Abstract
Inter-α-trypsin inhibitor heavy chain 5 (ITIH5) is supposed to be involved in extracellular matrix stability and thus may play a key role in the inhibition of tumor progression. The current study is the first to analyze in depth ITIH5 expression and DNA methylation, as well as its potential clinical impact in non-small-cell lung carcinoma (NSCLC). We examined ITIH5 mRNA expression in tumor and adjacent normal lung tissue specimens of NSCLC patients. In addition, methylation frequency of the ITIH5 promoter was investigated using methylation-specific PCR and pyrosequencing. Significance of our data was validated by independent data sets from The Cancer Genome Atlas and the Kaplan-Meier Plotter platform. Furthermore, ITIH5 protein expression was evaluated by immunohistochemistry utilizing a tissue microarray with 385 distinct lung tissue samples. Based on our tissue collections, ITIH5 mRNA expression was significantly decreased in NSCLC compared to normal lung tissue in line with an increased methylation frequency in lung cancer tissue. Independent TCGA data confirmed significant expression loss of ITIH5 in lung cancer concordant with ITIH5 promoter hypermethylation in NSCLC. Of interest, low ITIH5 mRNA expression was particularly found in the magnoid and squamoid ADC expression subtype, concordant with an unfavorable patients' outcome in squamoid as well as tobacco smoking ADC patients. In conclusion, ITIH5 may be a novel putative tumor suppressor gene in NSCLC with a potential molecular significance in the squamoid ADC subtype and further clinical impact for risk stratification of adenocarcinoma patients. In addition, ITIH5 may serve as a novel biomarker for prognosis of tobacco smoking ADC patients.
Keywords: biomarker; DNA-methylation, non-small-cell lung cancer (NSCLC); gene signature; ITIH5; intrinsic expression subtype
Abbreviations
- EGFR
Epidermal growth factor receptor
- ITIH
inter-α-trypsin inhibitor heavy chain.
Introduction
Lung cancer is the most common cause of global cancer-related mortality, leading to over a million deaths each year.1,2 Due to this high mortality rate, a thorough understanding of tumor biological and molecular processes is mandatory to enable individual prognosis and novel targeted therapies.
To date, targeted therapies for non-small-cell lung cancer (NSCLC), the most common bronchial tumor, have dramatically improved treatment for patients whose tumors harbor driver mutations or rearrangements resulting in constitutively active mutant signaling proteins, most commonly occurring in oncogenes such as EGFR, ALK, HER2, BRAF, PIK3CA, AKT1, ROS1, and MAP2K1.3 Therapy involving tyrosine kinase inhibitors tailored to the genetic background of individual tumors can lead to improved clinical benefit compared to cytotoxic chemotherapy. However, up to 25% of NSCLC patients harbor clinically challenging KRAS mutations, which are currently not treatable by kinase inhibitors, or in other potentially clinical relevant oncogenic drivers (33%), resulting in conventional chemotherapy treatment.3,4
Within the last year, microarray expression profiling of adenocarcinoma of the lung, the most frequent histological type of NSCLC, revealed intrinsic molecular subtypes encompassing diverse functional pathways and patients' outcomes.5-11 Using microarray data analysis, Hayes et al. originally defined 3 ADC gene expression subtypes, namely bronchioid, magnoid, and squamoid, dependent on transcriptomic similarities with histologically defined bronchioalveolar, large-cell, and squamous-cell carcinoma.6 Transcriptional expression profiles co-occur with distinct mutations and alterations in patient tumors.6,7 For instance, bronchioid ADC are generally of lower grade, have a higher proportion of EGFR mutations, occur predominantly in women and never-smokers, and have favorable overall survival.6,7 In contrast, magnoid and squamoid ADC harbor more KRAS mutations, occur more often in men and smokers, and have poorer overall survival.6,7 However, the clinical impact of these expression signatures is currently not well understood, thus limiting progress in targeted therapy development in ADC patients.7,12
Deciphering of novel biomarkers affecting molecular pathways within the distinct expression signatures of ADC is a critical challenge for efficient stratification of patients who will benefit from a particular therapy regimen improving patients' survival. In this context, the previously identified inter-α-trypsin inhibitor heavy chain 5 (ITIH5) could play a valuable biological and clinical role. Loss of ITIH5 expression in mammary, colon, and bladder tumors, due to frequent hypermethylation of the ITIH5 promoter, was shown to be associated with malignant progression and unfavorable patients' outcome, indicating a putative tumor suppressor function in breast,13 colon,14 and bladder cancer.15 Furthermore, the ITI heavy chains effectively stabilize the ECM.16 and have been shown to suppress processes such as tumor invasion.17 and metastasis,18 while the biological relevance of ITIH5 in NSCLC has not been investigated yet.
The present study is the first to show that expression of ITIH5 is clearly deregulated in NSCLC, providing evidence for a potential role as tumor suppressor particularly in adenocarcinoma of the lung. Moreover, an unfavorable patients' outcome in squamoid as well as tobacco smoking ADC patients, due to the epigenetic silencing of the ITIH5 gene promoter, point toward a possible clinical impact of ITIH5 in adenocarcinoma of the lung.
Results
ITIH5 expression is lost in non-small-cell lung carcinoma
In a recent study, we showed that ITIH5 promoter hypermethylation is the molecular cause for ITIH5 gene silencing in breast,13 colon,14 and bladder cancer,15 which was associated with unfavorable patients' outcome. To investigate the biological relevance of ITIH5 in lung cancer we initially analyzed mRNA expression in 10 NSCLC tumor and 11 normal lung tissues by real-time PCR. We verified a significant (P < 0.0001) loss of ITIH5 mRNA expression in tumor (median expression level: 0.0965) when compared to normal lung tissues (median expression level: 0.9003) (Fig. 1A). Available matched normal and tumor tissue pairs from the same patients (n = 3) confirmed expression loss in tumor compared to adjacent normal lung tissue.
Figure 1.
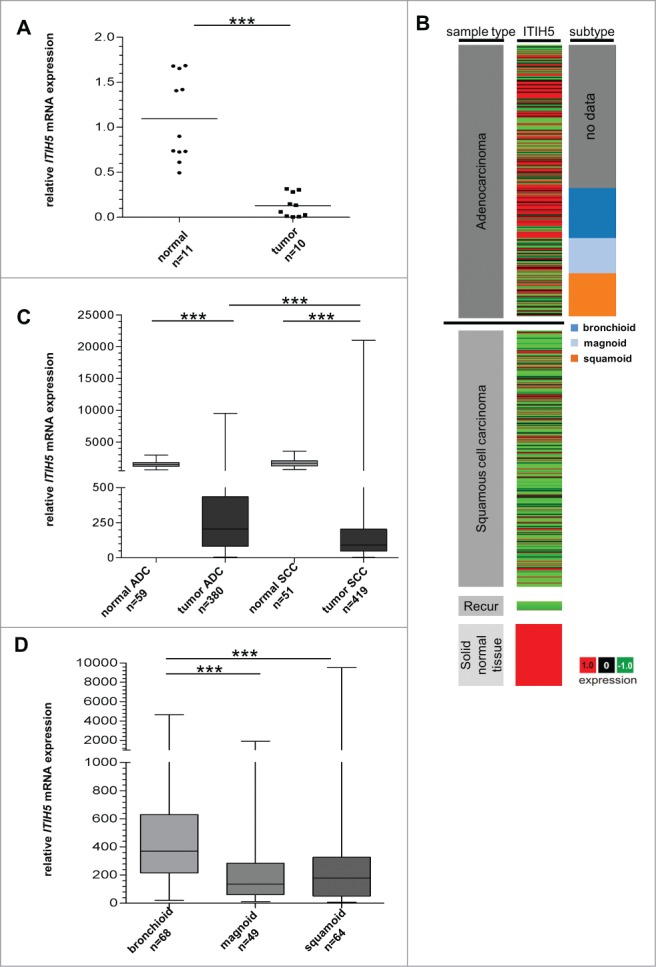
ITIH5 gene expression is lost in NSCLC cancer tissue. (A) ITIH5 mRNA expression is strongly decreased in NSCLC compared with normal lung tissue. Box plot analysis illustrates reduced ITIH5 mRNA expression in tumor tissue with a median expression level of 0.1402 compared to normal lung tissue (median expression level: 0.9003). Horizontal lines: grouped medians. ***P < 0.0001. (B) Tumor samples (based on TCGA Illumina HiSeq mRNA expression platform) are divided in adenocarcinoma (n = 380), squamous cell carcinoma (n = 419), recurrent (“Recur”) tumors (n = 2), and normal tissue samples (n = 110) (left panel). The middle panel illustrates relative values of ITIH5 mRNA expression: red (high expression) and green (low expression). Additionally, adenocarcinoma are stratified by expression subtype: bronchioid (n = 68), magnoid (n = 49), and squamoid (n = 64) (right panel). (C) Box plot analysis of ITIH5 mRNA expression in adenocarcinoma and squamous cell carcinoma compared to adjacent normal lung tissue. Horizontal lines: grouped medians. Boxes: 25–75% quartiles. Vertical lines: range, peak, and minimum. ***P < 0.0001. (D) Box plot analysis illustrates decreased expression of the ITIH5 gene in magnoid as well as squamoid adenocarcinoma. Horizontal lines: grouped medians. Boxes: 25–75% quartiles. Vertical lines: range, peak, and minimum. ***P < 0.0001.
To assess the significance of our data, we analyzed ITIH5 gene expression in a large dataset of an independent study. Using data of The Cancer Genome Atlas (TCGA) we verified a prevalent loss of ITIH5 gene expression in the major histological NSCLC subtypes, including ADC and SCC, when compared to adjacent normal lung tissue specimen (Fig. 1B and C). We found abundant ITIH5 mRNA expression in ADC (median expression level: 216) compared to SCC (median expression level: 94) (P < 0.0001) (Fig. 1C). Of interest, stratifying ADC patients by expression subtypes illustrated a significant decrease of ITIH5 mRNA expression, particularly in magnoid (median expression level: 136) and squamoid (median expression level: 178), contrary to bronchioid ADC (median FC: 370) (P < 0.0001) (Fig. 1D and Table 1). In addition, we could demonstrate a significant (P < 0.05) association of high ITIH5 mRNA expression concerning female and lower stage ADC patients (Table 1), while there were no significant correlations with any clinicopathological parameter in SCC patients.
Table 1.
Clinicopathological parameters in relation to ITIH5 mRNA expression in ADC of the lung using the TCGA data platform
| Variable |
ITIH5 mRNA expression |
||||
|---|---|---|---|---|---|
| Clinicopathological factors | na | low≤ 204b | high> 204b | P-valuec | r-valued |
| Age at diagnosis (median: 65 years) | |||||
| ≤ 65 years | 185 | 99 | 86 | 0.128 | 0.080 |
| > 65 years | 178 | 81 | 97 | ||
| gender | |||||
| Female | 205 | 91 | 114 | 0.018 | 0.121 |
| Male | 175 | 99 | 76 | ||
| Tumor size | |||||
| pT1–2 | 331 | 164 | 167 | 0.649 | −0.022 |
| pT3–4 | 47 | 25 | 22 | ||
| Lymph node status | |||||
| pN0 | 242 | 116 | 126 | 0.304 | −0.053 |
| pN1–3 | 129 | 71 | 58 | ||
| Tumor stage | |||||
| Stage I | 209 | 94 | 115 | 0.026 | −0.114 |
| Stage II-IV | 170 | 96 | 74 | ||
| Expression subtype | |||||
| Bronchioid | 68 | 15 | 53 | <0.001 | −0.297 |
| Magnoid | 49 | 35 | 14 | ||
| Squamoid | 64 | 36 | 28 | ||
| KRAS mutation status | |||||
| No mutation | 127 | 64 | 63 | 0.236 | 0.088 |
| mutation | 54 | 22 | 32 | ||
| EGFR mutation status | |||||
| No mutation | 158 | 77 | 81 | 0.392 | 0.064 |
| Mutation | 23 | 9 | 14 | ||
| ITIH5 methylation | |||||
| Low (≤47.4%) | 162 | 64 | 99 | 0.001 | −0.187 |
| High (>47.4%) | 170 | 98 | 71 | ||
Significant P-values marked in bold face.
Only patients with primary, invasive adenocarcinoma of the lung were included.
Median ITIH5 mRNA expression values.
Fisher's exact test at a 2-sided significance level of 0.05.
Pearson correlation coefficient.
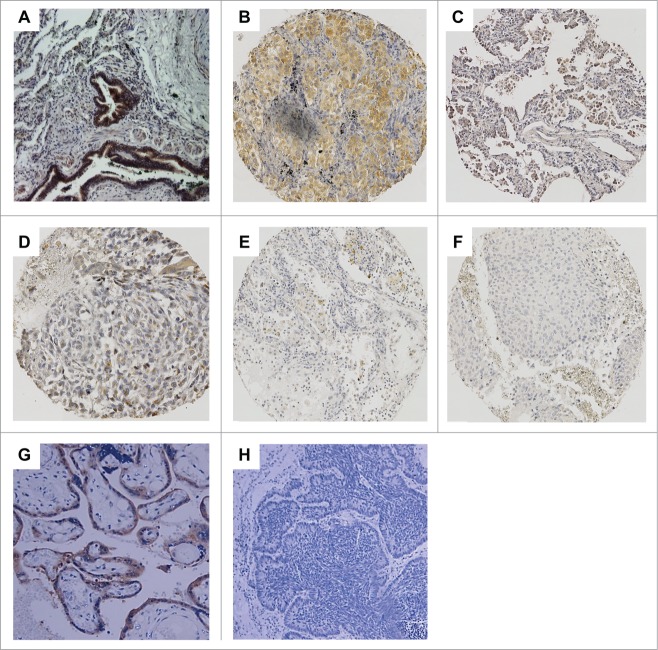
In concordance to the mRNA expression data, immunohistochemistry analysis based on a well-established polyclonal anti-ITIH5 antibody, confirmed abundant ITIH5 expression in epithelial cells of healthy respiratory tissue (Fig. 2A). As expected from the RNA expression pattern, NSCLC tumor cells showed decreased ITIH5 protein staining (Fig. 2B-D) or almost complete loss of ITIH5 protein (Fig. 2E and F). An association of ITIH5 protein expression with clinical data of the tissue microarray can be found in Table S1.
Figure 2.
Loss of ITIH5 protein expression in human lung cancer. (A) Strong ITIH5 expression in epithelial cells of normal respiratory tissue. (B–D) Moderate ITIH5 immunoreactivity in adenocarcinoma cells (B and C) and squamous cell carcinoma (D). (E) and (F) Very low staining in squamous cell carcinoma. (G) Strong ITIH5 protein expression in placenta tissue that served as positive control for ITIH5 staining. (H) Negative control of adenocarcinoma lung tissue. The application of primary antibody was omitted. Magnifications: (A, G, and H): × 200; (B, C, D, E, and F): × 100.
Decreased ITIH5 expression is caused by promoter hypermethylation, particularly in adenocarcinoma of the lung
Next, to prove if promoter methylation could be responsible for ITIH5 expression loss in lung cancer, we analyzed a set of 13 ADC and 14 SCC tissue samples by qualitative methylation-specific PCR (MSP). Methylation frequency of the ITIH5 gene promoter was 50% in SCC (i.e., 7 of 14 samples were methylated) and 54% in ADC (i.e., 7 of 13 samples were methylated). Considering the highest methylation value (6%) in normal lung tissue as the cut-off value, quantitative pyrosequencing in our independent cryoconserved tissue collection revealed a methylation frequency of 66% in ADC (i.e., 10 of 15 samples were methylated) as well as SCC (i.e., 4 of 6 samples were methylated) (Fig. 3A). Median methylation was significantly (P < 0.05) increased in ADC and SCC tissue samples compared to adjacent normal lung tissue, while ADC patients showed a higher median methylation (18%) in contrast to SCC patients (8.5%).
Figure 3.
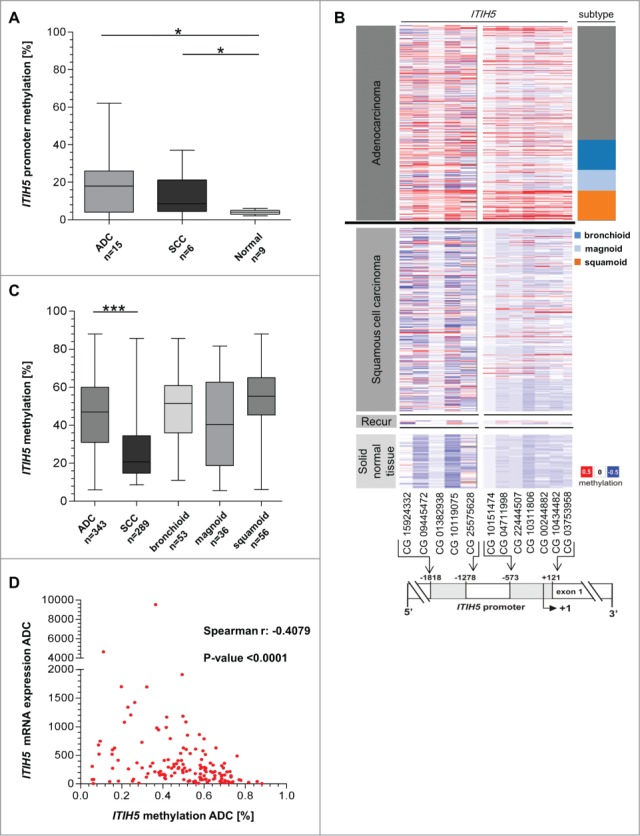
Expression of ITIH5 mRNA correlates with epigenetic inactivation particularly in adenocarcinoma. (A) Box plot diagram illustrates ITIH5 promoter hypermethylation between adenocarcinoma (ADC) (n = 15), squamous cell carcinoma (SCC) (n = 6), and normal lung (n = 9) specimens, based on quantitative pyrosequencing analysis. (B) DNA hypermethylation (Illumina HumanMethylation450 platform) of the ITIH5 promoter analyzed in lung cancer samples from the TCGA data portal. Tumor samples are divided in adenocarcinoma (n = 329), squamous cell carcinoma (n = 241), recurrent (“Recur”) tumors (n = 2), and normal tissue samples (n = 88) (left panel). The middle panel illustrates relative values of ITIH5 DNA hypermethylation for each CpG duplet: red (high methylation), white (mean methylation), and blue (low methylation). The relative positions of 12 analyzed CpG duplets (−1818 bp to +121 bp; 5′ to 3′) are indicated within a schematic map of the human ITIH5 promoter region. +1: ITIH5 transcription start site. The right panel shows the adenocarcinoma expression type bronchioid (n = 53), magnoid (n = 36), and squamoid (n = 56). (C) Box plot analysis demonstrates a significantly higher ITIH5 methylation level in all adenocarcinoma expression subtypes of the TCGA data portal compared to squamous cell carcinoma. Horizontal lines: grouped medians. Boxes: 25–75% quartiles. Vertical lines: range, peak and minimum, *P < 0.05, ***P < 0.001. (D) Scatter plot illustrates the association between expression (Illumina HiSeq mRNA expression platform) and DNA methylation status (Illumina HumanMethylation450 platform) of ITIH5 in 149 primary adenocarcinoma samples based on available TCGA data. Spearman correlation coefficient: r = −0.4053, P < 0.0001.
Based on the TCGA data set, CpG sites within the ITIH5 promoter that are closely located to the transcription start were commonly found methylated in primary lung cancer samples (Fig. 3B). In line with the data from our cryoconserved tissue collection, median methylation frequency of the ITIH5 gene promoter in the TCGA data were increased in ADC (including all ADC-specific expression subtypes) compared to SCC (Fig. 3C). To further examine the relation between ITIH5 mRNA expression loss and DNA-methylation, we performed a spearman correlation analysis between ITIH5 mRNA expression and methylation. This analysis revealed a higher inverse association between ITIH5 mRNA expression and DNA methylation in ADC (spearman r: −0.4053, P < 0.0001) (Fig. 3D) compared to SCC (spearman r: −0.2778, P < 0.0001).
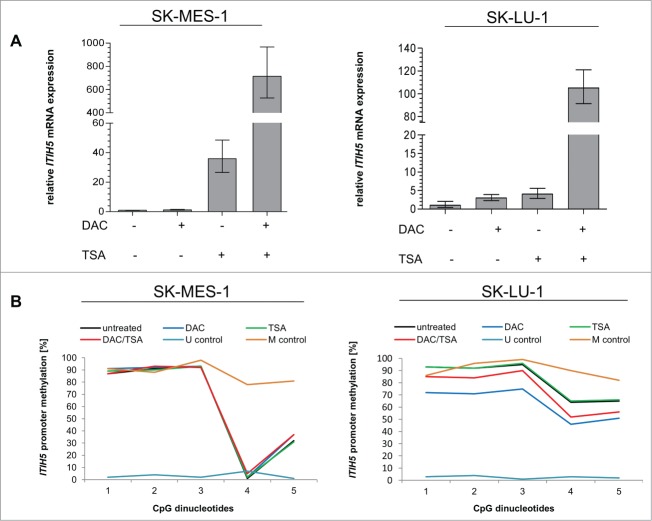
A functional association between ITIH5 promoter methylation and ITIH5 gene silencing was further supported by in vitro demethylation experiments using 2 different lung cancer cell lines lacking endogenous ITIH5 mRNA expression (SK-MES-1 (human squamous cell carcinoma) and SK-LU-1 (human adenocarcinoma). Real-time PCR analyses showed a clear re-expression of ITIH5 after 5-aza-2′-deoxycytidine (DAC) and trichostatin A (TSA) treatment in both cell lines compared to untreated cells (Fig. 4A), while methylation pattern in SK-MES-1 remained stable in contrast to SK-LU-1 cells (Fig. 4B).
Figure 4.
ITIH5 re-expression after in vitro demethylation. (A) Semi quantitative real-time PCR showing ITIH5 mRNA expression without and after treatment with 5-aza-2′-deoxycytidine (DAC) and Trichostatin A (TSA) in human SK-MES-1 and SK-LU-1 cells. Gain of ITIH5 mRNA expression is indicated as fold-change relative to each baseline expression. Expression of GAPDH served as a control for equal starting amounts of cDNA. Relative Y-axis scaling is related to non-treated SK-MES-1 and SK-LU-1 cells (set to one). SEM was derived from triplicate experiments. (B) Quantitative ITIH5 promoter methylation analysis of 5 CpG sites of SK-MES-1 and SK-LU-1 cells. Pyrogram of non-treated as well as DAC and TSA treated cells. Unmethylated and in vitro methylated DNA (Qiagen, Hilden, Germany) served as control.
Abundant ITIH5 expression predicts favorable outcome in patients diagnosed with squamoid ADC, with a pronounced clinical impact in tobacco smokers
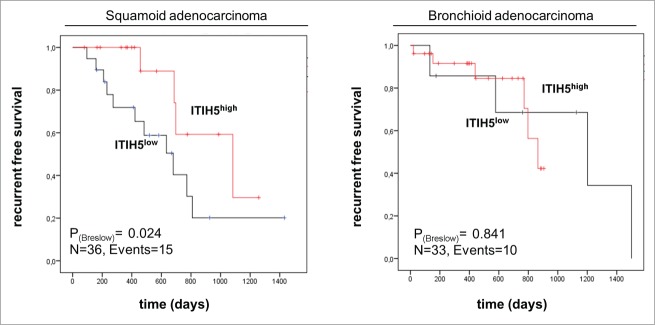
To reveal whether ITIH5 mRNA expression has an impact on patients' survival, a descriptive data analysis was performed with overall survival (OS) and recurrence free survival (RFS) data of the TCGA and the Kaplan-Meier Plotter (KMP) platform. RFS and OS data of the TCGA platform were compared between invasive ADC showing abundant ITIH5 mRNA expression (median expression ≥204) and SCC (median expression ≥90) by univariate statistics (Table 2). Concerning both ADC and SCC patients, KMP analysis revealed no prognostic impact of a strong ITIH5 mRNA expression concerning longer RFS or OS. However, with respect to a low ITIH5 mRNA expression in magnoid and squamoid ADC, we performed stratified univariate analysis within the expression subgroups. Interestingly, a favorable RFS could be demonstrated in the subgroup of squamoid ADC with abundant ITIH5 expression. Squamoid ADC patients with higher ITIH5 mRNA expression had an estimated median RFS of 2.96 years [95% confidence interval (CI): 1.36 – 4.57], compared to 1.86 years (95% CI: 1.07 – 2.65) in patients with decreased ITIH5 expression (P = 0.027) (Fig. 5). Next, we calculated a multivariate Cox regression model, including all factors potentially influencing the RFS in ADC, but statistical independency was missed. Of clinical relevance, evaluating data of the KMP platform, strong ITIH5 mRNA expression indicated a favorable outcome in tobacco smoking ADC patients (smoker at some point) (Fig. S1): Smoker patients with low ITIH5 expression had a worse OS compared to smoker patients showing high ITIH5 expression. In contrast, ITIH5 expression revealed no influence on patients' survival within the group of non-smoking adenocarcinoma patients.
Table 2.
Univariate analysis of clinicopathological factors regarding overall survival (OS) and recurrence-free (RFS) in ADC patients
| Variable | OS |
RFS |
||||
|---|---|---|---|---|---|---|
| Clinicopathological factors | na | Events | P-valueb | na | Events | P-valueb |
| Gender | ||||||
| Female | 113 | 28 | 0.168 | 113 | 27 | 0.715 |
| Male | 131 | 34 | 131 | 43 | ||
| Tumor size | ||||||
| pT1–2 | 214 | 52 | 0.004 | 214 | 59 | 0.024 |
| pT3–4 | 28 | 10 | 28 | 10 | ||
| Lymph node status | ||||||
| pN0 | 157 | 26 | 0.037 | 157 | 35 | 0.027 |
| pN1–3 | 80 | 35 | 80 | 4 | ||
| Tumor stage | ||||||
| Stage I | 133 | 19 | 0.001 | 133 | 29 | 0.030 |
| Stage II-IV | 110 | 43 | 110 | 41 | ||
| Expression subtype | ||||||
| Bronchioid | 33 | 10 | 0.642 | 33 | 10 | 0.014 |
| Magnoid | 20 | 8 | 20 | 11 | ||
| Squamoid | 37 | 15 | 37 | 15 | ||
| KRAS mutation status | ||||||
| No mutation | 66 | 26 | 0.605 | 66 | 26 | 0.067 |
| Mutation | 24 | 7 | 24 | 10 | ||
| EGFR mutation status | ||||||
| No mutation | 78 | 26 | 0.064 | 78 | 30 | 0.557 |
| Mutation | 12 | 7 | 12 | 6 | ||
| ITIH5 expression | ||||||
| Low (≤204) | 108 | 30 | 0.934 | 108 | 39 | 0.113 |
| High (>204) | 124 | 28 | 124 | 29 | ||
| ITIH5 methylation | ||||||
| Low (≤47.4%) | 112 | 29 | 0.403 | 112 | 38 | 0.018 |
| High (>47.4%) | 108 | 26 | 108 | 26 | ||
Only patients with primary, invasive adenocarcinoma of the lung were included.
Fisher's exact test at a 2-sided significance level of 0.05.
Figure 5.
Univariate survival analysis of ITIH5 mRNA expression according to the Kaplan–Meier Plotter (KMP) method reveals a favorable RFS in patients with squamoid adenocarcinoma. KMP analysis illustrating a prognostic value for ITIH5 in the clinically challenging group of patients with squamoid adenocarcinoma of the lung (which is not demonstrable in bronchioid adenocarcinoma patients). Red line: abundant ITIH5 expression (median ≥ 204); black line: weaker ITIH5 expression (median < 204). Vertical lines: censored cases.
To give a first insight into the close association of ITIH5 mRNA expression regarding the smoking-related mechanism of ADC patients, we re-analyzed a published transcriptomic microarray analysis consisting of 58 tissue samples of adenocarcinoma and 49 paired noninvolved lung tissue from current (n = 40), former (n = 36), and never smokers (n = 31).19 Again, array-based class comparison analysis revealed a significant (P < 0.0001) downregulation of ITIH5 mRNA expression in ADC cancer specimen compared to adjacent normal tissue samples (FC: 2.25). Besides, we identified more than 962 genes that are predominately downregulated in current compared to never smokers and 986 genes in former compared to never smokers, including ITIH5. Therewith, ITIH5 loss seems to be part of a common gene signature typical for the tobacco-smoking patients. A part of this signature, including ITH5, is shown as heatmap in Figure S2.
Discussion
Today, several lines of evidence suggest a potential role of ITIH5, a member of the ITIH family, in tumor biology, particularly in the prevention of tumor development and progression.13,20,21 We previously showed that loss of ITIH5 expression in breast,13 bladder,15 and colon cancer,14 caused by aberrant promoter hypermethylation, is associated with unfavorable prognosis. However, molecular relevance of ITIH5 in lung cancer remains elusive. The current study is the first to analyze in depth ITIH5 expression and DNA methylation, as well as its potential clinical impact toward NSCLC.
Initially, we verified, by both real-time PCR and immunohistochemistry, that ITIH5 was downregulated in human NSCLC tissue, suggesting that ITIH5 expression is lost in the course of tumor progression. To prove the accuracy of our results, we further analyzed independent ITIH5 mRNA expression data of the TCGA platform in NSCLC samples. In line, TCGA data analyses revealed a significantly decreased ITIH5 mRNA expression in NSCLC compared to normal lung tissue. Furthermore, TCGA data illustrated a significantly higher ITIH5 mRNA expression in ADC compared to SCC of the lung. To analyze the molecular cause of downregulation in ADC and SCC, we investigated the epigenetic configuration of the ITIH5 gene promoter, as it is known that the ITIH5 promoter sequence contains distinct CpG islands. Based on our cryoconserved tissue collection analyzed with quantitative pyrosequencing, we demonstrated a higher median methylation level in ADC compared to SCC tumor specimen. Moreover, hypermethylation of distinct CpG dinucleotides within the ITIH5 gene promoter discriminate significantly between lung cancer and normal tissue specimen. In line, TCGA data analyses confirmed the results of our independent cryoconserved tissue cohort by indicating a higher methylation frequency in ADC (median methylation: 47%) compared to SCC (median methylation: 20%). Concerning this data, one may hypothesize that ITIH5 promoter methylation is the leading molecular mechanism regulating ITIH5 gene expression in ADC, while in SCC further mechanisms may exist, resulting in low ITIH5 gene expression in unmethylated SCC. Indeed, in vitro demethylation of the human ADC cell line SK-LU-1 and the human SCC cell line SK-MES-1 revealed restoration of ITIH5 mRNA expression in both cell lines, but ITH5 promoter DNA methylation in SK-MES-1 cells remained stable. Additionally, we demonstrated a pronounced inverse correlation (r = −0.4079) of ITIH5 methylation and mRNA expression in ADC compared to SCC (r = −0.2778), strengthening promoter hypermethylation as the molecular cause of the ITIH5 loss in adenocarcinoma of the lung.
The predominant association of ITIH5 gene expression and promoter DNA methylation in ADC prompted us to take a deeper look into the putative impact of ITIH5 within the complex histological group of lung adenocarcinoma. In 2006, Hayes et al.6 described 3 major intrinsic molecular subtypes in ADC of the lung showing distinct gene expression patterns associated with significant morphologic and molecular heterogeneity.5,8-11 Clinical evaluation of these potentially intrinsic subtypes of ADC confers new opportunities for therapeutic strategies in the management of lung cancer patients, as it has successfully been demonstrated for the intrinsic breast cancer subtypes.22,23 Recently, new classification systems and developments at the molecular level using microarray expression profiling of ADC patients revealed further intrinsic molecular subtypes encompassing diverse functional pathways and patients' outcomes.24 Several studies confirmed an unfavorable prognosis of ADC patients with squamoid compared to those with bronchioid gene expression pattern.6 Contrary to the bronchioid subtype, a decreased ITIH5 mRNA expression was observed in magnoid and squamoid ADC. The low ITH5 mRNA expression in magnoid and squamoid ADC, in line with the different clinical characteristics of distinct expression subtypes, prompted us to perform a stratified univariate survival analysis in bronchioid compared to magnoid and squamoid ADC. Of clinical interest, we revealed a linkage of abundant ITIH5 mRNA expression with favorable RFS concerning the squamoid subtype. Because of its epigenetic mediated silencing, ITIH5 may be a potential novel tumor suppressor gene in ADC, particularly in the challenging clinical subgroups of squamoid and magnoid ADC, displaying an impact on tumor development.
Additionally, a subscribed molecular feature of squamoid adenocarcinoma of the lung is frequent KRAS mutation, while, in contrast, bronchioid ADC harbor mainly EGFR driver mutations.7 Interestingly, KMP data revealed a clear impact of abundant ITIH5 mRNA expression in tobacco-smoking patients, often showing multiple mutations, including a KRAS driver mutation,25,26 compared to non-smoking patients, who are known to have more frequent EGFR mutations. Hence, there is still a lack of suitable prognostic biomarkers for risk stratification in ADC patients showing KRAS mutation. Accordingly, all patients harboring KRAS mutation receive adjuvant chemotherapy, while the benefit is still controversial. In this context, ITIH5 may represent a novel prognostic biomarker with clinical utility in squamoid and KRAS mutated adenocarcinoma patients helping to estimate favorable patients' outcome. Of interest, based on in silico transcriptomic microarray analysis, we found a decreased ITIH5 expression level related to a common gene signature in tobacco-smoking ADC patients, providing strong evidence that ITIH5 loss is associated with smoking-related mechanism of ADC tumorigenesis. Nevertheless, the biological role of ITIH5 in the subtype- and smoking-related signaling pathway has to be unraveled in further studies.
In conclusion, these findings provide for the first time evidence that ITIH5 could act as a tumor suppressor gene in normal lung tissue. In addition, ITIH5 is potentially valuable as a prognostic biomarker for the clinically important group of patients with squamoid adenocarcinoma, whose disease management has to be adjusted to a personalized progression risk. Moreover, abundant ITIH5 expression might be an improved novel biomarker concerning tobacco-smoking ADC patients, with prognostic significance for patient survival. Further investigation of the contribution of ITIH5 to lung cancer progression concerning the potentially biological relevance in the ADC-specific intrinsic expression subtypes displaying distinct clinical characteristics may help to understand underlying pathways in more detail, finally helping to improve disease management.
Material and Methods
FFPE and cryoconserved patient samples
DNA methylation was analyzed using formalin-fixed, paraffin-embedded (FFPE) tissue collection, including ADC (n = 13) and SCC (n = 14) of the lung. In addition, an independent cryoconserved tissue collection for ITIH5 mRNA expression and DNA methylation analysis from patients with primary NSCLC, including ADC (n = 17) and SCC (n = 6), as well as adjacent normal tissue (n = 11), were obtained from the RWTH centralized biomaterial bank (RWTH cBMB; http://www.cbmb.rwth-aachen.de). All patients gave informed consent for retention and analysis of their tissue for research purposes (local ethical review board of the medical faculty of the RWTH Aachen, ref no. EK-206/09). Hematoxylin and eosin-stained sections were prepared for assessment of the percentage of tumor cells, only samples with >70% tumor cells were selected. An overview of the clinical characteristics of the patients is summarized in Tables S2 and S3.
In silico patient samples
Data from primary NSCLC tissues, including ADC and SCC of the lung, recurrent lung cancer tissues, and solid normal lung tissues were used from The Cancer Genome Atlas (TCGA), comprising patients data of 2 independent platforms: Illumina Infinium DNA methylation [HumanMethylation450 array and IlluminaHiSeq mRNA expression (ADC, n = 398; SCC, n = 419)]. An overview of the clinical characteristics of the patients is summarized in Table S4. In addition, data of the KMP portal was used to analyze a possible prognostic influence of abundant ITIH5 mRNA expression in ADC patients.27
Cell lines and reagents
The human adenocarcinoma cell line SK-LU-1 and the human squamous carcinoma cell line SK-MES-1 were obtained, tested, and authenticated from Cell Lines Service (Eppelheim, Germany) and were resuscitated before using in experiments. Used cell lines were regularly tested for mycoplasma infection using the PCR-based Venor® GeM Mycoplasma Detection Kit (Minerva Biolabs, Berlin, Germany).
Nucleic acid extraction and reverse transcription PCR
Genomic DNA from FFPE and cryoconserved NSCLC as well as normal lung tissue samples was isolated using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. By using TRIzol reagent (Invitrogen, Darmstadt, Germany) total cellular RNA from cell culture and tissue specimen was prepared. cDNA was synthesized using the reverse transcription system (Promega, Madison, WI) as previously described.13
Semiquantitative real-time PCR
cDNAs were amplified by semiquantitative real-time PCR using SYBR-Green PCR mix (Bio-Rad Laboratories, Munich, Germany) performed in an iCycler IQ5 (Bio-Rad Laboratories). Gene expression was quantified by the comparative CT method, normalizing CT-values to the housekeeping gene GAPDH, and calculating relative expression values.28 All primers used spanned at least one intron, and were described earlier.15 To ensure experiment accuracy, all reactions were performed in triplicate.
DNA bisulfite modification
The extracted tissue DNA was bisulfite-converted using the EZ DNA methylation kit (Zymo Research, Orange, CA, USA), as previously described.13
AZA/TSA treatment
A demethylation treatment of the lung cancer cell lines SK-LU-1 and SK-MES-1 was performed as previously described.29
Pyrosequencing
Pyrosequencing analysis of a distinct ITIH5 promoter region was performed by using the PyroMark PCR Kit (Qiagen) for initial fragment amplification. Afterwards, the PyroMark96 ID device and the PyroGoldSQA Reagent Kit (Qiagen) were implemented as previously described.30 The ITIH5 assays were designed by using the Pyromark Assay Design Software (Qiagen). Primers were described previously.15
ITIH5 immunohistochemistry
A lung cancer-specific tissue microarray (TMA) was kindly provided by Prof. I. Petersen (Institute of Pathology, University Hospital Jena) previously published by his working group.31 The TMA was constructed containing 380 lung cancer samples including SCC (n = 194), ADC (n = 83), non-small-cell lung cancer not otherwise specified (n = 47), lung cancer metastases (n = 29), neuroendocrine tumors (n = 10), large cell lung cancer (n = 7), and small-cell lung cancer (n = 10). Patients were operated between the years 1999 and 2002. Immunohistochemical staining was performed as previously described.15 with slight modifications. TMA slides were incubated with a polyclonal ITIH5 rabbit anti-human antibody (1:50) (Atlas Antibodies, Stockholm, Sweden). FFPE sections of non-cancerous placenta tissue served as positive control.
Statistical analysis
Statistical analysis was performed using SPSS 22.0 (SPSS, Chicago, IL) and GraphPad Prism 5.0 (GraphPad Software Inc., La Jolla, CA). The non-parametric Mann-Whitney U-test was used in order to compare ITIH5 mRNA expression between tumor and normal lung tissue, as well as ADC and SCC tumor specimen. Differences were considered statistically significant if the 2 sided P-values were equal or below 5% (≤0.05).
ITIH5 mRNA expression and DNA methylation pattern in human NSCLC samples and adjacent normal lung tissue were assessed using 2 independent own tissue collections, as well as an independent public data set (TCGA). Correlation of the ITIH5 mRNA expression (TCGA Illumina sequencing platform) and ITIH5 methylation data (TCGA HM450 platform) was performed by calculating a Spearman correlation coefficient. Recurrence-free survival (RFS) was measured from surgery until local or distant relapse and was censored for patients alive without evidence of relapse at the last follow-up. Multivariate Cox-regression analysis was carried out to test for an independent prognostic value of ITIH5 mRNA expression. Selection of the prognostic factors to be included in the multivariate model was based on statistical significance in univariate Breslow tests.
Re-analysis of gene expression using HG-U133A Affymetrix array to compare tobacco-smoking to never-smoking lung cancer patients.19 was performed using BRB-ArrayTools developed by Dr. Richard Simon and BRB-ArrayTools Development Team version 4.4.0 – Stable. In order to significantly identify genes differentially expressed between 2 classes the class comparison evaluation was used.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011; 61: 69-90 [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin 2014; 64: 9-29; PMID:24399786; http://dx.doi.org/ 10.3322/caac.21208 [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol 2011; 12: 175-80; PMID:21277552; http://dx.doi.org/ 10.1016/S1470-2045(10)70087-5 [DOI] [PubMed] [Google Scholar]
- 4.Pao W, Hutchinson KE. Chipping away at the lung cancer genome. Nat Med 2012; 18: 349-51; PMID:22395697; http://dx.doi.org/ 10.1038/nm.2697 [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al.. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA 2001; 98: 13790-5; PMID:11707567; http://dx.doi.org/ 10.1073/pnas.191502998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayes DN, Monti S, Parmigiani G, Gilks CB, Naoki K, Bhattacharjee A, et al.. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J Clin Oncol 2006; 24: 5079-90; PMID:17075127; http://dx.doi.org/ 10.1200/JCO.2005.05.1748 [DOI] [PubMed] [Google Scholar]
- 7.Wilkerson MD, Yin X, Walter V, Zhao N, Cabanski CR, Hayward MC, Socinski MA, Perou C, Meyerson M. Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PLoS One 2012; 7: e36530; PMID:22590557; http://dx.doi.org/ 10.1371/journal.pone.0036530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al.. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 2002; 8: 816-24; PMID:12118244 [DOI] [PubMed] [Google Scholar]
- 9.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, et al.. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A 2001; 98: 13784-9; PMID:11707590; http://dx.doi.org/ 10.1073/pnas.241500798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park YY, Park ES, Kim SB, Kim SC, Sohn BH, Chu IS, Jeong W, Mills GB, Byers LA, Lee JS. Development and validation of a prognostic gene-expression signature for lung adenocarcinoma. PLoS One 2012; 7: e44225; PMID:22970185; http://dx.doi.org/ 10.1371/journal.pone.0044225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeuchi T, Tomida S, Yatabe Y, Kosaka T, Osada H, Yanagisawa K, Mitsudomi T, Takahashi T. Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J Clin Oncol 2006; 24: 1679-88; PMID:16549822; http://dx.doi.org/ 10.1200/JCO.2005.03.8224 [DOI] [PubMed] [Google Scholar]
- 12.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst 2010; 102: 464-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veeck J, Chorovicer M, Naami A, Breuer E, Zafrakas M, Bektas N, Dürst M, Kristiansen G, Wild PJ, Hartmann A, et al.. The extracellular matrix protein ITIH5 is a novel prognostic marker in invasive node-negative breast cancer and its aberrant expression is caused by promoter hypermethylation. Oncogene 2008; 27: 865-76; PMID:17653090; http://dx.doi.org/ 10.1038/sj.onc.1210669 [DOI] [PubMed] [Google Scholar]
- 14.Kloten V, Rose M, Kaspar S, von SS, Knuchel R, Dahl E. Epigenetic inactivation of the novel candidate tumor suppressor gene ITIH5 in colon cancer predicts unfavorable overall survival in the CpG island methylator phenotype. Epigenetics. 2014; 9: 1290-301; PMID:25093535; http://dx.doi.org/ 10.4161/epi.32089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rose M, Gaisa NT, Antony P, Fiedler D, Heidenreich A, Otto W, Denzinger S, Bertz S, Hartmann A, Karl A, et al.. Epigenetic inactivation of ITIH5 promotes bladder cancer progression and predicts early relapse of pT1 high-grade urothelial tumours. Carcinogenesis 2014; 35: 727-36; PMID:24265292; http://dx.doi.org/ 10.1093/carcin/bgt375 [DOI] [PubMed] [Google Scholar]
- 16.Huang L, Yoneda M, Kimata K. A serum-derived hyaluronan-associated protein (SHAP) is the heavy chain of the inter α-trypsin inhibitor. J Biol Chem. 1993; 268: 26725-30; PMID:7504674 [PubMed] [Google Scholar]
- 17.Kobayashi H, Gotoh J, Hirashima Y, Fujie M, Sugino D, Terao T. Inhibitory effect of a conjugate between human urokinase and urinary trypsin inhibitor on tumor cell invasion in vitro. J Biol Chem. 1995; 270: 8361-6; PMID:7713945; http://dx.doi.org/ 10.1074/jbc.270.14.8361 [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi H, Shinohara H, Fujie M, Gotoh J, Itoh M, Takeuchi K, Terao T. Inhibition of metastasis of Lewis lung carcinoma by urinary trypsin inhibitor in experimental and spontaneous metastasis models. Int J Cancer 1995; 63: 455-62; PMID:7591248; http://dx.doi.org/ 10.1002/ijc.2910630326 [DOI] [PubMed] [Google Scholar]
- 19.Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et al.. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS.One 2008; 3: e1651; PMID:18297132; http://dx.doi.org/ 10.1371/journal.pone.0001651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oing C, Jost E, Dahl E, Wilop S, Brummendorf TH, Galm O. Aberrant DNA hypermethylation of the ITIH5 tumor suppressor gene in acute myeloid leukemia. Clin Epigenetics 2011; 2: 419-23; PMID:22704354; http://dx.doi.org/ 10.1007/s13148-011-0043-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veeck J, Breuer E, Rose M, Chorovicer M, Naami A, Bektas N, Alkaya S, von Serényi S, Horn F, Hartmann A, et al.. Novel prognostic marker in invasive breast cancer. ITIH5 expression is abrogated by aberrant promoter methylation. Pathologe 2008; 29 Suppll 2: 338-46; PMID:18810445; http://dx.doi.org/ 10.1007/s00292-008-1044-9 [DOI] [PubMed] [Google Scholar]
- 22.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al.. Molecular portraits of human breast tumours. Nature 2000; 406: 747-52; PMID:10963602; http://dx.doi.org/ 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- 23.Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, Davies SR, Snider J, Stijleman IJ, Reed J, et al.. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res 2010; 16: 5222-32; PMID:20837693; http://dx.doi.org/ 10.1158/1078-0432.CCR-10-1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warth A, Muley T, Meister M, Stenzinger A, Thomas M, Schirmacher P, Schnabel PA, Budczies J, Hoffmann H, Weichert W. The novel histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification system of lung adenocarcinoma is a stage-independent predictor of survival. J Clin Oncol 2012; 30: 1438-46; PMID:22393100; http://dx.doi.org/ 10.1200/JCO.2011.37.2185 [DOI] [PubMed] [Google Scholar]
- 25.Ahrendt SA, Decker PA, Alawi EA, Zhu Yr YR, Sanchez-Cespedes M, Yang SC, Haasler GB, Kajdacsy-Balla A, Demeure MJ, Sidransky D. Cigarette smoking is strongly associated with mutation of the K-ras gene in patients with primary adenocarcinoma of the lung. Cancer 2001; 92: 1525-30; PMID:11745231; http://dx.doi.org/ 10.1002/1097-0142(20010915)92:6%3c1525::AID-CNCR1478%3e3.0.CO;2-H [DOI] [PubMed] [Google Scholar]
- 26.Tam IY, Chung LP, Suen WS, Wang E, Wong MC, Ho KK, Lam WK, Chiu SW, Girard L, Minna JD, et al.. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res 2006; 12: 1647-53; PMID:16533793; http://dx.doi.org/ 10.1158/1078-0432.CCR-05-1981 [DOI] [PubMed] [Google Scholar]
- 27.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 2013; 8: e82241; PMID:24367507; http://dx.doi.org/ 10.1371/journal.pone.0082241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fink L, Seeger W, Ermert L, Hanze J, Stahl U, Grimminger F, Kummer W, Bohle RM. Real-time quantitative RT-PCR after laser-assisted cell picking. Nat Med 1998; 4: 1329-33; PMID:9809560; http://dx.doi.org/ 10.1038/3327 [DOI] [PubMed] [Google Scholar]
- 29.Veeck J, Niederacher D, An H, Klopocki E, Wiesmann F, Betz B, Galm O, Camara O, Dürst M, Kristiansen G, et al.. Aberrant methylation of the Wnt antagonist SFRP1 in breast cancer is associated with unfavourable prognosis. Oncogene 2006; 25: 3479-88; PMID:16449975; http://dx.doi.org/ 10.1038/sj.onc.1209386 [DOI] [PubMed] [Google Scholar]
- 30.Noetzel E, Rose M, Sevinc E, Hilgers RD, Hartmann A, Naami A, Knüchel R, Dahl f. Intermediate filament dynamics and breast cancer: aberrant promoter methylation of the Synemin gene is associated with early tumor relapse. Oncogene 2010; 29: 4814-25; PMID:20543860; http://dx.doi.org/ 10.1038/onc.2010.229 [DOI] [PubMed] [Google Scholar]
- 31.Kohler LH, Mireskandari M, Knosel T, Altendorf-Hofmann A, Kunze A, Schmidt A, Presselt N, Chen Y, Petersen I. FGFR1 expression and gene copy numbers in human lung cancer. Virchows Arch 2012; 461: 49-57; PMID:22648708; http://dx.doi.org/ 10.1007/s00428-012-1250-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.