Abstract
Leaf senescence is a complicated developmental process that involves degenerative changes and nutrient recycling. The progress of leaf senescence is controlled by various environmental cues and plant hormones, including ethylene, jasmonic acid, salicylic acid, abscisic acid, cytokinins, and strigolactones. The production of strigolactones is induced in response to nitrogen and phosphorous deficiency. Strigolactones also accelerate leaf senescence and regulate shoot branching and root architecture. Leaf senescence is actively promoted in a nutrient-poor soil environment, and nutrients are transported from old leaves to young tissues and seeds. Strigolactones might act as important signals in response to nutrient levels in the rhizosphere. In this review, we discuss the possible roles of strigolactones during leaf senescence.
Keywords: leaf senescence, phytohormones, nutrient deficiency, strigolactone, translocation
1. Introduction
Leaf senescence is an essential process observed in the final stage of leaf development [1]. In particular, age-dependent senescence in monocarpic plants precedes whole plant death at the end of its life cycle. During leaf development, leaf cells accumulate ribulose bisphosphate carboxylase (rubisco), chlorophyll-a/b binding protein (CAB), and other proteins involved in photosynthesis in the chloroplast, which contains more than 70% of the total leaf protein [2]. Photosynthesis fixes carbon using light energy and plays a central role in plant growth. When photosynthetic activity decreases in the lower (old) leaves, these leaves are no longer required and nutrients accumulated in them are recycled. In the early stage of leaf senescence, cellular metabolism and gene expression patterns change dramatically. The most important change is chloroplast breakdown, which is accompanied by chlorophyll degradation and progressive loss of chloroplast proteins, such as rubisco and CAB, and which causes leaf color changes from green to yellow. After chloroplast degeneration, plasma membrane integrity is lost as the final step of cell death, and cellular ionic substances leak out [3,4]. Some senescence-associated genes (SAGs) encode enzymes involved in protein, lipid, and nucleic acid degradation [5]. However, leaf senescence is not a passive and unregulated degeneration process. The degraded cellular compounds and minerals are transported out of the senescing leaf and back into the main plant body, where carbon, nitrogen, and mineral nutrients are redistributed to the growing parts, especially to young organs and seeds, and reused for synthetic processes [2].
Leaf senescence is not only controlled by the developmental stage but is also influenced by various internal and environmental cues, even in young plants. Major environmental cues include light intensity, temperature, drought, pathogen attack, and soil nutrient deficiency [2]. Plant hormones act as internal cues that influence plant development and responses to environmental stresses, and play an important role in the regulation of leaf senescence. In general, ethylene, jasmonic acid, and salicylic acid are involved in plant immune responses, such as responses to pathogen attacks and in wounding, and abscisic acid mediates plant osmotic response [6,7]. Cytokinins control nutrient remobilization between source and sink organs. Ethylene, jasmonic acid, salicylic acid, and abscisic acid act as positive regulators in leaf senescence, whereas cytokinins are potent inhibitors of leaf senescence [7]. In addition to these hormones, strigolactones (SLs, a class of plant hormones) appear to regulate leaf senescence because some SL-deficient and SL-insensitive mutants show delayed leaf senescence [8,9,10,11]. In this review, we discuss the physiological roles of SLs in leaf senescence.
2. SL Pathway
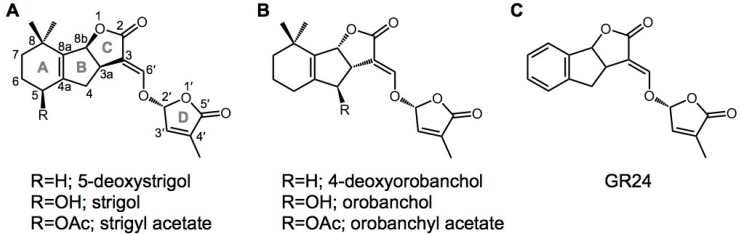
SLs are a group of terpenoid lactones that consist of a tricyclic lactone (ABC-ring) and hydroxymethyl butenolide (Figure 1). SLs were originally identified as seed germination stimulants in root parasitic plants such as Striga hermontica [12,13]. In 2005, SLs were characterized as inducers of hyphal branching in arbuscular mycorrhizal fungi [14]. Although SLs were well known as communication signals for parasitic and symbiotic interactions, they were later rediscovered as phytohormones for plant growth. Shoot branching inhibition by SLs was the first such discovery [15,16]. A series of mutants with enhanced shoot branching were isolated, including ramosus (rms) in pea, decreased apical meristem (dad) in petunia, more axillary growth (max) in Arabidopsis, and dwarf (d)/high tillering dwarf (htd) in rice. Grafting experiments with these mutants suggested that a root-derived graft-transmissible signal is involved in shoot branching inhibition [17]. Several RMS/DAD/MAX/D genes, mutations which cause the enhanced shoot branching phenotype, have been identified by molecular cloning. The genes MAX3/RMS5/D17/DAD3 and MAX4/RMS1/DAD1/D10 encode carotenoid cleavage dioxygenases 7 (CCD7) [18,19,20,21] and 8 (CCD8) [8,22,23,24], respectively, suggesting that the branch-inhibiting signal is derived from a carotenoid. MAX1 is a member of the cytochrome P450 superfamily, and acts downstream of CCD7 and CCD8 [25]. MAX2/RMS4/D3 is an F-box protein [21,26,27] and is the substrate-recognition subunit in the SKP1-CUL1-F-box-protein (SCF) ubiquitin E3 ligase complex, which targets proteins for proteasomal degradation [28]. Some F-box proteins are involved in the perception of plant hormones such as auxins, gibberellins, and jasmonic acid [28]. Endogenous SL levels are very low in ccd7, ccd8, and max1 mutants, and exogenously applied SLs inhibit enhanced shoot branching in these mutants, whereas the max2/rms4/d3 mutant is insensitive to SL treatment [15,16].
Figure 1.
Chemical structures of strigolactones (SLs) and the karrikin (KAR). Natural SLs of (A) the strigol type and (B) the orobanchol type are shown as representative SLs that have been identified from root exudates of various plant species; (C) the synthetic SL analog GR24 is widely used to evaluate the effects of SLs.
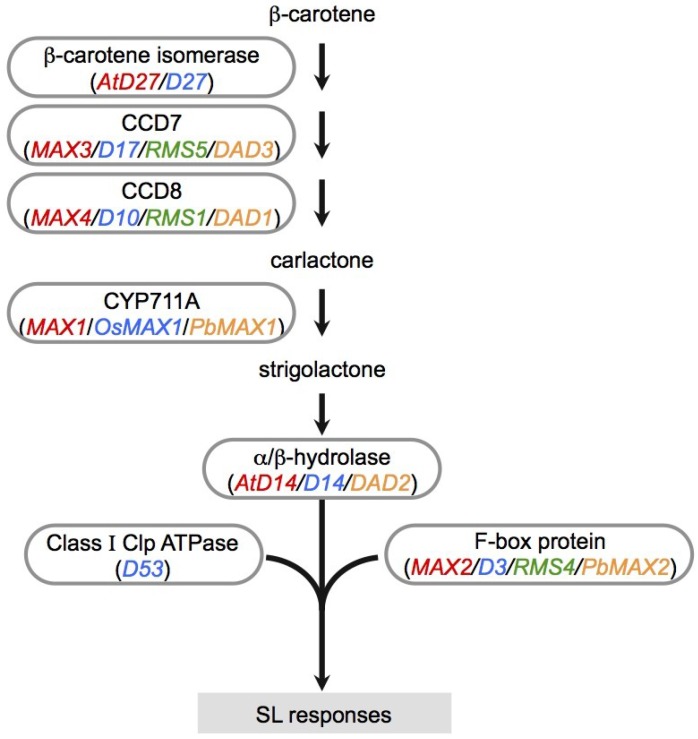
Later, another SL biosynthesis gene, D27, was identified in rice and Arabidopsis. D27 encodes an iron-containing protein localized in plastids; D27 does not have any conserved domains or homology to any known enzymes [29,30]. Recently, D27 was shown to be a β-carotene isomerase, and carlactone (CL), an SL-like carbon skeleton, was synthesized from β-carotene by three biosynthetic enzymes, D27, CCD7, and CCD8 in vitro (Figure 2) [31]. Seto et al. reported that endogenous CL was detectable in rice and Arabidopsis, and that 13C-labeled CL was converted to 13C-labeled SL, suggesting that CL is a biosynthetic precursor of SLs [32]. MAX1 catalyzes CL conversion to carlactonic acid upstream of SLs [33].
Figure 2.
SL biosynthesis and signaling pathway. The SL precursor carlactone is produced from β-carotene by three enzymes, β-carotene isomerase, CCD7, and CCD8. MAX1 is CYP711A, a member of the cytochrome P450 family, and acts as an oxidase for conversion of CL to SL. D14 (α/β-hydrolase) and F-box protein form an SCF-type ubiquitin ligase complex required for SL signaling. D53 is targeted for degradation by the SCF complex in SL signaling. Gene color coding: red, Arabidopsis; blue, rice; green, pea; orange, petunia.
In SL signaling, AtD14/DAD2/D14 and D53 function downstream of MAX1 as signaling components, as well as MAX2/RMS4/D3 (Figure 2). D14 belongs to the α/β-hydrolase family [34]. Crystal structure analysis of D14 and its orthologs from petunia (DAD2) and Arabidopsis (AtD14) demonstrated that the catalytic triad (Ser, His, Asp) is structurally conserved in these species [35,36,37]. Recently, a dominant SL-insensitive rice mutant, d53, was characterized [38,39]. The D53 protein interacts with D14, which binds GR24 (a synthetic SL analog; Figure 1) and is degraded through the D3-dependent proteasome pathway; D53 is thought to be a repressor of signaling downstream of SL [38,39]. Recently, it was reported that SLs control not only shoot branching, but also secondary growth, leaf senescence, and root growth [40,41,42,43,44,45].
3. Leaf Senescence Is Influenced by SL Signaling
Mutants with delayed leaf senescence in the dark were screened by using Arabidopsis seeds mutagenized with ethylmethane sulfonate or by T-DNA insertion, and some mutants were named oresara (ore, oresara means “long living” in Korean) [46]. Among the ore mutants, ore9 shows excessive shoot branching and delayed leaf senescence; ORE9 was later shown to be identical to MAX2 [9,27]. D3 is a MAX2/ORE9 ortholog in rice; the d3 mutant also shows delayed leaf senescence [10]. Not only the SL-insensitive mutants max2 and d3 but also SL biosynthesis mutants exhibit the delayed senescence phenotype in petunia and Lotus japonicus [8,47]. Not all SL mutants, however, show the delayed leaf senescence. No altered senescence phenotype has been reported for SL mutants/transgenics in pea or tomato [48,49,50]. To explore the effects of SLs on leaf senescence, GR24 was applied to leaf segments of rice and Arabidopsis incubated in the dark [44,45]. Leaf yellowing and membrane ion leakage from leaf segments were measured in the SL mutants as indicators of leaf senescence. Exogenously applied GR24 accelerated leaf senescence in the SL-deficient mutants d27, d17, and d10 of rice and max1, max3, and max4 of Arabidopsis, but appeared to have no effect on the SL-insensitive mutants d3 and d14 of rice and max2 and atd14 of Arabidopsis. SAGs are used as marker genes that are upregulated during both dark-induced and natural leaf senescence [51]. Among the SAGs, Yamada et al. [44] used the genes for α-keto acid dehydrogenase, which is a key enzyme in the catabolic pathway for branched-chain amino acids in mitochondria [52]; isocitrate lyase, which cleaves isocitrate to produce glyoxylate in the glyoxylate cycle [53]; and aspartic protease, which has been implicated in protein processing or degradation during plant senescence [54]. Although there were no significant differences in color, chlorophyll levels, or ion leakage between GR24 treatment and the control in wild-type leaves, SAG expression levels increased in wild-type plants and SL-deficient mutants after GR24 treatment, indicating that GR24 induces SAGs during senescence [44]. In senescing and fully senescent leaves, the transcript levels of SL biosynthesis-related genes MAX1, MAX3, and MAX4 are upregulated, suggesting that they play a role in leaf senescence [45]. SL levels in leaves are still unknown because they are too low to be measured. SLs might be produced in senescing leaves but not in young leaves. Carlactonic acid or methyl carlatonate, recently found SL-like compounds, might be synthesized in leaves and control the senescence [33]. It remains unknown whether SLs are synthesized in the leaf during senescence or SLs transported from roots promote leaf senescence.
Ethylene promotes leaf senescence. In Arabidopsis, five ethylene receptors, ETHYLENE RESISTANT1 (ETR1), ETR2, ETHYLENE INSENSITIVE4 (EIN4), ETHYLENE RESPONSE SENSOR1 (ERS1), and ERS2, have been identified; these receptors activate the serine/threonine kinase CTR1, a negative regulator of ethylene signaling [55]. Ethylene binding to the receptors inactivates CTR1, resulting in the translocation of the C-terminal fragment of the positive regulator EIN2 to the nucleus to stabilize the transcription factors EIN3 and ETHYLENE INSENSITIVE-LIKE1 (EIL1) [56]. EIN3 and EIL1 activate the expression of ethylene target genes. The ethylene-insensitive mutants ein2 and ein3 eil1 exhibit a strong delayed leaf senescence as well as SL mutants in the dark [45]. Both GR24 and ethylene treatments strongly promote leaf senescence in comparison with only ethylene treatment. These results suggest that SLs promote leaf senescence by enhancing the effects of ethylene. However, max1 ein2 double mutants show a stronger delayed leaf senescence phenotype than max1 or ein2 single mutants, suggesting that SLs partially promote leaf senescence through an ethylene-independent pathway.
In contrast, cytokinins inhibit leaf senescence. In Arabidopsis, the ahk2 ahk3 double mutants, which have defective cytokinin receptors, did not display the inhibition of dark-induced leaf senescence observed in wild type by exogenously applied cytokinin [57]. Delayed leaf senescence was shown in ore12, which has a recessive missense mutation in AHK3 [58]. In some plant species, SL mutants show delayed leaf senescence. However, the cytokinin level in xylem sap was rather reduced in SL mutants of Arabidopsis and pea, implying feedback regulation of xylem sap cytokinins [48,59,60]. In shoot branching regulation, strigolactones and cytokinins are thought to be transported acropetally through the xylem and act directly to control axillary bud outgrowth through joint regulation of a TCP (for TB1, CYCLOIDEA, PCF domain) transcription factor BRC1 [61,62]. In Arabidopsis, when wild-type and SL-deficient mutant max4 was grafted as scion and stock, respectively, leaves of wild type exhibited early senescence compared to that of max4 [45]. The main SLs to promote leaf senescence might be synthesized in leaves because max4 cannot produce SLs. To explore the interaction and transport further, it would be necessary to measure cytokinins and small amounts of SLs in leaves during senescence.
MAX2 is involved not only in SL, but also in karrikin (KAR) signaling in Arabidopsis [63,64]. KARs are a class of butenolide compounds; they promote seed germination and repress hypocotyl elongation. KARRIKIN INSENSITIVE2 (KAI2) is the AtD14 paralog required for KAR signaling [64]. KAI2 transduces the hormone signals to the SCF complex containing F-box protein MAX2 [65]. SUPPRESSOR OF MAX2 1 (SMAX1) is a possible suppressor in KAR signaling [66]. In dark-induced leaf senescence of kai2-3 and smax1, a delayed-senescence phenotype was not observed, suggesting that SL signaling specifically regulates leaf senescence but not KAR signaling [45,66].
4. Regulation of Leaf Senescence by SLs under Phosphate Deficiency
In various plants, SL levels are increased in response to the deficiency of nutrients such as nitrogen and phosphate (Pi) [16,67,68,69,70,71]. Conversely, nitrogen and Pi fertilization rapidly suppress SL production [72]. Pi is an important structural component of nucleic acids and membrane lipids and is also required for regulatory pathways involving phospholipid-derived signaling molecules or protein phosphorylation [73]. Thus, plant growth is generally suppressed by Pi deficiency. In rice, Pi deficiency was found to reduce shoot and tiller bud outgrowth [74]. Rice SL mutants were used to investigate whether SL elevated by Pi deficiency would inhibit tiller bud outgrowth. Under Pi deficiency, the levels of SL biosynthesis genes D10, D17, D27, and OsMAX1 (Os01g0700900) transcripts in roots are elevated, and endogenous SL is increased, inhibiting the tiller bud outgrowth in wild-type seedlings, but tiller bud outgrowth was observed in d mutants [75]. Similar results were obtained in Arabidopsis [76]. Thus, SLs are thought to be signal molecules that mediate plant adaptation to Pi deficiency. In addition, SLs produced in roots are secreted into the soil and induce symbiotic interactions with arbuscular mycorrhizal fungi in the rhizosphere to acquire limited Pi. Therefore, SLs have an important dual role in efficient Pi utilization and acquisition under Pi deficiency [77]. Pea showed enhanced mycorrahizal colonization in a Pi-deficient condition compared to a Pi-sufficient condition. The enhanced colonization was also confirmed in ccd8 mutants, indicating that SLs are not essential to show enhanced mycorrhizal colonaization under Pi deficiency [78].
To determine whether Pi deficiency would affect the SL responsiveness of leaf senescence in rice SL biosynthetic mutants, we examined the effect of exogenously applied GR24 on leaf senescence and found that GR24 responsiveness did not differ between Pi-sufficient and -deficient conditions in wild type and the d14 mutant [44]. Generally, leaf chlorophyll content decreases under Pi deficiency [79,80]. However, chlorophyll levels increase in SL-biosynthesis mutants and d3 under Pi deficiency [44]. As a result, stronger GR24 responsiveness was observed in SL-biosynthesis mutants under a Pi-deficient condition than a Pi-sufficient condition. Under Pi deficiency, Pi is redistributed from old leaves toward sink organs such as young leaves, growing roots, and developing seeds [81,82], which accelerates senescence of the old leaves. The SL signal is required for shoot branching inhibition and efficient progression of leaf senescence, suggesting that SLs might regulate phosphate allocation among plant tissues [83].
5. Nitrogen Metabolism and SLs
Nitrogen is required for the synthesis of nucleic acids, proteins, and secondary metabolites including nitrogen, and is a major limiting factor in plant productivity [73]. Under nitrogen deficiency, gene expression is repressed in amino acid synthesis, chlorophyll synthesis, photosynthetic light reaction, and Carvin cycle, and is induced in amino acid degradation. Levels of amino acids and nitrogen-containing compounds decrease, whereas starch and flavonoid levels increase. As mentioned above, SLs are produced in response to nitrogen deficiency in some species [67,70,71]. Plants use nitrate or ammonium ions (NH4+) as a primary nitrogen source. When NH4+ is assimilated in plant cells, glutamine synthase (GS) coupled with glutamate synthase (GOGAT) catalyzes the ATP-dependent addition of NH4+ to glutamate [84]. GOGAT generates two glutamate molecules from glutamine and 2-oxoglutalete in the presence of either ferredoxin or NADH. In rice, there are three cytosolic GS1 (OsGS1;1, OsGS1;2, and OsGS1;3) and two NADH-GOGATs (OsNADH-GOGAT1 and OsNADH-GOGAT2) [85]. In chloroplasts, GS2 and ferredoxin-GOGAT are responsible for the assimilation of NH4+ released during photorespiration [85]. OsGS1;2 and OsNADH-GOGAT1 are important for primary NH4+ assimilation in the roots. Tiller bud outgrowth in rice is severely reduced by the lack of OsGS1;2 [86]. Although a high SL content in rice roots is known to reduce the number of outgrowing tillers, the SL levels in gs1;2 and nadh-gogat1 are similar to those in wild type, indicating that the reduction of tiller bud outgrowth observed in gs1;2 is independent of the SL level. On the other hand, OsGS1;1 and OsNADH-GOGAT2 were expressed in mature rice leaves [87]. OsGS1;1 disruption causes a severe reduction of growth and grain filling [88]. Therefore, OsGS1;1 and OsNADH-GOGAT2 appear to be important in the remobilization of nitrogen during natural leaf senescence in rice. However, no information is available on the relationship between GS1;1 and SLs during leaf senescence.
6. Perspectives
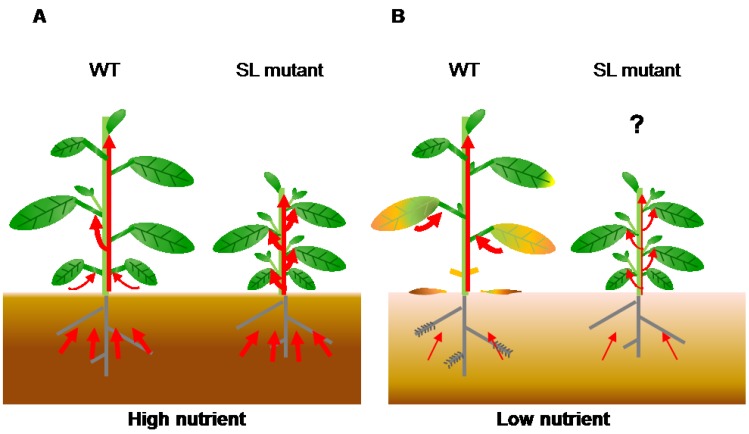
Plant growth is controlled by a complicated cross-talk of plant hormones. Recently, the interactions between SLs and other hormones have been uncovered in plant development [89] and stress responses [90,91]. However, the role of these interactions in leaf senescence is still unknown. Our hypothesis is that plants grown in soil with sufficient concentrations of nutrients (such as nitrogen and Pi) receive enough nutrients from the roots, and axillary buds can grow because SL levels are low (Figure 3). In contrast, plants grown in nutrient-deficient soil produce high SL levels to inhibit axillary bud outgrowth and accelerate leaf senescence for utilization of limited nutrients (Figure 3). To make up for nutrient deficiency, nutrients accumulated in old leaves are transported to young tissues and seeds. Because SL mutants show enhanced shoot branching and delayed leaf senescence, they may have reduced nutrient remobilization, resulting in a reduction of seed production. These findings indicate that SLs might affect crop grain yield through leaf senescence and shoot branching regulation. To address this question, it is necessary to compare the levels of primary metabolites (such as sugars and amino acids) and seed production in wild-type plants and SL mutants in the future.
Figure 3.
Transport of nutrients during leaf senescence under nutrient deficiency. (A) In nutrient-sufficient soil, plants absorb enough nutrients for growth and seed production. Wild-type plants show axillary bud outgrowth and the leaves remain green during leaf development because the SL levels are low. In SL mutants, the number of branches increases and nutrients absorbed from soil are evenly distributed between old and young organs because these plants have no SL signaling; (B) In nutrient-deficient soil, plants cannot absorb enough nutrients. In the wild type, increased SL levels inhibit shoot branching and accelerate leaf senescence to use limited nutrients. Nutrients are transported from old tissues to young tissues and seeds. SL mutants show enhanced shoot branching and delayed leaf senescence, which might reduce nutrient remobilization. Red arrows indicate flow of nutrients.
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research on Innovative Areas and for Scientific Research (C) from Japan’s Ministry of Education, Culture, Sports, Science and Technology (Nos. 23119523 and 26450144), by the Inoue Enryo Memorial Foundation for Promoting Sciences from Toyo University.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Noodén L.D. The phenomena of senescence and aging. In: Leopold A.C., Noodén L.D., editors. Senescence and aging in plants. Academic Press; San Diego, CA, USA: 1988. pp. 1–50. [Google Scholar]
- 2.Lim P.O., Kim H.J., Nam H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- 3.Buchanan-Wollaston V. The molecular biology of leaf senescence. J. Exp. Bot. 1997;48:181–199. doi: 10.1093/jxb/48.2.181. [DOI] [Google Scholar]
- 4.Gan S., Amasino R.M. Making sense of senescence (Molecular genetic regulation and manipulation of leaf senescence) Plant Physiol. 1997;113:313–319. doi: 10.1104/pp.113.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchanan-Wollaston V., Earl S., Harrison E., Mathas E., Navabpour S., Page T., Pink D. The molecular analysis of leaf senescence: A genomics approach. Plant Biotechnol. J. 2003;1:3–22. doi: 10.1046/j.1467-7652.2003.00004.x. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H., Zhou C. Signal transduction in leaf senescence. Plant Mol. Biol. 2013;82:539–545. doi: 10.1007/s11103-012-9980-4. [DOI] [PubMed] [Google Scholar]
- 7.Jibran R., Hunter D.A., Dijkwel P.P. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 2013;82:547–561. doi: 10.1007/s11103-013-0043-2. [DOI] [PubMed] [Google Scholar]
- 8.Snowden K.C., Simkin A.J., Janssen B.J., Templeton K.R., Loucas H.M., Simons J.L., Karunairetnam S., Gleave A.P., Clark D.G., Klee H.J. The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell. 2005;17:746–759. doi: 10.1105/tpc.104.027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woo H.R., Chung K.M., Park J.H., Oh S.A., Ahn T., Hong S.H., Jang S.K., Nam H.G. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell. 2001;13:1779–1790. doi: 10.1105/tpc.13.8.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan H., Saika H., Maekawa M., Takamure I., Tsutsumi N., Kyozuka J., Nakazono M. Rice tillering dwarf mutant dwarf3 has increased leaf longevity during darkness-induced senescence or hydrogen peroxide-induced cell death. Genes Genet. Syst. 2007;82:361–366. doi: 10.1266/ggs.82.361. [DOI] [PubMed] [Google Scholar]
- 11.Hamiaux C., Drummond R.S., Janssen B.J., Ledger S.E., Cooney J.M., Newcomb R.D., Snowden K.C. DAD2 is an alpha/beta hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 2012;22:2032–2036. doi: 10.1016/j.cub.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Cook C.E., Whichard L.P., Turner B., Wall M.E., Egley G.H. Germination of witchweed (Striga lutea Lour.): Isolation and properties of a potent stimulant. Science. 1966;154:1189–1190. doi: 10.1126/science.154.3753.1189. [DOI] [PubMed] [Google Scholar]
- 13.Cook C.E., Whichard L.P., Wall M.E., Egley G.H., Coggon P., Luhan P.A., McPhail A.T. Germination stimulants. 2. The structure of strigol—A potent seed germination stmulant for witchweed (Striga lutea Lour.) J. Am. Chem. Soc. 1972;94:6198–6199. doi: 10.1021/ja00772a048. [DOI] [Google Scholar]
- 14.Akiyama K., Matsuzaki K., Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. 2005;435:824–827. doi: 10.1038/nature03608. [DOI] [PubMed] [Google Scholar]
- 15.Gomez-Roldan V., Fermas S., Brewer P.B., Puech-Pages V., Dun E.A., Pillot J.P., Letisse F., Matusova R., Danoun S., Portais J.C., et al. Strigolactone inhibition of shoot branching. Nature. 2008;455:189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- 16.Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., Magome H., Kamiya Y., Shirasu K., Yoneyama K., et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455:195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- 17.Mouchel C.F., Leyser O. Novel phytohormones involved in long-range signaling. Curr. Opin. Plant Biol. 2007;10:473–476. doi: 10.1016/j.pbi.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Booker J., Auldridge M., Wills S., McCarty D., Klee H., Leyser O. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 2004;14:1232–1238. doi: 10.1016/j.cub.2004.06.061. [DOI] [PubMed] [Google Scholar]
- 19.Drummond R.S.M., Martinez-Sanchez N.M., Janssen B.J., Templeton K.R., Simons J.L., Quinn B.D., Karunairetnam S., Snowden K.C. Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 is involved in the production of negative and positive branching signals in petunia. Plant Physiol. 2009;151:1867–1877. doi: 10.1104/pp.109.146720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou J., Zhang S., Zhang W., Li G., Chen Z., Zhai W., Zhao X., Pan X., Xie Q., Zhu L. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 2006;48:687–698. doi: 10.1111/j.1365-313X.2006.02916.x. [DOI] [PubMed] [Google Scholar]
- 21.Johnson X., Brcich T., Dun E.A., Goussot M., Haurogne K., Beveridge C.A., Rameau C. Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol. 2006;142:1014–1026. doi: 10.1104/pp.106.087676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arite T., Iwata H., Ohshima K., Maekawa M., Nakajima M., Kojima M., Sakakibara H., Kyozuka J. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 2007;51:1019–1029. doi: 10.1111/j.1365-313X.2007.03210.x. [DOI] [PubMed] [Google Scholar]
- 23.Simons J.L., Napoli C.A., Janssen B.J., Plummer K.M., Snowden K.C. Analysis of the DECREASED APICAL DOMINANCE genes of petunia in the control of axillary branching. Plant Physiol. 2007;143:697–706. doi: 10.1104/pp.106.087957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorefan K., Booker J., Haurogne K., Goussot M., Bainbridge K., Foo E., Chatfield S., Ward S., Beveridge C., Rameau C., Leyser O. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 2003;17:1469–1474. doi: 10.1101/gad.256603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Booker J., Sieberer T., Wright W., Williamson L., Willett B., Stirnberg P., Turnbull C., Srinivasan M., Goddard P., Leyser O. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell. 2005;8:443–449. doi: 10.1016/j.devcel.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Ishikawa S., Maekawa M., Arite T., Onishi K., Takamure I., Kyozuka J. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 2005;46:79–86. doi: 10.1093/pcp/pci022. [DOI] [PubMed] [Google Scholar]
- 27.Stirnberg P., van de Sande K., Leyser H.M. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development. 2002;129:1131–1141. doi: 10.1242/dev.129.5.1131. [DOI] [PubMed] [Google Scholar]
- 28.Lechner E., Achard P., Vansiri A., Potuschak T., Genschik P. F-box proteins everywhere. Curr. Opin. Plant Biol. 2006;9:631–638. doi: 10.1016/j.pbi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Lin H., Wang R.X., Qian Q., Yan M.X., Meng X.B., Fu Z.M., Yan C.Y., Jiang B., Su Z., Li J.Y., Wang Y.H. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell. 2009;21:1512–1525. doi: 10.1105/tpc.109.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waters M.T., Brewer P.B., Bussell J.D., Smith S.M., Beveridge C.A. The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol. 2012;159:1073–1085. doi: 10.1104/pp.112.196253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alder A., Jamil M., Marzorati M., Bruno M., Vermathen M., Bigler P., Ghisla S., Bouwmeester H., Beyer P., Al-Babili S. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science. 2012;335:1348–1351. doi: 10.1126/science.1218094. [DOI] [PubMed] [Google Scholar]
- 32.Seto Y., Sado A., Asami K., Hanada A., Umehara M., Akiyama K., Yamaguchi S. Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc. Natl. Acad. Sci. USA. 2014;111:1640–1645. doi: 10.1073/pnas.1314805111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abe S., Sado A., Tanaka K., Kisugi T., Asami K., Ota S., Kim H.I., Yoneyama K., Xie X., Ohnishi T., et al. Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc. Natl. Acad. Sci. USA. 2014;111:18084–18089. doi: 10.1073/pnas.1410801111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arite T., Umehara M., Ishikawa S., Hanada A., Maekawa M., Yamaguchi S., Kyozuka J. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 2009;50:1416–1424. doi: 10.1093/pcp/pcp091. [DOI] [PubMed] [Google Scholar]
- 35.Kagiyama M., Hirano Y., Mori T., Kim S.Y., Kyozuka J., Seto Y., Yamaguchi S., Hakoshima T. Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells. 2013;18:147–160. doi: 10.1111/gtc.12025. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura H., Xue Y.L., Miyakawa T., Hou F., Qin H.M., Fukui K., Shi X., Ito E., Ito S., Park S.H., et al. Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun. 2013;4:2613. doi: 10.1038/ncomms3613. [DOI] [PubMed] [Google Scholar]
- 37.Zhao L.H., Zhou X.E., Wu Z.S., Yi W., Xu Y., Li S., Xu T.H., Liu Y., Chen R.Z., Kovach A., et al. Crystal structures of two phytohormone signal-transducing alpha/beta hydrolases: Karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 2013;23:436–439. doi: 10.1038/cr.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang L., Liu X., Xiong G., Liu H., Chen F., Wang L., Meng X., Liu G., Yu H., Yuan Y., et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature. 2013;504:401–405. doi: 10.1038/nature12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou F., Lin Q., Zhu L., Ren Y., Zhou K., Shabek N., Wu F., Mao H., Dong W., Gan L., et al. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature. 2013;504:406–410. doi: 10.1038/nature12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agusti J., Herold S., Schwarz M., Sanchez P., Ljung K., Dun E.A., Brewer P.B., Beveridge C.A., Sieberer T., Sehr E.M., et al. Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc. Natl. Acad. Sci. USA. 2011;108:20242–20247. doi: 10.1073/pnas.1111902108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapulnik Y., Delaux P.M., Resnick N., Mayzlish-Gati E., Wininger S., Bhattacharya C., Sejalon-Delmas N., Combier J.P., Becard G., Belausov E., et al. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta. 2011;233:209–216. doi: 10.1007/s00425-010-1310-y. [DOI] [PubMed] [Google Scholar]
- 42.Kapulnik Y., Resnick N., Mayzlish-Gati E., Kaplan Y., Wininger S., Hershenhorn J., Koltai H. Strigolactones interact with ethylene and auxin in regulating root-hair elongation in Arabidopsis. J. Exp. Bot. 2011 doi: 10.1093/jxb/erq464. [DOI] [PubMed] [Google Scholar]
- 43.Ruyter-Spira C., Kohlen W., Charnikhova T., van Zeijl A., van Bezouwen L., de Ruijter N., Cardoso C., Lopez-Raez J.A., Matusova R., Bours R., et al. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: Another belowground role for strigolactones? Plant Physiol. 2011;155:721–734. doi: 10.1104/pp.110.166645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada Y., Furusawa S., Nagasaka S., Shimomura K., Yamaguchi S., Umehara M. Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta. 2014;240:399–408. doi: 10.1007/s00425-014-2096-0. [DOI] [PubMed] [Google Scholar]
- 45.Ueda H., Kusaba M. Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis. Plant Physiol. 2015 doi: 10.1104/pp.15.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oh S.A., Park J.H., Lee G.I., Paek K.H., Park S.K., Nam H.G. Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J. 1997;12:527–535. doi: 10.1046/j.1365-313X.1997.00527.x. [DOI] [PubMed] [Google Scholar]
- 47.Liu J., Novero M., Charnikhova T., Ferrandino A., Schubert A., Ruyter-Spira C., Bonfante P., Lovisolo C., Bouwmeester H.J., Cardinale F. CAROTENOID CLEAVAGE DIOXYGENASE 7 modulates plant growth, reproduction, senescence, and determinate nodulation in the model legume Lotus japonicus. J. Exp. Bot. 2013;64:1967–1981. doi: 10.1093/jxb/ert056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beveridge C.A., Murfet I.C., Kerhoas L., Sotta B., Miginiac E., Rameau C. The shoot controls zeatin riboside export from pea roots. Evidence from the branching mutant rms4. Plant J. 1997;11:339–345. doi: 10.1046/j.1365-313X.1997.11020339.x. [DOI] [Google Scholar]
- 49.Vogel J.T., Walter M.H., Giavalisco P., Lytovchenko A., Kohlen W., Charnikhova T., Simkin A.J., Goulet C., Strack D., Bouwmeester H.J., et al. SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. Plant J. 2010;61:300–311. doi: 10.1111/j.1365-313X.2009.04056.x. [DOI] [PubMed] [Google Scholar]
- 50.Kohlen W., Charnikhova T., Lammers M., Pollina T., Toth P., Haider I., Pozo M.J., de Maagd R.A., Ruyter-Spira C., Bouwmeester H.J., et al. The tomato CAROTENOID CLEAVAGE DIOXYGENASE8 (SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytol. 2012;196:535–547. doi: 10.1111/j.1469-8137.2012.04265.x. [DOI] [PubMed] [Google Scholar]
- 51.Lee R.H., Wang C.H., Huang L.T., Chen S.C. Leaf senescence in rice plants: cloning and characterization of senescence up-regulated genes. J. Exp. Bot. 2001;52:1117–1121. doi: 10.1093/jexbot/52.358.1117. [DOI] [PubMed] [Google Scholar]
- 52.Fujiki Y., Sato T., Ito M., Watanabe A. Isolation and characterization of cDNA clones for the e1beta and E2 subunits of the branched-chain alpha-ketoacid dehydrogenase complex in Arabidopsis. J. Biol. Chem. 2000;275:6007–6013. doi: 10.1074/jbc.275.8.6007. [DOI] [PubMed] [Google Scholar]
- 53.Lu Y., Wu Y.R., Han B. Anaerobic induction of isocitrate lyase and malate synthase in submerged rice seedlings indicates the important metabolic role of the glyoxylate cycle. Acta Biochim. Biophys. Sin. 2005;37:406–414. doi: 10.1111/j.1745-7270.2005.00060.x. [DOI] [PubMed] [Google Scholar]
- 54.Roberts I.N., Caputo C., Criado M.V., Funk C. Senescence-associated proteases in plants. Physiol. Plant. 2012;145:130–139. doi: 10.1111/j.1399-3054.2012.01574.x. [DOI] [PubMed] [Google Scholar]
- 55.Merchante C., Alonso J.M., Stepanova A.N. Ethylene signaling: Simple ligand, complex regulation. Curr. Opin. Plant Biol. 2013;16:554–560. doi: 10.1016/j.pbi.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 56.Yoo S.D., Cho Y., Sheen J. Emerging connections in the ethylene signaling network. Trends Plant Sci. 2009;14:270–279. doi: 10.1016/j.tplants.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riefler M., Novak O., Strnad M., Schmulling T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell. 2006;18:40–54. doi: 10.1105/tpc.105.037796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim H.J., Ryu H., Hong S.H., Woo H.R., Lim P.O., Lee I.C., Sheen J., Nam H.G., Hwang I. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2006;103:814–819. doi: 10.1073/pnas.0505150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Foo E., Morris S.E., Parmenter K., Young N., Wang H.T., Jones A., Rameau C., Turnbull C.G.N., Beveridge C.A. Feedback regulation of xylem cytokinin content is conserved in pea and arabidopsis. Plant Physiol. 2007;143:1418–1428. doi: 10.1104/pp.106.093708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beveridge C.A., Symons G.M., Murfet I.C., Ross J.J., Rameau C. The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signal(s) Plant Physiol. 1997;115:1251–1258. [Google Scholar]
- 61.Braun N., de Saint Germain A., Pillot J.P., Boutet-Mercey S., Li X., Antoniadi I., Dalmais M., Maia-Grondard A., Lesignor C., Bouteiller N., et al. The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 2011;158:225–238. doi: 10.1104/pp.111.182725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dun E.A., de Saint Germain A., Rameau C., Beveridge C.A. Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol. 2012;158:487–498. doi: 10.1104/pp.111.186783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nelson D.C., Scaffidi A., Dun E.A., Waters M.T., Flematti G.R., Dixon K.W., Beveridge C.A., Ghisalberti E.L., Smith S.M. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2011;108:8897–8902. doi: 10.1073/pnas.1100987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waters M.T., Nelson D.C., Scaffidi A., Flematti G.R., Sun Y.K., Dixon K.W., Smith S.M. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development. 2012;139:1285–1295. doi: 10.1242/dev.074567. [DOI] [PubMed] [Google Scholar]
- 65.Waters M.T., Smith S.M. KAI2- and MAX2-mediated responses to karrikins and strigolactones are largely independent of HY5 in Arabidopsis seedlings. Mol. Plant. 2013;6:63–75. doi: 10.1093/mp/sss127. [DOI] [PubMed] [Google Scholar]
- 66.Stanga J.P., Smith S.M., Briggs W.R., Nelson D.C. SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol. 2013;163:318–330. doi: 10.1104/pp.113.221259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoneyama K., Xie X., Kusumoto D., Sekimoto H., Sugimoto Y., Takeuchi Y., Yoneyama K. Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta. 2007;227:125–132. doi: 10.1007/s00425-007-0600-5. [DOI] [PubMed] [Google Scholar]
- 68.Yoneyama K., Yoneyama K., Takeuchi Y., Sekimoto H. Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites. Planta. 2007;225:1031–1038. doi: 10.1007/s00425-006-0410-1. [DOI] [PubMed] [Google Scholar]
- 69.Lopez-Raez J.A., Charnikhova T., Gomez-Roldan V., Matusova R., Kohlen W., de Vos R., Verstappen F., Puech-Pages V., Becard G., Mulder P., et al. Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol. 2008;178:863–874. doi: 10.1111/j.1469-8137.2008.02406.x. [DOI] [PubMed] [Google Scholar]
- 70.Yoneyama K., Xie X., Kim H.I., Kisugi T., Nomura T., Sekimoto H., Yokota T., Yoneyama K. How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta. 2012;235:1197–1207. doi: 10.1007/s00425-011-1568-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jamil M., Charnikhova T., Cardoso C., Jamil T., Ueno K., Verstappen F., Asami T., Bouwmeester H.J. Quantification of the relationship between strigolactones and Striga hermonthica infection in rice under varying levels of nitrogen and phosphorus. Weed Res. 2011;51:373–385. doi: 10.1111/j.1365-3180.2011.00847.x. [DOI] [Google Scholar]
- 72.Yoneyama K., Xie X., Kisugi T., Nomura T., Yoneyama K. Nitrogen and phosphorus fertilization negatively affects strigolactone production and exudation in sorghum. Planta. 2013;238:885–894. doi: 10.1007/s00425-013-1943-8. [DOI] [PubMed] [Google Scholar]
- 73.Amtmann A., Armengaud P. Effects of N, P, K and S on metabolism: New knowledge gained from multi-level analysis. Curr. Opin. Plant Biol. 2009;12:275–283. doi: 10.1016/j.pbi.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 74.Luquet D., Zhang B.G., Dingkuhn M., Dexet A., Clement-Vidal A. Phenotypic plasticity of rice seedlings: Case of phosphorus deficiency. Plant Prod. Sci. 2005;8:145–151. doi: 10.1626/pps.8.145. [DOI] [Google Scholar]
- 75.Umehara M., Hanada A., Magome H., Takeda-Kamiya N., Yamaguchi S. Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol. 2010;51:1118–1126. doi: 10.1093/pcp/pcq084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kohlen W., Charnikhova T., Liu Q., Bours R., Domagalska M.A., Beguerie S., Verstappen F., Leyser O., Bouwmeester H., Ruyter-Spira C. Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol. 2011;155:974–987. doi: 10.1104/pp.110.164640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Czarnecki O., Yang J., Weston D.J., Tuskan G.A., Chen J.G. A dual role of strigolactones in phosphate acquisition and utilization in plants. Int J. Mol. Sci. 2013;14:7681–7701. doi: 10.3390/ijms14047681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Foo E., Yoneyama K., Hugill C., Quittenden L., Reid J. Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Mol. Plant. 2013;6:76–87. doi: 10.1093/mp/sss115. [DOI] [PubMed] [Google Scholar]
- 79.Park M.R., Baek S.-H., de los Reyes B.G., Yun S.J., Hasenstein K.H. Transcriptome profiling characterizes phosphate deficiency effects on carbohydrate metabolism in rice leaves. J. Plant Physiol. 2012;169:193–205. doi: 10.1016/j.jplph.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 80.Cai H., Chu Q., Yuan L., Liu J., Chen X., Chen F., Mi G., Zhang F. Identification of quantitative trait loci for leaf area and chlorophyll content in maize (Zea mays) under low nitrogen and low phosphorus supply. Mol. Breed. 2012;30:251–266. doi: 10.1007/s11032-011-9615-5. [DOI] [Google Scholar]
- 81.Raghothama K.G. Phosphate transport and signaling. Curr. Opin. Plant Biol. 2000;3:182–187. doi: 10.1016/S1369-5266(00)00062-5. [DOI] [PubMed] [Google Scholar]
- 82.Bucher M., Rausch C., Daram P. Molecular and biochemical mechanisms of phosphorus uptake into plants. J. Plant Nutr. Soil Sci. 2001;164:209–217. doi: 10.1002/1522-2624(200104)164:2<209::AID-JPLN209>3.0.CO;2-F. [DOI] [Google Scholar]
- 83.Umehara M. Strigolactone, a key regulator of nutrient allocation in plants. Plant Biotechnol. 2011;28:429–437. doi: 10.5511/plantbiotechnology.11.1109a. [DOI] [Google Scholar]
- 84.Lea P.J., Miflin B.J. Alternative route for nitrogen assimilation in higher plants. Nature. 1974;251:614–616. doi: 10.1038/251614a0. [DOI] [PubMed] [Google Scholar]
- 85.Yamaya T., Kusano M. Evidence supporting distinct functions of three cytosolic glutamine synthetases and two NADH-glutamate synthases in rice. J. Exp. Bot. 2014;65:5519–5525. doi: 10.1093/jxb/eru103. [DOI] [PubMed] [Google Scholar]
- 86.Ohashi M., Ishiyama K., Kusano M., Fukushima A., Kojima S., Hanada A., Kanno K., Hayakawa T., Seto Y., Kyozuka J., et al. Lack of cytosolic glutamine synthetase1;2 in vascular tissues of axillary buds causes severe reduction in their outgrowth and disorder of metabolic balance in rice seedlings. Plant J. 2015;81:347–356. doi: 10.1111/tpj.12731. [DOI] [PubMed] [Google Scholar]
- 87.Tabuchi M., Abiko T., Yamaya T. Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.) J. Exp. Bot. 2007;58:2319–2327. doi: 10.1093/jxb/erm016. [DOI] [PubMed] [Google Scholar]
- 88.Tabuchi M., Sugiyama K., Ishiyama K., Inoue E., Sato T., Takahashi H., Yamaya T. Severe reduction in growth rate and grain filling of rice mutants lacking OsGS1;1, a cytosolic glutamine synthetase1;1. Plant J. 2005;42:641–651. doi: 10.1111/j.1365-313X.2005.02406.x. [DOI] [PubMed] [Google Scholar]
- 89.Al-Babili S., Bouwmeester H.J. Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 2015;66:161–186. doi: 10.1146/annurev-arplant-043014-114759. [DOI] [PubMed] [Google Scholar]
- 90.Bu Q., Lv T., Shen H., Luong P., Wang J., Wang Z., Huang Z., Xiao L., Engineer C., Kim T.H., et al. Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant Physiol. 2014;164:424–439. doi: 10.1104/pp.113.226837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ha C.V., Leyva-Gonzalez M.A., Osakabe Y., Tran U.T., Nishiyama R., Watanabe Y., Tanaka M., Seki M., Yamaguchi S., Dong N.V., et al. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc. Natl. Acad. Sci. USA. 2014;111:851–856. doi: 10.1073/pnas.1322135111. [DOI] [PMC free article] [PubMed] [Google Scholar]