Summary
Chlamydia is a medically important bacterium that infects eukaryotic cells. Temporal expression of chlamydial genes during the intracellular infection is proposed to be regulated by changes in DNA supercoiling levels. To understand how chlamydial supercoiling levels are regulated, we purified and analyzed three putative Chlamydia trachomatis topoisomerases. As predicted by sequence homology, CT189/190 are the two subunits of DNA gyrase, whereas CT643 is a topoisomerase I. CT660/661 have been predicted to form a second DNA gyrase, but the reconstitute holoenzyme decatenated and relaxed DNA, indicating that the proteins are subunits of topoisomerase IV. Promoter analysis showed that each topoisomerase is transcribed from its own operon by the major chlamydial RNA polymerase. Surprisingly, all three topoisomerase promoters had higher activity from a more supercoiled DNA template. This supercoiling-responsivesness is consistent with negative feedback control of topoisomerase I and topoisomerase IV expression, which is typical of other bacteria. However, activation of the chlamydial gyrase promoter by increased supercoiling is unorthodox compared with the relaxation-induced transcription of gyrase in other bacteria. We present a model in which supercoiling levels during the intracellular chlamydial developmental cycle are regulated by unusual positive feedback control of the gyrase promoter and the temporal expression of three topoisomerases.
Introduction
Chlamydia is a pathogenic bacterium that causes the majority of infectious disease cases reported to the CDC each year (CDC, 2014). Chlamydia trachomatis is the most common cause of bacterial sexually transmitted infection in the USA (Batteiger and Tan, 2014). This organism also causes trachoma, a preventable form of blindness. C. pneumoniae causes community-acquired pneumonia (Hammerschlag et al., 2014). Although chlamydial species and strains produce infections with different clinical manifestations, they all share a similar intracellular lifestyle characterized by a biphasic developmental cycle (Moulder, 1991). The infectious chlamydial form, called an elementary body (EB), initiates the intracellular infection by binding and entering a host eukaryotic cell. By 2–8 h post infection (hpi), the EB converts into a reticulate body (RB), which is a larger, intracellular but non-infectious form that replicates by binary fission. After 8 to 10 rounds of division, RBs differentiate into EBs for release to start a new round of infection.
Chlamydial genes are expressed during this developmental cycle in three main temporal classes (Shaw et al., 2000; Belland et al., 2003; Nicholson et al., 2003; Abdelrahman and Belland, 2005). Early genes are selectively expressed during the first few hours of the intracellular infection by an unknown mechanism. Midcycle genes, which are the large majority of chlamydial genes, are transcribed during RB replication. Late genes are a small group of genes that are first expressed or upregulated at about 16–24 hpi, when RBs are converting into EBs. Late gene expression is proposed to be negatively regulated by a transcription factor called EUO, which binds and represses promoters of late genes (Rosario and Tan, 2012; Rosario et al., 2014). In contrast, chlamydial gene expression in midcycle is proposed to be activated by higher levels of negative DNA supercoiling that are a feature of this stage of the developmental cycle (Niehus et al., 2008). This model is supported by the finding that promoters of representative midcycle genes are transcribed at higher levels from more supercoiled templates (Niehus et al., 2008; Case et al., 2010). In addition, a subset of early genes that are upregulated in midcycle also have supercoiling-dependent promoters (Case et al., 2010; Cheng and Tan, 2012). DNA supercoiling co-ordinates the expression of 7–15% of the genome in other bacteria (Champoux, 2001; Gmuender et al., 2001; Niehus et al., 2004; Peter et al., 2004; Ferrandiz et al., 2010; Schroder et al., 2014), but it has the potential to be the global regulator of gene expression in Chlamydia by controlling the expression of the majority of genes in midcycle.
Bacterial DNA supercoiling levels are homeostatically regulated by enzymes called DNA topoisomerases. DNA gyrase, which is a heterotetrameric protein composed of GyrA and GyrB subunits, introduces negative supercoils into DNA (Nollmann et al., 2007). Its action is counterbalanced by topoisomerase I, which relaxes negatively supercoiled DNA (Tse and Wang, 1980; Viard and de la Tour, 2007). Topoisomerase IV, which is a heterotetramer composed of two ParC and two ParE subunits, catalyzes DNA decatenation and relaxation (Kato et al., 1992; Bebear et al., 1998; Barnes et al., 2003; Schmutz et al., 2004). DNA supercoiling levels in bacteria are subject to negative feedback control because the promoters of topoisomerase genes are transcribed in a supercoiling-dependent manner. For example, increased DNA supercoiling upregulates the activity of the topoisomerase I promoter, leading to more expression of this DNA relaxation enzyme. In a similar vein, higher supercoiling levels downregulate the promoter for the gyrase genes, leading to decreased expression of DNA gyrase (Menzel and Gellert, 1983; 1987b; Tse-Dinh, 1985).
The Chlamydia genome is predicted to encode three DNA topoisomerases, based on sequence similarity to known bacterial topoisomerases. In C. trachomatis, ct189 and ct190 are predicted to encode the two subunits of DNA gyrase and have been designated as gyrA_1 and gyrB_1, respectively, whereas ct0643 encodes a putative topoisomerase I (Stephens et al., 1998). C. trachomatis ct660 and ct661 were designated as gyrA_2 and gyrB_2 in the original genome annotation, based on the prediction that they encode a second chlamydial DNA gyrase (Stephens et al., 1998). However, these genes have been noted to share sequence similarity with parC and parE, which encode topoisomerase IV in other bacteria (Dessus-Babus et al., 1998), raising questions about their true function. Of the three putative chlamydial topoisomerases, only C. pneumoniae gyrA_1 and gyrB_1 have been shown to encode an active enzyme (Ameyama et al., 2003).
In this study, we performed functional studies on the three predicted C. trachomatis topoisomerases, measuring their ability to alter the superhelical density of a plasmid DNA template. We also examined the transcriptional regulation of the genes that encode these topoisomerases. Our findings support a model in which the temporal expression and feedback control of DNA topoisomerases regulate DNA supercoiling levels during the chlamydial developmental cycle.
Results
To investigate the enzymes that regulate DNA supercoiling levels in C. trachomatis, we purified five proteins that have been identified as putative topoisomerases or topoisomerase subunits. Proteins were individually expressed as epitope-tagged recombinant proteins in E. coli, purified by affinity chromatography and checked to ensure that they did not contain nuclease activity. They were then tested individually, or in combination, for topoisomerase activity with in vitro assays of DNA supercoiling, relaxation and decatenation.
ct189and ct190 encode the two subunits of C. trachomatis DNA gyrase
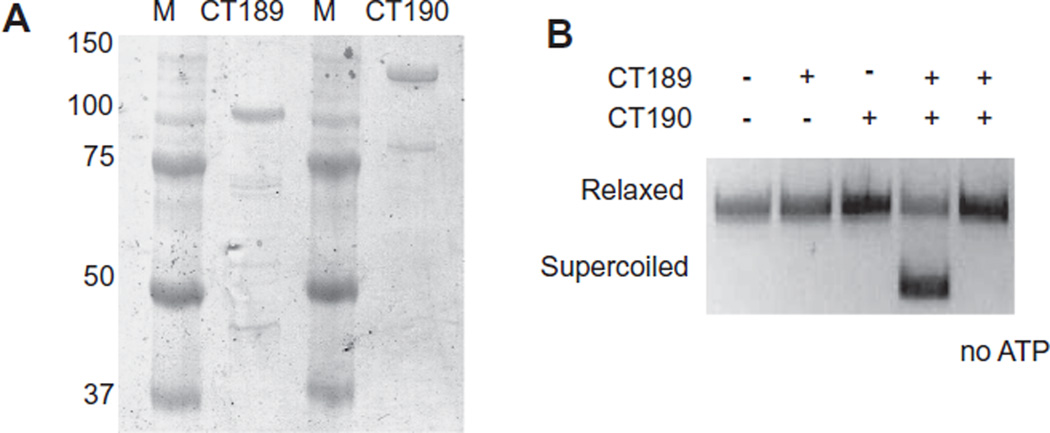
Chlamydia trachomatis ct189 and ct190 have been presumptively named gyrA and gyrB, respectively, because of sequence similarity to these two gyrase subunits in other bacteria. We cloned the two genes into individual expression vectors in order to express GyrA with a C-terminal histidine tag and GyrB as a fusion protein with maltose binding protein (MBP) at the N-terminus. The two recombinant proteins were purified to near homogeneity (Fig. 1A).
Fig. 1. ct189 and ct190 encode functional DNA gyrase subunits.
A. SDS PAGE of purified DNA gyrase subunits: CT189 (GyrA) and CT190 (GyrB) stained with Coomassie brilliant blue. The size of protein markers (M) in kDa is indicated to the left.
B. CT189 (GyrA) and CT190 (GyrB) proteins reconstitute an ATP-dependent supercoiling activity. Relaxed plasmid was incubated with CT189 (100 ng) and/or CT190 (50 ng) in the presence or absence of 2 mM ATP.
We tested if GyrA and GyrB had gyrase activity in an in vitro DNA supercoiling assay. Neither protein by itself altered DNA supercoiling levels, but the combination of GyrA and GyrB converted a relaxed plasmid substrate into its supercoiled form (Fig. 1B). This supercoiling activity was dependent on the addition of ATP (Fig. 1B) and a divalent cation (Fig. S1), which are properties of bacterial DNA gyrase. The reconstituted enzyme showed a strong preference for Mg2+, because there was no detectable supercoiling activity when the divalent cation in the reaction buffer was Mn2+, Ca2+ or Zn2+ (Fig. S1). DNA supercoiling activity was inhibited in a concentration-dependent manner by novobiocin and ciprofloxacin (Fig. S1), which are known pharmacologic inhibitors of bacterial DNA gyrase. Our reconstituted recombinant C. trachomatis DNA gyrase was completely inhibited by 100 µg ml−1 of novobiocin or 200 µg ml−1 of ciprofloxacin. These findings support the designations of ct189 and ct190 as gyrA and gyrB, respectively, and we will use these gene names for the rest of the manuscript.
ct643 encodes the C. trachomatis topoisomerase I
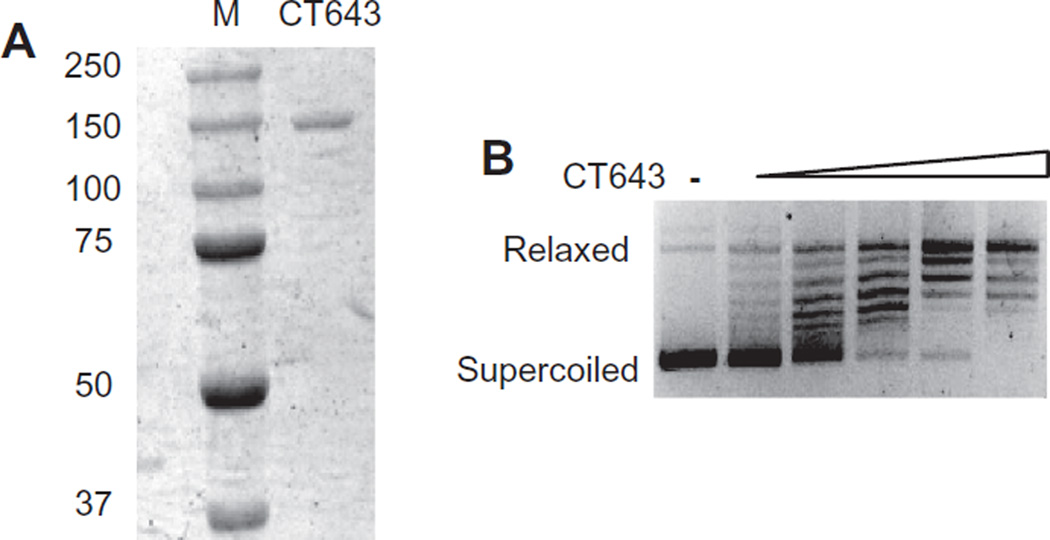
Chlamydia trachomatis ct643 is the predicted orthologue of topA, which encodes the single subunit of DNA topoisomerase I in other bacteria. We cloned ct643 into the pMAL-c5X vector and expressed C. trachomatis TopA in E. coli as a fusion protein with MBP at its N-terminus (Fig. 2A). The recombinant protein was enzymatically active and induced the concentration-dependent relaxation of negatively supercoiled plasmid DNA in an in vitro DNA relaxation assay (Fig. 2B). Five hundred micrograms of purified TopA was able to induce relaxation of 200 ng of supercoiled plasmid after a 30 min incubation at 37°C. This relaxation activity required a divalent cation, which is a property of topoisomerase I from other bacteria. The relaxation activity of our chlamydial enzyme was greatest with Ca2+, but also present with Mn2+ and to a lesser extent Mg2+ (Fig. S2). In contrast there was no detectable activity in the presence of Zn2+. Enzymatic activity did not require addition of ATP but was slightly inhibited at increased ATP concentrations and was inhibited by spermidine (Fig. S2), which are properties of bacterial type I topoisomerases (Srivenugopal and Morris, 1985). However, the chlamydial enzyme differed from other bacterial topoisomerase I by being sensitive to 50 mM NaCl (Fig. S2). These biochemical studies provide evidence that C. trachomatis ct643 encodes the chlamydial topoisomerase I.
Fig. 2. ct643 encodes a functional topoisomerase I.
A. SDS PAGE gel electrophoresis of purified CT643 (TopA) stained with Coomassie brilliant blue. The size of protein markers (M) in kDa is indicated to the left.
B. CT643 relaxes supercoiled DNA. Supercoiled plasmid was incubated with increasing concentrations of CT643.
ct660 and ct661 encode the two subunits of C. trachomatis DNA topoisomerase IV
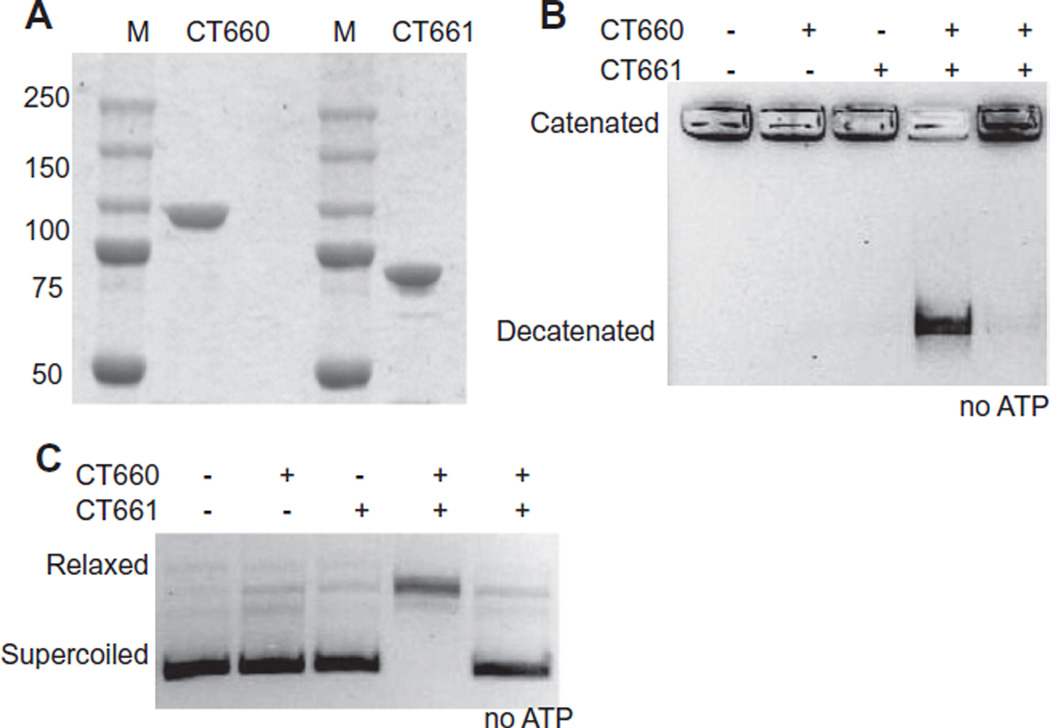
Unlike C. trachomatis DNA gyrase and topoisomerase I, there has been disagreement about the type of topoisomerase predicted to be encoded by C. trachomatis ct660 and ct661. Based on sequence similarity, these genes have been predicted to encode either a DNA gyrase or a topoisomerase IV, which are enzymes with opposite effects on DNA supercoiling levels (Dessus-Babus et al., 1998). To resolve this issue, we expressed CT660 and CT661 as recombinant proteins and measured their enzymatic effect on DNA topology. We cloned ct661 into pEXp5CT to express CT661 with a C-terminal histidine tag. We cloned ct660 into pMAL-c5X and expressed CT660 as an N-terminal MBP-tagged fusion protein. Protein purity was confirmed by SDS-PAGE (Fig. 3A).
Fig. 3. ct660 and ct661 encode functional DNA topoisomerase IV subunits.
A. SDS PAGE of purified topoisomerase IV subunits CT660 (ParC) and CT660 (ParE) stained with Coomassie brilliant blue. The size of protein markers (M) in kDa is indicated to the left.
B. CT660 and CT661 proteins reconstitute an ATP-dependent decatenation activity. kDNA was incubated with 75 ng CT660 and/or 75 ng CT661 in the presence or absence of 2 mM ATP. The catenated kDNA remains in the wells of the agarose gel; the position of decatenated DNA is indicated.
C. CT660 and CT661 proteins reconstitute an ATP-dependent relaxation activity. Supercoiled plasmid was incubated with 75 ng CT660 and/or 75 ng CT661 in the presence or absence of 2 mM ATP.
We first examined if CT660 and CT661 had enzymatic activity in a decatenation assay that measures the ATP-dependent unlinking of kinetoplast DNA (kDNA) into DNA minicircles (Fig. 3B). With either CT660 or CT661 alone, kDNA remained intact and failed to migrate from the well into the gel. In contrast, the combination of CT660 and CT661 caused decatenation of kDNA, as shown by migration of the released minicircle DNA into the gel. This decatenation activity was ATP dependent (Fig. 3B), and there was a preference for Mg2+ and Mn2+ over Ca2+ or Zn2+ as the required divalent cation (Fig. S3).
We also tested the enzymatic activity of CT660 and CT661 on DNA topology. Both subunits were required to relax supercoiled plasmid DNA in our in vitro relaxation activity assay (Fig. 3C). The reconstituted CT660 and CT661 enzyme required ATP (Fig. 3B) and a divalent cation for DNA relaxation, with Mg2+ preferred over Mn2+ and Ca2+ (Fig. S3). The relative preference for divalent cations (Mg2+ > Mn2+ > Ca2+, and no activity with Zn2+) was similar for both the decatenation and relaxation activities of CT660/CT661. In contrast, CT660 and CT661 did not convert relaxed DNA into its supercoiled form in our in vitro DNA supercoiling assay (data not shown), indicating that these proteins do not encode a DNA gyrase.
To verify the topoisomerase activity of CT660/CT661, we tested its susceptibility to known inhibitors of bacterial topoisomerase IV. Novobiocin and ciprofloxacin each inhibited the decatenation and DNA relaxation activities in a concentration-dependent manner (Fig. S3). At a concentration of 1 mg ml−1, novobiocin fully inhibited both topoisomerase IV activities. With ciprofloxacin, 1 mg ml−1 completely inhibited decatenation, whereas 0.5 mg ml−1 was required to block relaxation activity. Taken together, these results indicate that CT660 and CT661 are the two subunits of a functional C. trachomatis topoisomerase IV. Based on these findings, we propose to name ct660 as parC and ct661 as parE, which is the conventional naming of the two topoisomerase IV subunits in other bacteria.
Identification of promoters regions for the topoisomerase genes
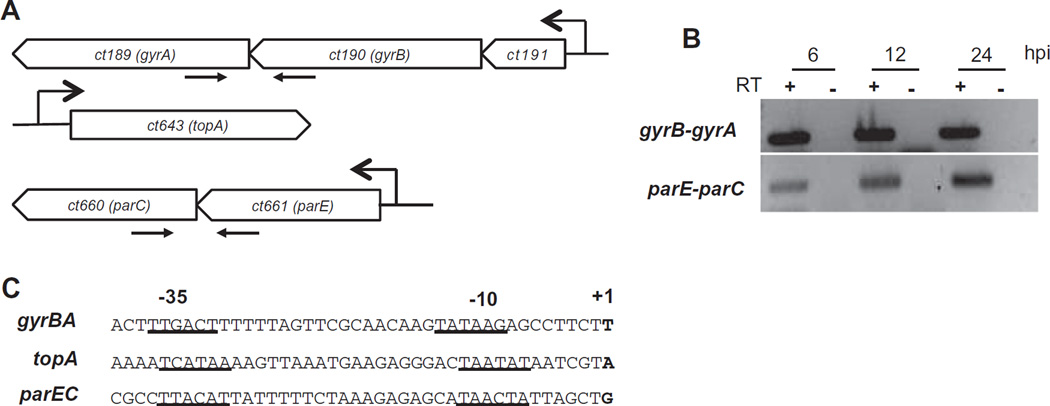
To examine how the expression of the five topoisomerase genes is regulated in C. trachomatis, we first identified the transcriptional unit for each gene (Fig. 4A). The genes for DNA gyrase (gyrB and gyrA) are grouped together as a pair, as is the case for topoisomerase IV (parE and parC), whereas the single topoisomerase I gene, topA, is at its own separate location. gyrB and gyrA appear to be transcribed from an operon because they are contiguous, and their intergenic region is small and lacks an identifiable terminator (Dessus-Babus et al., 1998). We verified that gyrB-gyrA is transcribed as a polycistronic message by using RT-PCR to amplify the overlapping region between the two genes from a cDNA fragment. We used a similar RT-PCR approach to amplify the overlapping region between parE-parC, indicating that parE and parC are also co-transcribed from an operon (Fig. 4B). These operon structures indicate that chlamydial gyrase and topoisomerase IV are each expressed as a single transcriptional unit.
Fig. 4. Transcriptional organization of C. trachomatis topoisomerase genes.
A. Schematic diagram of the topoisomerase genes, identified by name and number. The promoter for each transcription unit is indicated by an arrow with an open arrowhead. The pairs of smaller arrows represent primers used for the RT-PCR assay.
B. Co-transcription of gyrB-gyrA and of parE-parC. cDNA products shown in the gene diagram were amplified by RT-PCR from total RNA extracted from C. trachomatis-infected cells at 6, 12 or 24 hpi.
C. Alignment of putative topoisomerase promoters. The transcription start sites were mapped by 5′-RACE. Predicted −35 and −10 promoter elements are underlined. gyrBA= promoter of DNA gyrase operon, which also includes ct191; topA = promoter of DNA topoisomerase I gene; parEC = promoter of DNA topoisomerase IV operon.
We identified the promoters for topA, gyrBA and parEC by mapping their respective transcription start sites with 5′-RACE. The transcription start site for gyrBA promoter was located upstream of ct191, which is the gene immediately upstream of gyrB (Fig. 4A). ct191 encodes a hypothetical Chlamydia-specific protein of unknown function. By sequence inspection, and comparison with the preferred chlamydial σ66 promoter (Tan et al., 1998; Schaumburg and Tan, 2003; Tan, 2012), we located putative σ66 promoters upstream of each of the three transcription start sites (Fig. 4C).
Promoters for C. trachomatis topoisomerases genes are supercoiling sensitive
We investigated whether the chlamydial topoisomerase promoters are sensitive to DNA supercoiling, as they are in other bacteria. The three topoisomerase promoters were each cloned on a plasmid-based transcription template and verified to be transcribed by C. trachomatis σ66 RNA polymerase in vitro (data not shown), as well as by E. coli σ70 RNA polymerase (Fig. 5). We then tested each topoisomerase promoter with an in vitro supercoiling sensitivity transcription assay, in which topoisomers of the transcription plasmid were generated over a range of DNA supercoiling levels and individually tested for promoter activity (Niehus et al., 2008).
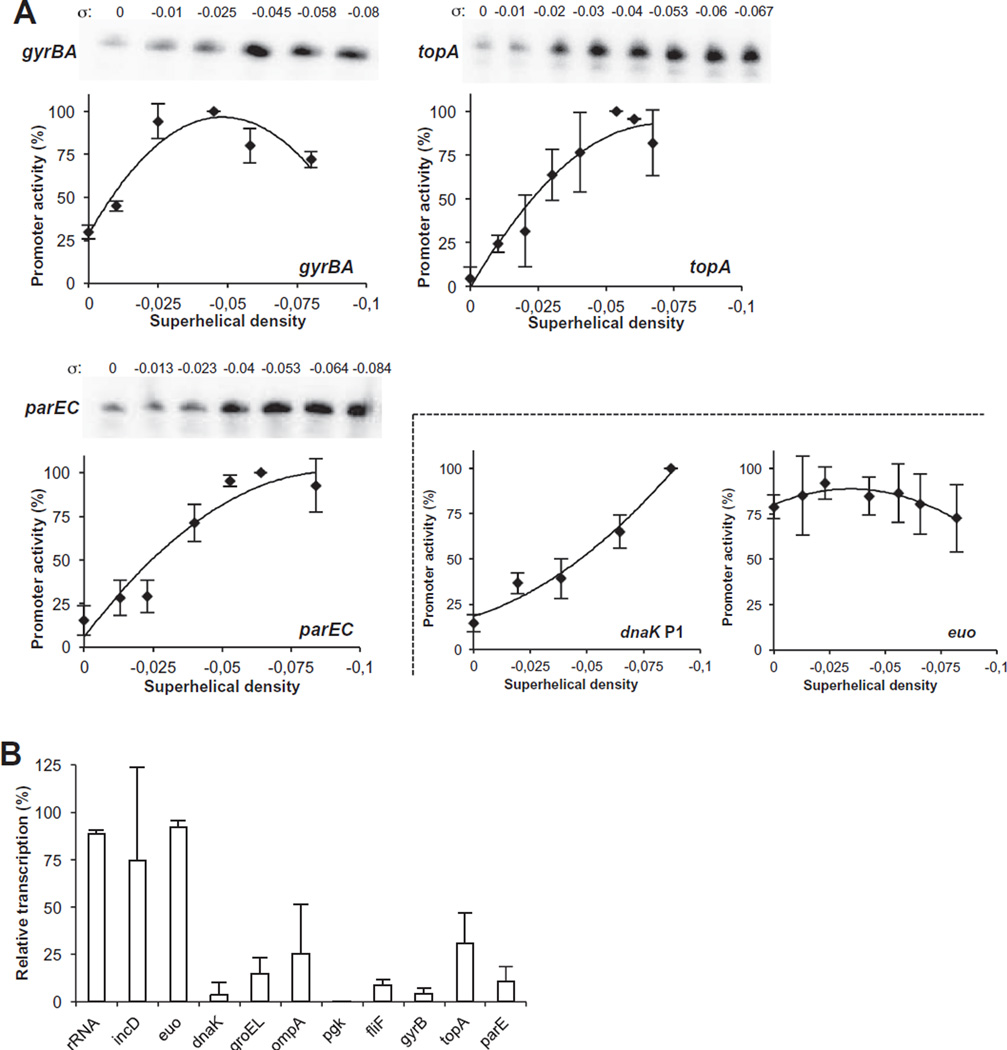
Fig. 5. DNA topoisomerase promoters are supercoiling sensitive.
A. In vitro supercoiling sensitivity assay of the chlamydial topoisomerase promoters. For each topoisomerase promoter cloned on a transcription plasmid, a series of topoisomers was generated over a range of superhelical densities (σ). Each topoisomer was transcribed by E. coli RNA polymerase in vitro. Relative promoter activity was calculated as a percentage of the maximal promoter activity (defined as 100%) over the range of superhelical densities tested. All reactions were performed as three independent experiments, and the error bars represent standard deviations. Comparison graphs showing the supercoiling response for a supercoiling-insensitive (euo) and a supercoiling-sensitive (dnaK P1) promoter are adapted from Cheng and Tan, 2012.
B. Supercoiling-response assay in Chlamydia-infected cells treated with novobiocin. The effects on transcription of selected chlamydial genes were measured by qRT-PCR. rRNA, incD and euo are early genes whose promoters have been shown to be supercoiling-insensitive in vitrodnak groEL ompApgk and fliF are early and midcycle genes whose promoters have been shown to be supercoiling sensitive in vitro (Case et al., 2010; Niehus et al., 2008; Cheng and Tan, 2012). Relative transcription was calculated as the percentage of the transcripts per chlamydial genome copy for novobiocin-treated cells compared with untreated samples. Results of three independent experiments are reported. Error bars indicate the standard deviation.
The gyrBA, topA and parEC promoters were all supercoiling-responsive. Each promoter was transcribed at a low level from a relaxed DNA template and at higher levels from more supercoiled topoisomers (Fig. 5A). The topA promoter was the most supercoiling-responsive, with a 21-fold range of promoter activity over the range of superhelical densities tested, whereas the parEC and gyrBA promoters had a 5.8- and 6.6-fold range of promoter activity respectively. These results indicate that there is negative feedback control of topoisomerase I and IV expression, with increased production of these DNA relaxation enzymes when supercoiling levels are high. However, the increased activity of the gyrBA promoter at higher supercoiling levels suggests that this promoter can be controlled by positive feedback control. This unusual response is different from E. coli and Mycobacterium smegmatis, in which gyrBA transcription is induced by DNA relaxation (Menzel and Gellert, 1983; Gellert and Nash, 1987; Unniraman and Nagaraja, 1999).
To verify that the chlamydial topoisomerase promoters are supercoiling-responsive, we used an inhibitor of DNA gyrase and topoisomerase IV to alter supercoiling levels in chlamydiae grown in cell culture (Fig. 5B). We treated Chlamydia-infected cells with novobiocin for 30 min at 16 hpi, which is before the peak in chlamydial supercoiling at about 18–24 h (Niehus et al., 2008), and measured transcript levels of selected genes by quantitative RT-PCR. In validation experiments, novobiocin treatment decreased transcription of dnaK, groEL, ompA, pgk and fliF, which each have a supercoiling-sensitive promoter (Niehus et al., 2008; Case et al., 2010; Cheng and Tan, 2012), but did not affect rRNA, euo and incD, early genes with supercoiling-insensitive promoters (Cheng and Tan, 2012). Transcription of all three chlamydial topoisomerases was decreased by novobiocin treatment, with transcript levels of gyrB, topA and parE reduced to 4.5%, 30.7% and 11% of untreated levels respectively. The results for topA and parE were consistent with the decreased transcription of topoisomerase I and IV in Haemophilus influenzae, Escherichia coli and Streptococcus treated with novobiocin (Gmuender et al., 2001; Peter et al., 2004; Ferrandiz et al., 2010). However, the response of gyrA was unusual because novobiocin increased transcription of the gyrase gene in these other bacteria (Menzel and Gellert, 1983; 1987b; Tse-Dinh, 1985). Thus, both our in vitro supercoiling sensitivity assay and this novobiocin treatment study of intracellular chlamydiae indicate that the chlamydial gyrase promoter has the unorthodox property of being activated by higher supercoiling levels.
C. trachomatis topoisomerase promoters are not controlled by the late regulator EUO
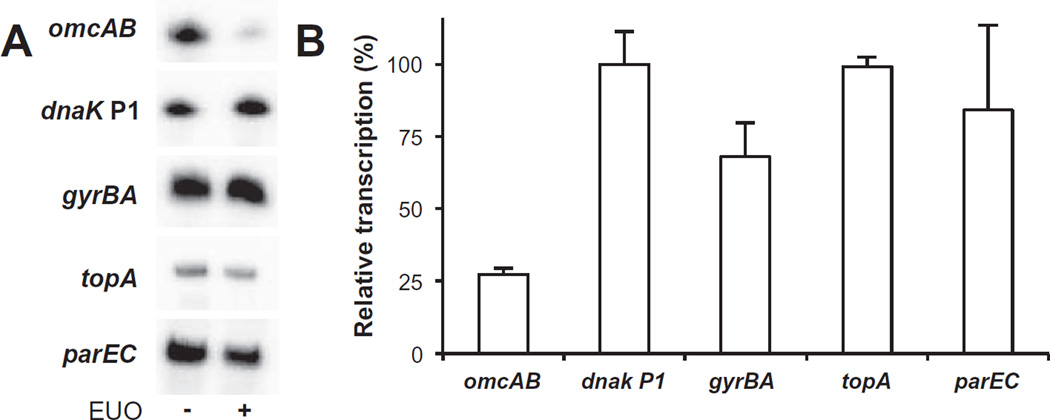
We also examined if the chlamydial topoisomerase genes are regulated by the transcription factor EUO. EUO selectively represses late promoters, and thus we anticipated that it would regulate parC and parE, which have been reported to have a late expression pattern (Belland et al., 2003), but not the early genes, gyrB and gyrA, or the midcycle gene topA. In an in vitro transcription assay EUO altered the transcriptional activity of the omcAB promoter, a known EUO target. However, EUO did not alter the in vitro activity of the topA, gyrBA or parEC promoters (Fig. 6). These results indicate that EUO does not regulate the three transcriptional units that encode the three topoisomerase enzymes in C. trachomatis. Thus, even though parC and parE have been categorized as late genes (Belland et al., 2003), the parEC promoter is supercoiling-responsive and EUO-independent, which are properties of a midcycle chlamydial promoter.
Fig. 6. Topoisomerase genes are not regulated by the master late regulator EUO.
A. In vitro transcription assay showing that EUO did not repress the three topoisomerase promoters. For each gene, a transcription templates containing the promoter region was transcribed with E. coli RNA polymerase in the absence or presence of EUO. The EUO target omcAB promoter and the EUO-independent dnak P1 promoter were used as positive and negative controls respectively.
B. Graph of the effect of EUO on transcriptional activity. For each promoter, transcription in the presence of EUO was normalized to baseline transcription in the absence of EUO (defined as 100%). Values are from the average of at least three independent experiments with standard deviation indicated by the error bar.
Discussion
In this study, we show that C. trachomatis expresses three DNA topoisomerases that can regulate unconstrained DNA supercoiling levels. In functional studies, we verified the counterbalancing activities of DNA gyrase to increase negative DNA supercoiling and topoisomerase I to relax DNA. The enzymatic activities of these proteins have been predicted in the genome annotation (Belland et al., 2003) and has been previously shown for DNA gyrase from the related species, C. pneumoniae (Ameyama et al., 2003). A third topoisomerase has been proposed to be either a second DNA gyrase (Belland et al., 2003) or a topoisomerase IV (Dessus-Babus et al., 1998), leaving doubts about its function in Chlamydia. We have resolved this issue by showing that CT660 and CT661 are the two subunits of a topoisomerase IV that decatenates DNA, and thus is likely to be involved in DNA replication. We thus propose that they should be named ParC and ParE respectively. In addition, we showed that this chlamydial enzyme, like other bacterial topoisomerase IV enzymes, is able to relax DNA in an ATP-dependent manner. C. trachomatis thus has a single DNA gyrase to increase DNA supercoiling but two topoisomerases that can relax DNA (Table 1).
Table 1.
Comparison of C. trachomatis and E. coli topoisomerase properties.
|
C. trachomatis |
E. coli |
|||||
|---|---|---|---|---|---|---|
| Enzyme | Gyrase | Topo I | Topo IV | Gyrase | Topo I | Topo IV |
| Gene | ct189 (gyrA) | ct643 (topA) | ct660 (parC) | gyrA | topA | parC |
| ct190 (gyrB) | ct661 (parE) | gyrB | parE | |||
| Size | 94 kD | 97 kD | 55 kD | 97 kD | 97 kD | 84 kD |
| 90 kD | 68 kD | 90 kD | 70 kD | |||
| Temporal expression | Early | Midcycle | Midcycle | Constitutive | Constitutive | Constitutive |
| Effect of supercoiling on transcription |
upregulation | upregulation | upregulation | downregulation | upregulation | upregulation |
| Dependence on ATP | + | − | + | + | − | + |
| Catalytic activity | Insertion of (−) supercoils |
Relaxation | Relaxation and Decatenation |
Insertion of (−) supercoils |
Relaxation | Relaxation and Decatenation |
These three DNA topoisomerases are present in most other bacteria (Forterre et al., 2007), but there are differences in how their genes are regulated in Chlamydia. A distinctive feature is that each chlamydial topoisomerase is expressed from its own transcriptional unit. In other Gram-negative bacteria, the genes for the two gyrase subunits are at different locations in the genome. Although the topoisomerases IV genes in E. coli and Salmonella typhimurium are adjacent, they are separated by a few kilobases (Huang, 1996). A potential advantage of the operon organization of Chlamydia gyrBA and parEC is that it allows each pair of subunits to be co-ordinately transcribed, which may be important for the temporal expression of the topoisomerase.
We found that the promoters for the chlamydial topoisomerase genes were transcribed at higher levels from more negatively supercoiled DNAtemplates. Supercoiling-responsiveness is a general property of topoisomerase promoters in other bacteria as part of a negative feedback mechanism for homeostatically regulating DNA supercoiling levels (Dorman and Corcoran, 2009). The chlamydial topA promoter conforms to this model because it was upregulated by higher DNA supercoiling levels, which would promote topoisomerase I expression and induce DNA relaxation (Tse-Dinh, 1985). Surprisingly, the C. trachomatis gyrBA promoter was also activated by higher levels of DNA supercoiling, which is a unique property that has not been described for a gyrase promoter. Instead in E. coli, S. pneumoniae, Mycobacterium and Streptomycces, transcription of the gyrase genes is reduced when supercoiling levels increase (Menzel and Gellert, 1987a; Thiara and Cundliffe, 1989; Unniraman and Nagaraja, 1999; Ferrandiz et al., 2010).
We demonstrated the unusual response of the chlamydial gyrase promoter to DNA supercoiling levels with supercoiling response assays performed in vitro and also in Chlamydia-infected cells treated with novobiocin. The in vitro transcription studies were performed with E. coli σ70 RNA polymerase because we could not obtain high enough transcription levels for quantification with Chlamydia σ66 RNA polymerase. This issue was mainly technical, related to the limited amounts of RNA polymerase that we are able to purify from Chlamydia-infected cells and compounded by the weak intrinsic activity of the chlamydial topoisomerase promoters. However, in previous studies, we have shown that the supercoiling response pattern for chlamydial promoters was similar for Chlamydia σ66 and E. coli σ70 RNA polymerase (Niehus et al., 2008). Thus, in Chlamydia, the topoisomerase promoters appear to be under feedback control, but the chlamydial gyrase promoter has the unusual property of being regulated by positive feedback.
Another feature of the chlamydial topoisomerase genes is that they have different temporal expression patterns during the intracellular developmental cycle. In the Belland microarray study, DNA gyrase was transcribed early, by 3 hpi, and topoisomerase I was transcribed as a midcycle gene by 8 hpi (Belland et al., 2003). The topoisomerase IV genes, parC and parE, were transcribed by 16 hpi and have been classified as late genes in microarray studies (Belland et al., 2003; Nicholson et al., 2003). However, our promoter analysis indicates that the parCE promoter is regulated as a midcycle promoter because it was supercoiling-dependent, in contrast to late promoters, and was not regulated by EUO, the master regulator of late gene expression (Schaumburg and Tan, 2003; Rosario et al., 2014). We also detected topoisomerase IV transcription at 12 hpi by RT-PCR, which is more consistent with a midcycle gene. These expression studies show that there is a delay between the early expression of DNA gyrase and the expression of topoisomerase I and topoisomerase IV in midcycle.
DNA supercoiling has been proposed to play an important role in temporal gene regulation during the chlamydial developmental cycle (Niehus et al., 2008; Cheng and Tan, 2012). In early reports, chlamydial supercoiling levels were shown to change during the intracellular infection (Solbrig et al., 1990; Barry et al., 1993). Niehus and collaborators then determined the temporal pattern by showing that DNA supercoiling levels are low at early times, rise to a peak in midcycle and then decrease to their initial levels at late times and in EBs (Niehus et al., 2008). This increased DNA supercoiling in midcycle has been proposed to drive the transcription of midcycle genes (Niehus et al., 2008; Case et al., 2010) and a subset of early genes that have supercoiling-dependent promoters (Case et al., 2010; Cheng and Tan, 2012). However, until now, there has been little understanding of how DNA supercoiling itself is regulated during the intracellular chlamydial infection.
Bacteria use a number of mechanisms to regulate topoisomerase expression and activity to control DNA supercoiling levels. In E. coli, the promoters for DNA gyrase and topoisomerase I are controlled by a nucleoid-associated protein called Fis (Cameron et al., 2011), but there is no Fis orthologue in Chlamydia. There are two chlamydial nucleoid-associated proteins, called the histone-like proteins hctA and hctB, but they bind and condense DNA in EBs, and not during the majority of the developmental cycle (Grieshaber et al., 2006a,b). DNA supercoiling can also be controlled by the ATP/ADP concentration ratio because gyrase activity is dependent on ATP while being inhibited by ADP (Westerhoff et al., 1988; Hsieh et al., 1991a,b). In Chlamydia, however, ATP levels accumulate to high levels in EBs (Tipples and McClarty, 1993), making it unlikely that there is a low ATP/ADP ratio to inhibit DNA gyrase at late times and in EBs. In addition, ATP levels may not have a selective effect on chlamydial DNA gyrase because topoisomerase IV is also ATP dependent.
We propose a model in which the changes in chlamydial DNA supercoiling levels are due to the regulated expression of the three topoisomerases during the developmental cycle. Early in the intracellular infection, DNA supercoiling levels are low but increase to a peak in midcycle (Niehus et al., 2008). We postulate that this initial stage of the developmental cycle is analogous to an E. coli topA mutant because there is expression of the chlamydial DNA gyrase prior to detectable transcription of chlamydial topoisomerase I and IV. topA mutants have highly negatively supercoiled DNA due to the inability to relax transcription-induced negative supercoiling (Stupina and Wang, 2005). We also predict that rising chlamydial supercoiling levels will activate the supercoiling-dependent gyrase promoter, amplifying gyrase expression through positive feedback.
After DNA supercoiling levels peak in midcycle, they decline to their original low levels (Niehus et al., 2008). This observation indicates that there must be a mechanism to prevent a runaway effect from positive feedback activation of the gyrase promoter. We propose that high DNA supercoiling levels in midcycle induce expression of topoisomerase I and then topoisomerase IV via their supercoiling-activated promoters. These DNA relaxation enzymes counterbalance and presumably exceed the effect of DNA gyrase so that supercoiling levels then decrease in the second half of the intracellular infection.
The unusual features of the chlamydial topoisomerase promoters described in this study play an important role in this model for the temporal control of chlamydial supercoiling levels. The DNA topoisomerases are constitutively expressed in other bacteria, but we postulate that the early expression of DNA gyrase, before the midcycle expression of topoisomerase I and IV, leads to an initial state of unbalanced gyrase activity. This temporal expression of chlamydial topoisomerases is facilitated by the organization of the two genes for DNA gyrase in their own transcriptional unit, as is also the case for the two genes for topoisomerase IV. Last but not least, the unusual activation of the chlamydial gyrase promoter by increased DNA supercoiling allows gyrase expression to be boosted by positive feedback. This response is in sharp contrast to a typical bacterial gyrase promoter, which is designed to be homeostatically downregulated by high supercoiling levels through negative feedback (Menzel and Gellert, 1987a).
A number of elements of this model await further experimental support. Although the sequential expression of the three chlamydial topoisomerase has been well described (Belland et al., 2003), it is not known how they are transcribed at different times even though all three have supercoiling-dependent promoters. It is also unclear if the combination of topoisomerase I and topoisomerase IV is necessary to reduce chlamydial supercoiling levels at later times. However, both enzymes have the capability to relax DNA, and the temporal correlation between topoisomerase I and topoisomerase IV expression and chlamydial DNA relaxation suggests that both enzymes may be involved. Topoisomerase I and topoisomerase IV also have different ATP and cation requirements, and thus even though they each mediate DNA relaxation, their activities may not be redundant.
This proposed role of the chlamydial topoisomerases in regulating DNA supercoiling levels may have implications for the treatment of chlamydial infections. The fluoroquinolones are a major class of antibiotics that interfere with bacterial DNA replication by inhibiting DNA gyrase and topoisomerase IV. In Chlamydia, the main target may be DNA gyrase because fluoroquinolone-resistance in C. trachomatis has only been linked with mutations in gyrA (Dessus-Babus et al., 1998). Furthermore, we found that C. trachomatis DNA gyrase was 2.5 to 5 times more sensitive to ciprofloxacin than topoisomerase IV in our in vitro studies. We predict that by inhibiting DNA gyrase, the fluoroquinolones will prevent changes in DNA supercoiling levels that regulate chlamydial gene expression. Thus, we propose that the fluoroquinolones may have a dual mechanism of action in Chlamydia by inhibiting DNA replication as well as progression of the developmental cycle.
Experimental procedures
Construction of topoisomerase subunits expressing plasmids
The ct189 (gyrA), ct190 (gyrB), ct643 (topA), ct660 (parC), ct661 (parE) genes were amplified from C. trachomatis serovar L2 (strain L2/434/Bu) genomic DNA using the primers listed in Table S1. ct190, ct643 and ct660 were ligated into SbfI/NcoI sites of the pMAL-c5X vector (New England Biolabs, NEB), whereas ct189 and ct661 were inserted into pEXp5CT/TOPO (Invitrogen). Correct DNA sequences were confirmed for each plasmid construct.
Expression and purification of the topoisomerase subunits
For MBP-tagged proteins, CT190, CT643 and CT660, E. coli BL21 (DE3) cells were transformed with the respective plasmid and grown in LB plus 100 µg ml−1 ampicillin, and 2 g l−1 glucose at 37°C. At an optical density at 600 nm (OD600) of 0.4 Isopropyl-β-D-thio-galactopyranoside (IPTG) was added to a final concentration of 0.3 mM. After incubation for 2 h at 37°C, cells were harvested, resuspended in lysis buffer (40 mM Tris [pH 8], 200 mM NaCl, 1 mM dithi-othreitol (DTT), 1 mM EDTA, 0.5 mg ml − 1 lysozyme) and disrupted by sonication. After centrifugation at 30 000 × g for 30 min at 4°C, the soluble fraction was loaded on an amylose column (NEB) and equilibrated with lysis buffer. Proteins were eluted in lysis buffer containing 10 mM maltose.
For His-tagged proteins, CT189 and CT661, E. coli T7 Express cells were transformed with the respective plasmid and grown in LB plus 100 µg ml−1 ampicillin at 37°C to an OD600 of 0.4. IPTG was added to a final concentration of 0.3 mM, and growth was continued overnight at 4°C. Cells were harvested, resuspended in 40 mM Tris [pH 8], 200 mM NaCl, 1 mM DTT and lysed by sonication. After centrifugation at 30 000x g for 30 min at 4°C, the soluble fraction was loaded onto a HisTrap FF column (GE healthcare) using an AKTÄ purifier system (Amersham Bioscience). Proteins were eluted over a 0–250 mM imidazole gradient.
Elution fractions containing the protein of interest, as determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), were pooled, dialysed overnight at 4°C against storage buffer (40 mM Tris [pH 8], 1 mM EDTA, 10% glycerol) and stored at 4°C.
Enzymatic assays
The DNA relaxation assay with topoisomerase I was performed in 40 mM HEPES-KOH [pH 8], 5 mM CaCl2, 1 mM DTT, BSA 50 µg ml−1 and 200 ng of negatively supercoiled pHOT-1 (TopoGEN) plasmid in a reaction volume of 20 µl. The samples were incubated for 30 min at 37°C, and the reactions were terminated by addition of Gel Loading Dye (NEB). The reaction products were electrophoresed on a 1% agarose gel without ethidium bromide (EtBr).
The DNA relaxation assay with topoisomerase IV subunits was performed in a similar manner except that the buffer contained 40 mM HEPES-KOH [pH 8], 100 mM K-glutamate, 10 mM DTT, 10 mM MgCl2, 50 µg ml−1 BSA, 2 mM ATP and 200 ng of negatively supercoiled pHOT-1 plasmid in a reaction volume of 20 µl.
For the decatenation assay, the topoisomerase IV subunits were mixed with 40 mM HEPES-KOH [pH 8], 100 mM K-glutamate, 10 mM DTT, 10 mM MgCl2, 50 µg ml−1 BSA, 2 mM ATP and 200 ng kinetoplast DNA (kDNA) (TopoGEN) in a reaction volume of 20 µl. Reaction mixes were incubated at 37°C for 30 min. The reactions were terminated and analyzed by electrophoresis as described above.
DNA supercoiling assay for the GyrA and GyrB proteins was performed in a buffer containing 40 mM HEPES-KOH [pH 8], 100 mM K-glutamate, 10 mM DTT, 10 mM MgCl2, 50 µg ml−1 BSA, 2 mM ATP and 100 ng relaxed pHOT-1 plasmid. Reaction mixes were incubated at 37°C for 30 min. The reactions were stopped and analyzed as described above.
Chlamydia cell culture
Chlamydia trachomatis serovar L2 (strain L2/434/Bu) was grown in L929 mouse fibroblast cells at 37°C with 5% CO2 in RPMI 1640 (Lonza) supplemented with 5% heat inactivated fetal bovine serum (FBS, Omega Scientific).
RT-PCR
L929 cells were infected with C. trachomatis at a multiplicity of infection (moi) of 3 and harvested at 6, 12 or 24 hpi. Total RNA was prepared using RNeasy plus Mini Kit (Qiagen) according to the manufacturer’s instructions. Residual DNA was removed by incubating the RNA with 20U RNase-free DNAse I (Roche) for 30 min at 37°C. cDNA synthesis was performed with AMV reverse transcriptase (Fisher Scientific) and random primers (Invitrogen) and amplified by PCR using AccuPower PCR premix (Bioneer). The primer sets used for RT-PCR are listed in Table S2.
Quantitative RT-PCR
L929 cells were infected with C. trachomatis at an moi of 3. At 16 hpi, 400 µg ml−1 novobiocin was added in the media. After 30 min incubation, the infected cells were collected. Total DNA was isolated using the QIAamp tissue kit (Qiagen) per the manufacturer’s protocol. Total RNA was prepared using RNeasy plus Mini Kit (Qiagen) according to the manufacturer’s instructions. Residual DNA was removed by incubating the RNA with 20U RNase-free DNAse I (Roche) for 30 min at 37°C. cDNA was synthesized and amplified by RT-PCR using the iScript one-step RT-PCR kit (Bio-Rad) on a Bio-Rad iCycler iQ per the manufacturer’s protocols.
qPCR and qRT-PCR data analysis
To normalize transcripts to genome copy number, standard curves were generated for each gene-specific primer pair (Bhaduri et al., 1998) by performing qPCR on C. trachomatis genomic DNA. For each sample, the Ct value obtained with 0.02% of total DNA was fit to the standard curve to determine the number of gene copies. qRT-PCR was performed for each RNA sample, and the Ct value obtained with 0.04% of total RNA (0.1% of total RNA for parE) was fit to the gene-specific standard curve to determine the number of transcript copies. The transcripts per genome copy were then calculated as the number of transcripts divided by the number of chlamydial genome copies measured with the same primer pair. The effect of novobiocin treatment was then determined by comparing the transcripts per genome copy of novobiocin-treated and untreated samples.
5′ Rapid amplification of cDNA ends (5’ RACE)
For each 5′ RACE reaction, 10 µg of RNA was treated with Terminator 5′-phosphate-dependent exonuclease (Epicentre) to remove processed transcripts. cDNA synthesis was carried out with AMV reverse transcriptase (Fisher Scientific) and random primers. A poly-dT tail was added to the 5′ end of the cDNA using terminal deoxynucleotidyltransferase (NEB). The RACE products were amplified by PCR with an adapter primer T602 (5′-CGCGAATTCCTCTTCTAGATGGGG GGGGGG-3′), which annealed to the poly-dT tail of the cDNA, and a gene specific primer (gyrB 5′-TAGCCTGAGTCATCCCATTAT-3′, parE 5′-CGGGAGCGAGATAAACTACA-3′; topA 5′-CGTGT AAATGCCGTCTTCAGC-3′). 5′-RACE PCR products were digested with EcoRI and subcloned into pGEM-7ZF (+) (Promega Biotech) between the EcoRI and SmaI sites. Plasmids were sequenced (Genewiz) to determine the 5′ end of each transcript.
Construction of in vitro transcription plasmids
Each promoter insert was cloned upstream of a promoter-less, G-less cassette transcription template, as previously described (Tan and Engel, 1996). Plasmid pMT1522 contains the promoter region of C. trachomatis gyrBA (positions −152 to +4). Plasmid pMT1539 contains the promoter region of topA (positions −138 to +4). Plasmid pMT1539 contains the promoter region of parEC (positions −119 to +4).
Generation of transcription plasmid topoisomers
For each transcription plasmid, a series of topoisomers over a range of superhelicities was generated, as previously described (Niehus et al., 2008). Ten micrograms of CsCl gradient purified plasmid DNA was treated for 3 h at 37°C with 8U of wheat germ Topoisomerase I (Promega) in a 40 µl mixture containing 50 mM Tris [pH 7.6], 0.1 mM EDTA, 1 mM DTT, 50 mM NaCl, 10% glycerol and concentrations of EtBr ranging from 0 to 140 µM. EtBr was removed by phenol-chloroform extraction, and the DNA was recovered by ethanol precipitation. The plasmid topoisomers were resolved on a 1.4% agarose gel containing EtBr. The average linking number difference (ΔLK) was determined by the band counting method of Keller (Keller, 1975). The average superhelical density (σ) was calculated using the equation σ =−10.5ΔLK/N, where N is the total number of base pairs in the plasmid.
In vitro transcription
For each promoter, a set of topoisomers of various superhelical densities was used as the transcription template in individual in vitro transcription reaction. Transcription reaction experiments were performed as described previously (Niehus et al., 2008). In a 10 µl reaction mixture, 0.5 U E. coli RNA polymerase holoenzyme (Epicentre) was used with 25 nM plasmid of each topoisomer. Radiolabeled transcripts were resolved on a urea-polyacrylamide gel and exposed to a phosphorimager screen. The screen was scanned with a Bio-Rad Personal FX scanner and quantified with ImageJ software. The relative promoter activity was calculated by defining the maximal promoter activity for the range of superhelicities tested as 100% and normalizing the promoter activity obtained for each topoisomer. Three measurements of relative promoter activity were obtained for each topoisomer, and a mean and a standard deviation were calculated.
For transcription assays examining regulation by EUO, plasmid DNA containing the transcription template was incubated with 2.5 µM C. trachomatis recombinant EUO (courtesy of Dr C. Rosario) at room temperature for 15 min. Transcription assays were initiated as described above.
Supplementary Material
Acknowledgements
We would like to thank Eike Neihus for identifying the topoisomerase promoters, and Brett Hanson, Kirsten Johnson, Jennifer Lee and Chris Rosario for critical reading of the manuscript. This work was supported by a grant from the NIH (Al44198) and a grant from Philippe Foundation, Inc (EO).
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Abdelrahman YM, Belland RJ. The chlamydial developmental cycle. FEMS Microbiol Rev. 2005;29:949–959. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Ameyama S, Shinmura Y, Takahata M. Inhibitory activities of quinolones against DNA gyrase of Chlamydia pneumoniae. Antimicrob Agents Chemother. 2003;47:2327–2329. doi: 10.1128/AAC.47.7.2327-2329.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes MH, LaMarr WA, Foster KA. DNA gyrase and DNAtopoisomerase of Bacillus subtilis: expression and characterization of recombinant enzymes encoded by the gyrA, gyrB and parC, parE genes. Protein Expr Purif. 2003;29:259–264. doi: 10.1016/s1046-5928(03)00068-8. [DOI] [PubMed] [Google Scholar]
- Barry CE, 3rd, Brickman TJ, Hackstadt T. Hc1-mediated effects on DNA structure: a potential regulator of chlamydial development. Mol Microbiol. 1993;9:273–283. doi: 10.1111/j.1365-2958.1993.tb01689.x. [DOI] [PubMed] [Google Scholar]
- Batteiger BE, Tan M. Chlamydia trachomatis (trachoma, genital infections, perinatal infections, and lym-phogranuloma venereum) In: Bennett JE, Dolin R, Mandell GL, editors. Mandell, Douglas, and Bennett’s: Principles and Practice of Infectious Diseases. Philadelphia, PA: Elsevier Inc; 2014. pp. 2154–2170. [Google Scholar]
- Bebear CM, Charron A, Bove JM, Bebear C, Renaudin J. Cloning and nucleotide sequences of the topoisomerase IV parC and parE genes of Mycoplasma hominis. Antimicrob Agents Chemother. 1998;42:2024–2031. doi: 10.1128/aac.42.8.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J, et al. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci USA. 2003;100:8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaduri T, Bagui TK, Sikder D, Nagaraja V. DNA topoisomerase I from Mycobacterium smegmatis. An enzyme with distinct features. J Biol Chem. 1998;273:13925–13932. doi: 10.1074/jbc.273.22.13925. [DOI] [PubMed] [Google Scholar]
- Cameron AD, Stoebel DM, Dorman CJ. DNA supercoiling is differentially regulated by environmental factors and FIS in Escherichia coli and Salmonella enterica. Mol Microbiol. 2011;80:85–101. doi: 10.1111/j.1365-2958.2011.07560.x. [DOI] [PubMed] [Google Scholar]
- Case ED, Peterson EM, Tan M. Promoters for Chlamydia type III secretion genes show a differential response to DNA supercoiling that correlates with temporal expression pattern. J Bacteriol. 2010;192:2569–2574. doi: 10.1128/JB.00068-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Summary of notifiable diseases – United States, 2012. MMWR. 2014;61:1–121. [PubMed] [Google Scholar]
- Champoux JJ. DNA topoisomerase I-mediated nicking of circular duplex DNA. Methods Mol Biol. 2001;95:81–87. doi: 10.1385/1-59259-057-8:81. [DOI] [PubMed] [Google Scholar]
- Cheng E, Tan M. Differential effects of DNA supercoiling on Chlamydia early promoters correlate with expression patterns in midcycle. J Bacteriol. 2012;194:3109–3115. doi: 10.1128/JB.00242-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessus-Babus S, Bebear CM, Charron A, Bebear C, de Barbeyrac B. Sequencing of gyrase and topoisomerase IV quinolone-resistance-determining regions of Chlamydia trachomatis and characterization of quinolone-resistant mutants obtained in vitro. Antimicrob Agents Chemother. 1998;42:2474–2481. doi: 10.1128/aac.42.10.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman CJ, Corcoran CP. Bacterial DNA topology and infectious disease. Nucleic Acids Res. 2009;37:672–678. doi: 10.1093/nar/gkn996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandiz MJ, Martin-Galiano AJ, Schvartzman JB, de la Campa AG. The genome of Streptococcus pneumoniae is organized in topology-reacting gene clusters. Nucleic Acids Res. 2010;38:3570–3581. doi: 10.1093/nar/gkq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Gellert M, Nash H. Communication between segments of DNA during site-specific recombination. Nature. 1987;325:401–404. doi: 10.1038/325401a0. [DOI] [PubMed] [Google Scholar]
- Gmuender H, Kuratli K, Di Padova K, Gray CP, Keck W, Evers S. Gene expression changes triggered by exposure of Haemophilus influenzae to novo-biocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 2001;11:28–42. doi: 10.1101/gr.157701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieshaber NA, Grieshaber SS, Fischer ER, Hackstadt T. A small RNA inhibits translation of the histone-like protein Hc1 in Chlamydia trachomatis. Mol Microbiol. 2006a;59:541–550. doi: 10.1111/j.1365-2958.2005.04949.x. [DOI] [PubMed] [Google Scholar]
- Grieshaber NA, Sager JB, Dooley CA, Hayes SF, Hackstadt T. Regulation of the Chlamydia trachomatis histone H1-like protein Hc2 is IspE dependent and IhtA independent. J Bacteriol. 2006b;188:5289–5292. doi: 10.1128/JB.00526-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschlag MR, Kohlhoff SA, Gaydos CA. Chlamydia pneumoniae. In: Bennett JE, Dolin R, Mandell GL, editors. Mandell, Douglas, and Bennett’s: Principles and Practice of Infectious Diseases. Philadelphia, PA: Elsevier Inc; 2014. pp. 2174–2182. [Google Scholar]
- Hsieh LS, Burger RM, Drlica K. Bacterial DNA supercoiling and [ATP]/[ADP]. Changes associated with a transition to anaerobic growth. J Mol Biol. 1991a;219:443–450. doi: 10.1016/0022-2836(91)90185-9. [DOI] [PubMed] [Google Scholar]
- Hsieh LS, Rouviere-Yaniv J, Drlica K. Bacterial DNA supercoiling and [ATP]/[ADP] ratio: changes associated with salt shock. J Bacteriol. 1991b;173:3914–3917. doi: 10.1128/jb.173.12.3914-3917.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WM. Bacterial diversity based on type II DNA topoisomerase genes. Annu Rev Genet. 1996;30:79–107. doi: 10.1146/annurev.genet.30.1.79. [DOI] [PubMed] [Google Scholar]
- Kato J, Suzuki H, Ikeda H. Purification and characterization of DNA topoisomerase IV in Escherichia coli. J Biol Chem. 1992;267:25676–25684. [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci USA. 1975;72:4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R, Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983;34:105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- Menzel R, Gellert M. Fusions of the Escherichia coli gyrA and gyrB control regions to the galactokinase gene are inducible by coumermycin treatment. J Bacteriol. 1987a;169:1272–1278. doi: 10.1128/jb.169.3.1272-1278.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R, Gellert M. Modulation of transcription by DNA supercoiling: a deletion analysis of the Escherichia coli gyrA and gyrB promoters. Proc Natl Acad Sci USA. 1987b;84:4185–4189. doi: 10.1073/pnas.84.12.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder JW. Interaction of Chlamydiae and host-cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson TL, Olinger L, Chong K, Schoolnik G, Stephens RS. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J Bacteriol. 2003;185:3179–3189. doi: 10.1128/JB.185.10.3179-3189.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehus E, Gressmann H, Ye F, Schlapbach R, Dehio M, Dehio C, et al. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol Microbiol. 2004;52:947–961. doi: 10.1111/j.1365-2958.2004.04006.x. [DOI] [PubMed] [Google Scholar]
- Niehus E, Cheng E, Tan M. DNA supercoiling-dependent gene regulation in Chlamydia. J Bacteriol. 2008;190:6419–6427. doi: 10.1128/JB.00431-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollmann M, Crisona NJ, Arimondo PB. Thirty years of Escherichia coli DNA gyrase: from in vivo function to single-molecule mechanism. Biochimie. 2007;89:490–499. doi: 10.1016/j.biochi.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Peter BJ, Arsuaga J, Breier AM, Khodursky AB, Brown PO, Cozzarelli NR. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004;5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario CJ, Tan M. The early gene product EUO is a transcriptional repressor that selectively regulates promoters of Chlamydia late genes. Mol Microbiol. 2012;84:1097–1107. doi: 10.1111/j.1365-2958.2012.08077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario CJ, Hanson BR, Tan M. The transcriptional repressor EUO regulates both subsets of Chlamydia late genes. Mol Microbiol. 2014:1097–1107. doi: 10.1111/mmi.12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaumburg CS, Tan M. Mutational analysis of the Chlamydia trachomatis dnaK promoter defines the optimal -35 promoter element. Nucleic Acids Res. 2003;31:551–555. doi: 10.1093/nar/gkg150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz E, Hennig S, Li SM, Heide L. Identification of a topoisomerase IV in actinobacteria: purification and characterization of ParYR and GyrBR from the coumermycin A1 producer Streptomyces rishiriensis DSM 40489. Microbiology. 2004;150:641–647. doi: 10.1099/mic.0.26867-0. [DOI] [PubMed] [Google Scholar]
- Schroder W, Bernhardt J, Marincola G, Klein-Hitpass L, Herbig A, Krupp G, et al. Altering gene expression by aminocoumarins: the role of DNA supercoiling in Staphylococcus aureus. BMC Genomics. 2014;15:291. doi: 10.1186/1471-2164-15-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw EI, Dooley CA, Fischer ER, Scidmore MA, Fields KA, Hackstadt T. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol Microbiol. 2000;37:913–925. doi: 10.1046/j.1365-2958.2000.02057.x. [DOI] [PubMed] [Google Scholar]
- Solbrig MV, Wong ML, Stephens RS. Developmental-stage-specific plasmid supercoiling in Chlamydia trachomatis. Mol Microbiol. 1990;4:1535–1541. doi: 10.1111/j.1365-2958.1990.tb02064.x. [DOI] [PubMed] [Google Scholar]
- Srivenugopal KS, Morris DR. Differential modulation by spermidine of reactions catalyzed by type 1 prokaryotic and eukaryotic topoisomerases. Biochemistry. 1985;24:4766–4771. doi: 10.1021/bi00339a009. [DOI] [PubMed] [Google Scholar]
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- Stupina VA, Wang JC. Viability of Escherichia coli topA mutants lacking DNA topoisomerase I. J Biol Chem. 2005;280:355–360. doi: 10.1074/jbc.M411924200. [DOI] [PubMed] [Google Scholar]
- Tan M. Temporal gene regulation during the chlamydial development cycle. In: Tan M, Bavoil PM, editors. Intracellular Pathogens I: Chlamydiales. Washington, DC: ASM Press; 2012. pp. 149–169. [Google Scholar]
- Tan M, Engel JN. Identification of sequences necessary for transcription in vitro from the Chlamydia trachomatis rRNA P1 promoter. J Bacteriol. 1996;178:6975–6982. doi: 10.1128/jb.178.23.6975-6982.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Gaal T, Gourse RL, Engel JN. Mutational analysis of the Chlamydia trachomatis rRNA P1 promoter defines four regions important for transcription in vitro. J Bacteriol. 1998;180:2359–2366. doi: 10.1128/jb.180.9.2359-2366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiara AS, Cundliffe E. Interplay of novobiocin-resistant and -sensitive DNA gyrase activities in self-protection of the novobiocin producer, Streptomyces sphaeroides. Gene. 1989;81:65–72. doi: 10.1016/0378-1119(89)90337-5. [DOI] [PubMed] [Google Scholar]
- Tipples G, McClarty G. The obligate intracellular bacterium Chlamydia trachomatis is auxotrophic for three of the four ribonucleoside triphosphates. Mol Microbiol. 1993;8:1105–1114. doi: 10.1111/j.1365-2958.1993.tb01655.x. [DOI] [PubMed] [Google Scholar]
- Tse Y, Wang JC. E. coli and M. luteus DNA topoisomerase I can catalyze catenation of decatenation of double-stranded DNA rings. Cell. 1980;22:269–276. doi: 10.1016/0092-8674(80)90174-9. [DOI] [PubMed] [Google Scholar]
- Tse-Dinh YC. Regulation of the Escherichia coli DNA topoisomerase I gene by DNA supercoiling. Nucleic Acids Res. 1985;13:4751–4763. doi: 10.1093/nar/13.13.4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unniraman S, Nagaraja V. Regulation of DNA gyrase operon in Mycobacterium smegmatis: a distinct mechanism of relaxation stimulated transcription. Genes Cells. 1999;4:697–706. doi: 10.1046/j.1365-2443.1999.00296.x. [DOI] [PubMed] [Google Scholar]
- Viard T, de la Tour CB. Type IA topoisomerases: a simple puzzle? Biochimie. 2007;89:456–467. doi: 10.1016/j.biochi.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Westerhoff HV, O’Dea MH, Maxwell A, Gellert M. DNA supercoiling by DNA gyrase. A static head analysis. Cell Biophys. 1988;12:157–181. doi: 10.1007/BF02918357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.