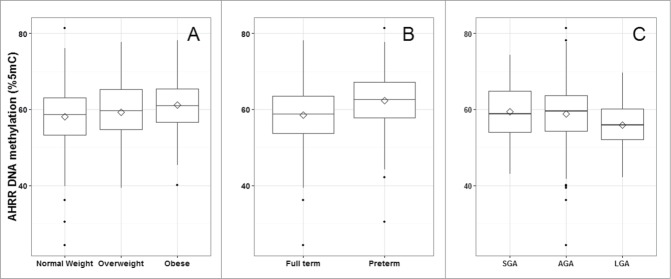
Mean and standard deviation (SD) methylation values (%5-methylcytosine) for the 3 AHRR CpG sites were 58.9 (7.8), 83.8 (6.2), and 64.3 (8.3) and were correlated with one another (Pearson correlations: r = 0.83 for CpG1 and CpG2; r = 0.61 for CpG2 and CpG3, r = 0.53 for CpG1 and CpG3; P < 0.0001) (Fig. 1). Bivariate analysis revealed that maternal BMI and education <12 years vs. 12 years, infant sex, gestational age, and birth weight-for-gestational age were all associated with AHRR DNA methylation (Table 2). Maternal education was marginally associated with AHRR DNA methylation. Maternal age, household smoke exposure, and parity were not associated with offspring AHRR DNA methylation. Plots of AHRR DNA methylation with maternal BMI, gestational age, and birth weight-for-gestational age reveal that the associations were similar among all 3 CpG sites, although weakest for CpG3 (Fig. 2). Representative plots of AHRR CpG1 with BMI categories (normal weight, overweight, and obese), preterm vs. full term birth and SGA, AGA, and LGA demonstrate that the direction of the associations is the same whether continuous or categorical variables are used to predict AHRR DNA methylation (Fig. 3).
Figure 1.
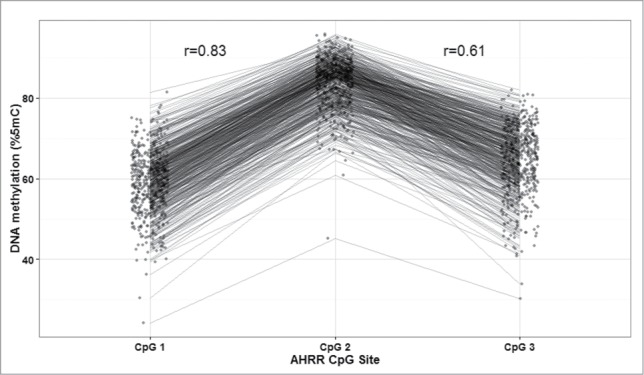
Aryl-hydrocarbon receptor repressor (AHRR) DNA methylation (%5-methylcytocines) at 3 CpG sites in cord blood DNA and their correlations, PROGRESS birth cohort, Mexico City, n = 512.
Table 2.
Associations of offspring AHRR DNA methylation with maternal and infant characteristics, PROGRESS birth cohort, Mexico City, 2007–2010
| Single variable models1 |
Multivariable-adjusted model2 |
|||||
|---|---|---|---|---|---|---|
| β (%5mC) | 95% CI | P value | β (%5mC) | 95% CI | P value | |
| Maternal characteristics | ||||||
| Maternal age (per year) | −0.02 | (−0.09, 0.05) | 0.5 | −0.02 | (−0.09, 0.05) | 0.5 |
| Maternal BMI (per kg/m2) | 0.13 | (0.05, 0.21) | 0.002 | 0.14 | (0.06, 0.22) | 0.00093 |
| Maternal education <12 years (vs. 12) | 0.89 | (0.00, 1.78) | 0.05 | 0.77 | (−0.09, 0.05) | 0.08 |
| Maternal education >12 years (vs. 12) | −0.12 | (−1.10, 0.85) | 0.8 | −0.20 | (−1.16, 0.75) | 0.7 |
| Multiparous (vs. primiparous) | −0.14 | (−0.88, 0.61) | 0.7 | −0.33 | (−1.09, 0.43) | 0.4 |
| Household smoke exposure (vs. no) | 0.42 | (−0.32, 1.2) | 0.3 | 0.36 | (−0.37, 1.09) | 0.3 |
| Infant characteristics | ||||||
| Gestational age (per week) | −0.43 | (−0.78, −0.08) | 0.02 | −0.86 | (−1.06, −0.65) | <0.00013 |
| Birth weight-for-gestational age (per SD) | −0.49 | (−0.88, −0.10) | 0.01 | −0.97 | (−1.26, −0.58) | <0.00013 |
| Male (vs. female) | 0.87 | (0.12, 1.6) | 0.02 | 0.48 | (−0.25, 1.21) | 0.2 |
Single variable models (n = 512), generalized estimating equations with robust variance used to account for correlations across 3 AHRR CpG sites.
Multivariable model including all variables listed (n = 507) due to missing values (n) for the following variables: birth weight (n = 1), infant sex (n = 1), parity (n = 2), household smoke defined as household member in home who smokes (n = 1).
P value robust to Bonferroni adjustment for multiple gene comparison (6 genes analyzed = 0.05/6 = 0.008) and adjustment to account for the 8 potential predictors (age, BMI, education, parity, smoke exposure, gestational age, birth weight-for-gestational age, and sex) (0.001/8 = 0.001).
Figure 2.
Aryl-hydrocarbon receptor repressor (AHRR) DNA methylation (%5-methylcytocine) and (A) maternal BMI; (B) infant gestational age; and (C) birth weight-for gestational age, PROGRESS birth cohort, Mexico City, n = 512.
Figure 3.
Distributions of aryl-hydrocarbon receptor repressor (AHRR) CpG 1 DNA methylation (%5-methylcytocines) among categories of (A) maternal BMI; (B) full-term vs. preterm births; and (C) birth weight-for-gestational age, PROGRESS birth cohort, Mexico City, n = 512. SGA, small-for-gestational age; AGA, appropriate-for-gestational age; LGA, large-for-gestational age.
Multivariable generalized estimating equation models mutually adjusting for maternal age, BMI, education, parity, household smoke exposure, gestational age, birth weight-for-gestational age, and infant sex revealed that only maternal BMI, gestational age, and birth-weight-for gestational age were significantly associated with AHRR DNA methylation in offspring cord blood DNA (Table 2). For every 1 kg/m2 increase in maternal BMI, AHRR DNA methylation was 0.14% higher (P = 0.0009). For every week increase in gestational age, AHRR DNA methylation was 0.86% lower (P < 0.0001). Per 1 unit higher Z score (or SD) in birth weight-for-gestational age, AHRR DNA methylation was 0.97% lower (P < 0.0001).
In similarly adjusted multivariable generalized estimating equation models using categorical predictors, we found that infants born to obese and overweight mothers had higher AHRR DNA methylation [2.1% (95% CI: 1.1, 3.2) and 0.9% (95% CI: 0.08, 1.7), respectively, vs. normal weight] (Table 3). Preterm (vs. full term) infants had higher AHRR DNA methylation [3.1% (95% CI: 2.0, 4.3)]. LGA (vs. AGA) infants had lower AHRR DNA methylation [−3.7% (95% CI: −6.2, −1.2)].
Table 3.
Multivariable associations of offspring aryl-hydrocarbon receptor repressor (AHRR) DNA methylation with categories of maternal BMI, preterm vs. term birth, and categories of fetal growth, PROGRESS birth cohort, Mexico City, n = 507
| β (AHRR DNA methylation, %5mC) | 95% CI | P value | |
|---|---|---|---|
| Model 1 | |||
| Overweight vs. normal weight | 0.91 | (0.08, 1.74) | 0.03 |
| Obese vs. normal weight | 2.14 | (1.01, 3.23) | 0.0001 |
| Model 2 | |||
| Preterm vs. full term | 3.15 | (1.97, 4.33) | <0.0001 |
| Model 3 | |||
| SGA vs. AGA | 1.11 | (0.20, 2.02) | 0.02 |
| LGA vs. AGA | −3.72 | (−6.20, −1.23) | 0.003 |
All models adjusted for maternal age, parity, household smoke exposure, infant sex using generalized estimating equations with robust variance.
Model 1 additionally adjusted for gestational age at delivery and birth weight-for-gestational age. Overweight defined as 2nd trimester body mass index (BMI) >27 and <32 kg/m2. Obese defined as a 2nd trimester BMI >32 kg/m2.
Model 2 additionally adjusted for maternal BMI and birth weight-for-gestational age. Preterm defined as <37 completed weeks of gestation and full term defined as >37 weeks of gestation.
Model 3 additionally adjusted for maternal BMI and gestational age at delivery. SGA, small-for-gestational age and LGA, large-for-gestational age defined as <10th and >90th percentile of birth weight-for-gestational age based on Fenton,22 respectively.
We performed several sensitivity analyses to ensure our findings were robust. First because cell type can be associated with DNA methylation,23 we re-ran the analysis in the subset of mother-infant pairs for whom cord blood leukocyte subtype differential counts were available (n = 405). Associations were unchanged and thus we chose final models that did not adjust for leukocyte subtypes to maintain the sample size of 507 (Table S2). Secondly, we analyzed the association of folic acid intake in the subset of women who had this data available (n = 65). Mean (SD) folic acid intake was 528 (237) micrograms per day. In the subset of women with folic acid intake data, associations, while less statistically significant, were of similar magnitudes and directions as in the larger cohort. Further, while folic acid intake was negatively associated with AHRR DNA methylation, it did not confound the associations (Table S3). We were concerned about batch effects because one laboratory extracted and stored DNA for the first 267 samples and a second laboratory extracted and stored DNA from the next 245 samples. Indeed, laboratory site was associated with AHRR DNA methylation values (P = 0.002). We performed 2 analyses to determine if storage site affected our findings. First we added laboratory site as a term in our models, which did not affect our findings (Table S4). Second, we performed stratified analyses to ensure that the associations were similar for participants whose samples were stored in one laboratory vs. the other. Our findings were robust to these 2 sensitivity analyses, suggesting that laboratory storage site did not confound the associations we observed. We also eliminated the single current smoker in primary analysis and our results were unchanged (data not shown). Our final sensitivity analysis addressed our choice to use measured BMI in the 2nd trimester instead of pre-pregnancy BMI based on self-reported pre-pregnancy weight and measured height during pregnancy. We re-ran our analysis using pre-pregnancy BMI and were reassured when associations with AHRR DNA methylation were the same. The β coefficient when we used pre-pregnancy BMI was 0.136 (95% CI: 0.055, 0.217) (P = 0.0009), while the β coefficient when we used measured 2nd trimester BMI was 0.137 (95% CI: 0.056, 0.218)(P = 0.0009).
Discussion
The aryl-hydrocarbon receptor (AHR) protein mediates the toxicity of xenobiotics including 2,3,7,8-tetrachlorodibenzo-p-dioxin.14 The AHR is a cytoplasmic receptor, similar to steroid receptor, in that that the ligand-receptor unit acts as a transcription factor for Phase 1 metabolic enzymes. Ligands include flavonoids and indoles commonly found in foods, as well as aromatic xenobiotic compounds such as dioxin. Once bound to a ligand (such as a dioxin), it is transferred to the nucleus and heterodimerizes with its protein partner, AHR nuclear translocator (ARNT). This complex then binds xenobiotic response elements (XRE) of AHR, thereby inducing transcription of target genes, including CYP1A1, CYP1A2, CYP1B1, and AHRR. The protein of interest for this study, AHRR, inhibits AHR function by disrupting the AHR/ARNT complex. Dioxin is among a number of planar aromatic hydrocarbons that act as ligands for AHR, including polybrominated diphenyl ethers24 and polycyclic aromatic hydrocarbons.25 In addition to its own XRE, the AHRR regulatory region contains a nuclear factor kappa B (NFKB) binding site that works as an inducible enhancer. NFKB and XRE sequences can act cooperatively on the gene transcription machinery,26 suggesting that inflammation could play a role in AHRR production. AHRR plays a role as a tumor suppressor and AHRR gene silencing has been shown to occur through DNA methylation.27 Taken together, AHRR DNA methylation has become a target for human epidemiologic studies as a potential marker of disruption of AHR activity. Several studies of smoking have demonstrated differential DNA methylation of AHRR between smokers and non-smokers.13 Others have demonstrated that these same methylation sites differed in the offspring of women who smoked during pregnancy, compared to offspring of non-smokers.12,28
Prior studies demonstrating associations between smoking and AHRR DNA methylation used epigenome-wide methylation assays designed to discover previously unknown associations.12,13 Given the results of these prior studies demonstrating that AHRR DNA methylation is modifiable during pregnancy,12 we chose AHRR as a candidate gene to conduct a hypothesis-driven study of perinatal predictors of AHRR DNA methylation in cord blood. We chose these CpG sites within the gene because of their location within the promoter of the gene. Methylation in this area would be expected to contribute to gene silencing of AHRR. Joubert et al. reported AHRR hypomethylation among smokers in prior studies. In contrast, we found higher methylation among offspring of obese women. However, close inspection of the data in Joubert et al. revealed that 1 of the 4 differentially methylated CpG sites was more highly methylated among infants whose mothers were smokers.12 Additionally, the magnitude of AHRR DNA methylation difference between mothers with undetectable vs. high cotinine levels at each of their 4 significant sites was of similar magnitude (∼4%) as associations with our categorical predictors (2–3%). We did not analyze the same CpG sites as the smoking-associated CpG sites reported in prior studies but instead chose to maximize the CpG sites in the AHRR promoter region that our pyrosequencing assay would capture. Furthermore, even if the same sites had been interrogated, different exposures might affect methylation in opposite directions.
Our observed association of higher maternal BMI with higher offspring AHRR DNA methylation may occur for several reasons. First, it is possible that the fetal tissues of overweight/obese women in our cohort may be exposed to different levels of unmeasured xenobiotics that cross the placenta. For example, dioxin-like polychlorinated biphenyls (PCBs) have been shown to be positively associated with BMI in women.29 Obese mothers may lead to fetal cell ligand-AHR binding thereby inducing AHRR gene expression. We do not have data in ours study of gene expression. However, obesity is a known systemic inflammatory state. Chronic inflammation induces cytokines that can induce NFKB,30 which in turn can also induce AHRR.26 It is currently unknown how higher levels of AHRR gene expression might trigger methylation, but potentially there could be a negative feedback loop. The negative associations of AHRR DNA methylation with gestational age and birth weight-for-gestational age might share a similar etiology of DNA methylation variation, given that both preterm deliveries31 and poor fetal growth may result from inflammatory states.32
The three associations with offspring AHRR DNA methylation and maternal BMI, infant gestational age and infant birth weight-for-gestational age are intriguing because each of these factors has been shown to be associated with long-term offspring metabolic syndrome including obesity and hypertension.33,34 How fetal life programs outcome later in life remains an area of intense investigation as epigenetic processes, including DNA methylation, may link fetal exposures to adult disease.35,36 Animal models of IUGR support this hypothesis. Animals with IUGR are not only at risk for developing central obesity and Type II diabetes mellitus37 but also have distinct DNA methylation profiles.38 We are currently following infants into early childhood and will be able to test the hypothesis that AHRR gene DNA methylation in humans is associated with long-term metabolic health and obesity in the future.
Strengths of our study include its prospective and hypothesis-driven design. We have detailed covariate data allowing for adjustment for potential confounding variables. One limitation of our study is the estimation of gestational age using last menstrual period because ultrasounds are not routinely performed as part of prenatal care for Instituto Mexicano del Seguro Social (IMSS) beneficiaries. However, we did obtain ultrasounds in a subset of 98 women participating in a prior cohort enrolled in the same setting by the same field staff and found a high correlation between ultrasound estimated gestational age and last menstrual period (LMP) estimated age (0.89, P < 0.0001). Furthermore, error in gestational age assessment is likely to be non-differential and thus unlikely to lead to Type I error. Limitations of gene-specific pyrosequencing assays include the inability to screen for other DNA methylation sites beyond the candidates that we have chosen to analyze. We also lack RNA or protein to determine functional biologic relevance of AHRR DNA methylation. We analyzed 3 CpG sites within AHRR and, thus, cannot determine whether AHRR DNA methylation in other regulatory regions for this gene might have different levels of association. However, other DNA methylation marks in proximity to one another in general are highly correlated,39 as was the case with the 3 sites we chose. Cord blood DNA might not be the target tissue of interest for some long-term cardiometabolic outcomes and, thus, is often considered a surrogate tissue. However, in this study, inflammation may link obesity, preterm birth, and poor fetal growth to long-term adverse sequelae making leukocytes a reasonable tissue to study. Despite these limitations, our findings add to a growing body of literature that suggests that the intrauterine environment may leave epigenetic marks. Furthermore, studies of maternal smoking and offspring AHRR DNA methylation should carefully consider effects of maternal BMI and infant birth weight and gestational age. Determining the persistence and long-term health effects of these marks remain worthwhile future directions of the field of perinatal epigenetics.
In conclusion, in our prospective birth cohort, we found significant associations of maternal BMI, infant gestational age, and birth weight-for-gestational age with offspring AHRR DNA methylation in umbilical cord blood DNA. Future work to determine the long-term consequences of AHRR DNA methylation later in childhood is warranted.
Methods
Cohort enrollment
We performed a prospective birth cohort study, Programming Research in Obesity, GRowth Environment and Social Stress (PROGRESS), in Mexico City from 2007–2011. Details of enrollment are published elsewhere.40 Briefly, pregnant women receiving prenatal care through the Mexican Social Security System (IMSS) in Mexico City were enrolled between 12 and 24 weeks gestation after they provided written informed consent. All study activities were approved by the Institutional Review Boards of the participating institutions.
Variable ascertainment
Study staff obtained demographic data through verbally administered questionnaires and interviews. Women self-reported pre-pregnancy weight and study staff weighed and measured the height of mothers upon enrollment. Because we noted some error in maternal report of pre-pregnancy weight with differences of over 10 kg between self-reported pre-pregnancy weights, we opted to calculate BMI using the measured weights obtained in the second trimester. However, when we categorized BMI, we added 2 kg to traditional cut-points to account for normal weight gain in the first trimester of pregnancy.41 We categorized BMI as normal (BMI ≤27 kg/m2), overweight (BMI >27 to ≤32 kg/m2), and obese (BMI >32 kg/m2). Pearson correlation between maternal pre-pregnancy BMI and calculated second trimester BMI was 0.89. Because ultrasounds were not routinely performed as standard of care, gestational age was based on LMP and by a standardized physical examination to determine gestational age at birth.42 When the physical examination assessment of gestational age differed by more than 3 weeks from the gestational age based on LMP, the physical exam was used in lieu of the LMP-determined gestational age. We dichotomized gestational age categories into preterm (<37 completed weeks of gestation) and full term (≥37 completed weeks of gestation). Birth weight was abstracted from the infants' hospital chart. Birth weight-for-gestational age Z scores were calculated using an international reference developed by Fenton and Kim, allowing us to analyze fetal growth as a continuous variable (per standard deviation (SD) increment).22 We considered infants small-for-gestational age (SGA) if their birth weights for gestational age were below the 10th percentile and large-for-gestational age (LGA) if their birth weights for gestational age were above the 90th percentile. Household smoke exposure was derived from any positive report from several questions asked in the second and third trimester about household members who smoke inside the home or outside the home. We divided education into 3 categories (<12 years, 12 years, or >12 years). We dichotomized women as multiparous or primiparous after delivery of the participating infant in the study, based on whether the women reported a prior live-born infant. We collected leukocyte differential counts from cord blood to adjust for percentage of monocytes, lymphocytes, and granulocytes. We also administered a food frequency questionnaire for folic acid intake to explore whether methyl donor intake was associated with offspring DNA methylation.
AHRR DNA methylation analysis
We obtained umbilical cord blood at the time of delivery and analyzed DNA methylation for 531 of the 948 infants born into the study. Most missing samples were due to births occurring late at night or in the very early morning hours. We have published details about extraction and DNA bisulfite treatment of these samples elsewhere.40 The first 260 whole blood samples were stored in PAXgene™ (PreAnalytiX GmbH, Hombrechtikon Switzerland) tubes and extracted using a QIAamp DNA Blood Kit (QIAGEN). The DNA was then stored at −80°C prior to bisulfite treatment. The next 271 samples were extracted by conventional Phenol–chloroform method after red cell lysis by a second laboratory. The second laboratory stored the DNA at 4°C. One μg of genomic DNA subsequently underwent bisulfite treatment using EZ DNA Methylation-Gold Kit (Zymo Research, CA, USA). The pyrosequencing assay was designed based upon promoter regions defined by SwitchGear Genomics43 (SwitchGear Genomics, Carlsbad, CA). We use a MethPrimer online tool44 (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi) to design PCR primers for use with pyrosequencing and bisulfite-converted DNA. Pyrosequencing primers were designed not to overlap with any single-nucleotide polymorphism (SNP) or repeated elements.45 The forward PCR primer was GGTAGTTATTTAGTTAAGTTTTTTTT and the reverse 5′ end biotin labeled primer was TTCACTCTAAATACTAAAACATTTC. PCR cycling condition was 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s for 40 cycles. PCR products were purified and sequenced by PSQ Q96 MD pyrosequencing System (QIAGEN), as previously described.46 The sequencing primer for pyrosequencing was TAAGTTTTTTTTTGA and the sequence entry was C/TGGGTTTTGAGGTTGTC/TGTTATAG/AAGC/TGGTAGA. Three CpG sites were analyzed including those at chr5:420,208–420,235 (GRCh37/hg19) (Fig. S1).
Of the 531 assays run, 517 were successful in PCR and pyrosequencing reactions at all 3 CpGs of AHRR. We further excluded the pyrosequencing data from 11 individuals after inspection of each pyrogram as those showed a high background peak. As quality controls, we placed 33 duplicate genomic DNA samples for technical replicates to estimate the internal plate variation and pyrosequencing run variation. The Pearson correlation range was from 0.91 to 0.95 at each of the CpG sites. We removed individuals without BMI, birth weight or gestational age (n = 8) from the analysis leaving 512 samples for inclusion in the analysis. Fully adjusted models included 507 participants with complete covariate data.
Statistical analysis
Visual inspection revealed DNA methylation values appeared normally distributed. After univariate descriptive analyses, we performed bivariate analyses to compare mean AHRR methylation among categories of maternal BMI, preterm vs. term births, and among birth weight-for-gestational age categories (SGA, AGA, and LGA). Subsequently, we fit multivariable generalized estimating equation models, adjusted for potential confounding variables, with an exchangeable correlation structure and robust standard errors to account for the non-independence of the 3 proximal CpGs as a repeated measure of AHRR DNA methylation. In this way, we estimated the associations of maternal and infant covariates with a marginal model for AHRR DNA methylation across the 3 sites. We fit separate models with continuous and categorical BMI, gestational age, and birth weight for gestational age as independent variables. To ensure results were not confounded by potential batch effects by DNA extraction and storage methods we introduced a laboratory site term into adjusted models. We also fit stratified methods to ensure that associations were the same within batch as they were in the overall analysis.
Because cell type can affect DNA methylation patterns,23 in a subset of 405 individuals for whom we had complete leukocyte differential data, we re-ran the analysis to evaluate the effect of associations of maternal and infant characteristics with AHRR DNA methylation adjusting for cell type. A very small proportion of individuals (65) from the end of cohort enrollment also had food-frequency data, allowing for adjustment for folic acid intake in a subset of the population because methyl donor intake might be associated with offspring DNA methylation.47 We also re-analyzed our data excluding the one current smoker in our cohort.
As part of the parent study, we also analyzed the DNA methylation of 5 other candidate genes. Because of our a priori hypothesis that AHRR would be associated with other risk factors for adverse perinatal and long-term cardio-metabolic outcomes, we report here the AHRR results. However, we did screen for associations with the other chosen genes for each association using multivariable linear regression with mean DNA methylation as the dependent variable (Table S1).
We used SAS, version 9.3, Cary NC. We also used R, (version 3.1.3:R Core Team (2015). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/) and specifically package “gee” (Vincent J Carey. Ported to R by Thomas Lumley and Brian Ripley. Note that maintainers are not available to give advice on using a package they did not author. (2012). gee: Generalized Estimation Equation solver. R package version 4.13–18. http://CRAN.R-project.org/package=gee).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We acknowledge the American British Cowdray (ABC) Hospital in Mexico City for the use of their facilities.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
Funding
This work was supported in part by NIH/NIEHS: K23ES022242, P30ES000002; P30ES023515; R01ES013744, R01ES020268; R01ES014930, R01ES021357; K99ES023450 and the Klarman Scholars Program at Beth Israel Deaconess Medical Center. This study was also supported and partially funded by the National Institute of Public Health/Ministry of Health of Mexico. Dr. Tamayo y Ortiz was funded by the Mexican Council for Science and Technology; CONACyT.
References
- 1.World Health Organization Obesity and overweight fact sheet [updated 2015 January; cited 2015 January 23]. Available from: http://www.who.int/mediacentre/factsheets/fs311/en/.
- 2.Catalano PM, Farrell K, Thomas A, Huston-Presley L, Mencin P, de Mouzon SH, Amini SB. Perinatal risk factors for childhood obesity and metabolic dysregulation. Am J Clin Nutr 2009; 90:1303-13; PMID:19759171; http://dx.doi.org/ 10.3945/ajcn.2008.27416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oken E, Gillman MW. Fetal origins of obesity. Obes Res 2003; 11:496-506; PMID:12690076; http://dx.doi.org/ 10.1038/oby.2003.69 [DOI] [PubMed] [Google Scholar]
- 4.Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med 2008; 359:61-73; PMID:18596274; http://dx.doi.org/ 10.1056/NEJMra0708473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barouki R, Gluckman PD, Grandjean P, Hanson M, Heindel JJ. Developmental origins of non-communicable disease: implications for research and public health. Environ Health 2012; 11:42; PMID:22715989; http://dx.doi.org/ 10.1186/1476-069X-11-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science 2001; 293:1089-93; PMID:11498579; http://dx.doi.org/ 10.1126/science.1063443 [DOI] [PubMed] [Google Scholar]
- 7.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet 2005; 14(Spec No 1):R47-58; PMID:15809273; http://dx.doi.org/ 10.1093/hmg/ddi114 [DOI] [PubMed] [Google Scholar]
- 8.Harrod CS, Reynolds RM, Chasan-Taber L, Fingerlin TE, Glueck DH, Brinton JT, Dabelea D. Quantity and timing of maternal prenatal smoking on neonatal body composition: the Healthy Start study. J Pediatr 2014; 165:707-12; PMID:25063722; http://dx.doi.org/ 10.1016/j.jpeds.2014.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savitz DA, Murnane P. Behavioral influences on preterm birth: a review. Epidemiology 2010; 21:291-9; PMID:20386169; http://dx.doi.org/ 10.1097/EDE.0b013e3181d3ca63 [DOI] [PubMed] [Google Scholar]
- 10.Dior UP, Lawrence GM, Sitlani C, Enquobahrie D, Manor O, Siscovick DS, Friedlander Y, Hochner H. Parental smoking during pregnancy and offspring cardio-metabolic risk factors at ages 17 and 32. Atherosclerosis 2014; 235:430-7; PMID:24937467; http://dx.doi.org/ 10.1016/j.atherosclerosis.2014.05.937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monick MM, Beach SR, Plume J, Sears R, Gerrard M, Brody GH, Philibert RA. Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am J Med Genet Part B Neuropsychiatr Genet 2012; 159B:141-51; PMID:22232023; http://dx.doi.org/ 10.1002/ajmg.b.32021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joubert BR, Haberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, Huang Z, Hoyo C, Midttun O, Cupul-Uicab LA, et al.. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect 2012; 120:1425-31; PMID:22851337; http://dx.doi.org/ 10.1289/ehp.1205412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philibert RA, Beach SR, Lei MK, Brody GH. Changes in DNA methylation at the aryl hydrocarbon receptor repressor may be a new biomarker for smoking. Clin Epigenet 2013; 5:19; PMID:24120260; http://dx.doi.org/ 10.1186/1868-7083-5-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: multitasking in the immune system. Ann Rev Immunol 2014; 32:403-32; PMID:24655296; http://dx.doi.org/ 10.1146/annurev-immunol-032713-120245 [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Chen Q, Tsai HJ, Wang G, Hong X, Zhou Y, Zhang C, Liu C, Liu R, Wang H, et al.. Maternal preconception body mass index and offspring cord blood DNA methylation: exploration of early life origins of disease. Environ Mol Mutagen 2014; 55:223-30; PMID:24243566; http://dx.doi.org/ 10.1002/em.21827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabano S, Colapietro P, Cetin I, Grati FR, Zanutto S, Mando C, Antonazzo P, Pileri P, Rossella F, Larizza L, et al.. Epigenetic modulation of the IGF2/H19 imprinted domain in human embryonic and extra-embryonic compartments and its possible role in fetal growth restriction. Epigenetics 2010; 5:313-24; PMID:20418667; http://dx.doi.org/ 10.4161/epi.5.4.11637 [DOI] [PubMed] [Google Scholar]
- 17.Haworth KE, Farrell WE, Emes RD, Ismail KM, Carroll WD, Hubball E, Rooney A, Yates AM, Mein C, Fryer AA. Methylation of the FGFR2 gene is associated with high birth weight centile in humans. Epigenomics 2014; 6:477-91; PMID:25431941; http://dx.doi.org/ 10.2217/epi.14.40 [DOI] [PubMed] [Google Scholar]
- 18.Rangel M, dos Santos JC, Ortiz PH, Hirata M, Jasiulionis MG, Araujo RC, Ierardi DF, Franco Mdo C. Modification of epigenetic patterns in low birth weight children: importance of hypomethylation of the ACE gene promoter. PloS One 2014; 9:e106138; PMID:25170764; http://dx.doi.org/ 10.1371/journal.pone.0106138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoyo C, Daltveit AK, Iversen E, Benjamin-Neelon SE, Fuemmeler B, Schildkraut J, Murtha AP, Overcash F, Vidal AC, Wang F, et al.. Erythrocyte folate concentrations, CpG methylation at genomically imprinted domains, and birth weight in a multiethnic newborn cohort. Epigenetics 2014; 9:1120-30; PMID:24874916; http://dx.doi.org/ 10.4161/epi.29332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engel SM, Joubert BR, Wu MC, Olshan AF, Haberg SE, Ueland PM, Nystad W, Nilsen RM, Vollset SE, Peddada SD, et al.. Neonatal genome-wide methylation patterns in relation to birth weight in the Norwegian Mother and Child Cohort. Am J Epidemiol 2014; 179:834-42; PMID:24561991; http://dx.doi.org/ 10.1093/aje/kwt433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parets SE, Conneely KN, Kilaru V, Fortunato SJ, Syed TA, Saade G, Smith AK, Menon R. Fetal DNA methylation associates with early spontaneous preterm birth and gestational age. PloS One 2013; 8:e67489; PMID:23826308; http://dx.doi.org/ 10.1371/journal.pone.0067489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenton TR, Kim JH. A systematic review and meta-analysis to revise the Fenton growth chart for preterm infants. BMC Pediatr 2013; 13:59; PMID:23601190; http://dx.doi.org/ 10.1186/1471-2431-13-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 2012; 13:86; PMID:22568884; http://dx.doi.org/ 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren XM, Guo LH. Molecular toxicology of polybrominated diphenyl ethers: nuclear hormone receptor mediated pathways. Environ Sci Processes Impacts 2013; 15:702-8; PMID:23467608; http://dx.doi.org/ 10.1039/c3em00023k [DOI] [PubMed] [Google Scholar]
- 25.Billiard SM, Hahn ME, Franks DG, Peterson RE, Bols NC, Hodson PV. Binding of polycyclic aromatic hydrocarbons (PAHs) to teleost aryl hydrocarbon receptors (AHRs). Comp Biochem Physiol Part B Biochem Mol Biol 2002; 133:55-68; PMID:12223212; http://dx.doi.org/ 10.1016/S1096-4959(02)00105-7 [DOI] [PubMed] [Google Scholar]
- 26.Baba T, Mimura J, Gradin K, Kuroiwa A, Watanabe T, Matsuda Y, Inazawa J, Sogawa K, Fujii-Kuriyama Y. Structure and expression of the Ah receptor repressor gene. J Biol Chem 2001; 276:33101-10; PMID:11423533; http://dx.doi.org/ 10.1074/jbc.M011497200 [DOI] [PubMed] [Google Scholar]
- 27.Zudaire E, Cuesta N, Murty V, Woodson K, Adams L, Gonzalez N, Martinez A, Narayan G, Kirsch I, Franklin W, et al.. The aryl hydrocarbon receptor repressor is a putative tumor suppressor gene in multiple human cancers. J Clin Invest 2008; 118:640-50; PMID:18172554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richmond RC, Simpkin AJ, Woodward G, Gaunt TR, Lyttleton O, McArdle WL, Ring SM, Smith AD, Timpson NJ, Tilling K, et al.. Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: findings from the Avon Longitudinal Study of Parents and Children (ALSPAC). Hum Mol Genet 2015; 24:2201-17; PMID:25552657; http://dx.doi.org/ 10.1093/hmg/ddu739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knutsen HK, Kvalem HE, Haugen M, Meltzer HM, Brantsaeter AL, Alexander J, Papke O, Liane VH, Becher G, Thomsen C. Sex, BMI and age in addition to dietary intakes influence blood concentrations and congener profiles of dioxins and PCBs. Mol Nutr Food Res 2011; 55:772-82; PMID:21280203; http://dx.doi.org/ 10.1002/mnfr.201000243 [DOI] [PubMed] [Google Scholar]
- 30.Quilley J. Oxidative stress and inflammation in the endothelial dysfunction of obesity: a role for nuclear factor kappa B? J Hypertens 2010; 28:2010-1; PMID:20844369; http://dx.doi.org/ 10.1097/HJH.0b013e32833e24cb [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McElrath TF, Hecht JL, Dammann O, Boggess K, Onderdonk A, Markenson G, Harper M, Delpapa E, Allred EN, Leviton A, et al.. Pregnancy disorders that lead to delivery before the 28th week of gestation: an epidemiologic approach to classification. Am J Epidemiol 2008; 168:980-9; PMID:18756014; http://dx.doi.org/ 10.1093/aje/kwn202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amarilyo G, Oren A, Mimouni FB, Ochshorn Y, Deutsch V, Mandel D. Increased cord serum inflammatory markers in small-for-gestational-age neonates. J Perinatol 2011; 31:30-2; PMID:20410909; http://dx.doi.org/ 10.1038/jp.2010.53 [DOI] [PubMed] [Google Scholar]
- 33.Skilton MR, Viikari JS, Juonala M, Laitinen T, Lehtimaki T, Taittonen L, Kahonen M, Celermajer DS, Raitakari OT. Fetal growth and preterm birth influence cardiovascular risk factors and arterial health in young adults: the Cardiovascular Risk in Young Finns Study. Arterioscler Thromb Vasc Biol 2011; 31:2975-81; PMID:21940950; http://dx.doi.org/ 10.1161/ATVBAHA.111.234757 [DOI] [PubMed] [Google Scholar]
- 34.Barker DJ, Winter PD, Osmond C, Margetts B, Simmonds SJ. Weight in infancy and death from ischaemic heart disease. Lancet 1989; 2:577-80; PMID:2570282; http://dx.doi.org/ 10.1016/S0140-6736(89)90710-1 [DOI] [PubMed] [Google Scholar]
- 35.Osborne-Majnik A, Fu Q, Lane RH. Epigenetic mechanisms in fetal origins of health and disease. Clin Obstet Gynecol 2013; 56:622-32; PMID:23787712; http://dx.doi.org/ 10.1097/GRF.0b013e31829cb99a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banister CE, Koestler DC, Maccani MA, Padbury JF, Houseman EA, Marsit CJ. Infant growth restriction is associated with distinct patterns of DNA methylation in human placentas. Epigenetics 2011; 6:920-7; PMID:21758004; http://dx.doi.org/ 10.4161/epi.6.7.16079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simmons R. Perinatal programming of obesity. Semin Perinatol 2008; 32:371-4; PMID:18929161; http://dx.doi.org/ 10.1053/j.semperi.2008.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seki Y, Williams L, Vuguin PM, Charron MJ. Minireview: Epigenetic programming of diabetes and obesity: animal models. Endocrinology 2012; 153:1031-8; PMID:22253432; http://dx.doi.org/ 10.1210/en.2011-1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al.. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet 2006; 38:1378-85; PMID:17072317; http://dx.doi.org/ 10.1038/ng1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burris HH, Braun JM, Byun HM, Tarantini L, Mercado A, Wright RJ, Schnaas L, Baccarelli AA, Wright RO, Tellez-Rojo MM. Association between birth weight and DNA methylation of IGF2, glucocorticoid receptor and repetitive elements LINE-1 and Alu. Epigenomics 2013; 5:271-81; PMID:23750643; http://dx.doi.org/ 10.2217/epi.13.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.In: Rasmussen KM, Yaktine AL, eds. The National Academies Collection: Reports funded by National Institutes of Health Weight Gain During Pregnancy: Reexamining the Guidelines Washington (DC): National Academies Press; 2009. [PubMed] [Google Scholar]
- 42.Capurro H, Konichezky S, Fonseca D, Caldeyro-Barcia R. A simplified method for diagnosis of gestational age in the newborn infant. J Pediatr 1978; 93:120-2; PMID:650322; http://dx.doi.org/ 10.1016/S0022-3476(78)80621-0 [DOI] [PubMed] [Google Scholar]
- 43.Hartzell DD, Trinklein ND, Mendez J, Murphy N, Aldred SF, Wood K, Urh M. A functional analysis of the CREB signaling pathway using HaloCHIP-chip and high throughput reporter assays. BMC Genomics 2009; 10:497; PMID:19860899; http://dx.doi.org/ 10.1186/1471-2164-10-497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002; 18:1427-31; PMID:12424112; http://dx.doi.org/ 10.1093/bioinformatics/18.11.1427 [DOI] [PubMed] [Google Scholar]
- 45.Barrow TM, Byun HM. Single nucleotide polymorphisms on DNA methylation microarrays: precautions against confounding. Epigenomics 2014; 6:577-9; PMID:25531251; http://dx.doi.org/ 10.2217/epi.14.55 [DOI] [PubMed] [Google Scholar]
- 46.Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med 2009; 179:572-8; PMID:19136372; http://dx.doi.org/ 10.1164/rccm.200807-1097OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dominguez-Salas P, Moore SE, Baker MS, Bergen AW, Cox SE, Dyer RA, Fulford AJ, Guan Y, Laritsky E, Silver MJ, et al.. Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat Commun 2014; 5:3746; PMID:24781383; http://dx.doi.org/ 10.1038/ncomms4746 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.