Abstract
The importance of epistasis — non-additive interactions between alleles — in shaping population fitness has long been a controversial topic, hampered in part by lack of empirical evidence1,2,3,4. Traditionally, epistasis is inferred based on non-independence of genotypic values between loci for a given trait. However epistasis for fitness should also have a genomic footprint5,6,7. To capture this signal, we have developed a simple approach that relies on detecting genotype ratio distortion (GRD) as a signal for epistasis, and we confirm experimentally that instances of GRD represent loci with epistatic fitness effects. In applying this method to a large panel of Drosophila melanogaster recombinant inbred lines8, 9, we conservatively estimate that any two haploid genomes in this study are expected to harbor 1.15 pairs of incompatible alleles. This observation has important implications for speciation genetics, as it indicates that the raw material to drive reproductive isolation is segregating contemporaneously within species and does not necessarily require, as proposed by the Dobzhansky–Muller model, the emergence of incompatible mutations independently derived and fixed in allopatry. The relevance of our result extends beyond speciation, as it demonstrates that epistasis is widespread but that it may often go undetected due to lack of statistical power or lack of genome-wide scope of the experiments.
The role of epistasis in shaping genetic variation and contributing to observable differences within and between populations has been the focus of much debate1,2,3. In complex trait genetics, the additive paradigm used in genome-wide association (GWA) studies10 has recently been challenged by mounting evidence highlighting the importance of non-additive interactions between alleles4. While the debate has been centered on the relative contribution of epistasis to the genetic variance, we still have a poor grasp of the extent to which epistatis affects the mean genotypic values of traits, an important step towards understanding the genetic basis of complex trait and the organization of molecular pathways5. Although epistasis is widely accepted to underlie the genetic basis of speciation, many details of this phenomenon remain poorly understood2,3,5. In particular, the evolutionary origins of the alleles that cause reproductive isolation are largely unidentified. Therefore, the importance of epistasis in shaping fitness within and between populations remains an important question in evolutionary biology.
Our understanding of the contribution of epistasis and the molecular details underlying non-additive genetic interactions is limited largely by the scarcity of available data. Although the idea that populations may harbor alleles with epistatic fitness effects has existed in the literature for some time, very few examples have been dissected at the genetic level (except for individual cases6,11). Furthermore, as yet, no systematic surveys have been conducted in diploid out-crossing species that are sufficiently powered to detect small fitness effects or to finely map interacting loci.
The traditional approach used to detect epistasis by statistical means relies on the observation of non-additivity of genotypic values between loci for a given phenotype. However, epistasis for fitness should have a genomic signature, regardless of our ability to measure a given phenotype5,6,7. In particular, one expects that unfavorable allelic combinations will be under-represented, and this should precipitate a deviation from Mendelian proportions among unlinked incompatible alleles (detected by performing a screen for statistical association between alleles at loci that are not physically linked; Supplemental Methods). Hereafter we refer to such deviations as Genotype-Ratio-Distortion (GRD). In natural populations an exhaustive search for GRD is computationally intractable, statistically underpowered, or both6. By contrast, model organisms allow us to create experimental populations, in which the amount of genetic variation and recombination can be controlled, thereby amplifying the signature of epistasis in a background of reduced dimensionality.
Here we apply tests of epistasis to the Drosophila Synthetic Population Resource (DSPR)8, 9 (Extended Data Figure 1). To create the DSPR, two sets of eight highly inbred strains of diverse geographic origins were independently crossed in a round-robin design. Each set was duplicated and maintained for 50 generations in large freely-mating population cages (generating 4 panels A-1,2 and B-1,2). Subsequently, approximately 400 recombinant inbred lines (RILs) in each of four independent panels were created through 20 generations of sib-mating. After inbreeding, each RIL was genotyped at densely spaced markers, allowing a description of each RIL’s genome as a genetic mosaic of the eight founding lines originally crossed (Extended Data Figure 1). The 50 generations of recombination and the large number of RILs within a panel provides replication over random allelic permutations. This replication is essential to attain statistical power for the detection of small effect epistasis.
We first excluded the possibility that residual population structure within the DSPR created association among alleles in the absence of epistasis by performing principle component analysis (Extended Data Figure 2, Supplemental Methods). Subsequently, we identified 22 pairs of epistatically interacting alleles in the DSPR (Figure 1, Extended Data Table 1, Extended Data Figure 3). Importantly, of the 44 incompatible alleles, 27 appear to be shared between two or more strains (Extended Data Table 1). This indicates that incompatible alleles are segregating at polymorphic frequencies in natural populations, and are not a result of inbreeding or long-term maintenance at small population size. Based on the frequencies in the founder strains, we estimate that any pairwise combination of founders has, on average, 1.15 pairs of epistatically interacting alleles. This is probably an underestimate, both because our statistical approach is conservative and because selectively disfavored allelic combinations may be purged by selection during the free-recombination phase of the DSPR.
Figure 1. Locus pairs showing significant Genotype Ratio Distortion across the DSPR lines of Drosophila.
The outer circle represent each chromosome arm. Each link represents a locus pair showing significant two-locus GRD. Yellow, blue and red links correspond respectively to RIL panel A-2, B-1 and B-2 (5% FDR corrected P < 0.05).
We next sought to confirm the predicted effect on reproductive fitness and to identify the underlying phenotype of two pairs of incompatible haplotypes (Figure 2-a,c). Using the original founder strains that contributed the putatively interacting alleles, we performed experimental crosses, and in both cases, we discovered that the negative interaction is caused by the minor alleles at each locus (Figure 2b,d; Extended Data Figure 1). Specifically, in the case of one incompatibility between chromosomes 2 and 3, males that are homozygous for both incompatible alleles produce on average 74% fewer offspring compared to all other allelic combinations (P = 5.51121E-09 LRT, Figure 2b, Extended Data Figure 4). No significant effect was detected in females for any combination of genotypes. Using the same approach we validated a second instance of GRD, selected in the low range of effect size, between a haplotype on chromosome X and 3 (Extended Data Figure 5). We again observe a significant decrease (22%) in F2 male fertility (P = 8.25e-5 LRT, Figure 2b, Extended Data Figure 4), suggesting that GRD is a reliable signature of epistasis. The ‘faster-males’ theory 2,12 and subsequent experimental confirmations [reviewed in 13] predicts that male infertility will evolve more rapidly than other forms of post-zygotic reproductive isolation. Although we only have phenotypic data for our confirmed examples, the fact that both implicate male fertility as the underlying phenotype suggests that this effect may extend to within-species fitness epistasis.
Figure 2. From missing genotypes to epistasis.
a. GRD signature between all genotyped loci on chromosomes 2R and 3R in RIL panel B-2. b. Average productivity of each genotypic class recovered from 318 F2 single-pair mating (progeny counts are F3). As predicted from the GRD signal (in a.), haplotypes tagged by SNPs 2R:4806926 and 3R:5870973 show strong negative epistasis for the aa;bb genotypes, P = 5.51121E-09 LRT (indicated by the red bar) c. GRD between loci on chromosomes 3L and X in RIL panel A-2. d. Average productivity of each genotypic class recovered from 401 F2 single pair mating. Haplotypes tagged by SNPs 3L: 11510853 and X: 16483812 show strong negative epistasis for the minor alleles on each haplotype aa;bb, P = 8.25e-5 LRT (indicated by the red bar).
The DSPR was intercrossed for sufficiently many generations (50+) that little linkage disequilibrium remains; hence this approach allows us to narrow down likely candidate genes associated with epistatic interaction for male fecundity. In total, there are three genes within the haplotype on chromosome arm 2R (~40 kb). notopleural (np) is at the peak of this region, a gene expressed in mature sperm14 with alleles that are known to affect viability and sterility 15. Notably, the human orthologue of np is associated with sperm-dysfunction in humans16. The interacting haplotype on chromosome arm 3R contains only two genes. In the center of this region is Cyp12e1, a P450-cytochrome associated with electron transport in the mitochondria17. Interestingly, Cyp12e1 harbors a non-synonymous mutation in a highly conserved protein domain. Mitochondrial dysfunction is commonly associated with male sterility in humans, plants, and D. melanogaster18, and therefore seems a plausible candidate phenotype.
To confirm that these observations were not specific to the Drosophila DSPR, we used the same method to screen for GRD in two additional RIL panels: The MAGIC panel in Arabidopsis19 and the NAM panel in maize20. We found 7 instances of GRDs in Arabidopsis and 5 in maize (Table S2). Although we have not validated these results, they suggest that GRD is present in other species as well.
Although the contribution of epistasis to variation in fitness is controversial in some fields21, the Dobzhansky-Muller incompatibility (DMI) model22, 2 is a widely accepted guiding principle for biologists studying of the genetic basis of intrinsic, postzygotic reproductive isolation. Largely motivated by this model, which predicts that alleles causing hybrid incompatibility are derived and fixed after population divergence, much empirical work in speciation genetics has been dedicated to mapping DMIs between species that diverged relatively long ago on an evolutionarily time scale2, 1 (Extended Data Figure 5). However, it is unclear if these known examples of so-called ‘speciation genes’1, 2, 23 are an accurate representation of the earliest events in speciation, which have the greatest biological significance2. Even species that have diverged for only ~250,000 years have evolved complete male sterility an estimated 15 times over24. A reasonable interpretation of this evidence may concede that known ‘speciation genes’ are unlikely to be the same as those that initially contributed to reproductive isolation, but that these examples are instructive as to the properties of those genes2 –logic that closely mirrors our own.
Our central finding, that fitness epistasis is widespread within natural populations indicates that the raw material to drive reproductive isolation is segregating contemporaneous within species and does not necessarily require, as proposed by the DMI model22, the emergence of genetically incompatible mutations independently derived and fixed in allopatric lineages23. We therefore need to explore the possibility that reproductive isolation could be achieved through divergence in frequencies of numerous preexisting, polymorphic, small-effect incompatibilities26, 27, 28. The implications of these results go beyond understanding the role of intra-specific incompatibility in the context of speciation. Our work shows that epistasis for fitness related traits has a detectable genomic footprint and supports the idea that latent incompatibilities often exists between segregating variation within populations, only to be released when divergent lineages hybridize. This discovery highlights the importance of understanding the contribution of epistasis to observable phenotypic differences within and between populations.
Methods Summary
We genotyped the RILs of the DSPR by requiring that each putative variant be supported by a minimum of five reads. All sites wherein two or more alleles are supported by five reads were discarded. We confirmed that the RIL panels were free of cryptic population structure by performing principal component analysis (Extended Data Figure 2). We next excluded sites wherein fewer than 150 individuals have a supported genotype, where the minor allele was present in fewer than 10 individuals, or where more than 15% percent of individuals with data had heterozygous genotypes. Following this, we assessed statistical significance for non-independence between pairwise combinations of alleles using a χ2 test, and applied a 5% false discovery rate to correct for multiple testing. To reduce type 1 error, we restricted our search to inter-chromosomal comparisons and required that each putative instance of GRD be consistent with signal from adjacent variants (see Supplemental Methods).
To confirm the predictions of the GRD scan, we first crossed the two DSPR founder strains that contributed the predicted interacting alleles. We then intercrossed the F1 progeny to produce F2 offspring. Virgin F2 females were then individually and randomly mated to a single F2 male. After mating for 4 days, the F2 pairs were individually genotyped at known variable sites near the interacting alleles. We recorded the number of progeny of each pair to assay productivity. We used TaqMan kits to perform qPCR on the F2 parents, and performed numerous statistical analyses5,29 to quantify epistatic effects as a product of genotypes at the two sites (see Supplemental Methods).
Extended Data
Extended data Figure 1. Description of the DSPR and validation scheme.
a. Geographic distribution of the DSPR founding strains (in orange panel A and in red panel B). b. Construction of the recombinant inbred lines. For each panel each founder strains were crossed in a round-robin design (Line 1 ♀ × Line 2 ♂, Line 2 ♀ × Line 3 ♂,…, Line 8 ♀ × Line 1 ♂) to produce F1s, the F1 were then allowed to mate free to produce an F2 population. In each panel A and B, these F2 population were split into two independent population to create panels A1, A2 and B1, B2. Each was allowed to recombine freely for 50 generations, in very large population. After 50 generation, for each replicate panel, about 400 isofemale lines were inbred for 25 generations to create the 4 panels of RIL used in this study. c. Crossing scheme used to validate epistatic effects. A pair of founder segregating incompatible allele was selected and crossed to produce F1‘s, we then intercrossed the F1 progeny to produce a large F2 population, segregating all possible allelic combinations between alleles at loci 1 and 2. We then counted the progeny each pair produced by intercrossing a large number of F2’s which were later genotyped at sites near to the predicted interacting loci.
Extended data Figure 2. Principal component analysis of each three DSPR RILs panel.

In Green panel A-2, blue panel B-1 and red panel B-2. Showing no evidence of population structure.
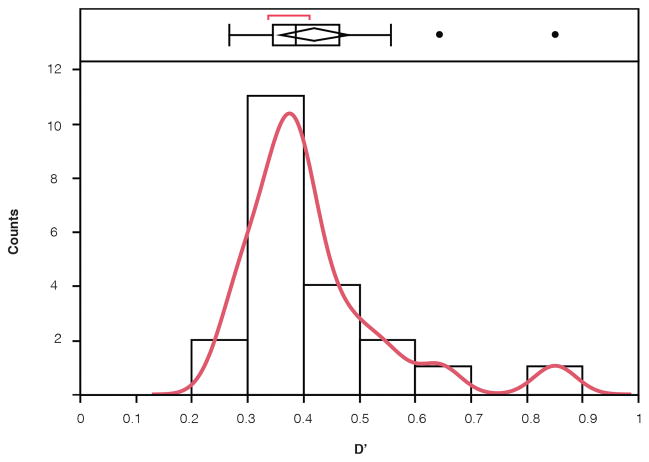
Extended data Figure 3. D′ distribution for significant GRD.
(plotted across DSPR panels).
Extended data Figure 4. Epistasis plot for each validated instance of GRD.

a. GRD between chromosomes 2R and 3R (tagged by SNPs 2R:4806926, on the X axis and 3R:5870973, colored lines) shows strong negative epistasis due to the low fitness of the aa;bb genotype. The additive-by-additive genetic effect is equal to −13.75 (sensu Phillips et al5 and Cheverud29). b. GRD between chromosomes 3L and X (tagged by SNPs 3L: 11510853, on the X axis and X: 16483812, colored lines) also shows negative epistasis. Here the additive-by-additive genetic effect equals −5.94.
Extended data Figure 5. The accumulation of post-zygotic reproductive isolation through time (log-scaled X-axis).
Approximate divergence times of commonly studied Drosophila species are indicated by blue stars, and the red star indicates a reasonable expectation for divergence times of stocks used to found the DSPR (~10,000 years). The red-line indicates a very approximate “speciation threshold”, and indicates that many species pairs that are commonly studied exceed this threshold significantly.
Extended data Table 1.
List of all significant inter-chromosomal GRD identified in the DSPR
| SNP based analysis | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Panel | Chromosome 1 | Position 1 | Chromosome 2 | Position 2 | Number of RILs counted | 1st Major Allele | 1st Minor Allele | 2dn Major Allele | 2dn Minor Allele | 1st Major allele freq | 1st Minor allele freq | 2dn Major allelefreq | 2dn Major allele freq | Major-Major frequency | Major-Minor frequency | Minor-Major frequency | Minor- Minor frequency | d | d′ | r | Chi-square | p-value |
| B-2 | 2L | 2767815 | 3R | 4492436 | 391 | C | A | A | T | 0.91 | 0.09 | 0.92 | 0.08 | 0.86 | 0.05 | 0.06 | 0.03 | 0.02 | 0.27 | 0.25 | 23.70 | 5.85E-07 |
| B-2 | 2L | 8027605 | X | 13053319 | 418 | C | T | C | T | 0.94 | 0.06 | 0.88 | 0.12 | 0.85 | 0.10 | 0.03 | 0.03 | 0.02 | 0.38 | 0.25 | 26.89 | 1.12E-07 |
| B-2 | 2L | 10869984 | 3R | 10633352 | 177 | C | G | T | indel | 0.94 | 0.06 | 0.95 | 0.05 | 0.92 | 0.03 | 0.03 | 0.02 | 0.02 | 0.41 | 0.39 | 26.77 | 1.18E-07 |
| B-2 | 2L | 21657908 | 3R | 5870973 | 444 | A | C | G | A | 0.78 | 0.22 | 0.85 | 0.15 | 0.70 | 0.08 | 0.15 | 0.07 | 0.04 | 0.34 | 0.26 | 30.69 | 1.56E-08 |
| B-2 | 2R | 4806926 | 3R | 5870973 | 443 | G | A | G | A | 0.50 | 0.50 | 0.85 | 0.15 | 0.49 | 0.01 | 0.36 | 0.14 | 0.06 | 0.85 | 0.36 | 58.05 | 1.30E-14 |
| B-2 | 2R | 8464341 | X | 5753834 | 457 | A | G | T | A | 0.89 | 0.11 | 0.77 | 0.23 | 0.72 | 0.17 | 0.05 | 0.06 | 0.03 | 0.39 | 0.25 | 29.29 | 3.22E-08 |
| B-2 | 2R | 20512785 | X | 19647595 | 436 | T | G | A | C | 0.64 | 0.36 | 0.59 | 0.41 | 0.33 | 0.32 | 0.27 | 0.09 | −0.06 | −0.40 | −0.24 | 25.79 | 1.98E-07 |
| B-2 | 3L | 9627942 | X | 13622563 | 263 | G | T | A | G | 0.94 | 0.06 | 0.94 | 0.06 | 0.89 | 0.04 | 0.04 | 0.02 | 0.02 | 0.31 | 0.31 | 24.99 | 3.00E-07 |
| B-2 | 3R | 20437352 | X | 19127400 | 385 | C | T | A | T | 0.90 | 0.10 | 0.95 | 0.05 | 0.87 | 0.03 | 0.08 | 0.02 | 0.02 | 0.36 | 0.26 | 26.13 | 1.65E-07 |
|
| ||||||||||||||||||||||
| B-1 | 2L | 66907 | 3L | 11787066 | 256 | A | G | C | T | 0.92 | 0.08 | 0.89 | 0.11 | 0.85 | 0.07 | 0.04 | 0.04 | 0.03 | 0.39 | 0.33 | 27.30 | 9.03E-08 |
| B-1 | 2L | 22131178 | 3R | 5598460 | 375 | A | G | A | T | 0.75 | 0.25 | 0.90 | 0.10 | 0.71 | 0.04 | 0.19 | 0.06 | 0.04 | 0.48 | 0.28 | 28.94 | 3.85E-08 |
| B-1 | 2L | 2896002 | X | 11823233 | 274 | A | G | T | G | 0.95 | 0.05 | 0.93 | 0.07 | 0.90 | 0.05 | 0.03 | 0.02 | 0.02 | 0.36 | 0.31 | 26.88 | 1.12E-07 |
| B-1 | 2L | 10065007 | 3R | 11588046 | 283 | C | A | T | G | 0.92 | 0.08 | 0.87 | 0.13 | 0.83 | 0.08 | 0.04 | 0.04 | 0.03 | 0.45 | 0.35 | 35.10 | 1.61E-09 |
| B-1 | 2L | 4140219 | X | 11514794 | 361 | G | A | T | indel | 0.94 | 0.06 | 0.95 | 0.05 | 0.91 | 0.03 | 0.04 | 0.02 | 0.02 | 0.33 | 0.30 | 33.13 | 4.43E-09 |
| B-1 | 2R | 3232234 | 3R | 5598051 | 356 | G | A | G | A | 0.75 | 0.25 | 0.92 | 0.08 | 0.72 | 0.03 | 0.19 | 0.06 | 0.04 | 0.56 | 0.29 | 30.98 | 1.34E-08 |
| B-1 | 2R | 14543771 | 3R | 22691609 | 355 | A | G | T | C | 0.85 | 0.15 | 0.93 | 0.07 | 0.81 | 0.04 | 0.12 | 0.04 | 0.03 | 0.41 | 0.27 | 26.33 | 1.49E-07 |
| B-1 | 3R | 13807981 | X | 8763898 | 151 | T | C | C | T | 0.91 | 0.09 | 0.88 | 0.12 | 0.84 | 0.07 | 0.04 | 0.05 | 0.04 | 0.51 | 0.45 | 30.06 | 2.17E-08 |
| B-1 | 3R | 18284739 | X | 14686047 | 378 | G | T | G | A | 0.94 | 0.06 | 0.92 | 0.08 | 0.88 | 0.06 | 0.04 | 0.02 | 0.02 | 0.30 | 0.25 | 23.46 | 6.62E-07 |
|
| ||||||||||||||||||||||
| A-2 | 2L | 19531958 | X | 12584624 | 326 | G | T | C | G | 0.94 | 0.06 | 0.94 | 0.06 | 0.91 | 0.04 | 0.03 | 0.02 | 0.02 | 0.35 | 0.34 | 37.94 | 3.74E-10 |
| A-2 | 3L | 11510853 | X | 16483812 | 354 | A | T | G | A | 0.92 | 0.08 | 0.91 | 0.09 | 0.86 | 0.06 | 0.05 | 0.03 | 0.02 | 0.28 | 0.27 | 26.16 | 1.62E-07 |
| A-2 | 3R | 23793328 | X | 14472525 | 64 | A | T | C | T | 0.84 | 0.16 | 0.84 | 0.16 | 0.80 | 0.05 | 0.05 | 0.11 | 0.08 | 0.64 | 0.64 | 26.58 | 1.31E-07 |
| A-2 | 2L | 16549805 | 3L | 10566820 | 236 | C | T | A | G | 0.92 | 0.08 | 0.92 | 0.08 | 0.87 | 0.05 | 0.05 | 0.03 | 0.03 | 0.37 | 0.37 | 32.37 | 6.55E-09 |
| HMM based analysis | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Panel | Chromosome 1 | Position 1 | Chromosome 2 | Position 2 | Number of RILs counted | 1st Major Allele | 1st Minor Allele | 2dn Major Allele | 2dn Minor Allele | Incompat. Panel-1 A | Incompat. Panel-1 B | Incompat. Panel-2 A | Incompat. Panel-2 B | Maximum/Chi-square | Maximum P-value |
| B-2 | 2L | 2767815 | 3R | 4492436 | 391 | C | A | A | T | 2,3 | 6,7,8 | 1,5 | 2,3,4,6,7 | 22.285 | 1.22E-06 |
| B-2 | 2L | 8027605 | X | 13053319 | 418 | C | T | C | T | 2,7 | 1,3,6,8 | 5,6 | 1,2,3,7 | 28.552 | 4.71E-08 |
| B-2 | 2L | 10869984 | 3R | 10633352 | 177 | C | G | T | indel | 1,3 | 6,7 | 2 | 3,4,5,6,7 | 14.007 | 9.69E-05 |
| B-2 | 2L | 21657908 | 3R | 5870973 | 444 | A | C | G | A | 1,2,7 | 3,6,8 | 2 | ,3,4,5,6, | 31.207 | 1.19E-08 |
| B-2 | 2R | 4806926 | 3R | 5870973 | 443 | G | A | G | A | 3 | 1,2,6,7,8 | 2 | ,3,4,5,6, | 62.734 | 1.20E-15 |
| B-2 | 2R | 8464341 | X | 5753834 | 457 | A | G | T | A | 1,3,8 | 2,5,6 | 1,4,5 | 2,3,7 | 37.070 | 5.84E-10 |
| B-2 | 2R | 20512785 | X | 19647595 | 436 | T | G | A | C | 4,8 | 2,3,5,6,7 | 3,5 | 1,2,4,6 | 28.937 | 3.86E-08 |
| B-2 | 3L | 9627942 | X | 13622563 | 263 | G | T | A | G | 2,3,5 | 1,6,7,8 | 5,7 | 1,3,6 | 17.625 | 1.41E-05 |
| B-2 | 3R | 20437352 | X | 19127400 | 385 | C | T | A | T | 3,5 | 1,4,6 | 8 | ,2,4,5,6, | 10.122 | 7.95E-04 |
|
| |||||||||||||||
| B-1 | 2L | 66907 | 3L | 11787066 | 256 | A | G | C | T | 7 | 1,2,3,6,8 | 5 | 2,3,4,6,7 | 16.936 | 2.04E-05 |
| B-1 | 2L | 22131178 | 3R | 5598460 | 375 | A | G | A | T | 1,2,3,6 | 4,7,8 | 1,2,3,6 | 4,7,8 | 28.154 | 5.79E-08 |
| B-1 | 2L | 2896002 | X | 11823233 | 274 | A | G | T | G | 6,7,8 | 2,3,4,5 | 2,8 | 1,3,5,6,7 | 27.374 | 8.67E-08 |
| B-1 | 2L | 10065007 | 3R | 11588046 | 283 | C | A | T | G | 1,7 | 2,3,4,6 | 2,8 | 1,2,3,6,7 | 29.341 | 3.13E-08 |
| B-1 | 2L | 4140219 | X | 11514794 | 361 | G | A | T | indel | 3 | 2,4,6,7,8 | 3,5,6 | 1,2,7,8 | 44.297 | 1.44E-11 |
| B-1 | 2R | 3232234 | 3R | 5598051 | 356 | G | A | G | A | 4,7,8 | 1,2,3,6 | 4,7,8 | 1,2,3,6 | 30.049 | 2.17E-08 |
| B-1 | 2R | 14543771 | 3R | 22691609 | 355 | A | G | T | C | 2,6 | 1,3,5,8 | 4 | 2,3,5,6,7 | 26.540 | 1.34E-07 |
| B-1 | 3R | 13807981 | X | 8763898 | 151 | T | C | C | T | 3,4 | 1,2,6,7,8 | 5,7,8 | 1,3,6 | 18.243 | 1.02E-05 |
| B-1 | 3R | 18284739 | X | 14686047 | 378 | G | T | G | A | 2,7 | 1,4,6,8 | 5 | 1,3,6,8 | 29.620 | 2.71E-08 |
|
| |||||||||||||||
| A-2 | 2L | 19531958 | X | 12584624 | 326 | G | T | C | G | 1 | 2,4,5,7 | 5 | ,2,3,4,6, | 42.031 | 4.59E-11 |
| A-2 | 3L | 11510853 | X | 16483812 | 354 | A | T | G | A | 4 | 2,3,5,6 | 5 | 1,3,4,6,7 | 34.180 | 2.58E-09 |
| A-2 | 3R | 23793328 | X | 14472525 | 64 | A | T | C | T | 5 | 3,4,6,7 | 1,5 | 2,3,4,6,7 | 12.913 | 1.74E-04 |
| A-2 | 2L | 16549805 | 3L | 10566820 | 236 | C | T | A | G | 1,8 | 2,4,5,7 | 4 | 3,5 | 20.660 | 2.86E-06 |
Supplementary Material
Figure 3. Model for unlinked loci with segregating pairs of incompatible alleles.
The dendrograms on the left and the right represent the genealogies of two haplotypes segregating within a species. The blue dot and the red rectangle indicate the origins of incompatible mutations on each respective genealogy. On the left, derived blue alleles are incompatible with derived red alleles on the right. These genealogies yield the individuals shown in the center; wherein each line segment corresponds to a chromosome and each colored square indicates the derived incompatible allele. Importantly, these incompatible allele pairs are polymorphic in this sample of individuals, thus individuals who inherit both incompatible alleles have lower fitness than those with either none or only a single incompatibility.
Acknowledgments
We are grateful to S Macdonald and E King for creating the DSPR and sharing the RIL and founder strains with us. We thank C Jones, B de Bivort, T Sackton, SD Kocher, J Grenier, and NE Soltis and three anonymous reviewers for helpful comments and discussions. We thank Xi Shi for technical assistance. This work was supported by grants: NIH GM065169 and GM084236 to DLH, HD059060 to AGC, Harvard Society of Fellows Fellowship and Harvard Milton Funds to JFA. RBCD is supported by a Harvard Prize Fellowship.
Abbreviations
- DMI
Dobzhansky-Muller incompatibility
- DSPR
Drosophila synthetic population resource
- GRD
Genotype-Ratio-Distortion
- RADseq
restriction-site-associated DNA sequencing
- RIL
recombinant inbred line
Footnotes
Authors contribution: R.B.C.D., J.F.A., conducted bioinformatics and statistical analyses, R.B.C.D., J.F.A., J.Z., performed experiments, J.Z. carried out molecular work; A.G.C., D.L.H., gave analytical and conceptual advice throughout the project; J.F.A conceived of the project.
Authors declare no competing interests.
All code used and generated for this study is available upon request.
References
- 1.Presgraves DC. The molecular evolutionary basis of species formation. Nature Rev Genet. 2010;11(3):175–180. doi: 10.1038/nrg2718. [DOI] [PubMed] [Google Scholar]
- 2.Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- 3.Cutter AD. The polymorphic prelude to Bateson–Dobzhansky–Muller incompatibilities. TREE. 2011;27(4):209–218. doi: 10.1016/j.tree.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Carlborg O, Haley CS. Epistasis: too often neglected in complex trait studies? Nature Rev Genet. 2004;5:618–625. doi: 10.1038/nrg1407. [DOI] [PubMed] [Google Scholar]
- 5.Phillips PC. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nature Rev Genet. 2008;9(11):855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bomblies K, et al. Autoimmune response as a mechanism for a Dobzhansky-Muller- type incompatibility syndrome in plants. PLoS biol. 2007;5(9):e236. doi: 10.1371/journal.pbio.0050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Payseur BA, Hoekstra HE. Signatures of reproductive isolation in patterns of single nucleotide diversity across inbred strains of house mice. Genetics. 2005;171:1905– 1916. doi: 10.1534/genetics.105.046193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King EG, Macdonald SJ, Long AD. Properties and power of the Drosophila Synthetic Population Resource for the routine dissection of complex traits. Genetics. 2012;191(3):935–949. doi: 10.1534/genetics.112.138537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King EG, et al. Genetic dissection of a model complex trait using the Drosophila Synthetic Population Resource. Gen Resch. 2012;22(8):1558–1566. doi: 10.1101/gr.134031.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuk Or, et al. The mystery of missing heritability: genetic interactions create phantom heritability. PNAS. 2012;109(4):1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bikard D, et al. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science. 2009;323(5914):623–626. doi: 10.1126/science.1165917. [DOI] [PubMed] [Google Scholar]
- 12.Palopoli MF, Wu CI. Genetics of hybrid male sterility between Drosophila sibling species: a complex web of epistasis is revealed in interspecific studies. Genetics. 1994;138(2):329–341. doi: 10.1093/genetics/138.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu CI, Johnson NA, Palopoli MF. Haldane’s rule its legacy: Why are there so many sterile males? TREE. 1996;11(7):281–284. doi: 10.1016/0169-5347(96)10033-1. [DOI] [PubMed] [Google Scholar]
- 14.WasBrough RW, et al. The Drosophila melanogaster sperm proteome-II (DmSP-II) J Proteomics. 2010;73:2171–2185. doi: 10.1016/j.jprot.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Dockendorff TC, Robertson SE, Faulkner DL, Jongens TA. Genetic characterization of the 44D-45B region of the Drosophila melanogaster genome based on an F2 lethal screen. Mol Gen Genet. 2000;263(1):137–143. doi: 10.1007/s004380050040. [DOI] [PubMed] [Google Scholar]
- 16.Netzel-Arnett S, et al. The glycosylphosphatidylinositol-anchored serine protease PRSS21 (testisin) imparts murine epididymal sperm cell maturation and fertilizing ability. Biol reprod. 2009;81(5):921–932. doi: 10.1095/biolreprod.109.076273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasai S, Tomita T. Male specific expression of a cytochrome P450 (Cyp312a1) in Drosophila melanogaster. Biochem Biophys Res Commun. 2003;300(4):894–900. doi: 10.1016/s0006-291x(02)02950-9. [DOI] [PubMed] [Google Scholar]
- 18.Meiklejohn CD, Montooth KL, Rand DM. Positive and negative selection on the mitochondrial genome. TIG. 2007;23(6):259–263. doi: 10.1016/j.tig.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Kover PX, et al. multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS gen. 2009;5(7):e1000551. doi: 10.1371/journal.pgen.1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMullen MD, et al. Genetic properties of the maize nested association mapping population. Science. 2009;325(5941):737–740. doi: 10.1126/science.1174320. [DOI] [PubMed] [Google Scholar]
- 21.Hill WG, Goddard ME, Visscher PM. Data and Theory Point to Mainly Additive Genetic Variance for Complex Traits. PLoS Genet. 2008;4(2):e1000008. doi: 10.1371/journal.pgen.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobzhansky T. Genetics and the Origin of Species. Columbia University Press; New York: 1937. [Google Scholar]
- 23.Orr HA, Turelli M. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution. 2001;55(6):1085–1094. doi: 10.1111/j.0014-3820.2001.tb00628.x. [DOI] [PubMed] [Google Scholar]
- 24.Presgraves DC, Stephan W. Pervasive adaptive evolution among interactors of the Drosophila hybrid inviability gene, Nup96. MBE. 2007;24(1):306–314. doi: 10.1093/molbev/msl157. [DOI] [PubMed] [Google Scholar]
- 25.Tao Yun, et al. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. I. Differential accumulation of hybrid male sterility effects on the X and autosomes. Genetics. 2003;164(4):1383–1398. doi: 10.1093/genetics/164.4.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fitzpatrick BM. Hybrid dysfunction: Population genetic and quantitative genetic perspectives. Am Nat. 2008;171(4):491–498. doi: 10.1086/528991. [DOI] [PubMed] [Google Scholar]
- 27.Demuth JP, Wade MJ. On the theoretical and empirical framework for studying genetic interactions within and among species. Am Nat. 2005;165(5):524–536. doi: 10.1086/429276. [DOI] [PubMed] [Google Scholar]
- 28.Reed LK, Markow TA. Early events in speciation: polymorphism for hybrid male sterility in Drosophila. PNAS. 2004;101(24):9009–9012. doi: 10.1073/pnas.0403106101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheverud JM, Routman EJ. Epistasis and its contribution to genetic variance components. Genetics. 1995;139(3):1455–1461. doi: 10.1093/genetics/139.3.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.