Abstract
Background
This study meta-analyzed data on the possible association of the miR-196a2 C>T (rs11614913) and miR-499 A>G (rs3746444) polymorphisms with risk of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC).
Methods
Databases in PubMed, EMBASE, Web of Science, China BioMedicine, and Google Scholar were systematically searched to identify relevant studies. Meta-analyses were performed to examine the association of the miR-196a2 C>T and miR-499 A>G polymorphisms with HBV-related HCC risk. Odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated.
Results
A total of 13 studies involving 3,964 cases and 5,875 healthy controls were included. Random-effect meta-analysis showed that the T allele and TT genotype of miR-196a2 C>T were associated with significantly lower HBV-related HCC risk (allelic model, OR =0.84, 95% CI =0.71–0.99, P=0.04; homozygous model, OR =0.68, 95% CI =0.47–0.98, P=0.04). In contrast, miR-499 A>G showed no significant association with HBV-related HCC risk in either overall pooled analysis or ethnic subgroup analysis according to any of the four genetic models. Based on analysis of ethnic subgroups, neither miR-196a2 C>T nor miR-499 A>G was significantly associated with risk of HBV-related HCC in Chinese population.
Conclusion
The polymorphism miR-196a2 C>T, but not miR-499 A>G, may be associated with decreased HBV-related HCC risk. These conclusions should be verified in large, well-designed studies.
Keywords: microRNA, single nucleotide polymorphisms, hepatitis B virus related, meta-analysis, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is one of the most common causes of cancer-related mortality worldwide.1 It is well known that chronic hepatitis B virus (HBV) is a primary risk factor for HCC, especially in Asian populations and particularly Chinese,2 but only a fraction of infected patients develop HCC during their lifetime. This suggests that genetic factors play a role in tumor development.
MicroRNAs (miRNAs) are short, noncoding RNA molecules 18–25 nucleotides long. Bioinformatics studies indicate that a single miRNA may bind to as many as 200 gene targets, and miRNAs have been proposed to regulate the expression of approximately one-third of protein-coding messenger RNAs. miRNAs posttranscriptionally suppress target gene expression, usually by binding to the 3′-untranslated region of messenger RNA, in a broad range of organisms in both physiological and disease contexts. This suppression plays important roles in regulating diverse biological processes, including development, cell cycle, proliferation, differentiation, apoptosis, and response to stress.3,4 Recent studies further suggest that miRNAs play a critical role as tumor suppressors and oncogenes.5,6
The miRNA polymorphisms miR-196a2 C>T (rs11614913) and miR-499 A>G (rs3746444) have been linked to risk of certain types of cancer.7,8 Numerous case–control studies have investigated a potential association between miR-196a2 C>T and risk of HBV-related HCC. Most of these studies have associated miR-196a2 C>T with decreased risk of HBV-related HCC,9–12 while others have failed to detect any significant association.13–15 Studies of a possible association between miR-499 A>G and risk of HBV-related HCC have given similarly inconsistent results.15–21 Meta-analyses have attempted to synthesize the available evidence on these associations,22,23 but the results are inconclusive, primarily because of the small sample sizes involved.
Thus, we conducted an updated meta-analysis of all relevant literature to provide more comprehensive and reliable insights into possible associations of miR-196a2 C>T and miR-499 A>G with risk of HBV-related HCC.
Methods
Search strategy
The databases in PubMed, EMBASE, Web of Science, China BioMedicine, and Google Scholar were systematically searched to identify all clinical and experimental case–control studies examining associations of miR-196a2 C>T and/or miR-499 A>G with risk of HBV-related HCC. The last round of searching was conducted on January 3, 2016. The following search terms and strings were used: miR-196a2, miRNA-196a2, microRNA-196a2, rs11614913, miR-499, miRNA-499, microRNA-499, or rs3746444; each of the aforementioned terms in combination with polymorphism, SNP, variant, variants, variation, genotype, genetic or mutation; and each of the aforementioned terms in combination with hepatocellular carcinoma or liver cancer. Reference lists in identified articles and reviews were also manually searched to identify additional eligible studies.
Inclusion criteria
To be included in this meta-analysis, studies had to 1) use a case–control design to assess the association of miR-196a2 C>T (rs11614913) and/or miR-499 A>G (rs3746444) with risk of HBV-related HCC in humans; 2) be available as full text; and 3) report sufficient data for estimating an odds ratio (OR) with 95% confidence interval (CI), including genotype frequencies. If multiple publications reported data for the same study population, only the study with the largest sample size was included.
Data extraction
Two authors (S-LZ and J-HZ) independently extracted the following data from included studies: first author’s family name, year of publication, ethnicity, country of origin, genotyping methods, P describing Hardy–Weinberg equilibrium (HWE) in controls, numbers and genotypes of cases and controls, and frequencies of genotypes in cases and controls. Discrepancies were resolved by consensus. Only those studies meeting the predetermined inclusion criteria were included.
Statistical analysis
Using previously described methods,24–26 we calculated the unadjusted OR with 95% CI to assess the strength of the association of miR-196a2 C>T and miR-499 A>G with risk of HBV-related HCC based on genotype frequencies in cases and controls. The significance of pooled ORs was determined using the Z-test, with P<0.05 defined as the significance threshold. A meta-analysis was conducted using a fixed-effect model when P>0.10 for the Q-test, indicating lack of heterogeneity among studies; otherwise, a random-effect model was used. It was performed over the course of the entire study in all participants, as well as for subgroups of Asian participants in general and of Chinese participants in particular. All statistical tests for this meta-analysis were performed in Review Manager 5.2 (Cochrane Collaboration, London, UK).
Publication bias was assessed using Begg’s funnel plot and Egger’s weighted regression, with P<0.05 considered statistically significant. Both these tests were performed using Stata 12.0 (StataCorp LP, College Station, TX, USA).
Results
Description of studies
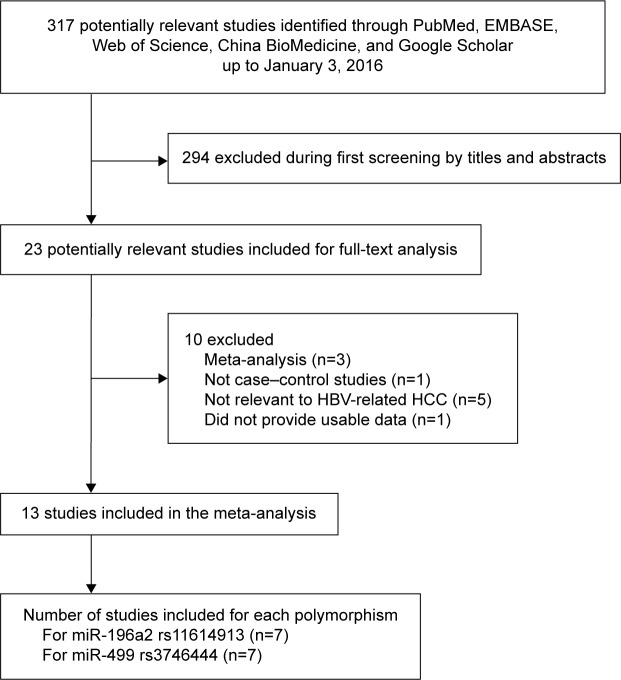
Figure 1 is a flow diagram illustrating the search and study selection criteria. A total of 317 potentially relevant publications up to January 3, 2016, were systematically identified in the databases of PubMed, EMBASE, Web of Science, China BioMedicine, and Google Scholar. Of these, we excluded 294 studies based on review of titles and abstracts. An additional ten studies were excluded because they were meta-analyses (n=3), not relevant to HBV-related HCC (n=5), not case–control studies (n=1), or did not provide usable data (n=1). In the end, 13 studies9–21 were included in this meta-analysis based on our search strategy and inclusion criteria. Their characteristics are summarized in Table 1. Seven studies9–15 evaluated the association between miR-196a2 C>T and risk of HBV-related HCC in a total of 2,693 cases and 3,594 controls. Seven studies15–21 evaluated the association between miR-499 A>G and risk of HBV-related HCC in a total of 1,271 cases and 2,281 controls. The miR-196a2 C>T study by Akkız et al,11 involving 105 cases and 185 controls, involved a Caucasian population. The other 12 studies9,10,12–21 involved Asian populations. The distribution of genotypes in controls was consistent with HWE (P>0.05) in all but six studies.9,10,18–21
Figure 1.
Flowchart of study selection.
Abbreviations: HBV, hepatitis B virus; HCC, hepatocellular carcinoma.
Table 1.
Characteristics of studies in the meta-analysis
| First author | Year | Ethnicity | Country | Testing methods | P for HWE | Cases–controls | No of cases | Allele frequencies of controls (n [%]) | No of controls | Allele frequencies of cases (n [%]) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miR-196a2 C>T (rs11614913) | CC | CT | TT | C | T | CC | CT | TT | C | T | ||||||
| Qi et al13 | 2010 | Asian | People’s Republic of China | PCR-LDR | 0.87 | 361/391 | 82 | 179 | 100 | 343 (47.5) | 379 (52.5) | 92 | 197 | 102 | 381 (48.7) | 401 (51.3) |
| Akkız et al11 | 2011 | Caucasian | Turkey | PCR-RFLP | 0.49 | 105/185 | 46 | 48 | 11 | 140 (66.7) | 70 (33.3) | 58 | 87 | 40 | 203 (54.9) | 167 (45.1) |
| Kim et al15 | 2012 | Asian | Korea | PCR-RFLP | 0.36 | 127/201 | 24 | 70 | 33 | 118 (46.5) | 136 (53.5) | 45 | 107 | 49 | 197 (49.0) | 205 (51.0) |
| Han et al14 | 2013 | Asian | People’s Republic of China | Q-PCR | 0.31 | 1,017/1,009 | 207 | 505 | 305 | 919 (45.2) | 1,115 (54.8) | 220 | 485 | 304 | 925 (45.8) | 1,093 (54.2) |
| Zhang et al12 | 2013 | Asian | People’s Republic of China | MassARRAY | 0.25 | 771/995 | 171 | 376 | 224 | 718 (46.6) | 824 (53.4) | 165 | 502 | 328 | 832 (41.8) | 1,158 (58.2) |
| Kou et al9 | 2014 | Asian | People’s Republic of China | Duplex PCR | <0.001 | 159/532 | 56 | 85 | 18 | 197 (61.9) | 121 (38.1) | 125 | 304 | 103 | 554 (52.1) | 510 (47.9) |
| Zhou et al10 | 2014 | Asian | People’s Republic of China | PCR-RFLP | 0.02 | 153/281 | 57 | 80 | 16 | 194 (63.4) | 112 (36.6) | 66 | 160 | 55 | 292 (52.0) | 270 (48.0) |
| miR-499 A>G (rs3746444) | AA | AG | GG | A | G | AA | AG | GG | A | G | ||||||
| Kim et al15 | 2012 | Asian | Korea | PCR-RFLP | 0.278 | 127/201 | 91 | 34 | 2 | 216 (85.0) | 38 (15.0) | 120 | 74 | 7 | 314 (78.1) | 88 (21.9) |
| Xiang et al17 | 2012 | Asian | People’s Republic of China | PCR-RFLP | 0.284 | 73/100 | 27 | 30 | 16 | 84 (57.5) | 62 (42.5) | 54 | 36 | 10 | 144 (72) | 56 (28) |
| Shan et al21 | 2013 | Asian | People’s Republic of China | PCR-RFLP | 0.005 | 71/185 | 54 | 14 | 3 | 122 (85.9) | 20 (14.1) | 123 | 48 | 14 | 294 (79.5) | 76 (20.5) |
| Zou and Zhao18 | 2013 | Asian | People’s Republic of China | PCR-RFLP | 0.005 | 71/185 | 54 | 14 | 3 | 122 (85.9) | 20 (14.1) | 123 | 48 | 14 | 294 (79.5) | 76 (20.5) |
| Chu et al16 | 2014 | Asian | People’s Republic of China | PCR-RFLP | 0.321 | 80/337 | 54 | 22 | 4 | 130 (81.3) | 30 (18.7) | 281 | 55 | 1 | 617 (91.5) | 57 (8.5) |
| Ma et al20 | 2014 | Asian | People’s Republic of China | MassARRAY | <0.001 | 760/969 | 558 | 189 | 13 | 1,305 (85.9) | 215 (14.1) | 765 | 179 | 25 | 1,709 (88.2) | 229 (11.8) |
| Wang et al19 | 2014 | Asian | People’s Republic of China | PCR-RFLP | <0.001 | 89/304 | 59 | 18 | 12 | 136 (76.4) | 42 (23.6) | 218 | 62 | 24 | 498 (81.9) | 110 (18.1) |
Abbreviations: HWE, Hardy–Weinberg equilibrium; LDR, ligation detection reaction; PCR, polymerase chain reaction; Q, quantitative; RFLP, restriction fragment length polymorphism.
Quantitative data synthesis
Total population
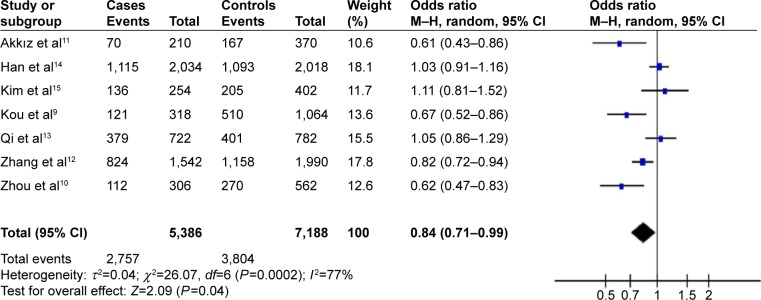
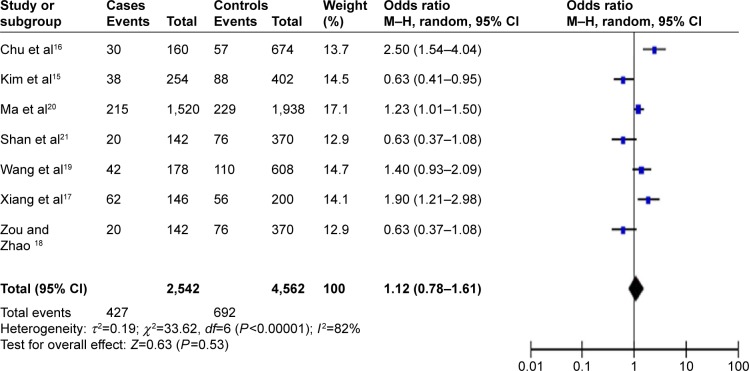
In the total population of 2,693 cases and 3,594 controls in which miR-196a2 C>T was examined, random-effect meta-analysis showed that the T allele and TT genotype were associated with significantly lower HBV-related HCC risk (allelic model, OR =0.84, 95% CI =0.71–0.99, P=0.04, Figure 2; homozygous model, OR =0.68, 95% CI =0.47–0.98, P=0.04). In the total population of 1,271 cases and 2,281 controls in which miR-499 A>G was examined, no association with HBV-related HCC risk was observed in the allelic model (OR =1.12, 95% CI =0.78–1.61, P=0.53, Figure 3), recessive model (OR =1.16, 95% CI =0.58–2.31, P=0.68), dominant model (OR =0.90, 95% CI =0.61–1.32, P=0.60), or homozygous model (OR =1.19, 95% CI =0.56–2.54, P=0.65). Table 2 summarizes the meta-analysis results for the two polymorphisms across the total population.
Figure 2.
Forest plot describing the association of miR196a2 C>T polymorphism with risk of hepatitis B virus-related hepatocellular carcinoma (T-allele vs C-allele).
Abbreviations: CI, confidence interval; df, degrees of freedom; M–H, Mantel–Haenszel.
Figure 3.
Forest plot describing the association of miR-499 A>G polymorphism with risk of hepatitis B virus-related hepatocellular carcinoma (G-allele vs A-allele).
Abbreviations: CI, confidence interval; df, degrees of freedom; M–H, Mantel–Haenszel.
Table 2.
Overall meta-analysis of the association of the polymorphisms miR-196a2 C>T and miR-499 A>G with risk of hepatitis B virus-related hepatocellular carcinoma
| Genotype comparison | OR (95% CI) | Z (P-value) | Heterogeneity of study design
|
Analysis model | ||
|---|---|---|---|---|---|---|
| χ2 | df (P-value) | I2 (%) | ||||
| miR-196a2 C>T (rs11614913) in total population (2,693 cases and 3,594 controls) | ||||||
| Allelic model (T-allele vs C-allele) | 0.84 (0.71–0.99) | 2.09 (0.04) | 26.07 | 6 (<0.001) | 77 | Random |
| Recessive model (TT vs CT + CC) | 0.81 (0.65–1.01) | 1.88 (0.06) | 15.53 | 6 (0.02) | 61 | Random |
| Dominant model (CC vs CT + TT) | 1.23 (0.95–1.59) | 1.59 (0.11) | 23.57 | 6 (<0.001) | 75 | Random |
| Homozygous model (TT vs CC) | 0.68 (0.47–0.98) | 2.08 (0.04) | 28.08 | 6 (<0.001) | 79 | Random |
| miR-196a2 C>T (rs11614913) in Asian population (2,693 cases and 3,594 controls) | ||||||
| Allelic model (T-allele vs C-allele) | 0.87 (0.74–1.03) | 1.62 (0.10) | 21.43 | 5 (<0.001) | 77 | Random |
| Recessive model (TT vs CT + CC) | 0.86 (0.70–1.05) | 1.46 (0.14) | 11.43 | 5 (0.04) | 56 | Random |
| Dominant model (CC vs CT + TT) | 1.23 (0.94–1.62) | 1.50 (0.13) | 20.66 | 5 (<0.001) | 76 | Random |
| Homozygous model (TT vs CC) | 0.74 (0.51–1.07) | 1.62 (0.11) | 23.48 | 5 (<0.001) | 79 | Random |
| miR-196a2 C>T (rs11614913) in Chinese population (2,461 cases and 3,208 controls) | ||||||
| Allelic model (T-allele vs C-allele) | 0.84 (0.70–1.01) | 1.87 (0.06) | 19.71 | 4 (<0.001) | 80 | Random |
| Recessive model (TT vs CT + CC) | 0.83 (0.66–1.04) | 1.61 (0.11) | 10.84 | 4 (0.03) | 63 | Random |
| Dominant model (CC vs CT + TT) | 1.32 (0.97–1.75) | 1.78 (0.07) | 18.72 | 4 (<0.001) | 79 | Random |
| Homozygous model (TT vs CC) | 0.68 (0.45–1.01) | 1.89 (0.06) | 21.75 | 4 (<0.001) | 82 | Random |
| miR-499 rs3746444: A>G in total/Asian population (1,271 cases and 2,281 controls) | ||||||
| Allelic model (G-allele vs A-allele) | 1.12 (0.78–1.61) | 0.63 (0.53) | 33.62 | 6 (<0.001) | 82 | Random |
| Recessive model (GG vs AG + AA) | 1.16 (0.58–2.31) | 0.41 (0.68) | 17.35 | 6 (0.008) | 65 | Random |
| Dominant model (AA vs AG + GG) | 0.90 (0.61–1.32) | 0.53 (0.60) | 27.20 | 6 (<0.001) | 78 | Random |
| Homozygous model (GG vs AA) | 1.19 (0.56–2.54) | 0.45 (0.65) | 20.14 | 6 (0.003) | 70 | Random |
| miR-499 rs3746444: A>G in Chinese population (1,144 cases and 2,080 controls) | ||||||
| Allelic model (G-allele vs A-allele) | 1.24 (0.86–1.78) | 1.17 (0.24) | 23.95 | 5 (<0.001) | 79 | Random |
| Recessive model (GG vs AG + AA) | 1.30 (0.62–2.73) | 0.69 (0.49) | 15.90 | 5 (0.007) | 69 | Random |
| Dominant model (AA vs AG + GG) | 0.80 (0.55–1.17) | 1.15 (0.25) | 17.51 | 5 (0.004) | 71 | Random |
| Homozygous model (GG vs AA) | 0.97 (0.44–2.15) | 0.08 (0.94) | 14.56 | 5 (0.01) | 66 | Random |
Abbreviations: CI, confidence interval; df, degrees of freedom; OR, odds ratios.
Asian population
Stratifying study participants by ethnicity defined a subpopulation of 2,588 Asian cases and 3,409 Asian controls in which miR-196a2 C>T was examined. No association was observed between miR-196a2 C>T and risk of HBV-related HCC in an allelic model (OR =0.87, 95% CI =0.74–1.03, P=0.10), recessive model (OR =0.86, 95%CI =0.70–1.05, P=0.14), dominant model (OR =1.23, 95% CI =0.94–1.62, P=0.13), or homozygous model (OR =0.74, 95% CI =0.51–1.07, P=0.11). The entire population of the seven studies examining miR-499 A>G were Asian, and all were included in the Asian subgroup for meta-analysis of this polymorphism. No association was found between miR-499 A>G and HBV-related HCC risk in the four genetic models. Table 2 summarizes the results of meta-analyses of both polymorphisms in the total/Asian population.
Chinese population
Stratifying the study population by ethnicity defined a subgroup of 2,461 Chinese cases and 3,208 Chinese controls in which miR-196a2 C>T was examined. This polymorphism was not associated with HBV-related HCC risk in the allelic model (OR =0.84, 95% CI =0.70–1.01, P=0.06), recessive model (OR =0.83, 95% CI =0.66–1.04, P=0.11), dominant model (OR =1.32, 95% CI =0.97–1.75, P=0.07), or homozygous model (OR =0.68, 95% CI =0.45–1.01, P=0.06). Similarly, among 1,144 Chinese cases and 2,080 Chinese controls, there was no significant association between miR-499 A>G and risk of HBV-related HCC in the allelic model (OR =1.24, 95% CI =0.86–1.78, P=0.24), recessive model (OR =1.30, 95% CI =0.62–2.73, P=0.49), dominant model (OR =0.80, 95% CI =0.55–1.17, P=0.25), or homozygous model (OR =0.97, 95% CI =0.44–2.15, P=0.94). Table 2 summarizes the results of meta-analyses for the two polymorphisms in the Chinese population.
Sensitivity analysis
Sensitivity analysis of the miR-196a2 C>T meta-analysis was carried out by excluding the study by Zhang et al12 or the one by Han et al,14 both of which appeared to heavily influence the results. The results from the allele model were different after excluding the study by Zhang et al;12 the new OR was 0.83 (95% CI =0.68–1.03, P=0.09). Excluding the study by Han et al14 led to a statistically significant result in the recessive genetic model (OR =0.75, 95% CI =0.56–0.99, P=0.04) and in the dominant genetic model (OR =1.38, 95% CI =1.07–1.78, P=0.01). The results from the dominant genetic model were different after excluding the study by Zhang et al;12 the new OR was 0.67 (95% CI =0.42–1.07, P=0.09). Therefore, our meta-analysis results should be interpreted with caution.
Sensitivity analysis of the miR-499 A>G meta-analysis was carried out by excluding the study with a large weight.20 This did not significantly alter the pooled ORs from any of the four genetic models (data not shown), implying that our results based on all seven studies of miR-499 A>G are robust.
Publication bias
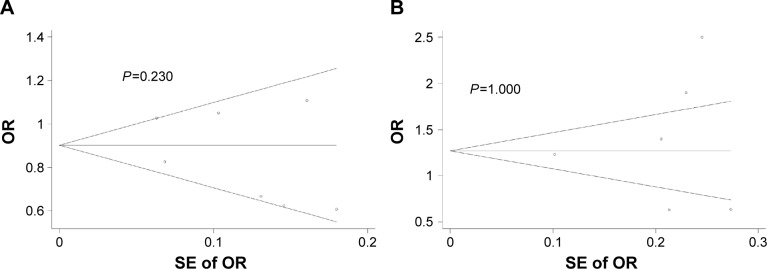
Begg’s funnel plot and Egger’s test were performed to detect potential publication bias in this meta-analysis. No obvious asymmetry was observed in Begg’s funnel plots based on the allelic models (Figure 4A and B). Similarly, Egger’s tests showed no potential publication bias in the allelic models for miR-196a2 C>T (T-allele vs C-allele: t=−1.07, P=0.332) or miR-499 A>G (G-allele vs A-allele: t=0.08, P=0.940).
Figure 4.
(A) Begg’s funnel plot to assess risk of publication bias in analysis of the association between miR-196a2 C>T polymorphism and HBV-related HCC risk according to the allelic model (T-allele vs C-allele). (B) Begg’s funnel plot to assess risk of publication bias in analysis of the association between miR-499 A>G polymorphism and HBV-related HCC risk according to the allelic model (G-allele vs A-allele). Pseudo 95% confidence interval.
Abbreviations: OR, odds ratio; SE, standard error; HBV, hepatitis B virus; HCC, hepatocellular carcinoma.
Discussion
miRNAs play important roles in almost all kinds of cancer, where they modulate key processes during tumorigenesis such as metastasis, apoptosis, proliferation, or angiogenesis.27 Many studies have examined single-nucleotide polymorphisms in precursor and mature miRNAs and their potential associations with HCC risk and progression.9–21 Several meta-analyses have explored the association between miRNA polymorphisms and cancer,8,22,23,28 but their results have been largely inconsistent and few have looked specifically at HBV-related HCC. Therefore, we meta-analyzed the available literature to provide more comprehensive and reliable estimates of the association of two common functional polymorphisms in miRNAs, miR-196a2 C>T and miR-499 A>G, with risk of HBV-related HCC.
Meta-analysis by Xu et al23 found a significant relationship between the miR-196a2 C>T polymorphism and HBV-related HCC risk in Caucasians, but not in the total population. Similarly, meta-analysis of five case–control studies by Su et al,22 involving 2,381 cases and 2,781 controls, showed a significant association between miR-196a2 C>T polymorphism and risk of HBV-related HCC in Caucasians, but not in the total population. Both these meta-analyses contained only one study in a Caucasian population: the study by Akkız et al11 also included in the present meta-analysis. The present work contains more case–control studies than those two previous meta-analyses, and it involved 2,693 cases and 3,394 controls in which miR-196a2 C>T was examined. In contrast to the two previous meta-analyses, our data indicated an association between miR-196a2 C>T and decreased risk of HBV-related HCC in the allelic and homozygous models. This suggests that miR-196a2 polymorphism may reduce the risk of HBV-related HCC. These results are consistent with most case–control studies.9–12 Nevertheless, these findings should be verified in larger studies, given that our results were only marginally significant in the allelic and homozygous models (both P=0.04). In addition, we failed to detect any other significant relationships between miR-196a2 C>T and risk of HBV-related HCC in ethnic subgroup analysis. Larger sample sizes may uncover additional significant correlations. Indeed, we did not examine a Caucasian subgroup because only one relevant study, the one by Akkız et al,11 was identified in the literature. Detailed and well-designed studies involving larger, multiethnic samples are needed, particularly with non-Asian participants.
The present work included more case–control studies of miR-499 A>G than previous meta-analyses,22,23 though all were from Asian populations. Similar to previous meta-analyses, we failed to find any association between miR-499 A>G and risk of HBV-related HCC in the four genetic models. These results were the same for the Asian population in general as well as among Chinese in particular.
The findings in this systematic review were limited by the designs of the included studies. First, the healthy controls in the included studies were not uniformly defined, such that it was impossible to determine which controls were population-based and which were hospital-based. Second, the P-value of HWE was <0.05 in two studies of miR-196a2 C>T9,10 and four studies of miR-499 A>G18–21 (Table 1), suggesting that these study populations were not representative of the broader target population. Nevertheless, we decided to retain these studies in the meta-analysis because excluding them did not significantly affect the pooled ORs (data not shown); in particular, excluding them did not affect the lack of association between miR-499 A>G and risk of HBV-related HCC. Third, the results may be affected by confounding factors, such as hepatitis C infection status, tumor status, sex, or age, but most studies either did not report these baseline data or aggregated them in different ways, making it impossible to include them in the meta-analysis. Fourth, methods used to test for polymorphisms were not uniform and they varied in sensitivity and specificity, which may reduce the robustness of the meta-analysis. Lastly, this meta-analysis included only one study from a non-Asian population; large, well-designed studies in Caucasian and African populations are warranted to reevaluate these associations.
Conclusion
In conclusion, this meta-analysis suggests that miR-196a2 polymorphism may reduce the risk of HBV-related HCC, but we failed to replicate this result in ethnic subgroups, reflecting the small number of studies. We also failed to find any significant association between miR-499 A>G and risk of HBV-related HCC. These findings highlight the need for further detailed and well-designed studies involving larger, multiethnic samples to clarify the role of miR-196a2 C>T and miR-499 A>G in HBV-related HCC. Future studies should also explore gene–gene and gene–environment interactions that may mediate the association of these polymorphisms with disease risk.
Acknowledgments
This research was supported by the National Science and Technology Major Special Project (2012ZX10002010001009), the Self-Raised Scientific Research Fund of the Ministry of Health of Guangxi Province (Z2015621, Z2014241), and the Guangxi University of Science and Technology Research Fund (KY2015LX056).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel R, Miller K, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–886. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Kang Z, Li Y, He X, et al. Quantitative assessment of the association between miR-196a2 rs11614913 polymorphism and cancer risk: evidence based on 45,816 subjects. Tumour Biol. 2014;35(7):6271–6282. doi: 10.1007/s13277-014-1822-3. [DOI] [PubMed] [Google Scholar]
- 8.Chen C, Yang S, Chaugai S, Wang Y, Wang DW. Meta-analysis of Hsa-mir-499 polymorphism (rs3746444) for cancer risk: evidence from 31 case-control studies. BMC Med Genet. 2014;15(1):126. doi: 10.1186/s12881-014-0126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kou JT, Fan H, Han D, et al. Association between four common microRNA polymorphisms and the risk of hepatocellular carcinoma and HBV infection. Oncol Lett. 2014;8(3):1255–1260. doi: 10.3892/ol.2014.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou B, Dong L, Jing X, et al. Association between miR-146aG. C and miR-196a2C> T polymorphisms and the risk of hepatocellular carcinoma in a Chinese population. Tumour Biol. 2014;35(8):7775–7780. doi: 10.1007/s13277-014-2020-z. [DOI] [PubMed] [Google Scholar]
- 11.Akkız H, Bayram S, Bekar A, Akgöllü E, Ulger Y. A functional polymorphism in pre-microRNA-196a-2 contributes to the susceptibility of hepatocellular carcinoma in a Turkish population: a case–control study. J Viral Hepat. 2011;18(7):e399–e407. doi: 10.1111/j.1365-2893.2010.01414.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Wang R, Ma Y, et al. Association between single nucleotide polymorphisms in miRNA196a-2 and miRNA146a and susceptibility to hepatocellular carcinoma in a Chinese population. Asian Pac J Cancer Prev. 2013;14(11):6427–6431. doi: 10.7314/apjcp.2013.14.11.6427. [DOI] [PubMed] [Google Scholar]
- 13.Qi P, Dou TH, Geng L, et al. Association of a variant in MIR 196A2 with susceptibility to hepatocellular carcinoma in male Chinese patients with chronic hepatitis B virus infection. Hum Immunol. 2010;71(6):621–626. doi: 10.1016/j.humimm.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 14.Han Y, Pu R, Han X, et al. Associations of pri-miR-34b/c and pre-miR-196a2 polymorphisms and their multiplicative interactions with hepatitis B virus mutations with hepatocellular carcinoma risk. PLoS One. 2013;8(3):e58564. doi: 10.1371/journal.pone.0058564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim WH, Min KT, Jeon YJ, et al. Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. Gene. 2012;504(1):92–97. doi: 10.1016/j.gene.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 16.Chu YH, Hsieh M, Chiou H, et al. MicroRNA gene polymorphisms and environmental factors increase patient susceptibility to hepatocellular carcinoma. PLoS One. 2014;9(2):e89930. doi: 10.1371/journal.pone.0089930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang Y, Fan S, Cao J, Huang S, Zhang LP. Association of the microRNA-499 variants with susceptibility to hepatocellular carcinoma in a Chinese population. Mol Biol Rep. 2012;39(6):7019–7023. doi: 10.1007/s11033-012-1532-0. [DOI] [PubMed] [Google Scholar]
- 18.Zou HZ, Zhao YQ. Positive association between miR-499A. G and hepatocellular carcinoma risk in a Chinese population. Asian Pac J Cancer Prev. 2013;14(3):1769–1772. doi: 10.7314/apjcp.2013.14.3.1769. [DOI] [PubMed] [Google Scholar]
- 19.Wang XH, Wang FR, Tang YF, Zou HZ, Zhao YQ. Association of miR-149C. T and miR-499A. G polymorphisms with the risk of hepatocellular carcinoma in the Chinese population. Genet Mol Res. 2014;13(3):5048–5054. doi: 10.4238/2014.July.4.20. [DOI] [PubMed] [Google Scholar]
- 20.Ma Y, Wang R, Zhang J, et al. Identification of miR-423 and miR-499 polymorphisms on affecting the risk of hepatocellular carcinoma in a large-scale population. Genet Test Mol Biomarkers. 2014;18(7):516–524. doi: 10.1089/gtmb.2013.0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shan YF, Huang YH, Chen ZK, et al. miR-499A> G rs3746444 and miR-146aG. C expression and hepatocellular carcinoma risk in the Chinese population. Genet Mol Res. 2013;12(4):5365–5371. doi: 10.4238/2013.November.7.11. [DOI] [PubMed] [Google Scholar]
- 22.Su G, Yang J, Hu L, Fu Z, Zhao Z, Yang W. Correlation between single nucleotide polymorphisms in microRNAs and hepatitis B virus-related hepatocellular carcinoma risk. J Sci Appl: Biomed. 2015;3(3):45–55. [Google Scholar]
- 23.Xu Y, Li L, Xiang X, et al. Three common functional polymorphisms in microRNA encoding genes in the susceptibility to hepatocellular carcinoma: a systematic review and meta-analysis. Gene. 2013;527(2):584–593. doi: 10.1016/j.gene.2013.05.085. [DOI] [PubMed] [Google Scholar]
- 24.Zhong JH, You XM, Gong WF, et al. Epidermal growth factor gene polymorphism and risk of hepatocellular carcinoma: a meta-analysis. PLoS One. 2012;7(3):e32159. doi: 10.1371/journal.pone.0032159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong JH, Xiang BD, Ma L, You XM, Li LQ, Xie GS. Meta-analysis of microsomal epoxide hydrolase gene polymorphism and risk of hepatocellular carcinoma. PLoS One. 2013;8(2):e57064. doi: 10.1371/journal.pone.0057064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong JH, Zhang ZM, Li LQ. mEH Tyr113His polymorphism and the risk of ovarian cancer development. J Ovarian Res. 2010;6(1):40. doi: 10.1186/1757-2215-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacFarlane L, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics. 2010;11(7):537. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun H, Li Q, Yang T, Wang W. Quantitative assessment of the association between microRNA-499 rs3746444 A/G polymorphism and cancer risk. Tumour Biol. 2014;35(3):2351–2358. doi: 10.1007/s13277-013-1407-6. [DOI] [PubMed] [Google Scholar]