Abstract
Bacteria use type IV secretion systems (T4SS) to translocate macromolecular substrates destined for bacterial, plant or human target cells. The T4SS are medically important, contributing to virulence-gene spread, genome plasticity and the alteration of host cellular processes during infection. The T4SS are ancestrally related to bacterial conjugation machines, but present-day functions include (i) conjugal transfer of DNA by cell-to-cell contact, (ii) translocation of effector molecules to eukaryotic target cells, and (iii) DNA uptake from or release to the extracellular milieu. Rapid progress has been made toward identification of type IV secretion substrates and the requirements for substrate recognition.
Bacteria have evolved type IV secretion systems (T4SS) to transfer DNA or protein macromolecules to a wide array of target-cell types [1,2]. Originally, the T4SS nomenclature referred to the virB–D4-encoded translocation system of Agrobacterium tumefaciens and two closely related systems encoded by the transfer (tra) region of the IncN plasmid pKM101 and the ptl operon of Bordetella pertussis [3]. In the past decade, the type IV family expanded considerably in number, partly because many new VirB–D4-type systems were discovered by genetic screens or genome sequencing. The criteria were also relaxed, so that now we define the T4SS as translocation systems ancestrally related to any conjugation system of Gram-negative or -positive bacteria [1,4]. However, to distinguish functional translocation machines from the many fragments of mobile elements characteristically found in bacterial genomes, mutagenesis of a putative type IV system should yield a phenotype at least consistent with a translocation defect.
In an earlier review, we suggested sub-classifying the T4SS on the basis of ancestral lineage [5]. Thus, we designated the VirB–D4-type systems as type IVA, and a second subfamily represented by the plasmid ColIb-P9 Tra and Legionella pneumophila Dot–Icm systems as type IVB. This leaves open the possibility of further subclassifying systems such as the Gram-positive plasmid transfer or the Gram-negative or Gram-positive conjugative transposons that are essentially unrelated to the VirB–D4 or ColIb-P9 systems. Although this scheme remains a useful way of depicting how the different T4SS might have evolved, this review emphasizes the distinctive biological activities of this secretion family by a grouping scheme based on function. We will summarize recent progress on two questions of central interest. First, what are the defining properties of type IV substrates? Second, how are secretion substrates recruited to the secretory apparatus? For detailed discussions of other features of T4SS, such as machine biogenesis, architecture and the consequences of inter-kingdom substrate transfer, readers are referred to several excellent reviews [2,6–9].
The versatile type IV family
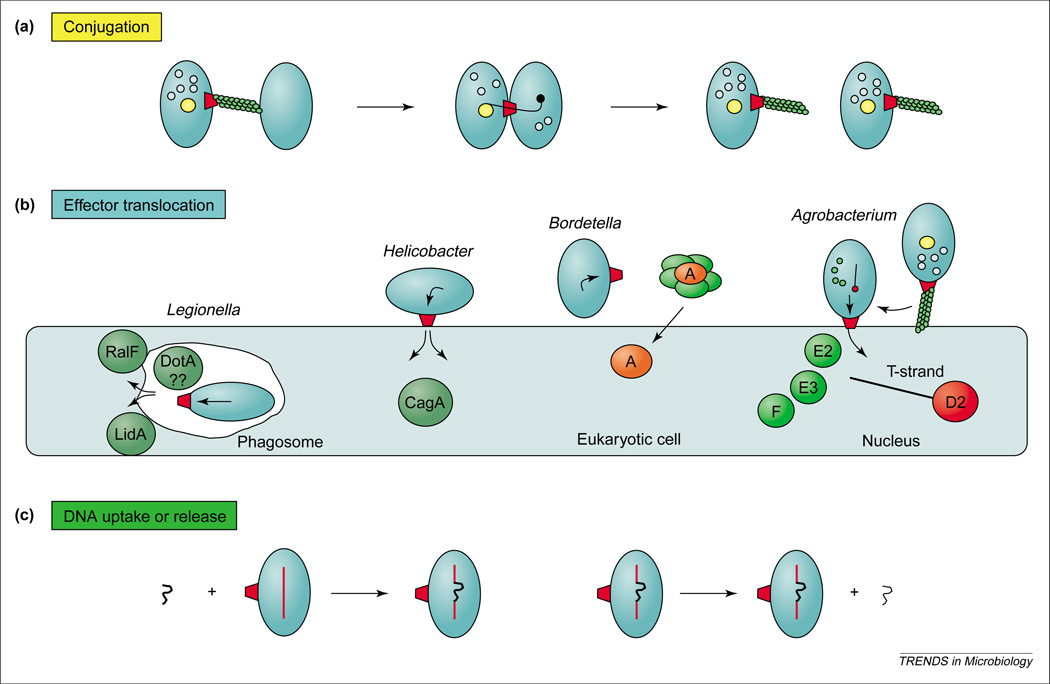
The type IV family members can be grouped as (i) conjugation systems mediating DNA transfer to recipient cells, (ii) ‘effector translocator’ systems that transfer molecules termed effectors to eukaryotic cells during infection, and (iii) ‘DNA uptake or release’ systems mediating exchange of DNA with the milieu (Figure 1). By definition, the conjugation systems deliver DNA substrates by establishing direct physical contact with target cells. Examples include the well-studied A. tumefaciens T-DNA transfer system and the F, RP4 and R388 plasmid transfer (Tra) systems (Figure 2). Although the conjugation systems are known mainly for their role in disseminating DNA among bacterial populations, they can also translocate protein substrates independently of DNA [10]. A subset of these systems can also transfer DNA and protein substrates to a range of eukaryotic cell types, including plant, fungal and human [11–14]. The conjugation systems comprise the largest type IV subgroup, and numerous systems share common ancestries with the A. tumefaciens VirB–D4 (IVA) or the ColIB-P9 (IVB) T4SS [8,15].
Figure 1.
The type IV secretion systems (T4SS) translocate DNA and protein substrates by cell-contact-dependent and –independent mechanisms. Three functional subfamilies include the (a) conjugation systems composed of a transfer channel (red trapezoid) and, for Gram-negative bacteria, an extracellular pilus, (b) ‘effector translocators’ dedicated to the transfer of effector molecules during infection, and (c) DNA uptake or release systems that translocate DNA independently of target-cell contact. Thick and thin squiggly lines are double- and single-stranded DNA, respectively.
Figure 2.
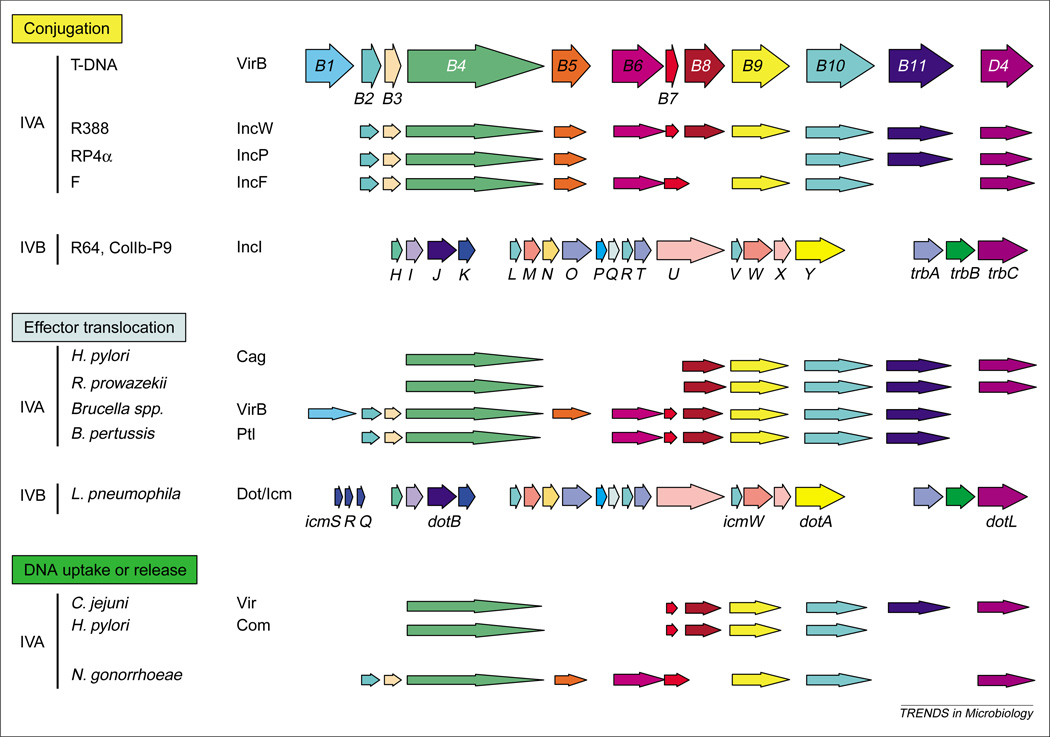
Conservation of type IV secretion genes. The Agrobacterium tumefaciens VirB–D4 reference system is a type IVA secretion system composed of 11 VirB proteins (Mpf) and the VirD4 T4CP (type IV coupling protein), whereas the Dot–Icm, R64 and ColIb-P9 systems are representatives of the type IVB subclass. Dot–Icm proteins discussed in the text or related to VirB proteins are identified. Genes encoding protein homologs are shown. These are not necessarily in the order found in the respective genomes. Homologs of the VirB4 ATPase and VirB7 through to VirB10 are postulated to comprise an ancestral ‘core’ structure to which function-specifying subunits or protein subassemblies were added to evolve the present-day family.
Most of the type IV effector translocator systems inject their substrates directly into the eukaryotic cytosol, as also shown for the type III secretion systems (T3SS) [16]. This type of translocation is now recognized as a dominant virulence mechanism of the phytopathogen A. tumefaciens, and of several medically important pathogens, including Helicobacter pylori, L. pneumophila, Brucella spp. and Bartonella spp. (Figures 1,2). We also include in this subfamily the Ptl system of B. pertussis, although this system exports its protein substrate, pertussis toxin (PT), independently of host-cell contact [17]. Presently, at least 10 type IVA and several type IVB effector translocators are thought to be essential for infection of eukaryotic hosts [8,15].
The ‘DNA uptake and release’ systems, presently with three members, translocate DNA substrates across the cell envelope to or from the extracellular milieu (Figures 1,2). Neisseria gonorrhoeae uses a system encoded by the gonococcal genetic island (GGI) to export DNA [18,19]. Intriguingly, recent studies have established that this system is very closely related to the conjugation system of the E. coli F plasmid. Because the gonococci are naturally competent, it is thought that the GGI-encoded T4SS have evolved to supply exogenous DNA to neighboring gonococci, creating the potential for genetic variation of virulence factors such as pili and surface proteins [19]. Also very remarkably, two other members of this subfamily, identified in Campylobacter jejuni and H. pylori, translocate DNA in the opposite direction. These ‘competence’ systems have also probably evolved to promote genetic variation to enhance cell survival and invasion of the eukaryotic host [20,21].
Building blocks of the type IV secretion machines
Distinct protein complexes carry out the three early stages of conjugal DNA transfer: DNA processing, substrate recruitment to the secretory apparatus and substrate translocation. The DNA replication and transfer (Dtr) proteins process DNA substrates by assembling at the origin-of-transfer (oriT) sequence of a mobile DNA element, forming the relaxosome. One relaxosome component, the relaxase, nicks the strand of DNA (T-strand) destined for export and then remains covalently bound to its 5′ end [22,23]. Next, a homomultimer of a protein, termed the coupling protein or T4CP as it is referred to here, is thought to recruit the DNA to the transfer apparatus through interactions with relaxosome components [6,24,25]. Recent structural studies suggest the T4CP assembles as a homohexamer with an overall structure strikingly similar to F1-ATPase [26,27]. This structure, additional biochemical properties and sequence similarities with two known DNA translocases, SpoIIIE and FtsK, suggests that in addition to substrate recruitment the T4CPs might function as inner membrane DNA translocases [6,28].
The Mpf (mating pair formation) proteins direct substrate transfer by assembling as a transenvelope structure, known as the ‘mating pore’ [22]. For almost every T4SS, an ATPase homologous to A. tumefaciens VirB11 assembles, probably as a homomultimer, at the cytoplasmic face of the inner membrane. For one homolog, H. pylori HP0525, a crystal structure presents as a homohexameric double-stacked ring, formed by self-association of N- and C-terminal domains of the protomers [29,30]. The VirB11 ATPases are thought to interact with other Mpf proteins and undergo ATP-dependent conformational changes required for machine morphogenesis or substrate transfer [6,30–32]. For the A. tumefaciens VirB–D4 system, the membrane-spanning Mpf complex is composed of the VirB proteins, a subset of which form a stable ‘core’ subcomplex: the inner membrane VirB4 ATPase, VirB8 and VirB10 proteins, and the outer-membrane-associated VirB7 lipoprotein and VirB9 protein [22,33–36]. A subset of Tra proteins encoded by the E. coli F plasmid transfer system also assembles as a stable transenvelope complex, possibly structurally analogous to the VirB core [37]. Finally, the conjugation systems of Gram-negative bacteria produce extracellular pili that are readily detectable by electron microscopy and recovered from culture supernatants or by shearing of cells [38]. The conjugative pili, as well as other recently described extracellular filaments of effector translocator systems [39–41], appear to function predominantly as attachment organelles.
Type IV secretion substrates: identification and characteristics
DNA substrates
The T4SS are unique among the bacterial secretion machines in their capacity to export DNA substrates. The oriT is clearly the signature sequence for the DNA substrate as this is the only cis-sequence required for transfer. However, recent work establishes that it is the relaxosome, more specifically the relaxase bound at oriT, that mediates DNA-substrate contact with a specific subunit, the T4CP, of a cognate conjugation machine [23,25,42,43]. Also of interest, in addition to relaxase binding to the 5′ end of the T-strand, DNA transfer is 5′–3′ directional, and there is some evidence for translocation of at least one relaxase, A. tumefaciens VirD2, to target cells [13]. These observations suggest that the relaxase is essential for all three early stages of conjugation mentioned previously, and for the last stage possibly serving to pilot the T-strand across the cell envelope.
For other DNA-translocation systems, the F Tra-like DNA release system of N. gonorrhoeae is dependent on Dtr proteins and therefore probably also exports single-stranded DNA (J. Dillard, pers. commun.). It seems reasonable to predict that these proteins process DNA substrates at chromosomal oriT-like sequences to generate a relaxase–T-strand substrate. For the DNA-uptake systems, exogenous double-stranded (ds) circular-plasmid and linear-chromosomal DNA fragments serve as substrates [20,21]. In principle, these systems might translocate such substrates, but by analogy to other competence systems [44], it seems more probable that they first convert the dsDNA to a single-stranded molecule for translocation.
Protein substrates and screens for their identification
Recently, many investigations have focused on identifying protein substrates of type IV effector translocators. As summarized in Table 1, the outcome is an impressive array of creative screens and a rapidly expanding list of candidate substrates. Two of the more powerful assays test for activities of translocated proteins in the target cell. The first assay, genetic complementation, was originally used to demonstrate conjugal transfer of the ColIb-P9 Sog primase to recipient bacteria [10]. This approach also supplied evidence for protein translocation to plant cells by the A. tumefaciens VirB–D4 system. Accordingly, A. tumefaciens mutants defective for synthesis of a suspected protein effector (e.g. VirE2) were complemented either by mixed infection with a second agrobacterial strain producing the effector, or by production of the effector in a transgenic plant host [12]. For the second assay, a suspected effector is fused to a reporter protein whose activity is only detected upon transfer to the eukaryotic target cell. For example, the A. tumefaciens VirB–D4 system delivers the VirE2 and VirF effector proteins fused to Cre recombinase to plant and yeast cells, as monitored by Cre recombination at lox sites engineered into the eukaryotic cells [45]. The Cre reporter system was also used to demonstrate A. tumefaciens VirB–D4-dependent transfer of a novel effector, VirE3, to plant and yeast cells [46]. Other promising candidate substrates for T4SS of mammalian pathogens are being identified using YopP toxin and adenylate cyclase protein fusions (Table 1).
Table 1.
Type IV protein substrates and assays for identificationa
| Assay | T4SS | Substrate or Interaction | Refs |
|---|---|---|---|
| Genetic complementation: | |||
| dnaG Ts mutation | ColIb-P9 | Sog primase | [10] |
| Mixed infection | A. t. VirB–D4 | VirE2, VirF | [12] |
| Transgenic plants | A. t. VirB–D4 | VirE2, VirF | [12] |
| Reporter protein fusions: | |||
| Cre recombinase | A. t. VirB–D4 | VirE2, VirF, VirE3 |
[45] |
| [46] | |||
| YopP | Brucella spp. | ?? | D. O′Callaghanb |
| Adenylate cyclase | L. p. Dot–IcM | ?? | J. Vogelc |
| Complex formation with T4SS: | |||
| In vitro with purified protein | RP4 Tra | TraG–TraI | [24] |
| RP4 Tra | TraG–pBHR1 | [42] | |
| F Tra | TraD–TraM | [43] | |
| Bacterial two-hybrid | RP4 Tra | TraG–Mob (pBHR1) | [42] |
| Cytology-based two-hybrid | A. t. VirB–D4 | VirD4–VirE2 | [48] |
| Substrate-dependent assembly | L. p. Dot–Icm | LidA | [47] |
| Complex formation with chaperone: | |||
| Biochemical–IP or pull-down | A. t. VirB–D4 | VirE1–VirE2 | [53,55] |
| L. p. Dot–Icm | IcmR–IcmQ | [56] | |
| IcmS–P130 | [56] | ||
| Yeast dihybrid | A. t. VirB–D4 | VirE1–VirE2 | [52–54] |
| L. p. Dot–Icm | IcmR–IcmQ | [56] | |
| IcmS–IcmW | [56] | ||
| Secretion: | |||
| Milieu | B. p. Ptl | PT | [17] |
| Milieu | L. p. Dot–Icm | DotA | [51] |
| Periplasm or milieu | A. t. VirB–D4 | VirE2, VirF | [64,65] |
| Translocation-dependent modification: | |||
| Tyrosine phosphorylation | H. p. Cag | CagA | [66] |
| Sequence-based predictions: | |||
| Homology to eukaryotic proteins associated with virulence | L. p. Dot–Icm | RalF | [59] |
| C-terminal type IV secretion signal | A. t. VirB–D4 | VirE3 | [46] |
Abbreviations: A. t., Agrobacterium tumefaciens; B. p., Bordetella pertussis; L. p., Legionella pneumophila; T4SS, Type IV secretion systems.
Data from D. O’Callaghan, unpublished.
Data from J. Vogel, unpublished.
Screens aimed at identifying interactions between secretion substrates and components of the T4SS have also shown considerable promise. In L. pneumophila, certain mutations of the DotL T4CP are lethal, suggesting impairment of assembly or activity of the Dot–Icm translocase. It was hypothesized that mutations in secreted effectors might also result in misregulation of type IV function. Therefore, a genetic screen was devised to identify mutations in genes that confer reduced viability in the presence of the secretory apparatus and are phenotypically silent in the absence of the apparatus. One identified protein, LidA (lowered viability in the presence of dot), is important for maintaining bacterial cell integrity in the presence of the Dot–Icm complex and was also shown to be deposited by a Dot–Icm-dependent mechanism on the surface of the phagosome during infection [47].
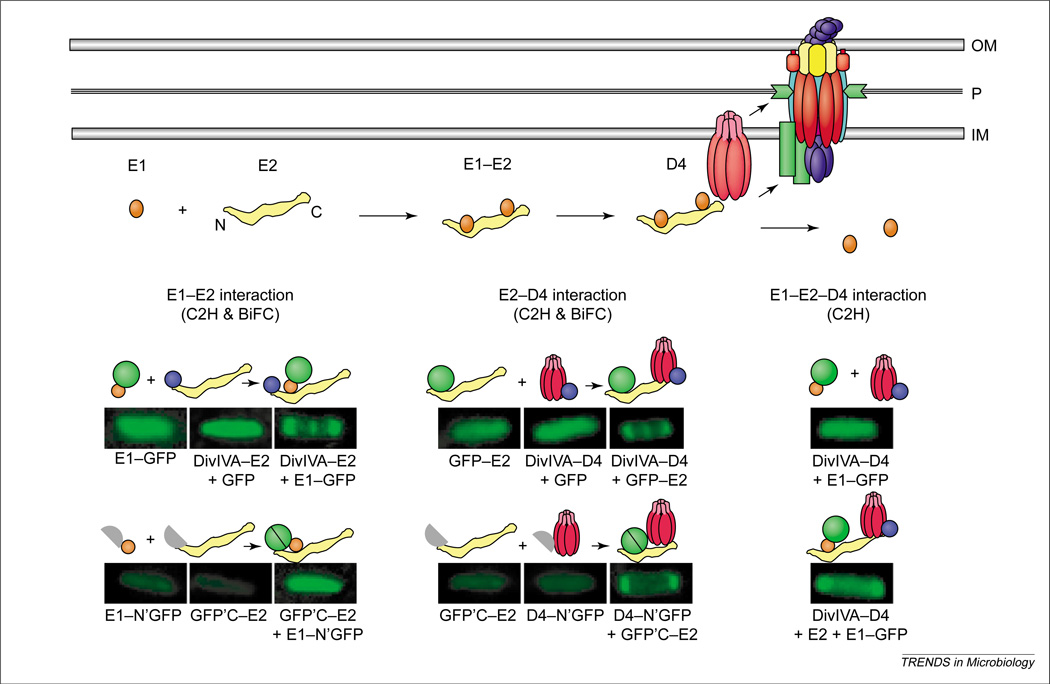
Additional screens have assayed specifically for interactions between suspected substrates and T4CPs (Table 1). For example, immunoprecipitation studies yielded evidence for complex formation between the A. tumefaciens VirE2 effector and the VirD4 T4CP [48]. In these studies, chemical crosslinking was essential for recovery of complexes, possibly to stabilize transient or weak-affinity interactions. Whereas two-hybrid screens in the heterologous yeast-host thus far have not proven successful for demonstrating substrate contacts with T4CPs, the bacterial adenylate cyclase two-hybrid assay identified a relaxase–T4CP interaction (Table 1) [42]. Recently, cytology-based two-hybrid screens have also been applied to studies of a substrate–T4CP interaction in bacteria. As illustrated in Figure 3, the cytology-based screens monitor recruitment of GFP-tagged forms of putative substrates to T4CP localized at discrete sites within the cell. In one application, it was shown that the VirE2 effector tagged with GFP is recruited to VirD4 T4CP [48], which localizes in wild-type A. tumefaciens at the cell poles [49]. Furthermore, it was shown that VirD4, when relocalized to the midcell by fusion to the cell division protein DivIVA [50], recruits GFP-VirE2 to the midcell [48]. Finally, an assay termed bimolecular fluorescence complementation (BiFC), which tests for reconstitution of fluorescent proteins from halves fused to putative partner proteins [51], supplied additional evidence for the VirD4–VirE2 interaction [48]. As described in more detail below, these cytological assays are extremely powerful because they permit studies of substrate–T4CP interactions in vivo, thus enabling definition of the genetic requirements for complex formation.
Figure 3.
Processing and recruitment of the VirE2 effector to the Agrobacterium tumefaciens VirB–D4 system. Top: Newly synthesized VirE2 forms a stabilizing complex with the secretion chaperone VirE1. Chaperone interactions with N-terminal and central domains prevent VirE2 self-aggregation and formation of premature complexes with other exported effectors, for example, the T-strand. A secretion signal located near the C-terminus of VirE2 mediates complex formation with the VirD4 T4CP (type IV coupling protein) without contributions from VirE1 or the VirB proteins. Lower: Genetic requirements for complex formation between VirE1 and VirE2, and between VirE2 and VirD4, as defined with novel cytology two-hybrid (C2H) and bimolecular fluorescence complementation (BiFC) assays. Fluorescence patterns of cells shown result from production of full- or half-length GFP fusion proteins listed below each image, and the patterns supply evidence for the interactions depicted above each image. Green circle, GFP; purple circle, DivIVA.
In type III secretion, translocation of several effectors is dependent on complex formation with secretion chaperones. On the assumption that type IV substrates might also interact with secretion chaperones, several studies have attempted to identify chaperone partner proteins. In act, this approach is validated by studies in A. tumefaciens, whereby VirE2 requires the VirE1 chaperone for translocation to plant cells and VirE1–VirE2 complex formation is readily demonstrated by several in vivo and in vitro screens [52–55]. In L. pneumophila, a sequence-based search for soluble proteins that bear properties reminiscent of the type III chaperones (small size, acidic pI and amphipathic C-terminus) identified two proteins, IcmR and IcmS, and subsequent interaction assays using GST-tagged derivatives identified several partner proteins including IcmQ, P130 and IcmW [56]. Finally, some T4SS might release substrates to the extracellular milieu either through a natural routing pathway, as in the case of the B. pertussis Ptl system for PT export [17], or because of a gating defect. One protein apparently secreted in a type IV-dependent manner is the L. pneumophila DotA protein, which is surprising because DotA was originally reported to be a polytopic inner membrane protein [57].
Properties of protein substrates and possible secretion signals
Clearly, the screens described above have identified many new possible type IV substrates. However, it is important to note that in several cases further confirmatory studies are needed. This is certainly true for L. pneumophila DotA given that there is no precedent for secretion of a polytopic inner membrane protein, and the presumptive secretion of DotA displays no biological effects. Additionally, DotA homologs are encoded by the transfer systems of IncI plasmids (Figure 2), making it more probable that DotA is part of the secretion apparatus itself. Similarly, further studies are needed to demonstrate bona fide secretion at some stage in the L. pneumophila infection cycle of other candidate Dot–Icm substrates identified by indirect means, such as the chaperone-interacting proteins [56]. Indeed, even for the well-characterized conjugation machines, to date there is no definitive evidence for the intercellular transfer of any relaxase. Therefore, the proposal that the relaxase pilots the T-strand through the mating channel remains a working model. Finally, below we cite an example of a biological phenomenon attributed to type IV secretion that might instead result from activation of a receptor-dependent signaling pathway by the secretory apparatus itself. According to this, it is conceivable that a candidate effector is in fact not a true secretion substrate, but is nevertheless dependent on the secretory apparatus for stabilization at the cell surface where it exerts its effects.
With these caveats in mind, do the candidate secretion substrates identified so far display any common features for targeting to the secretion machine? No universally conserved primary sequence motifs or physical characteristics are readily discernible. The substrates vary considerably in size, from ~22 kDa for A. tumefaciens VirF to ~145 kDa for H. pylori CagA. They also vary in subunit composition, from monomers to DNA–protein complexes or the 6-subunit PT holotoxin. Among the known substrates, the PT subunits are unique in possessing characteristic N-terminal secretion signals (see below). Intriguingly, some substrates, such as VirE2, are probably maintained in an unfolded or partially folded state through chaperone contacts, whereas others such as the relaxases are catalytic in the bacterium and therefore almost certainly folded before export. These general observations suggest there are different requirements for substrate processing and recruitment to the various T4SS.
For a given secretion system, however, different substrates might possess a conserved recognition signal. In support of this proposal, recent studies suggest that a motif located within the C-terminal region of several secretion substrates is important for recruitment to the VirD4 T4CP of A. tumefaciens. By the use of cytology-based two-hybrid screens, it was shown that VirE2 is recruited to VirD4 independently of VirB proteins (Mpf structure), the VirD2 relaxase (T-DNA processing reaction) and complex formation with its secretion chaperone, VirE1 [48]. This latter finding is especially noteworthy in view of the fact that chaperone–effector complex formation is essential for VirE2 stabilization and maintenance of a translocation-competent configuration [53,55]. VirE1 therefore acts early in the VirE2 translocation pathway, although it does not participate in substrate targeting. Moreover, analyses of VirE2 truncation derivatives showed that the C-terminal 100 residues of VirE2 are both necessary and sufficient for recruitment of GFP-VirE2 derivatives to VirD4, whereas small deletions or insertions at the extreme C-terminus impede complex formation [48]. Therefore, the C-terminus of VirE2 is important for recruitment to VirD4.
Two independent lines of study support a proposal that VirB–D4 substrates carry C-terminal recognition signals. First, by use of the genetic complementation assay (Table 1) it was shown that the C-terminal 18 residues of VirE2, although dispensable for effector function in the plant cell, are required for VirB–D4-dependent translocation across the A. tumefaciens envelope [58]. Second, by use of the Cre recombinase reporter system, it was shown that a C-terminal fragment of another substrate, VirF, mediates transfer of Cre recombinase to the plant cell by a VirB–D4-dependent mechanism [45]. Intriguingly, these investigators identified conserved Arg-Pro-Arg motifs near the C-termini of VirE2, VirE3 and VirF; the three identified secretion substrates of this system [45,46,58]. These residues or similar clusters of basic residues might be critical for establishment of productive substrate–T4CP contacts. Whether other type IV substrates carry recognition signals at their C-termini awaits further study, but the observation that the L. pneumophila RalF protein, a substrate of the Dot–Icm T4SS, carries a Sec7 homology domain at its extreme N-terminus is consistent with a C-terminal location for an export signal [15,59].
T4CP-mediated and alternative substrate recruitment mechanisms
As discussed above, T4CPs recruit DNA substrates through interactions with relaxases and auxiliary components of the relaxosome, and there is also increasing evidence that the T4CPs recruit protein substrates to the secretory apparatus. In view of a proposal that T4CPs function as general substrate-recruitment factors [48], it is noteworthy that, although T4CPs are ubiquitous components of conjugation machines, they are common to most but not all effector translocators. One exception, the B. pertussis Ptl system, appears to bypass the T4CP requirement by use of the general secretory pathway (GSP) for delivery of the PT subunits across the cytoplasmic membrane [17]. The Brucella spp. type IV systems also lack a T4CP and therefore might use the GSP or another mechanism for early stages of substrate recognition and translocation [9]. Also, the H. pylori Com system [21] and a VirB-mediated DNA-uptake system described for A. tumefaciens [60] both function independently of a T4CP. The C. jejuni Com system encodes a VirD4 homolog that is dispensable for virulence, but it is not yet reported whether this protein is required for DNA uptake [61].
There is also some evidence that a given type IV system might function by alternative, coupling protein-dependent and -independent pathways. In H. pylori, for example, the Cag system requires the VirD4 coupling protein for CagA translocation. However, a virD4 null-mutant still induces interleukin (IL)-8 secretion, prompting a proposal that this system can alternatively export an effector for IL-8 secretion by a T4CP-independent mechanism [62]. Further studies are needed to test this idea, however, because similar findings led to the alternative proposal that the Cag secretory apparatus might act directly through receptor-dependent activation of IL-8 secretion [63]. Additionally, it has been reported that the A. tumefaciens VirE2, VirE3 and VirF effectors accumulate at low levels in periplasmic fractions, forming associations with the periplasmic protein VirJ or its homolog AcvB. Intriguingly, these substrates can also be detected in the periplasmic fractions of virB and virD4 mutants [64,65]. Although further control studies are necessary and the biological relevance of these findings for virulence remains to be shown, it is still possible that a VirB–D4-independent pathway can translocate these substrates across the inner membrane.
The translocation route
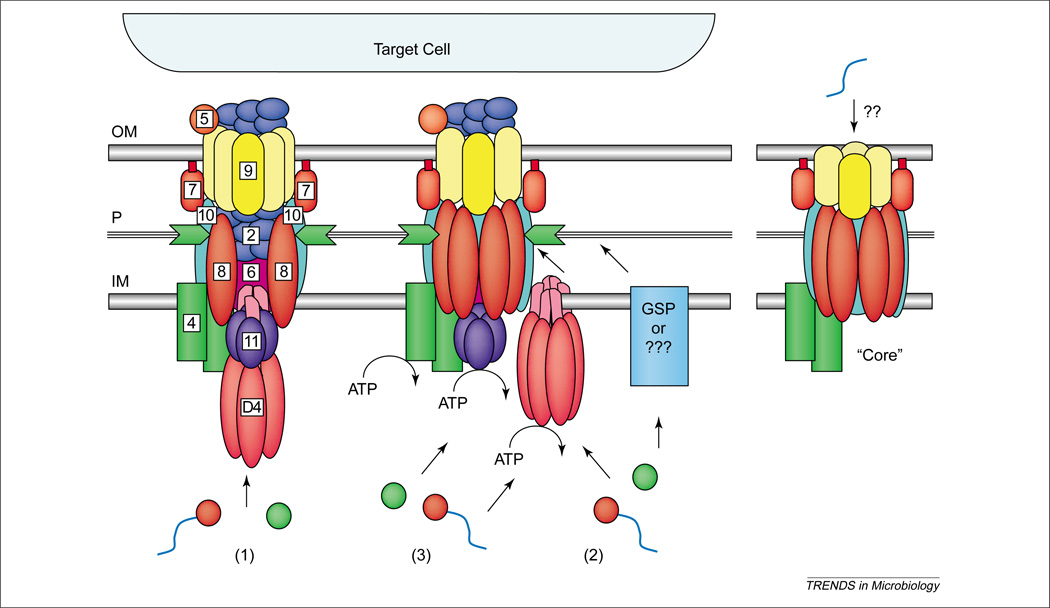
Once DNA and protein substrates are recruited to the secretory apparatus, how are they translocated across the cell envelope? Presently, there are three working models to describe the possible routing pathways and, interestingly, each makes specific and testable predictions regarding the contribution of the T4CP to translocation. As depicted in Figure 4, the first model envisioned for conjugation systems suggests that the Mpf proteins assemble as a transenvelope channel for substrate export in one step [22]. A strong prediction derived from this model is that the T4CP recruits DNA and protein substrates to the translocation apparatus, then coordinates its activity with a VirB11-like ATPase to drive substrate transfer through the Mpf channel. The second model represents a generalized version of the two-step routing pathway described for export of B. pertussis PT [17]. In step one, an inner membrane translocase delivers substrates across the inner membrane. In step two, the T4SS translocase, composed of Mpf proteins, delivers substrates across the outer membrane [60]. A prediction from this model is that, when present, the T4CP functions as an inner membrane translocase for both DNA and protein substrates, acting completely independently of the Mpf proteins.
Figure 4.
Possible architectures and substrate translocation routes for the type IV systems. A Mpf (mating pair formation) core structure might function as a competence machine with only minor evolutionary adaptation. Correspondingly, the addition of inner membrane translocases and, for some systems, a pilus biogenesis pathway, yields the DNA and protein translocation systems at the left. Three working models describe the possible machine architectures and translocation routes: (1) a one-step model using a transenvelope channel, (2) a two-step model using the T4CP or alternative translocase for subtrate transfer across the inner membrane and the Mpf complex for outer membrane transit, and (3) a different two-step model, the ‘shoot and pump’ model, whereby the T4CP recruits substrates and translocates DNA across the inner membrane, and delivers protein substrates to the Mpf protein export apparatus. Blue line, T-strand substrate; red circle, relaxase bound to the T-strand; green circle, protein substrate.
Finally, an alternative two-step model, termed the ‘shoot and pump’ model, also suggests there are two inner membrane translocases. However, in this case the T4CP functions as a DNA translocase, whereas the Mpf protein complex translocates protein substrates. In the periplasm, these pathways converge for Mpf-dependent export across the outer membrane [6]. This model does not exclude the proposed function of T4CP as a general recruitment factor, but rather postulates that upon recruitment the T4CP translocates DNA and delivers the protein substrate to the Mpf channel. Of these models, the ‘shoot and pump’ model is especially appealing, not only because it accommodates all experimental findings to date but it also nicely accounts for how the T4SS evolved to be so highly versatile [6]. Future studies defining the contribution of the T4CP to translocation and the nature of T4CP–Mpf contacts should establish which, if any, of these models best describes the type IV secretory pathways.
Conclusions
The most alluring feature of the T4SS as subjects for basic and applied studies is their extreme versatility. These systems recognize a wide array of DNA and protein substrates, translocate substrates by both cell-contact-dependent and -independent mechanisms, and deliver substrates to an exceptionally wide range of prokaryotic and eukaryotic taxa. During the past few years, important new information has emerged regarding the dynamics of substrate recognition and recruitment to the secretory apparatus, much of which emphasizes the prominent role of the T4CP in early stages of translocation. Equally importantly, recent studies have identified an entirely new type IV subfamily, the DNA uptake and release systems. Therefore, although progress is being made toward defining the mechanisms of action of T4SS on several fronts, we now face the fact that related systems also function in recognition and import of exogenous DNA. Furthermore, the discovery of an F Tra-like DNA release system dispels a 50-year old perception that a conjugation machine translocates DNA solely through direct physical contact with a target-cell. Clearly, the characterization of these new systems along with comparisons among the archetypal systems will unveil a deeper mechanistic understanding of the dynamics of type IV machine function. Concomitantly, the findings should foster development of therapeutic strategies aimed at incapacitating the T4SS of several medically important pathogens (see Questions for Future Research).
Questions for Future Research.
Does the T4CP function as a general recruitment factor for all type IV protein substrates?
What is the mechanism for substrate recruitment (in the cytoplasm or periplasm) by type IV systems that lack a T4CP?
Does the T4CP function as an inner membrane translocase for DNA or protein substrates? How does the T4CP and the Mpf machine physically and functionally interact?
What is the translocation route across the cell envelope for type IV substrates? For a given system, do alternative pathways exist?
How do the recently described type IV systems mediating DNA uptake or DNA release recognize, process and translocate DNA substrates?
Acknowledgments
We wish to acknowledge the important contributions of all investigators of type IV systems, and we apologize for omissions of data or citation in this review because of space limitations. We thank the members of the laboratory for helpful discussions and critiques of this review. We thank W. Margolin and members of his laboratory for guidance with and use of the Margolin microscopy facility. We gratefully acknowledge the financial support of the NIH for studies in this laboratory of the A. tumefaciens VirB–D4 type IV secretion system.
References
- 1.Christie PJ. Type IV secretion: Intercellular transfer of macromolecules by systems ancestrally-related to conjugation machines. Mol. Microbiol. 2001;40:294–305. doi: 10.1046/j.1365-2958.2001.02302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron C, et al. Bacterial secrets of secretion: Euro conference on the biology of type IV secretion processes. Mol. Microbiol. 2002;43:1359–1365. doi: 10.1046/j.1365-2958.2002.02816.x. [DOI] [PubMed] [Google Scholar]
- 3.Salmond GPC. Secretion of extracellular virulence factors by plant pathogenic bacteria. Annu. Rev. Phytopathol. 1994;32:181–200. [Google Scholar]
- 4.Grohmann E, et al. Conjugative plasmid transfer in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003;67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christie PJ, Vogel JP. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 2000;8:354–360. doi: 10.1016/s0966-842x(00)01792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Llosa M, et al. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 2002;45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 7.Lawley TD, et al. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 2003;224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 8.Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. doi: 10.1038/nrmicro753. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boschiroli ML, et al. Type IV secretion and Brucella virulence. Vet. Microbiol. 2002;90:341–348. doi: 10.1016/s0378-1135(02)00219-5. [DOI] [PubMed] [Google Scholar]
- 10.Wilkins BM, Thomas AT. DNA-independent transport of plasmid primase protein between bacteria by the I1 conjugation system. Mol. Microbiol. 2000;38:650–657. doi: 10.1046/j.1365-2958.2000.02164.x. [DOI] [PubMed] [Google Scholar]
- 11.Bates S, et al. IncP plasmids are unusually effective in mediating conjugation of Escherichia coli and Saccharomyces cerevisiae. J. Bacteriol. 1998;180:6538–6543. doi: 10.1128/jb.180.24.6538-6543.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ward DV, Zambryski PC. The six functions of Agrobacterium VirE2. Proc. Natl. Acad. Sci. U. S. A. 2001;98:385–386. doi: 10.1073/pnas.98.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu J, et al. The bases of crown gall tumorigenesis. J. Bacteriol. 2000;182:3885–3895. doi: 10.1128/jb.182.14.3885-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waters VL. Conjugation between bacterial and mammalian cells. Nat. Genet. 2001;29:375–376. doi: 10.1038/ng779. [DOI] [PubMed] [Google Scholar]
- 15.Sexton JA, Vogel JP. Type IVB secretion by intracellular pathogens. Traffic. 2002;3:178–185. doi: 10.1034/j.1600-0854.2002.030303.x. [DOI] [PubMed] [Google Scholar]
- 16.Blocker A, et al. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3027–3030. doi: 10.1073/pnas.0535335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burns DL. Type IV transporters of pathogenic bacteria. Curr. Opin. Microbiol. 2003;6:29–34. doi: 10.1016/s1369-5274(02)00006-1. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton HL, et al. Insertion-duplication mutagenesis of Neisseria : use in characterization of DNA transfer genes in the gonococcal genetic island. J. Bacteriol. 2001;183:4718–4726. doi: 10.1128/JB.183.16.4718-4726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dillard JP, Seifert HS. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol. Microbiol. 2001;41:263–277. doi: 10.1046/j.1365-2958.2001.02520.x. [DOI] [PubMed] [Google Scholar]
- 20.Bacon DJ, et al. Involvement of a plasmid in virulence of Campylobacter jejuni 81–176. Infect. Immun. 2000;68:4384–4390. doi: 10.1128/iai.68.8.4384-4390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofreuter D, et al. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 2001;41:379–391. doi: 10.1046/j.1365-2958.2001.02502.x. [DOI] [PubMed] [Google Scholar]
- 22.Christie PJ. The Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fekete RA, Frost LS. Mobilization of chimeric oriT plasmids by F and R100-1: role of relaxosome formation in defining plasmid specificity. J. Bacteriol. 2000;182:4022–4027. doi: 10.1128/jb.182.14.4022-4027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schroder G, et al. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 2002;184:2767–2779. doi: 10.1128/JB.184.10.2767-2779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamilton CM, et al. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J. Bacteriol. 2000;182:1541–1548. doi: 10.1128/jb.182.6.1541-1548.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomis-Ruth FX, et al. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature. 2001;409:637–641. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- 27.Hormaeche I, et al. Purification and properties of TrwB, a hexameric, ATP-binding integral membrane protein essential for R388 plasmid conjugation. J. Biol. Chem. 2002;277:46456–46462. doi: 10.1074/jbc.M207250200. [DOI] [PubMed] [Google Scholar]
- 28.Errington J, et al. DNA transport in bacteria. Nat. Rev. Mol. Cell Biol. 2001;2:538–545. doi: 10.1038/35080005. [DOI] [PubMed] [Google Scholar]
- 29.Yeo H-J, et al. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV system. Mol. Cell. 2000;6:1461–1472. doi: 10.1016/s1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
- 30.Savvides SN, et al. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 2003;22:1969–1980. doi: 10.1093/emboj/cdg223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sagulenko Y, et al. Role of Agrobacterium VirB11 ATPase inT-pilus assembly and substrate selection. J. Bacteriol. 2001;183:5813–5825. doi: 10.1128/JB.183.20.5813-5825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krause S, et al. Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J. Bacteriol. 2000;182:2761–2770. doi: 10.1128/jb.182.10.2761-2770.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das A, Xie Y-H. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 2000;182:758–763. doi: 10.1128/jb.182.3.758-763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward DV, et al. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11493–11500. doi: 10.1073/pnas.172390299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jakubowski S, et al. Agrobacterium tumefaciens VirB6 proteins participates in assembly of VirB7 and VirB9 complexes required for type IV secretion. J. Bacteriol. 2003;185:2867–2878. doi: 10.1128/JB.185.9.2867-2878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krall L, et al. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11405–11410. doi: 10.1073/pnas.172390699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris RL, et al. Evidence that F-plasmid proteins TraV, TraK and TraB assemble into an envelope-spanning structure in Escherichia coli. Mol. Microbiol. 2001;42:757–766. doi: 10.1046/j.1365-2958.2001.02667.x. [DOI] [PubMed] [Google Scholar]
- 38.Lai EM, Kado CI. The T-pilus of Agrobacterium tumefaciens. Trends Microbiol. 2000;8:361–369. doi: 10.1016/s0966-842x(00)01802-3. [DOI] [PubMed] [Google Scholar]
- 39.Rohde M, et al. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol. Microbiol. 2003;49:219–234. doi: 10.1046/j.1365-2958.2003.03549.x. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka J, et al. Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell. Microbiol. 2003;5:395–404. doi: 10.1046/j.1462-5822.2003.00286.x. [DOI] [PubMed] [Google Scholar]
- 41.Watarai M, et al. Formation of a fibrous structure on the surface of Legionella pneumophila associated with exposure of DotH and DotO proteins after intracellular growth. Mol. Microbiol. 2001;39:313–329. doi: 10.1046/j.1365-2958.2001.02193.x. [DOI] [PubMed] [Google Scholar]
- 42.Szpirer CY, et al. Interaction between the RP4 coupling protein TraG and the pBHR1 mobilization protein Mob. Mol. Microbiol. 2000;37:1283–1292. doi: 10.1046/j.1365-2958.2000.02077.x. [DOI] [PubMed] [Google Scholar]
- 43.Disque-Kochem C, Dreiseikelmann B. The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J. Bacteriol. 1997;179:6133–6137. doi: 10.1128/jb.179.19.6133-6137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dubnau D. DNA uptake in bacteria. Annu. Rev. Microbiol. 1999;53:217–244. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- 45.Vergunst AC, et al. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- 46.Schrammeijer B, et al. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucleic Acids Res. 2003;31:860–868. doi: 10.1093/nar/gkg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conover GM, et al. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 2003;48:305–321. doi: 10.1046/j.1365-2958.2003.03400.x. [DOI] [PubMed] [Google Scholar]
- 48.Atmakuri K, et al. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at the cell poles of Agrobacterium tumefaciens. Mol. Microbiol. 2003;49:1699–1713. doi: 10.1046/j.1365-2958.2003.03669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar RB, Das A. Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol. Microbiol. 2002;43:1523–1532. doi: 10.1046/j.1365-2958.2002.02829.x. [DOI] [PubMed] [Google Scholar]
- 50.Ding Z, et al. A novel cytology-based, two-hybrid screen for bacteria applied to protein–protein interaction studies of a type IV secretion system. J. Bacteriol. 2002;184:5572–5582. doi: 10.1128/JB.184.20.5572-5582.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu CD, et al. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 52.Sundberg CD, Ream W. The Agrobacterium tumefaciens chaperone-like protein, VirE1, interacts with VirE2 at domains required for single-stranded DNA binding and cooperative interaction. J. Bacteriol. 1999;181:6850–6855. doi: 10.1128/jb.181.21.6850-6855.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng W, et al. VirE1 is a specific molecular chaperone for the exported single-stranded-DNA-binding protein VirE2 in Agrobacterium. Mol. Microbiol. 1999;31:1795–1807. doi: 10.1046/j.1365-2958.1999.01316.x. [DOI] [PubMed] [Google Scholar]
- 54.Zhou X-R, Christie PJ. Mutagenesis of Agrobacterium VirE2 single-stranded DNA-binding protein identifies regions required for self-association and interaction with VirE1 and a permissive site for hybrid protein construction. J. Bacteriol. 1999;181:4342–4352. doi: 10.1128/jb.181.14.4342-4352.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao Z, et al. Activities of virE1 and the VirE1 secretion chaperone in export of the multifunctional VirE2 effector via an Agrobacterium type IV secretion pathway. J. Bacteriol. 2001;183:3855–3865. doi: 10.1128/JB.183.13.3855-3865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coers J, et al. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 2000;38:719–736. doi: 10.1046/j.1365-2958.2000.02176.x. [DOI] [PubMed] [Google Scholar]
- 57.Nagai H, Roy CR. The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. EMBO J. 2001;20:5962–5970. doi: 10.1093/emboj/20.21.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simone M, et al. The carboxy-terminus of VirE2 from Agrobacterium tumefaciens is required for its transport to host cells by the virB-encoded type IV transport system. Mol. Microbiol. 2001;41:1283–1293. doi: 10.1046/j.1365-2958.2001.02582.x. [DOI] [PubMed] [Google Scholar]
- 59.Nagai H, et al. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295:679–682. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- 60.Liu Z, Binns AN. Functional subsets of the VirB type IV transport complex proteins involved in the capacity of Agrobacterium tumefaciens to serve as a recipient in virB-mediated conjugal transfer of plasmid RSF1010. J. Bacteriol. 2003;185:3259–3269. doi: 10.1128/JB.185.11.3259-3269.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bacon DJ, et al. DNA sequence and mutational analyses of the pVir plasmid of Campylobacter jejuni 81–176. Infect. Immun. 2002;70:6242–6250. doi: 10.1128/IAI.70.11.6242-6250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Selbach M, et al. Functional analysis of the Helicobacter pylori cag pathogenicity island reveals both VirD4-CagA-dependent and VirD4-CagA-independent mechanisms. Infect. Immun. 2002;70:665–671. doi: 10.1128/iai.70.2.665-671.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fischer W, et al. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 2001;42:1337–1348. doi: 10.1046/j.1365-2958.2001.02714.x. [DOI] [PubMed] [Google Scholar]
- 64.Chen L, et al. Transferred DNA (T-DNA)-associated proteins of Agrobacterium tumefaciens are exported independently of virB. Proc. Natl. Acad. Sci. U. S. A. 2000;97:7545–7550. doi: 10.1073/pnas.120156997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pantoja M, et al. Agrobacterium type IV secretion is a two-step process in which export substrates associate with the virulence protein VirJ in the periplasm. Mol. Microbiol. 2002;45:1325–1335. doi: 10.1046/j.1365-2958.2002.03098.x. [DOI] [PubMed] [Google Scholar]
- 66.Backert S, et al. Helicobacter pylori type IV secretion, host cell signalling and vaccine development. Keio J. Med. 2002;51(Suppl 2):6–14. doi: 10.2302/kjm.51.supplement2_6. [DOI] [PubMed] [Google Scholar]