Abstract
Weedy invasive Cirsium spp. are widespread in temperate regions of North America and some of their biological control agents have attacked native Cirsium spp. A phylogenetic tree was developed from DNA sequences for the internal transcribed spacer and external transcribed spacer regions from native and non-native Great Plains Cirsium spp. and other thistles to determine if host specificity follows phylogeny. The monophyly of Cirsium spp. and Carduus within the tribe Cardinae was confirmed with native North American and European lineages of the Cirsium spp. examined. We did not detect interspecific hybridization between the introduced invasive and the native North American Cirsium spp. Selected host-biological control agent interactions were mapped onto the phylogenic tree derived by maximum likelihood analysis to examine the co-occurrence of known hosts with biological control agents. Within Cirsium-Cardueae, the insect biological control agents do not associate with host phylogenetic lines. Thus, more comprehensive testing of species in host-specificity trials, rather than relying on a single representative of a given clade may be necessary; because the assumption that host-specificity follows phylogeny does not necessarily hold. Since the assumption does not always hold, it will also be important to evaluate ecological factors to provide better cues for host specificity.
Keywords: biological control, Canada thistle, Cirsium, Cirsium arvense, phylogeny, thistles, weed
1. Introduction
The invasion history, genetic diversity associated with founding populations, and evolutionary relationships to proximal native species should be considered in developing biological control management strategies for weeds [1,2]; gaps in such knowledge have led to failures in biological control [3]. Indeed, assessment of genetic diversity in invasive plant populations can assist in predicting the effectiveness and longevity of herbicidal, biological, and other control measures [4,5,6,7].
Canada thistle (Cirsium arvense (L.) Scop.) is one of the world’s most serious weeds and is a highly invasive plant in temperate regions of North America [8,9]. The introduction of Canada thistle to North America is suspected to result from contaminated goods shipped from Europe [10]. In 1795, Vermont was the first state to identify it as a noxious weed [11]. Canada thistle is now classified as a noxious weed by 49 states/provincial governments [12] because it causes economic loss through reduced crop yield, deterred grazing, and habitat loss in natural areas [9,13]. Auxin-type herbicides provide some control of Canada thistle; however, the most effective control is obtained by integration of chemical, mechanical, cultural, and biological control methods [8,14]. For example, multiple applications of the native bacterial biological control agent Pseudomonas syringae pv. tagetis in conjunction with other control measures were necessary to produce infection and sufficient damage to control growth and seed production of Canada thistle [15].
Several insects have been introduced to North America as biological control agents for non-native thistles (Cirsium arvense, Cirsium vulgare, and Carduus nutans (=Carduus thoemeri). The root and stem weevil Hadroplontus litura (=Ceutorhynchus litura) was released in North America in 1971 specifically for C. arvense control, but this agent has had little or no impact on C. arvense populations [8,16]. The weevils Rhinocyllis conicus and Larinus planus were introduced with some success for control of C. nutans (musk thistle). However, these insect species have non-target effects. For example, L. planus attacks C. arvense and the native Cirsium undulatum, as well as other native North American species [17,18,19]. Unfortunately, the risks associated with the release of biological control agents are not typically fully evaluated with regard to the native flora [18]; the focus is generally non-target effects on economically important plants like crops and forages [8].
Kelch and Baldwin [20] examined genetic diversity and ecological variation of North American Cirsium to determine the timing of New World thistle diversification, particularly those of the California Floristic Province. The phylogenetic estimates used maximum likelihood analysis of external transcribed spacer (ETS) and the internal transcribed spacer (ITS) of nuclear ribosomal DNA (nrDNA) sequence. Single origins were indicated for old world Cirsium, New World Cirsium, and representatives from the California Floristic Province. They indicated further sampling within the Cardueae was required to substantiate that Cirsium and Carduus are monotypic genera; these species were attributed to a single genus in the past. Inclusion of additional Great Plains representatives of these species would assist in determining relationships among taxa that are not well-delimited morphologically and co-occur in areas where C. arvense infestations are greatest.
We examined genetic variation and population structure within and between C. arvense populations to develop a greater understanding of the biology and reproductive mechanism [21,22]. The level of diversity within populations was greater than expected for a clonally reproducing perennial, indicating high level of outcrossing between populations in North America. Here, we evaluate the relationship of invasive and native North American thistles. The interaction (e.g., hybridization, introgression) of non-native invasive and native endemic thistle species in the Great Plains of North America was also evaluated to identify any increased potential of non-target effects with biological control agents. The occurrences of some known biological control agents were mapped onto the resulting phylogeny to investigate the patterns of host specificity and preference.
2. Results
Direct sequencing of polymerase chain reaction (PCR) products was successful for all samples (Table 1). Pairwise sequence similarity of the ETS, ITS1, and ITS2 regions ranged from 100% within Cirsium flodmanii (populations SD7, TS, and 905) for the ingroup of Cirsium taxa to 51.2% between Centaurea rigida and Tagetes spp. when considering all species. Within C. arvense, the greatest sequence similarity was for C. flodmanii representatives (100%) and the least similarity was for Cirsium monocephalum (91.3%). In species with multiple populations, average pairwise similarity was C. arvense 98.3%, C. flodmanii 99.9%, Cirsium muticum 99.6%, C. pitcheri 99.4%, C. undulatum 99.4%, and C. vulgare 99.5%.
Table 1.
Species for phylogenetic analysis are indicated. Sequences generated for the current study are indicated by the population identification numbers with species (e.g., arvense IN1.1), corresponding collection information (Sources of TBS and MF (authors) and collaborator (J. Fant), or voucher number). Voucher specimens were deposited at the U.S. National Arboretum (NA) herbarium and the accession number is provided. Published sequences included in the analysis are indicated by the accession number of GenBank.
| Genus | Species | Source | Voucher at NA | ITS | ETS |
|---|---|---|---|---|---|
| Cirsium | arvense IN1.1 | TBS2004-13 | 48765 | JX867618 | JX867646 |
| Cirsium | arvense Itasca 1 | TBS2004-70 | 48793 | JX867619 | JX867647 |
| Cirsium | arvense Itasca 6 | TBS2004-71 | 48792 | JX867620 | JX867648 |
| Cirsium | arvense MN3 | TBS2004-33 | 48790 | JX867621 | JX867649 |
| Cirsium | arvense ND25s8 | TBS2004-72 | 48842 | JX867622 | JX867650 |
| Cirsium | arvense ND26s35 | TBS2004-58 | 48834 | JX867623 | JX867651 |
| Cirsium | arvense TS | TBS2004-30 | 48817 | JX867624 | JX867652 |
| Cirsium | canescens SD7 | TBS | ---- | JX867625 | JX867653 |
| Cirsium | canovirens | TBS2005-36 | 48883 | JX867626 | JX867654 |
| Cirsium | flodmanii 64 | TBS2004-64 | 48867 | JX867627 | JX867655 |
| Cirsium | flodmanii 905 | TBS2004-62 | 48861 | JX867628 | JX867656 |
| Cirsium | flodmanii SD7 | TBS2005-25 | 48870 | JX867629 | JX867657 |
| Cirsium | flodmanii TS | TBS2004-60 | 48872 | JX867630 | JX867658 |
| Cirsium | foliosum | TBS2005-33 | 48878 | JX867631 | JX867659 |
| Cirsium | hillii | Jeremie Fant, Chicago Bot Gard | JX867632 | JX867660 | |
| Cirsium | muticum | TBS2005-17 | 48873 | JX867633 | JX867661 |
| Cirsium | pitcheri | Jeremie Fant, Chicago Bot Gard | JX867634 | JX867662 | |
| Cirsium | undulatum 903.1 | TBS2004-59 | 48877 | JX867635 | JX867663 |
| Cirsium | undulatum 904.1 | TBS2004-59 | 48877 | JX867636 | JX867664 |
| Cirsium | undulatum SD | TBS sn. 27 July 2005 | 48876 | JX867637 | JX867665 |
| Cirsium | vulgare 1.1 | MF2 | 48794 | JX867638 | JX867666 |
| Cirsium | vulgare 2.1 | MF2 | 48794 | JX867639 | JX867667 |
| Cirsium | vulgare SD7 | TBS2005-26 | 48812 | JX867640 | JX867668 |
| Cirsium | andersonii | GenBank | AF443683 | AF443735 | |
| Cirsium | andrewsii | GenBank | AF443684 | AF443736 | |
| Cirsium | arvense clone 1 | GenBank | AF443680 | AF443734 | |
| Cirsium | arvense clone 2 | GenBank | AF443681 | AF443734 | |
| Cirsium | arvense clone 3 | GenBank | AF443682 | AF443734 | |
| Cirsium | brevistylum | GenBank | AF443685 | AF443737 | |
| Cirsium | calcareum | GenBank | AF443687 | AF443739 | |
| Cirsium | canovirens | GenBank | AF443688 | AF443740 | |
| Cirsium | canum | GenBank | AF443689 | AF443741 | |
| Cirsium | congdonii | GenBank | AF443690 | AF443742 | |
| Cirsium | cymosum | GenBank | AF443691 | AF443743 | |
| Cirsium | discolor | GenBank | AF443692 | AF443744 | |
| Cirsium | douglasii | GenBank | AF443686 | AF443738 | |
| Cirsium | eatonii | GenBank | AF443694 | AF443746 | |
| Cirsium | edule | GenBank | AF443711 | AF443763 | |
| Cirsium | ehrenbergii | GenBank | AF443726 | AF443778 | |
| Cirsium | faucium | GenBank | AF443725 | AF443777 | |
| Cirsium | fontinale var. obispoense | GenBank | AF443696 | AF443748 | |
| Cirsium | henryi | GenBank | AF443697 | AF443749 | |
| Cirsium | hydrophilum | GenBank | AF443698 | AF443750 | |
| Cirsium | jorullense | GenBank | AF443699 | AF443751 | |
| Cirsium | lineare | GenBank | AF443727 | AF443779 | |
| Cirsium | mohavense | GenBank | AF443700 | AF443752 | |
| Cirsium | monocephalum | GenBank | AF443701 | AF443753 | |
| Cirsium | monspessulanum | GenBank | AF443717 | AF443769 | |
| Cirsium | muticum | GenBank | AF443722 | AF443774 | |
| Cirsium | neomexicanum | GenBank | AF443718 | AF443770 | |
| Cirsium | occidentale var. venustum | GenBank | AF443703 | AF443755 | |
| Cirsium | occidentale | GenBank | AF443702 | AF443754 | |
| Cirsium | palustre | GenBank | AF443704 | AF443756 | |
| Cirsium | pitcheri | GenBank | AF443705 | AF443757 | |
| Cirsium | quercetorum | GenBank | AF443706 | AF443758 | |
| Cirsium | remotifolium | GenBank | AF443707 | AF443759 | |
| Cirsium | rhaphilepis | GenBank | AF443708 | AF443760 | |
| Cirsium | rhothophilum | GenBank | AF443709 | AF443761 | |
| Cirsium | rydbergii | GenBank | AF443710 | AF443762 | |
| Cirsium | scariosum | GenBank | AF443693 | AF443745 | |
| Cirsium | spinosissimum | GenBank | AF443720 | AF443772 | |
| Cirsium | subniveum | GenBank | AF443712 | AF443764 | |
| Cirsium | tioganum | GenBank | AF443721 | AF443773 | |
| Cirsium | tymphaeum | GenBank | AF443723 | AF443775 | |
| Cirsium | utahense | GenBank | AF443713 | AF443765 | |
| Cirsium | velatum | GenBank | AF443714 | AF443766 | |
| Cirsium | vulgare clone 1 | GenBank | AF443715 | AF443767 | |
| Cirsium | vulgare clone 2 | GenBank | AF443716 | AF443738 | |
| Cirsium | wheeleri | GenBank | AF443719 | AF443771 | |
| Carduus | acanthoides | MF3 | 48795 | JX867643 | JX867669 |
| Carduus | nutans | GenBank | AF443678 | AF443730 | |
| Carduus | nutans | TBS2005-14 | 48801 | JX867642 | JX867670 |
| Carduus | tenuiflorus | GenBank | AF44679 | AF4433731 | |
| Carthamus | oxyacanthus | GenBank | AJ867986-7 | AJ867985 | |
| Centaurea | rigidi | GenBank | AJ867989 | AJ867988 | |
| Cynara | scolymus | TBS | greenhouse grown | JX867643 | JX867671 |
| Helianthus | anuus | TBS | greenhouse grown | JX867644 | Not sequenced |
| Jurinea | narynensi | GenBank | AJ868001-2 | AJ868000 | |
| Onopordum | acaulon | GenBank | AF443676 | AF443728 | |
| Onopordum | illyricum | GenBank | AY78046 | AF4433729 | |
| Saussurea | riederi | GenBank | AJ868070-1 | AJ868069 | |
| Tagetes | spp. | TBS | greenhouse grown | JX867645 | JX867672 |
Eighty-nine sequences representing 59 species were analyzed for ITS1, ITS2, and ETS for the total combined analysis; the 5.8S nrDNA region was excluded due to missing data for several taxa in GenBank (Table 1). Partition homogeneity indicated (p = 0.01) the ITS and ETS data sets were not incongruent. When considering all samples, 414 out of 1,082 base pair alignments analyzed were parsimony informative and 154 were parsimony informative within the ingroup of Cirsium. Thirty-seven most parsimonious trees arose with a tree length of 1,574 steps in a heuristic search using tree bisection-reconnection (TBR) branch-swapping with random addition of 1,000 bootstrap replicates. Bootstrap analysis conducted using the above parameters indicated high support (>75%) for 22 clades and moderate support (50–75%) for an additional 13 clades. Most notably, there was high support for the genus Cirsium (82%) and Carduus (100%), as well as species clades: C. arvense (95%), C. vulgare (97%), C. flodmanii (98%), and C. undulatum (100%). Bootstrap support values greater than 50% were not obtained for unique North American or European Cirsium clades. Taxa did not form robust clades based upon geographical regions within North American endemic Cirsium, except for a clade of the endemic California taxa.
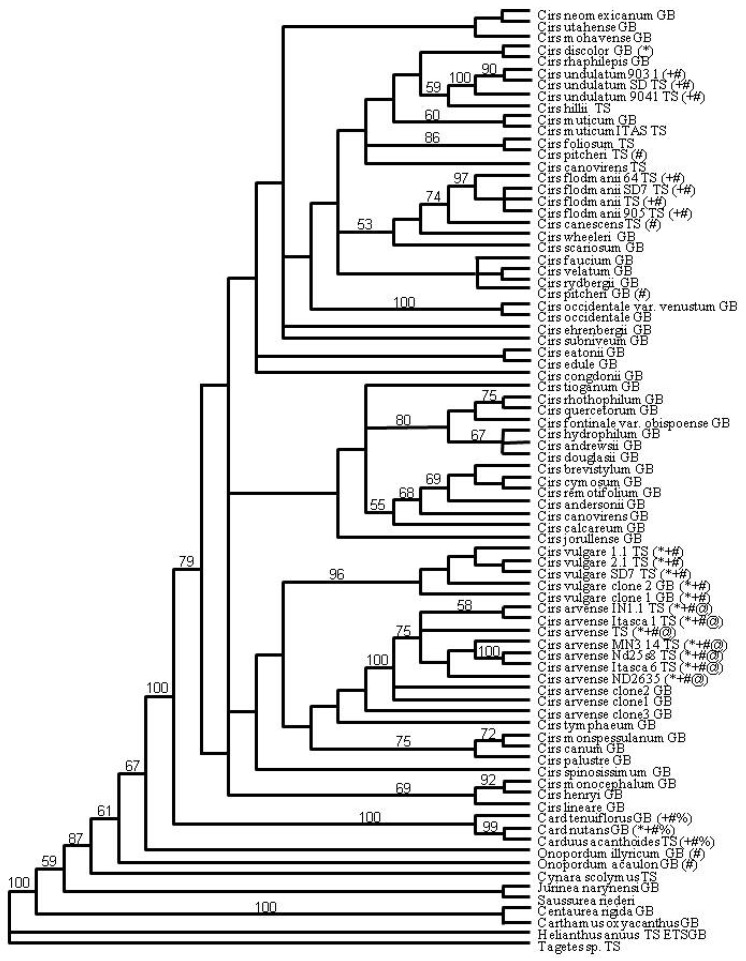
A greater resolution of species relationships was obtained using maximum likelihood analysis (Figure 1). Separate clades were obtained for the North American and the European Cirsium species in addition to species clades obtained from the bootstrap analyses. Known host-biological control agent interactions were mapped onto the phylogenic tree (Figure 1). Within Cirsium-Cardueae, the majority of biological control agents do not associate with the phylogenetic lines.
Figure 1.
Maximum likelihood analysis of ITS1, ITS2, and ETS. Biological control agents are labeled as follows Trichosirocalus horridus (*), Larinus planus (+), Rhinocyllis conicus (#), Puccinia carduorus (%), and Pseudomonas syringe (@). Cirs = Cirsium, Card = Carduus, TS = sequence generated here, GB = sequence available on GenBank. Bootstrap values (1,000 iterations) are indicated when support was greater than 50%.
3. Discussion
Cirsium and Carduus are monophyletic genera based on our analysis, supporting Kelch and Baldwin [20] and Garcia-Jacas et al. [23]. The genera are morphologically distinct with the plumouse pappus of Cirsium and the generally winged stems of Carduus [24]. Tribal relationships resolved in our investigation also support those of Garcia-Jacas et al. [23]. The Cardinae subtribe composed of the Carduus group, Onopordum, Cynara, Jurinea, and Sassurea did not resolve with >50% bootstrap support and is paraphyletic with Centaurea, consistent with analysis of ITS and matK [23].
Fine scale analyses within Cirsium did not resolve phylogeographic relationships; for example, the Great Plains species do not form a clade. A single origin is indicated for the North American taxa separating these from the European species, but bootstrap support was weak (<50%). This single origin is in general agreement with Kelch and Badwin [20]. The clade (80% bootstrap) of endemic California species is the only group reflecting geographical distribution; however, other species with broader distributions were not included in this clade. These narrow endemics are most likely of recent origin derived from taxa found west of the Rocky Mountains [20].
Representatives included from the Northern Great Plains (east of the Rocky Mountains, west of the Great Lakes and north of Nebraska) formed in clades based on species identity when multiple populations were sampled, but did not segregate as a geographic group. A loosely grouped (e.g., short branch lengths and lack of bootstrap support) set of Cirsium canescens, Cirsium canovirens, Cirsium discolor, C. flodmanii, Cirsium foliosum, Cirsium hillii, Cirsium pitcheri, Cirsium scariosum, C. undulatum, and Cirsium wheeleri resolved as paraphyletic with several Mexican taxa [25]. The majority of these taxa are thought to have originated in a species complex in the mountainous, western regions of North America [26].
Chromosomal numbers observed for Cirsium brevifolium (2n = 22), C. canovirens (2n = 34), C. flodmanii (2n = 22), C. pitcheri (2n = 34), C. undulatum var. tracyi (syn. C. tracyi 2n = 24), C. undulatum (2n = 26), and C. wheeleri (2n = 28) led to their placement in the series Undulata with a basal chromosome number of 2n = 34 and subsequent reduction during species diversification and expansion [26]. Cirsium altissimum (2n = 18), C. discolor (2n = 20), and C. muticum (2n = 20) were placed in separate series (Altissima) based on morphological characters and thought to be derived within an eastern, plains to rolling hills complex of taxa. Several distinctions between C. altissimum, C. brevifolium, and C. flodmanii to others in the Undulata series include the lack of mucilage on wet achenes and the presence of a yellow apical band on achenes. Considering the distribution of C. flodmanii and C. pitcheri, with no locations west of the Rocky Mountains, these species may be derived from an eastern complex of taxa that moved westward, as reflected in the lack of a well-supported clade of the Undulata series. Separation of these taxa into the defined series is not supported as paraphyly of the biogeographical groups. No gene flow between these species was indicated by the ITS and ETS sequences analyzed. Thus, we conclude that these species remain genetically distinct.
Morphological similarities and distributional overlap do not correspond to the phylogeny as C. canescens, C. discolor, C. flodmanii, and C. undulatum resolve as moderately to strongly supported unique species in separate clades based on the molecular analyses. These species are difficult to distinguish morphologically based on a gradation of leaf and stem pubescence, depth of leaf sinuses, and flower head shape [27]. Additionally, habitat preference and distribution also delimit these taxa. C. discolor is distributed further east (western Dakotas to the Atlantic) than the other taxa, with C. flodmanii (Michigan to Idaho) and C. canescens (Great Plains) in prairie habitats, and C. undulatum (Indiana, Texas to the Pacific) in dry grasslands [28].
Concerted evolution has been sufficient in the introduced and North American endemic species to homogenize ribosomal repeat region. Conspecifics formed independent clades with North American (C. muticum, C. flodmanii, and C. undulatum) and worldwide (C. arvense, C. vulgare) distributions. Concerted evolution of the nrDNA has occurred with the North American endemic species since their separation from Eurasian taxa during the Late Miocene (12 million years ago) [29]. The relatively recent introduction of the Eurasian C. arvense and C. vulgare (<300 years ago) to North America, in conjunction with the relatively low sequence divergence and high degree of concerted evolution of the nrDNA, supports continued gene flow within these species across North America or lack of lineage sorting. Strong support for clades consisting of representatives across the range for C. arvense and C. vulgare indicate a large source of genetic diversity in their ranges and potentially multiple introduction events consistent with Guggisberg et al. [10], at least for C. arvense.
The insect biological control agents do not follow the phylogenic relationships of hosts as judged by specificity of biological control options for Cirsium arvense, C. vulgare, and C. nutans (Figure 1). The weevil L. planus feeds upon C. arvense and C. palustre and other Cirsium, Carduus, Galactities, and Serratula species over its native range from Europe to North Africa [30]. L. planus is now found throughout the Great Plains of North America in areas with heavy infestations of invasive thistles. Its larvae develop within flower heads destroying florets leading to up to 95% suppression of seed production in C. arvense and Carduus, but also in the native thistles such as C. undulatum var. tracyi and C. flodmanii [17]. There is no correlation between host phylogenetic relationships and non-target effects as determined by phylogenetic mapping of thistle species affected by L. planus (Figure 1). Basically, L. planus is opportunistically feeding upon native species when there is an insufficient source of the targeted hosts like C. arvense.
Rhinocyllus conicus attacks seed heads of Carduus spp., Cirsium spp., and Silybum marianum within its native range in Europe [31]. Host-plant specificity tests in Europe for feeding, ovipositing, and better larval performance on C. nutans than on the Cirsium spp. influenced its selection as a biological control agent for C. nutans in North America [32]. Its introduction had mixed results [33,34]. Although the most efficient ovipositing and larval development in seeds heads occurred for C. nutans, C. arvense, and C. vulgare in North America, ovipositing was also discovered to occur in the native North American thistle species C. canescens, C. centaureae, C. flodmanii, C. pitcheri, Cirsium pulchellum, and C. undulatum [34]. The native thistles have greater pubescence and are genetically distant from the invasive species (Figure 1), yet non-target oviposit and feeding occurs. In the absence of the preferred host C. nutans, R. conicus fed on thistles with similar phenology and synchronous flowering times, which reduces seed set and population viability of the native thistle [32,34]. Prediction of non-target host selection for R. conicus would not have identified the native thistle C. canescens as a host plant based on plant morphology and the phylogenetic relationships (Figure 1). Likewise, prediction of non-target host selection for the foliar feeder Trichosirocalus horridus, which also was introduced from Europe into the U.S. in late 1960s as biological control agent for C. nutans [35], may not have identified the potential for foliar damage observed on North American native thistles C. altissimum, C. discolor, and C. carolinianum [36]. It is now known that various ecological factors like habitat preference of the biological control agent and geographical proximity to related plants provide better cues to potential alternative hosts [32,36,37,38].
Pathogens can be a particularly useful tool for weed control in natural areas that are rich in valued non-target species [39]. The fungal pathogen P. carduorum was evaluated as a biological control agent for Carduus spp. (musk thistles) [24]. P. carduorum collected from Turkey and Bulgaria was inoculated on three large flowered Carduus spp., twenty-four Cirsium spp., and C. scolymus. The Cirsium spp. selected for screening included a portion of the taxa that geographically overlapped the targeted Carduus thoemeri. In contrast with the above mentioned insect biological control agents, the strain of rust fungus tested by Bruckart et al. [24] coincides with plant host phylogenetic lines (Figure 1), as only Carduus spp. were susceptible. More extensive testing on seven rare, endangered, or threatened Cirsium spp. in California and extensive analysis using molecular marker data support that the rust strain only affected Carduus spp. [40,41].
Cirsium is a genus with a high affinity to form natural interspecific hybrids [42]. Fortunately, we did not detect interspecific hybridization between the introduced invasive and the native North American Cirsium spp. because such hybrids may provide a bridge for movement of host-specific biological control agents to expand their host range to non-target parental plants [43]. However, we did detect higher levels of variation within the invasive- relative to the native-Cirsium spp. This intraspecific genetic variation in the C. arvense may present challenges for identification of highly efficacious host-specific biological control agents. Molecular-based approaches that evaluate the phylogenetic or genetic diversity of invasive host plants and insect and pathogen biological control agents will be important for matching hosts and potential biological control agents [43]. Beyond the molecular-based pairing and the phylogenetic methods for delineating host range [44], it will be important to evaluate ecological factors to provide better cues to potential alternative hosts since some biological control agents do not follow host phylogenetic lines [37].
4. Experimental Section
4.1. Plant Material
Leaf material was collected from 15 to 43 individuals per population of Cirsium arvense (Table 1) and either dried with silica gel or kept at 4–8 °C until frozen at −80°C. Material from other Cirsium spp. (Table 1) was collected from 1 to 6 individuals per population and stored as described. Genomic DNA was extracted using the DNEasy kit (Qiagen Inc., Valencia, CA, USA). The DNA was quantified by spectrophotometry (Nanodrop Technologies, Wilmington, DE, USA).
Molecular markers for 26 samples representing eleven Great Plains Cirsium, two Carduus, and a Cynara (artichoke) species were sequenced to compile a matrix with published Cirsium sequences (Table 1). Multiple populations were included for species to examine variation between conspecifics. Populations with multiple species of Cirsium were included to determine if interspecific hybridization occurs between native and introduced species.
4.2. Amplification and Sequencing
Primers previously used for amplification of Cardueae (18S-ETS and ETS-Car1 and ITS1 and ITS4) [20] successfully amplified the regions for all samples. Reaction conditions were 1× Buffer E (Epicentre Biotechnologies, Madison, WI, USA), 0.5 mM primer, and 0.5 U Taq Polymerase (Promega Corp., Madison, WI, USA) with 10–25 ng genomic DNA. Amplification program parameters for ITS and ETS regions were those of Kelch and Baldwin [20]. Polymerase chain reaction (PCR) products were purified using a QiaQuick Gel Extraction kit (Qiagen Inc, Valencia, CA, USA) and quantified by spectrophotometery. Sequences were obtained using amplification primers (5 pmol) and 20–50 ng PCR product and sequenced with an ABI 3730 DNA Analyzer (Applied Biosystems, Inc., Foster City, CA, USA).
4.3. Data Analysis
Contiguous consensus sequences were compiled from double stranded DNA using Seqman (LaserGene, DNAStar, Madison, WI, USA). Alignments for the combined and independent data sets were produced in CLUSTALX, with gaps treated as missing data [45]. We deposited new DNA sequences in GenBank (Table 1). Phylogenetic analyses were conducted in PAUP*4.0b10 [46] with random stepwise addition of 100 iterations and tree bisection-reconnection branch-swapping. Partition homogeneity analysis in PAUP identified if the ITS and ETS were incongruent. The HKY85 model of evolution was used for maximum likelihood analysis [20]. Divergence between samples was calculated in PAUP as pairwise sequence similarity.
Several introduced and native biological control agents (e.g., insects and pathogens) were mapped onto the phylogeny to examine patterns of host preference [8,14,17,36,40]. The non-native, introduced insects were L. planus, R. conicus, and Trichosirocalus horridus, native pathogens were the rust pathogen Puccinia carduorum, and the bacterial pathogen P. syringae pv. tagetis.
5. Conclusions
From this research we conclude that there has not been interspecific hybridization between the introduced invasive such as Canada thistle and the native North American Cirsium spp. In addition, within Cirsium-Cardueae, the insect biological control agents do not associate with host phylogenetic lines. Thus, more comprehensive testing of species in host-specificity trials, rather than relying on a single representative of a given clade may be necessary; because the assumption that host-specificity follows phylogeny does not necessarily hold. Even if the assumption does not always hold, it is also important to evaluate ecological factors like habitat preference of the biological control agent and geographical proximity to related plants to provide better cues for host specificity [3,34].
Acknowledgment
Lab assistance was provided by Laura Kelley.
References
- 1.Sakai A.K., Allendorf F.W., Holt J.S., Lodge D.M., Molofsky J., With K.A., Baughman S., Cabin R.J., Cohen J.E., Elstrand N.C., et al. The population biology of invasive species. Ann. Rev. Ecol. Syst. 2001;32:305–332. doi: 10.1146/annurev.ecolsys.32.081501.114037. [DOI] [Google Scholar]
- 2.Simberloff D. How much information on population biology is needed to manage introduced species? Conserv. Biol. 2003;17:83–92. doi: 10.1046/j.1523-1739.2003.02028.x. [DOI] [Google Scholar]
- 3.Louda S.M., Rand T.A., Russell F.L., Arnett A.E. Assessment of ecological risks in weed biocontrol: Input from retrospective ecological analyses. Biol. Control. 2005;35:253–264. doi: 10.1016/j.biocontrol.2005.07.022. [DOI] [Google Scholar]
- 4.Hodgson J.M. The response of Canada thistle ecotypes to 2,4-D, amitrole, and intensive cultivation. Weed Sci. 1970;18:253–254. [Google Scholar]
- 5.Donald W.W. Management and control of Canada thistle (Cirsium arvense) Rev. Weed Sci. 1990;5:193–250. [Google Scholar]
- 6.Nissen S.J., Masters R.A., Lee D.J., Rowe M.L. DNA-based markers systems to determine genetic diversity of weedy species and their application to biocontrol. Weed Sci. 1995;44:504–513. [Google Scholar]
- 7.Lavergne S., Molofsky J. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc. Natl. Acad. Sci. USA. 2007;104:3883–3888. doi: 10.1073/pnas.0607324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McClay A.S. Canada thistle. In: van Driesche R., Blossey B., Hoddle M., Lyon S., Reardon R., editors. Biological Control of Invasive Plants in the Eastern United States. USDA Forest Service Publication FHTET-2002-04; USDA Forest Service, Morgantown, WV., USA: 2002. pp. 217–228. [Google Scholar]
- 9.Tiley G.E.D. Biological flora of the British Isles: Cirsium arvense (L.) Scop. J. Ecol. 2010;98:938–983. doi: 10.1111/j.1365-2745.2010.01678.x. [DOI] [Google Scholar]
- 10.Guggisberg A., Welk E., Sforza R., Horvath D.P., Anderson J.V., Foley M.E., Rieseberg L.H. Invasion history of North American Canada thistle, Cirsium arvense. J. Biogeogr. 2012;39:1919–1931. doi: 10.1111/j.1365-2699.2012.02746.x. [DOI] [Google Scholar]
- 11.Voss E.G. Michigan Flora: Part III Dicots (Pyrolaceae to Compositae) Cranbrook Institute; Bloomfield Hills, MI, USA: 1996. pp. 510–524. [Google Scholar]
- 12.Invaders Database System. [(accessed on 18 July 2012)]. Available online: http://invader.dbs.umt.edu/noxious_weeds/noxlist.asp/
- 13.Lym R.G., Duncan C.L. Canada thistle Cirsium arvense (L.) Scop. In: Duncan C.L., Clark J.K., editors. Invasive Plants of Range and Wildlands and Their Environmental, Economic, and Societal Impacts. Weed Science Society of America; Lawrence, KS, USA: 2005. pp. 69–83. [Google Scholar]
- 14.Hoeft E.V., Jordan N., Zhang J., Wyse D.L. Integrated cultural and biological control of Canada thistle in conservation tillage soybean. Weed Sci. 2001;49:642–646. doi: 10.1614/0043-1745(2001)049[0642:ICABCO]2.0.CO;2. [DOI] [Google Scholar]
- 15.Tichich R.P., Doll J.D. Field-based evaluation of a novel approach for infecting Canada thistle (Cirsium arvense) with Pseudomonas syringae pv. tagetis. Weed Sci. 2006;54:166–171. doi: 10.1614/WS-03-144R3.1. [DOI] [Google Scholar]
- 16.Cripps M.G., Gassmann A., Fowler S.V., Bourdôt G.W., McClay A.S., Edwards G.R. Classical biological control of Cirsium arvense: Lessons from the past. Biol. Control. 2011;57:165–174. doi: 10.1016/j.biocontrol.2011.03.011. [DOI] [Google Scholar]
- 17.Louda S.M., O’Brien C.W. Unexpected ecological effects of distributing the exotic weevil, Larinus planus (F), for the biological control of Canada thistle. Conserv. Biol. 2002;16:717–727. doi: 10.1046/j.1523-1739.2002.00541.x. [DOI] [Google Scholar]
- 18.Louda S.M., Pemberton R.W., Johnson M.T., Follett P.A. Nontarget effects—The Achilles’ Heel of biological control? Retrospective analyses to reduce risk associated with biocontrol introductions. Ann. Rev. Entomol. 2003;48:365–396. doi: 10.1146/annurev.ento.48.060402.102800. [DOI] [PubMed] [Google Scholar]
- 19.Rand T.A., Louda S.M. Exotic weed invasion increases the susceptibility of native plants to attack by a biocontrol herbivore. Ecology. 2004;85:1548–1554. [Google Scholar]
- 20.Kelch D.G., Baldwin B.G. Phylogeny and ecological radiation of New World thistles (Cirsium, Cardueae-Compositae) based on ITS and ETS rDNA sequence data. Mol. Ecol. 2003;12:141–151. doi: 10.1046/j.1365-294x.2003.01710.x. [DOI] [PubMed] [Google Scholar]
- 21.Slotta T.A.B., Rothhouse J., Horvath D.P., Foley M.E. Genetic Diversity of Canada thistle (Cirsium arvense) in North Dakota. Weed Sci. 2006;54:1080–1085. doi: 10.1614/WS-06-038R1.1. [DOI] [Google Scholar]
- 22.Slotta T.A.B., Foley M.E., Chao S., Hufbauer R.A., Horvath D.P. Assessing genetic diversity of Canada thistle (Cirsium arvense) in North America with microsatellites. Weed Sci. 2010;58:387–394. doi: 10.1614/WS-D-09-00070.1. [DOI] [Google Scholar]
- 23.Garcia-Jacas N., Garnatje T., Susanna A., Vilatersana R. Tribal and subtribal delimitation and phylogeny of the Cardueae (Asteraceae): A combined nuclear and chloroplast DNA analysis. Mol. Phylogenet. Evol. 2002;22:51–64. doi: 10.1006/mpev.2001.1038. [DOI] [PubMed] [Google Scholar]
- 24.Bruckart W.L., Poltis D.J., DeFago G., Rosenthal S.S., Supkoff D.M. Susceptibility of Carduus, Cirsium and Cynara scolymus species artificially inoculated with Puccinia carduorum from musk thistle. Biol. Control. 1996;6:215–221. doi: 10.1006/bcon.1996.0026. [DOI] [Google Scholar]
- 25.Kissinger D.G. Curculionidae of America North of Mexico, A Key to the Genera. Taxonomic Publications; South Lancaster, MA, USA: 1964. p. 143. [Google Scholar]
- 26.Moore R.J., Frankton C. Cytotaxonomy of Cirsium species of the Eastern United States, with a key to the eastern species. Can. J. Bot. 1969;47:1257–1275. doi: 10.1139/b69-177. [DOI] [Google Scholar]
- 27.Lym R.G., Christianson K.M. The Thistles of North Dakota, North Dakota State University Extension Service Bulletin; W-1120. North Dakota State University; Fargo, ND, USA: 1996. [Google Scholar]
- 28.Kartesz J.T. The Biota of North America Program (BONAP). North American Plant Atlas. [(accessed on 17 July 2012)]. Available online: http://www.bonap.org/MapSwitchboard.html/
- 29.Zwölfer H. Evolutionary and ecological relationships of the insect fauna of thistles. Ann. Rev. Entomol. 1988;33:103–122. [Google Scholar]
- 30.Volovnik S.V. Distribution and ecology of some species of Cleoninae (Coleoptera, Curculionidae). III. Genus Larinus Germ. Entomol. Rev. 1996;75:10–19. [Google Scholar]
- 31.Zwölfer H., Harris P. Biology and host specificity of Rhinocyllus conicus (Froel.) (Col., Curculionidae), a successful agent for biocontrol of the thistle, Carduus nutans L. Z. Angew. Entomol. 1984;97:36–62. [Google Scholar]
- 32.Arnett A., Louda S. Re-test of Rhinocyllus conicus host specificity and the prediction of ecological risk in biological control. Biol. Conserv. 2002;106:251–257. doi: 10.1016/S0006-3207(01)00251-8. [DOI] [Google Scholar]
- 33.Rees N.E. Impact of Rhinocyllus conicus on thistles in southwestern Montana. Environ. Entomol. 1977;6:839–842. [Google Scholar]
- 34.Louda S.M., Kendall D., Connor J., Simberloff D. Ecological effects of an insect introduced for the biological control of weeds. Science. 1997;277:1088–1090. doi: 10.1126/science.277.5329.1088. [DOI] [Google Scholar]
- 35.Ward R.H., Pienkowski R.L., Kok L.T. Host specificity of the first-instar of Ceuthorhynchidius horridus, a weevil for biological control of thistle. J. Econ. Entomol. 1974;67:735–737. [Google Scholar]
- 36.Wiggins G.J., Grant J.F., Lambdin P.L., Ranney J.W., Wilkerson J.B., van Manen F.T. Spatial prediction of habitat overlap of introduced and native thistles to identify potential areas of nontarget activity of biological control agents. Environ. Entomol. 2010;39:1866–1877. doi: 10.1603/EN10112. [DOI] [PubMed] [Google Scholar]
- 37.Anderson R.S. Weevils and plants: Phylogenetic versus ecological mediation of evolution of host plant associations in Curculioninae (Coleoptera: Curculionidae) Mem. Ent. Soc. Can. 1993;165:197–232. doi: 10.4039/entm125165197-1. [DOI] [Google Scholar]
- 38.Russell F.L., Louda S.M. Indirect interaction between two native thistles mediated by an invasive exotic floral herbivore. Oecologia. 2005;146:373–384. doi: 10.1007/s00442-005-0204-3. [DOI] [PubMed] [Google Scholar]
- 39.Barton J. Predictability of pathogen host range in classical biological control of weeds: An update. BioControl. 2012;57:289–305. doi: 10.1007/s10526-011-9401-7. [DOI] [Google Scholar]
- 40.Bruckart W.L., III. Supplemental risk evaluations and status of Puccinia carduorum for biological control of musk thistle. Biol. Control. 2005;32:348–355. doi: 10.1016/j.biocontrol.2004.10.013. [DOI] [Google Scholar]
- 41.Berner D.K., Bruckart W.L. Comparing predictions from mixed model equations with host range determinations from historical disease evaluation data of two previously released weed biological control pathogens. Biol. Control. 2012;60:207–215. [Google Scholar]
- 42.Bureš P., Wang Y.-F., Horová L., Suda J. Genome size variation in central European species of Cirsium (Compositae) and their natural hybrids. Ann. Bot. 2004;94:353–363. doi: 10.1093/aob/mch151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaskin J.F., Bon M.C., Cock M.J.W., Cristofaro M., Biase A.D., de Clerck-Floate R., Ellison C.A., Hinz H.L., Hufbauer R.A., Julien M.H., et al. Applying molecular-based approaches to classical biological control of weeds. Biol. Control. 2011;58:1–21. doi: 10.1016/j.biocontrol.2011.03.015. [DOI] [Google Scholar]
- 44.Wapshere A.J. Host specificity of phytophagous organisms and the evolutionary centres of plant genera or sub-genera. Entomophaga. 1974;19:301–309. doi: 10.1007/BF02371055. [DOI] [Google Scholar]
- 45.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swofford D.L. PAUP* Phylogenetic Analysis using Parsimony, ver. 4.0b10. Sinauer Associates; Sunderland, MA, USA: 2001. [Google Scholar]