Graphical abstract

Keywords: Lysine acetylation, HDACs, Inflammation, NF-κB p65, HDAC inhibitors, Lung disease
Abstract
The increasing number of patients suffering from chronic obstructive pulmonary disease (COPD) represents a major and increasing health problem. Therefore, novel therapeutic approaches are needed. Class I HDACs 1, 2 and 3 play key roles in the regulation of inflammatory gene expression with a particular pro-inflammatory role for HDAC 3. HDAC 3 has been reported to be an important player in inflammation by deacetylating NF-κB p65, which has been implicated in the pathology of COPD. Here, we applied the pharmacological HDAC 3-selective inhibitor RGFP966, which attenuated pro-inflammatory gene expression in models for inflammatory lung diseases. Consistent with this, a robust decrease of the transcriptional activity of NF-κB p65 was observed. HDAC 3 inhibition affected neither the acetylation status of NF-κB p65 nor histone H3 or histone H4. This indicates that HDAC 3 inhibition does not inhibit NF-κB p65 transcriptional activity by affecting its deacetylation but rather by inhibiting enzymatic activity of HDAC 3. Taken together, our findings indicate that pharmacological HDAC 3-selective inhibition by inhibitors such as RGFP966 may provide a novel and effective approach toward development of therapeutics for inflammatory lung diseases.
1. Introduction
Chronic obstructive pulmonary disease (COPD) is a major and increasing health problem that is a leading cause of death worldwide [1]. COPD is associated with chronic inflammatory responses, predominantly in small airways and lung parenchyma. For some patients the currently available anti-inflammatory therapy is not effective due to reduced responsiveness to corticosteroids [2]. Considering this, it is necessary to explore molecular mechanisms that influence inflammatory responses within the context of lung inflammation with the perspective to develop novel therapeutics.
Gene expression in inflammatory signaling is among others regulated by post-translational modifications of histone proteins, for example lysine acetylations. The enzymes regulating the balance between lysine acetylation and deacetylation are denoted as histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively [3], [4], [5]. Aberrant activities of either HATs or HDACs alter the expression of (disease-associated) genes and contribute to the development of diseases like cancer and chronic inflammation [6], [7], also including COPD.
Apart from histones also non-histone proteins, like the NF-κB p65 transcription factor, which has been implicated in the pathology of COPD, are modified by histone-modifying enzymes such as HDACs [8]. Acetylations of specific lysine residues in NF-κB p65 play a key role in the regulation of its transcriptional capacity, DNA binding ability and duration of action resulting in NF-κB p65 activation or deactivation [9], [10], [11]. Seven lysine residues of NF-κB p65 have been described to be subjected to acetylation, respectively, 122, 123, 218, 221, 310, 314 and 315. Acetylations at lysines 122 and 123 decrease DNA binding, whereas acetylations at lysines 218 and 221 increase binding to κB enhancers. While acetylation at lysine 310 is required for full transcriptional activity of NF-κB p65, it does not affect DNA binding or assembly with IκBα [12]. Acetylations at lysines 314 and 315 do not affect the general transcriptional activity of the NF-κB complex, but rather increase promoter selectivity [13]. These examples demonstrate that direct acetylation or deacetylation of specific lysine residues of NF-κB p65 plays a crucial role in the regulation of NF-κB-mediated gene expression.
Several studies have shed more light on the role of HDAC iso-enzymes in the NF-κB p65 pathway, notably HDAC 3. HDAC 3 has been reported to be an important player in inflammation by deacetylating NF-κB p65. For example, the positive regulatory role of HDAC 3 in IL-1-induced (human IL-8 or murine Cxcl2) gene expression involves binding to the NF-κB p65 subunit and concomitant deacetylation at various lysines [14]. In the same study, using mutagenesis it was demonstrated that acetylation at lysine 314 and 315, in addition to acetylation at lysine 122 and 123, negatively regulates NF-κB p65 activity. Moreover, it was shown that HDAC 3 is involved in the removal of the inhibitory NF-κB p65 acetylations at lysine 122, 123, 314 and 315. Therefore, HDAC 3 is an important positive regulator in IL-1-induced inflammatory signaling by deacetylating four specific lysines in the NF-κB p65 subunit.
Considering the importance of HDAC 3 in the NF-κB pathway, small molecules interfering with the enzymatic activity of HDAC 3 have great potential to regulate the acetylation and so the activity of NF-κB p65 thereby affecting inflammatory gene expression. Very recently a possible role has been described for HDAC 3 in LPS-mediated pulmonary endothelial barrier dysfunction and acute lung injury [15] indicating that HDAC 3-selective inhibition may in particular have potential as anti-inflammatory therapy in inflammatory lung diseases.
Ongoing efforts to develop HDAC inhibitors with improved selectivity provided the inhibitor RGFP966 [16], a selective and potent inhibitor of HDAC 3 with nanomolar potency and no effective inhibition of any other class I HDAC at low micromolar concentrations [17], [18], [19]. The development of this potent and HDAC-3-selective inhibitor sets the stage to explore this inhibitor in inflammatory diseases. Based on current literature on the activating role of HDAC 3 in pro-inflammatory gene expression via the NF-κB pathway, we hypothesize that selective pharmacological inhibition of HDAC 3 would inhibit pro-inflammatory gene expression in model systems for inflammatory lung diseases by inhibition of the activation of the NF-κB pathway.
In this study we explored the utility of the HDAC 3 inhibitor RGFP966 to alter the balance between pro- and anti-inflammatory gene expression in in vitro and ex vivo models for inflammatory lung diseases. Firstly, we investigated the effects of siRNA-mediated downregulation of HDACs 1, 2 and 3 on inflammatory gene expression in mouse RAW 264.7 macrophages. Next, we examined the effects of RGFP966 on attenuating inflammatory gene expression in cell-based model systems for inflammatory lung diseases using mouse RAW 264.7 macrophages, human bronchial epithelial (HBE) cells and human airway smooth muscle (hASM) cells. Subsequently, ex vivo cultured mouse precision-cut lung slices (PCLS) were subjected to HDAC 3 inhibition by RGFP966. In RAW 264.7 macrophages and mouse PCLS, RGFP966 robustly downregulated pro-inflammatory and upregulated anti-inflammatory gene expression. The mechanism of action of RGFP966 was further investigated in mouse RAW 264.7 macrophages with respect to NF-κB p65 transcriptional activity, inhibition of NF-κB p65 and histone acetylation, and intracellular localization of NF-κB p65 and HDACs 1–3.
2. Methods and materials
2.1. Cell culture
Mouse RAW 264.7 macrophages were obtained from the American Type Culture Collection (ATCC; Wesel, Germany) and cultured in plastic tissue culture plates or flasks (Costar Europe, Badhoevedorp, The Netherlands) at 37 °C under 5% CO2/95% air in Dulbecco’s Modification of Eagle’s Medium (DMEM) containing GlutaMAX™ (Gibco® by Life Technologies, Bleiswijk, The Netherlands) supplemented with 10% (v/v) heat inactivated fetal bovine serum (FBS; Invitrogen, Breda, The Netherlands), 2 mM additional GlutaMAX™ (Gibco® by Life Technologies), 100 U/ml penicillin (Gibco® by Life Technologies) and 100 μg/ml streptomycin (Gibco® by Life Technologies). RAW 264.7 macrophages were used between passage 5 and 16.
The immortalized human bronchial epithelial cell line (16HBE14o-; abbreviated as HBE) was kindly provided by Dr. D.C. Gruenert, University of Vermont, Burlington, Vermont, USA [20]. Cells were cultured in fibronectin/collagen-coated plastic tissue culture plates or flasks at 37 °C under 5% CO2/95% air in minimal essential medium (MEM) supplemented with 10% heat inactivated FBS, 2 mM l-glutamine (Gibco® by Life Technologies), 50 U/ml penicillin and 50 μg/ml streptomycin, as described previously [21]. HBE cells were used between passage 73 and 103. Cells were serum-starved in MEM supplemented with antibiotics prior to each experiment.
Human airway smooth muscle (hASM) cell lines, immortalized by human telomerase reverse transcriptase were kindly provided by Prof. Dr. R. Gosens (Department of Molecular Pharmacology, University of Groningen). The primary cultured hASM cells used to generate each immortalized cell line were prepared as described previously [22]. hASM cells were cultured in plastic tissue culture plates or flasks at 37 °C under 5% CO2/95% air in DMEM supplemented with 10% FBS, 50 U/ml penicillin and 50 μg/ml streptomycin. For all experiments, immortalized hASM cells derived from two to three different donors were used between passage 27 and 35. Prior to experimentation, cells were serum-starved in DMEM supplemented with antibiotics and ITS (5 μg/ml insulin, 5 μg/ml transferrin and 5 ng/ml selenium).
2.2. HDAC 1–3 downregulation by siRNA
In order to downregulate the expression of HDACs 1, 2 and 3, cells were subjected to HDAC 1–3 selective siRNAs as follows. One day prior to transfection, RAW 264.7 macrophages were seeded at 20,000 cells/cm2 to obtain identical cell density at the start of the experiment. siRNA transfection experiments were performed in a 12-well plate upon complexing 50 nM siRNA with 3.5 μl Lipofectamine® 2000 (LF2K, Life technologies) according to the manufacturer’s protocol. After 24 h RAW 264.7 macrophages were washed twice with ice-cold PBS and harvested for RNA isolation or Western blotting. Where indicated, cells were stimulated with 10 ng/ml lipopolysaccharide (LPS, Escherichia coli, serotype 0111:B4; Sigma–Aldrich, Zwijndrecht, The Netherlands) and 10 ng/ml interferon gamma (IFNγ, #315-05; PeproTech, Hamburg, Germany) for the last 4 h of the experiment. The following siRNAs were used: mouse HDAC 1 (s119559), HDAC 2 (s67417), HDAC 3 (s67421) and Negative Control siRNA (4390843). All siRNAs used were Silencer® Select Pre-Designed & Validated siRNAs purchased from Ambion® by Life Technologies.
2.3. Gene expression analysis by RT-qPCR
RAW 264.7 macrophages, human bronchial epithelial (HBE) cells and human airway smooth muscle (hASM) cells were incubated with 0.41 μM SAHA, 1 μM RGFP966 (RAW 264.7) or 2 μM RGFP966 (HBE and hASM) for 20 h and stimulated with LPS/IFNγ for the last 4 h of the experiment. Subsequently, cells were washed twice with ice-cold Dulbecco’s Phosphate-buffered Saline (DPBS; Gibco® by Life Technologies) and total RNA was isolated using the SV Total RNA Isolation kit (Promega, Leiden, The Netherlands). RNA from mouse precision-cut lung slices (PCLS) was isolated using the Maxwell® 16 LEV simplyRNA Tissue Kit (Promega). Both RNA isolation procedures were performed according to the manufacturer’s protocol. RNA integrity was determined by 28S/18S ratio detection on an 1% agarose gel, which was consistently found intact. RNA concentration (OD260) and purity (OD260/OD280) were measured by NanoDrop ND-1000 UV–Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). For gene expression analysis, RNA was reverse transcribed using the Reverse Transcription kit (#A3500, Promega). Subsequently, 10 ng of cDNA was applied for each RT-qPCR, which was performed on an ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands). The primers for TNFα (Mm00443258_m1 and Hs01113624_g1), iNOS (Mm00440502_m1 and Hs01075529_m1), IL-1β (Mm00434228_m1 and Hs01555410_m1), IL-6 (Mm00446190_m1 and Hs00985639_m1), IL-10 (Mm00439614_m1 and Hs00961622_m1), IL-12b (Mm00434174_m1 and Hs01011518_m1), HDAC 1 (Mm02391771_g1), HDAC 2 (Mm00515108_m1), HDAC 3 (Mm00515916_m1), Arg1 (Mm00475988_m1), Fizz1 (Mm00445109_m1), MRC1 (Mm00485148_m1) and GAPDH (Mm99999915_g1 and Hs02758991_g1) were purchased as Assay-on-Demand (Applied Biosystems). For each sample, the real-time PCR reactions were performed in duplicate and the averages of the obtained Ct values were used for further calculations. Data analysis was performed with the relative quantification manager software (Applied Biosystems). Gene expression levels were normalized to the expression of the reference gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which was not influenced by the experimental conditions resulting in the ΔCt value. Gene expression levels were calculated by the comparative Ct method (2−ΔΔCt) [23].
2.4. Protein expression analysis by Western blot
RAW 264.7 macrophages were washed twice with ice-cold DPBS and subsequently lysed in ice-cold cell lysis buffer (25 mM Hepes, 5 mM MgCl2, 5 mM EDTA, 0.5% Triton X-100; supplemented with 1 mM DTT, 1 mM sodium butyrate, and protease inhibitors (#88266; Thermo Scientific, Rockford, IL, USA)). Next, lysates were freeze-thawed (4×) and centrifuged (10 min, 13,000g) to remove cell debris. Protein concentrations were determined using the RC DC Protein Assay (Bio-Rad) according to the manufacturer’s protocol. Equal amounts of protein was loaded on a 10% polyacrylamide gel, separated by SDS–PAGE with a Mini-Protean II apparatus (Bio-Rad Laboratories, Veenendaal, The Netherlands), and transferred with a Trans-Blot Electrophoretic Transfer system (Bio-Rad Laboratories) onto a polyvinylidene difluoride membrane (PVDF; Bio-Rad Laboratories). The membrane was blocked at room temperature for 1 h in DPBS/0.1% Tween 20 (Sigma–Aldrich; solution referred to as PBST) containing 5% skimmed milk (Campina, Friesland, The Netherlands) and subsequently incubated overnight at 4 °C with the appropriate primary antibody in 5% BSA (Sigma–Aldrich) or 5% skimmed milk. The following primary antibodies and dilutions were used: NF-κB p65 (#8242, 1:2500), IκBα (#4812, 1:1000), PARP-1 (#9532, 1:1000), HDAC 1 (#5356, 1:1000), HDAC 2 (#5113, 1:1000), HDAC 3 (#3949, 1:1000 and #2632, 1:1000); all from Cell Signaling, Leiden, The Netherlands and anti-acetyl lysine (AB3879, 1:500; Millipore, Billerica, MA, USA). For loading control the lower part of the blot was incubated for 1 h at room temperature with monoclonal rabbit anti-β-actin (#4970, 1:10,000; Cell Signaling). Membranes were washed in PBST and incubated at room temperature for 1 h with peroxidase-conjugated secondary antibodies. The following secondary antibodies were used: goat anti-rabbit IgG/HRP (#P0448), rabbit anti-goat IgG/HRP (#P0449) and rabbit anti-mouse IgG/HRP (#P0260, all 1:2000; DakoCytomation, Glostrup, Denmark). The bands were visualized by the enhanced chemiluminescence method using the Western Lightning® Plus-ECL kit (PerkinElmer, Waltham, MA, USA), quantified by imaging (GeneSnap from SynGene, Frederick, MD, USA), and processed using ImageJ software (1.48d; National Institute of Health, USA).
2.5. HDAC inhibition assay
The assay was performed in triplicate as described before [24]. Briefly, the respective human recombinant HDAC enzymes (BPS Bioscience, San Diego, CA, USA) were incubated in absence and/or in presence of various concentrations RGFP966 and a pro-fluorogenic substrate at room temperature for 60 min. Next, the deacetylation reaction was stopped by the addition of the HDAC Stop Solution (6 mg/ml trypsin, 0.3 mM SAHA) in all wells and the plate was incubated at 37 °C for 20 min. The release of the fluorescent 7-amino-4-methylcoumarin was monitored by measuring the fluorescence at λem = 460 nm and λex = 390 nm using a Synergy H1 plate reader (BioTek Instruments, Winooski, VT, USA). The fluorescence value of the background wells was subtracted from the fluorescence of the positive control, blank and inhibitor wells. Nonlinear regression was used to fit the data to the log(inhibitor) vs. response curve using GraphPad Prism (GraphPad software 5.00, San Diego CA, USA).
2.6. Investigation of cellular toxicity by MTS assay
To investigate the influence of the HDAC 3-selective inhibitor RGFP966 on cell viability, RAW 264.7 macrophages, HBE cells and hASM cells were seeded in 96-well plates. To obtain identical cell density at the start of the experiments, RAW 264.7 macrophages were seeded at 25,000 cells/cm2, HBE cells and hASM cells were seeded at 70% confluency (based on surface area) and were serum-starved for 24 h prior incubation with RGFP966. Shortly before incubation with RGFP966, the medium was replaced by 100 μl fresh (if appropriate serum free) culture medium. Incubations with LPS and IFNγ were performed as described for HDAC 1–3 downregulation by siRNA. After 20 h of incubation with RGFP966, 20 μl of CellTiter 96 AQueous One Solution reagent (Promega) was added to each well and incubated at 37 °C for 1 h in the dark. The absorbance at 490 nm was measured using a Synergy H1 plate reader. LPS/IFNγ-stimulated cells without addition of RGFP966 were considered 100%.
2.7. Effect of RGFP966 on NF-κB p65 transcriptional activity by QUANTI-Blue assay
RAW-Blue™ cells (InvivoGen, San Diego, CA, USA) were originally derived from mouse RAW 264.7 macrophages, which stably express a secreted embryonic alkaline phosphatase (SEAP) gene inducible by NF-κB and AP-1 transcription factors. RAW-Blue cells were treated with RGFP966 for a total of 20 h from which the last 4 h was supplemented with 10 ng/ml LPS and 10 ng/ml INFγ. The secretion of SEAP into the medium was determined using the QUANTI-Blue™ detection medium (InvivoGen) according to the manufacturer’s protocol. Briefly, 50 μl of supernatant was added to 150 μl of QUANTI-Blue™ detection medium and incubated at 37 °C for 1–2 h in the dark. Subsequently, SEAP activity was assessed by measuring the absorbance at 630 nm using a Synergy H1 plate reader. Results were plotted as % of control.
2.8. Animals
C57bl/6 male mice (weight 24–28 g; age 8–10 weeks) were purchased from Harlan (Zeist, The Netherlands). Animals were maintained on mouse chow and tap water ad libitum in a humidity- and temperature-controlled room at 24 °C with a 12 h light/dark cycle. All experiments were performed according to the national guidelines and upon approval of the experimental procedures by the local Animal Care and Use committee of Groningen University, DEC number 6962A. Mice were randomly assigned to the experiments.
2.9. Precision-cut lung slices and treatment
Mouse precision-cut lung slices (PCLS) were prepared as previously described (Eleftheriadis et al. [25]). Briefly, male mice were anesthetized by subcutaneous injection of ketamine (75 mg/kg, Alfasan, Woerden, The Netherlands) and dexdomitor (0.5 mg/kg, Orion Pharma, Mechelen, Belgium). Subsequently, the trachea was cannulated and the animal was exsanguinated by cutting the jugular vein, after which the lungs were filled trough the cannula with 1.5 ml low melting-point agarose solution (1.5% final concentration). The lungs were placed on ice for 15 min to solidify the agarose for slicing. The lobes were separated and tissue cores were prepared of the individual lobes, after which the lobes were sliced at a thickness of 250 μm. Tissue slices were incubated at 37 °C in a humid atmosphere under 5% CO2/95% air. In order to remove the agarose and cell debris from the tissue, slices were washed every 30 min (four times in total).
PCLS were incubated in DMEM supplemented with sodium pyruvate (1 mM), MEM non-essential amino acid mixture (1:100; Gibco® by Life Technologies), gentamycin (45 μg/ml; Gibco® by Life Technologies), penicillin (100 U/ml), streptomycin (100 μg/ml) and amphotericin B (1.5 μg/ml; Gibco® by Life Technologies). Slices were cultured at 37 °C in a humidified atmosphere under 5% CO2/95% air in 12-well tissue culture plates, using 3 slices per well. Slices were treated with RGFP966 for 20 h at final concentrations of 1 and 10 μM, and where indicated, stimulated with 10 ng/ml LPS and 10 ng/ml IFNγ.
2.10. Assessment of tissue viability using lactate dehydrogenase
To assess the viability of the PCLS subjected to RGFP966, the amount of lactate dehydrogenase (LDH) released from the tissue slices into the incubation medium was analyzed. Maximal LDH release was determined by lysing 3 slices with 1% Triton X-100 for 30 min at 37 °C at the start of the experiments. Supernatants were stored at −80 °C. LDH release was determined using an assay form Roche Diagnostics (Mannheim, Germany), and was measured using a Hitachi automatic analyzer (Modular Analytics, Roche Diagnostics). LDH release from the PCLS into the incubation medium was plotted relative to maximal LDH release.
2.11. Confocal laser scanning microscopy
The effect of the RGFP966 on the nuclear translocation of NF-κB p65 and localization of HDAC 3 was evaluated by immunofluorescence. 20,000 cells/cm2 cells were seeded in Lab-Tek chambers (#177445; Nunc, Rochester, NY, USA) and subjected to 1 μM RGFP966 for 20 h. Cells were washed twice with ice-cold DPBS and fixed with 4% formaldehyde at room temperature for 20 min. Subsequently, cells were permeabilized with 0.25% Triton X‐100 in DPBS and blocked with DPBS/5% BSA for 30–60 min at room temperature (to minimize non-specific binding). NF-κB p65 was detected with rabbit monoclonal NF-κB p65 antibody (2 h incubation at 4 °C, 1:250; # 8242, Cell Signaling), followed by 1 h incubation with goat anti-rabbit AlexaFluor488 (10 μg/ml; A-11008, Molecular Probes, Leiden, The Netherlands). HDAC 3 was detected with mouse monoclonal HDAC 3 antibody (2 h incubation at 4 °C, 1:100; # 3949, Cell Signaling), followed by 1 h incubation with goat anti-mouse AlexaFluor546 (10 μg/ml; A-11018, Molecular Probes). All antibody incubation steps were carried out in 5% BSA/DPBS and between incubation steps slides were washed with DPBS/0.5% BSA/0.05% Tween 20. For negative control, samples were processed omitting the primary antibody. At the end of the incubation steps, nuclei were stained with Hoechst 33342 (Molecular Probes) and mounted using Aqua Poly/Mount medium (Polysciences, Warrington, PA, USA), air dried for 24 h, and stored in the dark at 4 °C. Fluorescence was examined using a confocal laser scanning microscope (CLSM) equipped with true confocal scanner (TCS; SP8 Leica, Heidelberg, Germany), using a 63× oil immersion lens. Sequential scans were obtained to avoid bleed through. AlexaFluor488 was excited using the 488 nm blue laser line, AlexaFluor546 was excited using the 552 nm green laser line and Hoechst 33342 using the 405 nm UV laser line. All images were recorded in the linear range, avoiding local saturation, at an image resolution of 2048 × 2048 pixels and with a pinhole size of 1 Airy unit. Presented images show a single z-scan. Images were further processed using ImageJ 1.48d.
2.12. Nuclear and cytosolic fractionation
RAW 264.7 macrophages were subjected to 1 μM RGFP966 for 20 h, stimulated with 10 ng/ml LPS and 10 ng/ml IFNγ for 4 h and subsequently lysed. Nuclear and cytosol fractions of RAW 264.7 macrophages were prepared using the NE-PER® Nuclear and Cytoplasmic Extraction kit (Thermo Scientific) according to the protocol of the manufacturer.
2.13. Cell lysis and immunoprecipitation of NF-κB p65
RAW 264.7 macrophages (175 cm2) were subjected to 1 μM RGFP966 for 20 h, stimulated with 10 ng/ml LPS and 10 ng/ml IFNγ for 1 h and subsequently lysed in lysis buffer (20 mM Tris–HCl pH 7.5, 120 mM NaCl, 1% IGEPAL CA-630, 2 mM EDTA; all from Sigma–Aldrich) supplemented with protease inhibitors (#88266; Thermo Scientific). Lysates were incubated at 4 °C with constant rotation for 4 h and cleared by centrifugation (14,000g, 10 min, 4 °C). Cleared lysates were incubated with mouse monoclonal NF-κB p65 antibody (#6956, 1:100 (v/v); Cell Signaling) overnight at 4 °C. Subsequently, lysates were incubated with 50 μl of Protein A/G PLUS-Agarose Immunoprecipitation Reagent (sc-2003; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) at constant rotation for 6 h at 4 °C. Protein A/G agarose bound immunocomplexes were precipitated by centrifugation (4000g, 5 min, 4 °C) and washed 3 times with ice-cold lysis buffer. The pellet was re-suspended in 100 μl of 1× SDS sample buffer, boiled (95 °C, 5 min), and cleared by centrifugation (14,000g, 1 min).
2.14. Histone extraction and Micro BCA™ protein assay
Histone extractions were performed as previously described in the literature with minor modifications [26]. After histone extractions the samples were diluted with Phosphate-Buffered Saline (PBS; PAA Laboratories GmbH, Austria) to determine the total protein concentration using the micro BCA protein assay according to the manufacturer’s instructions (Pierce, Rockford, USA, # 23235). Absorbance was measured with a Synergy H1 plate reader at 562 nm. A bovine serum albumin standard (2 mg/ml, Pierce, Rockford, USA, # 23209) was used to calibrate the assay.
2.15. Acetylation of histones with acetic anhydride-d6
For the reaction, 7 μg of histones were loaded on a 15% polyacrylamide gel and resolved by SDS-PAGE electrophoresis. After Coomassie staining, bands for histones H3 and H4 were excised from the gel. Then 50 μl of acetonitrile and 50 μl of ammonium bicarbonate buffer (100 mM) were added to destain the gel bands. Gel bands were then dried in acetonitrile. Subsequently, 5 μl of acetic anhydride-d6 was added, after which 100 μl of ammonium bicarbonate (1 M) was added immediately. Subsequently, samples were incubated at 37 °C for 15 min. Gel bands were then washed 3 times with H2O. The acetic anhydride-d6 reaction was then repeated. After the second reaction, gel bands were dried using acetonitrile and trypsin (Promega, Madison, Wisconsin, USA, #V511A) was added at a 1:20 ratio in ammonium bicarbonate (50 mM). Histones were digested at 37 °C for 16 h. Supernatants containing histone peptides were subjected to LC MS/MS analysis as described below.
2.16. NanoChip LC–MS/MS QTOF
A quadruple time-of-flight mass spectrometer (QTOF, Agilent 6510) with a liquid chromatography-chip cube (#G4240) electrospray ionization interface was coupled to a nanoLC system (Agilent 1200) composed of a nanopump (#G2226A), a capillary loading pump (#G1376A) and a solvent degasser (#G1379B). Injections were performed with an autosampler (#G1389A) equipped with an injection loop of 40 μl and a thermostated cooler maintaining the samples in the autosampler at 4 °C during the analysis (#G1377A Micro WPS). The instrument was operated under the MassHunter Data Acquisition software (Agilent Technologies, Santa Clara, USA, version B.04.00, B4033.3). A chip (ProtID-Chip-150 II 300A, #G4240-62006) with a 40 nl trap column and a 75 μm × 150 mm analytical column filled with Zorbax 300SB-C18, 5 μm (Agilent Technologies) was used for peptide separation.
The following mobile phases were used for LC separations: solvent A, ultrapure water (resistance 18.2 MΩ, Millipore) with 0.1% (v/v) formic acid (FA), and solvent B, acetonitrile (ACN) with 0.1% (v/v) FA. The samples were injected and trapped at a flow rate 4.0 μl/min in a solution of 3% (v/v) ACN and 0.1% (v/v) FA in ultrapure water. The flow rate for peptide separation was 0.25 μl/min. The following program was used for elution peptides: 0–3 min linear gradient from 3% to 10% solvent B; 3–65 min linear gradient to 35% solvent B; 65–75 min linear gradient to 50% solvent B; 75–80 min linear gradient to 90% solvent B; 80–82 min isocratic 90% solvent B; 82–82.5 min linear gradient to 3% eluent B; 82.5–90 min isocratic 3% solvent B.
The identification of peptides was based on data collected in auto MS/MS 2 GHz mode using the following settings; fragmentor: 175 V, skimmer: 65 V, OCT 1 RF Vpp: 750 V, precursor ion selection: medium (4 m/z), mass range: 200–2500 m/z, acquisition rate for MS: 2 spectra/s, for MS/MS 3 spectra/s; MS/MS range: 50–3000 m/z; ramped collision energy: slope 3.8, offset: 0, precursor setting: maximum 3 precursors/cycle; absolute threshold for peak selection was 1000; relative threshold was 0.01% of the most intense peak, active exclusion enabled after 1 selection, release of active exclusion after 0.6 min, precursors were sorted by abundance only. The MS/MS files were stored in centroid and profile mode. MS1 absolute threshold 50 and MS2 absolute threshold 35 were applied to account for detector noise. Static exclusion range 200–350 m/z for precursor selection was applied. Gas temperature (nitrogen) was 325 °C and gas flow was 5 l/min. The quantification of peptides was based on data collected in MS mode using the same settings except of the mass range 20–3000 m/z; acquisition rate 1 spectra/s. In both cases lock masses 1221.990 m/z and 299.294 (Agilent) were used to recalibrate spectra during the acquisition. The area of the manually extracted ion chromatograms (0.1 m/z tolerance) of the selected peptides was used for label-free quantification.
2.17. Database search
Tandem mass spectra were extracted, charge state deconvoluted and deisotoped by the MassHunter Qualitative Analysis software version B.05.00 (Agilent Technologies) and saved as .mgf files. All MS/MS data were analyzed using Phenyx (GeneBio, Geneva, Switzerland); version CYCLONE (2010.12.01.1)). The fragment ion mass tolerance of 0.30 Da and a parent ion tolerance of 400 ppm were selected for a database search. The oxidation of methionine (+15.99), light (+42.01) and heavy (+45.03) acetylation of lysine (K), heavy acetylation with light methylation (+59.045) of K, methylation (+14.01), dimethylation (+28.03), trimethylation (+42.04) of K, methylation of arginine (R) (+14.01), dimethylation of arginine (R) (+28.03) N-terminal acetylation (+42.01), deamidation (+0.98) of asparagine (N) and glutamine (Q) were specified in Phenyx as variable modifications. The database search was performed in 2 steps. First, Phenyx was setup to search the uniprot swissprot database (selected for Mus musculus, Release 2012_08 of 05-Sep-12, 16547 sequences) setting the digestion enzyme to trypsin with up to one allowed missed cleavage site to identify a set of histones in the sample. Second, database search against the selected protein sequences (P43276, P43277, P43274, P43275, P15864, P10853, Q64525, Q8CGP1, P0C0S6, P68433, P62806, Q8CGP6, Q8R1M2, Q64523, P27661, P10922, Q9D1P2, O55128) was performed setting the digestion enzyme to trypsin with up to five allowed missed cleavage sites.
2.18. Statistical analysis
Statistical analysis of the results was performed by a two-tailed unpaired Student’s t-test, assuming equal variances to compare two replicate groups. Analysis of differences between multiple replicate groups was analyzed with a one-way ANOVA followed by Bonferroni post hoc analysis. p values ⩽0.05 were considered to be significant. Data were analyzed with GraphPad Prism.
3. Results
3.1. Downregulation of HDAC 3 attenuates the LPS/IFNγ-induced expression of pro-inflammatory genes and upregulates anti-inflammatory gene expression in RAW 264.7 macrophages
Among the class I HDACs, activity of HDAC 3 has been described to play a key role in pro-inflammatory gene expression [27]. In order to establish the role of class I HDACs 1, 2 and 3 in our model we explored the gene expression of TNFα, iNOS, IL-1β, IL-6, IL-10 and IL-12b in mouse LPS/IFNγ-stimulated RAW 264.7 macrophages upon HDAC 1, 2 and 3 downregulation. HDAC 1, 2 and 3 gene expression was downregulated by 50–80%, as compared to control using specific siRNAs (Fig. 1A), which was corroborated by downregulation of HDAC 1–3 protein expression (Fig. 1B). We investigated the expression of pro- and anti-inflammatory genes in RAW 264.7 macrophages in response to inflammatory stimulation using LPS and INFγ to ensure classical activation toward M1 (pro-inflammatory) macrophages. LPS/IFNγ stimulation did not affect the gene expression levels of HDACs 1–3 in RAW 264.7 macrophages (data not shown).
Fig. 1.
Downregulation of HDAC 3 by siRNA reduced the mRNA expression of LPS/IFNγ-induced pro-inflammatory genes and upregulated the anti-inflammatory gene IL-10 in LPS/IFNγ-stimulated RAW 264.7 macrophages. 20 h after transfection with HDACs 1, 2 and 3 or negative control siRNA RAW 264.7 macrophages were stimulated with LPS/IFNγ for the last 4 h of the experiment. HDACs 1, 2 and 3 gene expression was expressed as fold change compared to control (LPS/IFNγ-treated) group (A). Data are presented as mean values ± SD of 4–9 independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001 compared to the control (LPS/IFNγ-treated) group. Immunoblot that corroborate downregulation of HDACs 1, 2 and 3 is shown in (B). The presented data set shows a representative blot of 3 independent experiments performed under inflammatory (LPS/IFNγ) conditions. Inflammatory gene expression was assessed by RT-qPCR and expressed as fold change compared to control (LPS/IFNγ-treated) group (C). Data are presented as mean values ± SD of 4–9 independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001 compared to the control (LPS/IFNγ-treated) group.
Downregulation of HDAC 1 in LPS/IFNγ-stimulated RAW 264.7 macrophages significantly reduced the expression of the pro-inflammatory gene IL-1β, whereas the expression of other genes remained largely unaltered (Fig. 1C). The expression of the pro-inflammatory genes was largely unaffected upon HDAC 2 downregulation, while HDAC 3 downregulation reduced the expression of the pro-inflammatory genes IL-1β, IL-6 and IL-12b up to 60%, and upregulated the expression of the anti-inflammatory gene IL-10 by 2-fold compared to control. The observed pro-inflammatory role for HDAC 3 is in line with recent literature [14], [27] as described in the introduction. Taken together, this justifies the investigation of a pharmacological HDAC 3-selective inhibitor in model systems to examine alleviation of inflammatory lung diseases.
3.2. The pharmacological HDAC 3-selective inhibitor RGFP966 attenuates the inflammatory gene expression in LPS/IFNγ-stimulated RAW 264.7 macrophages and mouse precision-cut lung slices
Having established HDAC 3 as an important HDAC iso-enzyme in (co)regulating inflammatory gene expression we moved on to study which molecular consequences are associated with HDAC 3 inhibition in model systems for pulmonary inflammation, using the pharmacological HDAC 3-selective inhibitor RGFP966. As a first step we verified the IC50 values of RGFP966 against the deacetylase activity of recombinant human HDACs 1–3 and 8 [18], [19]. RGFP966 potently and selectively inhibited HDAC 3 at nanomolar concentrations, while HDACs 1, 2 and 8 were not inhibited at such concentrations, (Fig. 2A) indicating a good level of selectivity for HDAC 3. As a reference, we included the well-studied pan-HDAC inhibitor SAHA for which we confirmed its lack of selectivity among class I HDAC iso-enzymes (Fig. 2B).
Fig. 2.
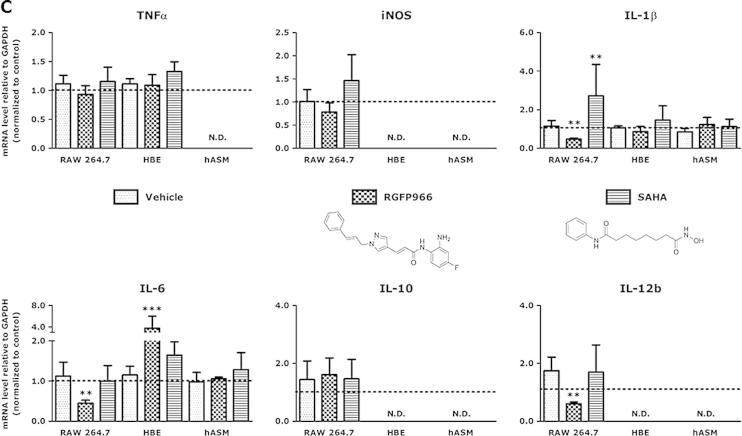
HDAC 3-selective inhibitor RGFP966 attenuates the expression of pro-inflammatory genes in RAW 264.7 macrophages. IC50 values of RGFP966 (A) and the pan-HDAC inhibitor SAHA (B) against the deacetylase activity of recombinant human class I HDACs 1, 2, 3 and 8 were determined. Inhibitory potency of RGFP966 and SAHA is shown in red. The log IC50 values are presented as mean values (μM) of 3 independent experiments ± SD. Subsequently, RAW 264.7 macrophages, human bronchial epithelial (HBE) cells and human airway smooth muscle (hASM) cells were incubated with RGFP966 and SAHA for 20 h and stimulated with LPS/IFNγ for the last 4 h of the experiment (C). Gene expression was assessed by RT-qPCR and expressed as fold change compared to control (LPS/IFNγ-treated) group. Data are presented as mean values ± SD of 3–5 independent experiments. **p < 0.01; ***p < 0.001 compared to vehicle. N.D. = not determined or not detected. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Next, RGFP966 was applied in cell-based model systems for inflammatory lung diseases using mouse RAW 264.7 macrophages, human bronchial epithelial (HBE) cells and human airway smooth muscle (hASM) cells that were subjected to LPS/IFNγ-stimulation. First, we investigated the cytotoxicity of RGFP966 in these cell types and observed no significant decrease in cell viability at the applied concentrations, as compared to control cells (data not shown). In LPS/IFNγ-stimulated RAW 264.7 macrophages treatment with RGFP966 did not change the expression of the genes TNFα, iNOS, and IL-10 but provided a significant downregulation of the expression of the pro-inflammatory genes IL-1β, IL-6 and IL-12b (Fig. 2C). This effect was specific for macrophages as in HBE cells and hASM cells the expression of the investigated genes was not affected except for pro-inflammatory gene IL-6, which was upregulated in HBE cells. For comparison, the pan-HDAC inhibitor SAHA upregulated IL-1β expression in RAW 264.7 macrophages demonstrating the differential effects observed for pan-HDAC inhibition compared to HDAC 3-selective inhibition (Fig. 2C). The observed differential effects on different cell types clearly demonstrates that the effects of HDAC inhibitors are cell type dependent and that HDAC 3 inhibition is particularly effective in repressing pro-inflammatory responses in macrophages.
To study the influence of RGFP966 treatment on the expression of LPS/IFNγ-induced cytokines in a more complex model system for inflammatory lung diseases we applied mouse precision-cut lung slices (PCLS) (Fig. 3A), which represent all cell types present in the in vivo situation and conserve the original architecture and matrix environment. Although PCLS do not have abundant inflammatory cells, tissue resident macrophages, epithelial cells and smooth muscle cells are present. First, the effect of RGFP966 treatment on the viability of PCLS was determined by measuring the release of lactate dehydrogenase (LDH) into the culture medium, which demonstrated no significant increase in LDH release upon RGFP966 treatment up to concentrations of ⩽10 μM (data not shown). Next, we examined the expression of pro-inflammatory genes TNFα, iNOS, IL-1β, IL-6, IL-12b, and the anti-inflammatory gene IL-10 in PCLS (either untreated or stimulated with LPS/IFNγ for 4 h), which confirmed robust induction of their expression (data not shown). Upon RGFP966 treatment, the expression of the anti-inflammatory gene IL-10 was upregulated by 2-fold in PCLS (Fig. 3B). This was accompanied by upregulation of the Fizz1 gene (Fig. 3C), which is a marker gene for the anti-inflammatory M2 phenotype of macrophages [28]. In addition, the expression of pro-inflammatory genes iNOS and IL-12b was significantly downregulated, and in contrast to our previous observations in RAW 264.7 macrophages, expression of the pro-inflammatory gene IL-1β was increased by more than 2-fold in PCLS (Fig. 3B). The expression of the investigated inflammatory genes in PCLS remained unaltered upon treatment with the pan-HDAC inhibitor SAHA (Fig. 3B). Taken together, since IL-10 has anti-inflammatory properties and is connected with the anti-inflammatory M2 macrophage phenotype, the observed changes in IL-10 expression are indicative for a potential anti-inflammatory effect of RGFP966 in inflammatory lung diseases, which is corroborated by the observed increased Fizz1 expression, and the attenuation of iNOS and IL-12b expression.
Fig. 3.
Effect of pharmacological HDAC 3 inhibition on the (pro)inflammatory gene expression profile in mouse precision-cut lung slices. Schematic representation of the experimental setup and timeline (A). Mouse precision-cut lung slices (PCLS) were subjected to RGFP966 and SAHA for 20 h and stimulated with LPS/IFNγ for the last 4 h of the experiment and subsequently lysed. Inflammatory gene expression was assessed by real-time PCR and expressed as fold change compared to control (LPS/IFNγ-treated) group (B). Effect of HDAC 3 inhibition on M2 marker gene expression in PCLS is shown in (C). Data are presented as mean values ± SD; n = 3–5. *p < 0.05; **p < 0.01 compared to vehicle. N.D. = not determined for SAHA.
3.3. RGFP966 attenuates LPS/IFNγ-induced NF-κB p65 transcriptional activity but does not affect NF-κB p65 acetylation, nuclear translocation, IκBα activation, or the overall histone acetylation status in RAW 264.7 macrophages
Inflammatory responses are frequently driven by well-defined stimulus-regulated transcription factors such as NF-κB p65. NF-κB p65 activation is required for the (in)direct expression of e.g., TNFα, iNOS, IL-1β, IL-6, IL-8, IL-10 and IL12b [29]. A number of biochemical studies have shown that HDAC 3 is involved in the deacetylation of the NF-κB p65 subunit, leading to activation of its transcriptional activity [14], [30]. Therefore, we investigate whether the observed attenuation of pro-inflammatory responses by the HDAC 3-selective inhibitor RGFP966 is connected to changes in NF-κB p65 transcriptional activity and lysine acetylation.
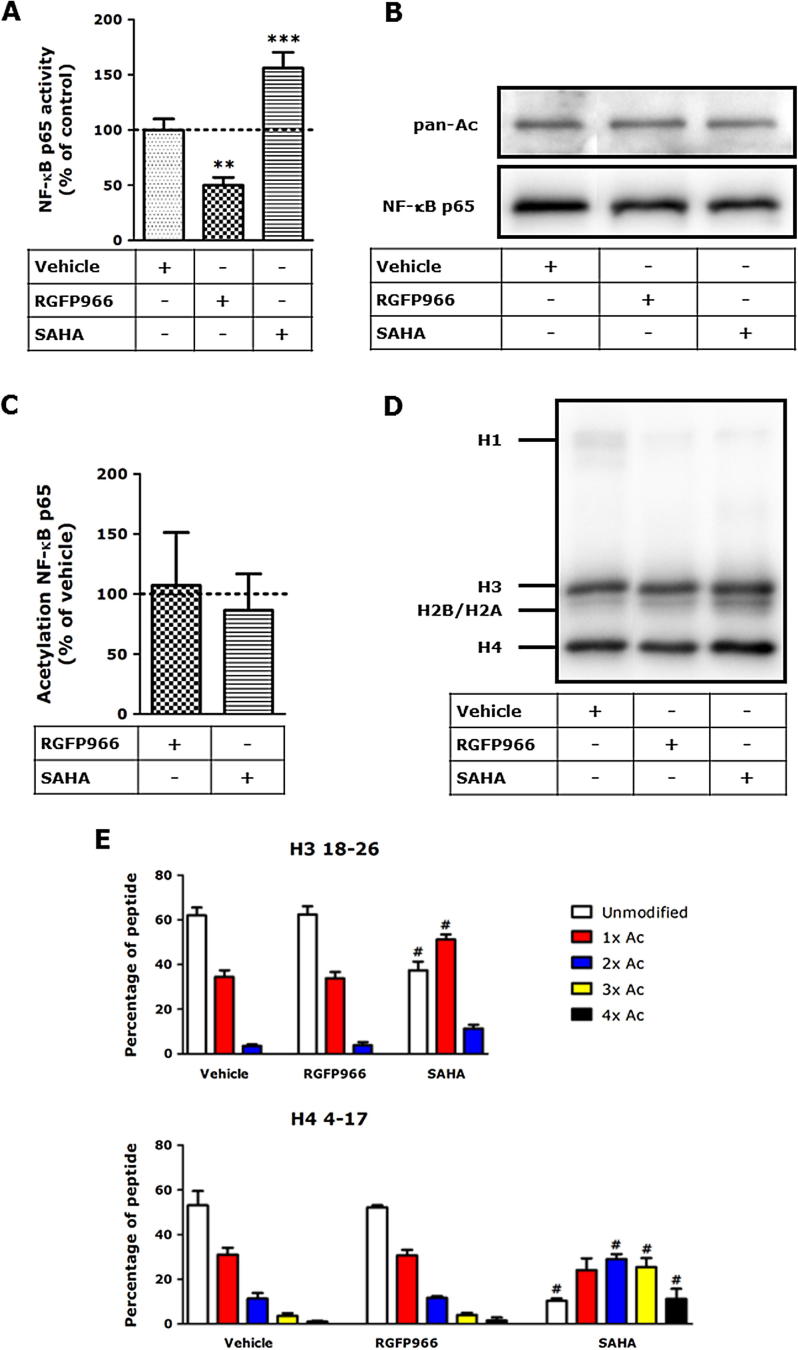
First, the transcriptional activity of NF-κB p65 was assessed using a reporter gene assay. As shown in Fig. 4A, RGFP966 provided a robust reduction of the transcriptional activity of NF-κB p65. In contrast, the pan-HDAC inhibitor SAHA increased the transcriptional activity of NF-κB p65. Subsequently, changes in the acetylation status of NF-κB p65 upon RGFP966 treatment were investigated using Western blot (Fig. 4B) and quantified by densitometric analysis (Fig. 4C). Using this method, no changes in the total acetylation of NF-κB p65 in RAW 264.7 macrophages were observed upon treatment with the HDAC 3-selective inhibitor RGFP966, indicating that HDAC 3 inhibition does not inhibit NF-κB p65 transcriptional activity by its deacetylation but rather by other mechanisms such as inhibition of co-activation of its transcriptional activity.
Fig. 4.
HDAC 3-selective inhibitor RGFP966 attenuates LPS/IFNγ-induced NF-κB p65 activity but affects neither the NF-κB p65 acetylation nor the histone acetylation status in RAW 264.7 macrophages. RAW-Blue cells were subjected to RGFP966 and SAHA for 20 h and stimulated with LPS/IFNγ for the last 4 h of the experiment. In the absence of an inflammatory stimulus, treatment of RAW-Blue cells with HDAC inhibitors did not affect the secretion of embryonic alkaline phosphatase (SEAP; data not shown), whereas treatment of RAW-Blue cells with RGFP966 followed by a pro-inflammatory stimulus significantly reduced the secretion of SEAP (A). Data are presented as mean values ± SD of 3–4 independent experiments. ***p < 0.001, compared to vehicle-treated group. The effect of HDAC 3 inhibition on total NF-κB p65 acetylation was assessed by (B) immunoblotting and quantified by densitometric analysis (C). RAW 264.7 macrophages were incubated with HDAC inhibitors for 20 h and subsequently lysed. For the detection of NF-κB p65 acetylation, lysates were incubated and immunoprecipitated with 5 μg of mouse monoclonal NF-κB p65-specific antibody as described. Data are presented as mean values ± SD expressed as fold change compared to control (vehicle-treated) group of 4 independent experiments. Histones were separated on SDS–PAGE (D), subsequently histones H3 and H4 were excised from the gel. Gel pieces were treated with acetic anhydride d6, followed by trypsin digestion. Resulting peptides were subjected to LC–MS/MS analysis (E). RGFP966 treatment did not affect the acetylation status of histone H3 (res. peptide 18–26: KQLATKAAR) and histone H4 (res. peptide 4–17: GKGGKGLGKGGAKR), whereas SAHA increased acetylation status of both peptides. Data are presented as mean values ± SD of 3–5 independent experiments. No significant differences were observed between untreated and vehicle treated cells (data not shown).
Next to NF-κB p65 acetylation, histone lysine acetylation was analyzed. Histones were extracted from RAW 264.7 macrophages and the acetylation levels of histone H3 and H4 were assessed by Western blot. RAW 264.7 macrophages subjected to HDAC 3 inhibitor RGFP966 showed no alteration in histone acetylation compared to vehicle-treated cells (Fig. 4D). In order to assess the changes in lysine acetylation on specific residues, histone acetylation of histone H3 and H4 was analyzed using mass spectrometry. To enable quantification, a previously described approach was employed in which the histones were fully acetylated using deuterated acetic acid anhydride ((CD3CO)2O) as a reagent [31], [32], [33], [34]. Using this method deuterated acetyl groups were installed on non-acetylated lysine residues. Subsequent tryptic digestion results in C-terminal cleavage at arginine residues, generating peptide fragments that are subjected to LC–MS/MS analysis. Based on the mass difference between deuterated and non-deuterated lysine acetylations the endogenous lysine acetylation ratio can be determined. Using this method a comprehensive overview of histone H3 and histone H4 acetylations was obtained (histone H3 peptide 18–26: KQLATKAAR and histone H4 peptide 4–17: GKGGKGLGKGGAKR are depicted in Fig. 4E). These and other peptides of histone H3 and histone H4 did not show changes in lysine acetylation status upon RGFP966 treatment, whereas SAHA treatment resulted in significant increases in histone lysine acetylation. This indicates that HDAC 3-selective inhibition does not induce global changes in histone lysine acetylations as observed for the pan-HDAC inhibitor SAHA (Fig. 4E).
In order to explore the influence of pharmacological HDAC 3 inhibition on NF-kB-mediated signaling further the effect of RGFP966 on nuclear translocation was investigated using fluorescence microscopy. No alterations in NF-κB p65 nuclear localization were observed upon RGFP966 treatment (Fig. 5A), which was confirmed by cellular fractionation experiments in which the nuclear and cytosolic fractions were separated and blotted for NF-κB p65 (Fig. 5B and C). In addition, to confirm proper LPS/IFNγ-stimulation and to determine the levels of IκBα in the presence of RGFP966, IκBα expression was assessed by Western blotting (Fig. 5D). These results demonstrate that HDAC 3-selective inhibition by RGFP966 does not affect IκBα activation. Taken together, these results indicate that HDAC 3-selective inhibitor RGFP966 pharmacologically attenuates NF-κB p65 transcriptional activity, but does not affect NF-κB p65 acetylation, histone H3 and H4 acetylation, IκBα activation or NF-κB p65 nuclear translocation in RAW 264.7 macrophages.
Fig. 5.
RGFP966 does not affect the nuclear translocation of NF-kB p65 in RAW 264.7 macrophages. After 20 h incubation with RGFP966 followed by 1 h LPS/IFNγ stimulation, RAW 264.7 macrophages were prepared for immunofluorescence microscopy (A). Green signal represents NF-kB p65 protein, while the blue signal visualizes the Hoechst-stained nuclei. RGFP966 did not affect the nuclear translocation of NF-kB p65 compared to (vehicle-treated) control group. The presented data set shows representative images of 3 independent experiments, original magnification 400×. All images were taken under identical instrumental conditions. In addition, the effect of RGFP966 on NF-kB p65 translocation in LPS/IFNγ-stimulated RAW 264.7 macrophages was analyzed by immunoblotting cell fractions (B) and quantified by densitometric analysis (C). PARP-1 and β-actin were used as internal controls. Data are presented as mean values ± SD expressed as fold change compared to control (LPS/IFNγ-treated) group of 4 independent experiments. RAW 264.7 macrophages were stimulated with 10 ng/ml LPS/IFNγ for 15, 30 and 60 min and subsequently lysed. In parallel, to confirm proper LPS/IFNγ-stimulation and to determine the levels of IκBα in the presence of RGFP966, IκBα expression was assessed by Western blotting (D). β-Actin was used as loading control. The presented data set shows a representative blot of 3 independent experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. HDAC 1–3 localization, expression and HDAC 3 – NF-κB p65 interaction in RGFP966-mediated attenuation of the LPS/IFNγ-induced inflammatory response in RAW 264.7 macrophages
Recently, it was demonstrated that HDAC inhibitors are able to affect HDAC expression levels and localization, besides attenuating HDAC activity [35], [36]. This prompted us to determine the expression levels of HDACs 1, 2 and 3 and to investigate their localization in LPS-induced RAW 264.7 macrophages.
The mRNA levels of HDACs 1, 2 and 3 were not significantly affected by RGFP966 in RAW 264.7 macrophages (Fig. 6A), whereas the HDAC 1 and HDAC 2 protein levels were slightly, though significantly, reduced upon RGFP966 treatment (Fig. 6B and C). In addition, neither mRNA nor protein levels of NF-κB p65 and IκBα were affected by RGFP966 in RAW 264.7 macrophages (Fig. 6A). In contrast, in PCLS, both mRNA and protein levels of HDACs 1, 2 and 3 were not affected by RGFP966, however, the IκBα level was significantly upregulated (data not shown). Moreover, differences in HDAC expression as reflected by, for example, the different basal mRNA expression of HDACs 1, 2 and 3 in cells and PCLS, might guide alterations of (pro)inflammatory gene expression.
Fig. 6.
HDAC 3-selective inhibitor RGFP966 reduces the protein expression of HDAC 1 and HDAC 2 in RAW 264.7 macrophages. RAW 264.7 macrophages were incubated with RGFP966, stimulated for 4 h with LPS/IFNγ and subsequently harvested. Gene expression of NF-kB p65, IκBα, HDAC 1, HDAC 2 and HDAC 3 was analyzed by real-time qPCR (A). Data are presented as mean values ± SD; n = 3–4. In parallel, the effect of RGFP966 on total NF-kB p65 and HDAC 1–3 protein expression in LPS/IFNγ-stimulated RAW 264.7 macrophages was analyzed by immunoblotting (B) and quantified by densitometric analysis (C). Protein levels were normalized against β-actin and control (LPS/IFNγ-treated) cells were set at 100%. Data are presented as mean values ± SD of 4 independent experiments and a representative blot is shown in B. *p < 0.05; **p < 0.01 compared to vehicle.
In order to evaluate if RGFP966 treatment influences NF-kB p65 activation by alteration of the localization of HDACs the influence of this inhibitor on the localization of NF-κB p65 and HDACs 1, 2 and 3 was investigated. The localization of HDACs 1–3 was studied by fluorescence microscopy (Fig. 7A) and Western blotting of nuclear and cytosolic fractions (Fig. 7B). As shown in Fig. 7A and B, no differences in HDAC 1, 2 and 3 localization were observed in RGFP966-treated RAW 264.7 macrophages compared to vehicle control.
Fig. 7.
RGFP966 does not affect the localization of HDAC 3 and HDAC 3 – NF-κB p65 interaction in LPS/IFNγ-stimulated RAW 264.7 macrophages. After 20 h incubation with RGFP966 followed by 1 h LPS/IFNγ stimulation, RAW 264.7 macrophages were prepared for immunofluorescence microscopy (A). The green signal represents NF-kB p65 protein, while the blue signal visualizes the Hoechst-stained nuclei. RGFP966 did not affect the nuclear localization of HDACs 1–3 compared to (vehicle-treated) control group. The presented data set shows representative images of 4 independent experiments. Images of the cells were taken using confocal laser scanning microscopy and all images were taken with identical instrumental conditions, original magnification 630×. In addition, the effect of RGFP966 on HDAC 1–3 localization in LPS/IFNγ-stimulated RAW 264.7 macrophages was analyzed by immunoblotting cell fractions (data not shown) and quantified by densitometric analysis (B). PARP-1 and β-actin were used as internal controls. Control (vehicle-treated) cells were set at 100%. Data are presented as mean values ± SD of 3–4 independent experiments. The HDAC 3 – NF-κB p65 interaction in LPS/IFNγ-stimulated RGFP966-treated RAW 264.7 macrophages was investigated by immunoprecipitation of NF-κB p65 followed by immunoblotting for HDAC 3, which was quantified by densitometric analysis (C). Protein levels were normalized against NF-κB p65. Data are presented as mean values ± SD of 3 independent experiments and a representative blot is shown. *p < 0.05 compared to vehicle. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Considering that pharmacological HDAC 3-selective inhibition reduced the NF-κB p65 transcriptional activity while leaving the NF-κB p65 acetylation and nuclear localization of NF-κB p65 and HDAC 3 unaltered suggests that HDAC 3 inhibition affects the NF-κB p65 transcriptional activity as a co-activator and not as a NF-κB p65 modifying enzyme in our model i.e., LPS/IFNγ-stimulated RAW 264.7 macrophages. Therefore, the interaction of HDAC 3 with NF-κB p65 was investigated by immunoprecipitation of NF-κB p65 and Western blot for HDAC 3 (Fig. 7C). As shown in Fig. 7C the interaction of HDAC 3 with NF-κB p65 was not affected in RGFP966-treated RAW 264.7 macrophages compared to vehicle-treated RAW 264.7 macrophages. Taken together, these results demonstrate that NF-κB p65 transcriptional activity is influenced by HDAC 3 enzymatic activity, while its acetylation, nuclear translocation as well as the localization and expression of HDACs 1, 2 and 3 remains unaltered in LPS/IFNγ-stimulated mouse RAW 264.7 macrophages.
4. Discussion
Class I HDACs 1, 2 and 3 play key roles in the regulation of pro-inflammatory gene expression in immune cells with a particular pro-inflammatory role for HDAC 3 [4], [14]. HDAC 3 has been reported to be an important player in inflammation by deacetylating NF-κB p65, which has been implicated in the pathology of COPD. Therefore, we investigated the application of a pharmacological HDAC 3-selective inhibitor in model systems for inflammatory lung diseases, to explore potential therapeutic applications.
As a first step we investigated the influence of HDACs 1, 2 and 3 on attenuating (pro)inflammatory gene expression in mouse RAW 264.7 macrophages by siRNA-mediated downregulation of HDACs 1, 2 and 3. Downregulation of HDAC 1 significantly reduced the expression of IL-1β, whereas the expression of other genes remained unaffected (Fig. 1C). Contrary to our findings, Serrat et al. demonstrated that downregulation of HDAC 1 (using siRNA) in bone marrow-derived macrophages led to a reduction of LPS-induced gene expression of iNOS and IL-6, but not to a reduction of LPS-induced IL-1β or TNFα [37]. Opposite to these anti-inflammatory effects, HDAC 1 has been described as a co-repressor of NF-κB p65-mediated gene expression [38]. In addition, it has been reported that HDAC 1 downregulation leads to a prolonged NF-κB p65 phosphorylation and nuclear localization [39]. Taken together, these studies demonstrate a versatile role for HDAC 1 in inflammation.
HDAC 2 downregulation in RAW 264.7 macrophages resulted in an increased expression of TNFα, iNOS and IL-6 indicating an anti-inflammatory role for HDAC 2, which is in line with literature (Fig. 1C). Recently it was demonstrated that HDAC 2 represses expression of IL-6 in dendritic cells [40]. Moreover, this enzyme has been shown to play a role in chronic obstructive pulmonary disease (COPD). HDAC 2 levels were significantly reduced in the lungs of patients with COPD with expression levels being reported at less than 5% of that in control lungs [41]. Similarly, it was reported that HDAC 2 levels were reduced in smoking asthmatics compared to non-smoking asthmatics [42]. Furthermore, transfection of HDAC 2 in alveolar macrophages has been shown to restore corticosteroid resistance [43]. Taken together, these studies demonstrate versatile roles for HDAC 2 in different types of immune cells or in different disease models, and call for a more profound investigation of the role of HDAC 2 in inflammation.
HDAC 3 downregulation reduced the expression of IL-1β, IL-6 and IL-12b in RAW 264.7 macrophages and increased the expression of IL-10 (Fig. 1C). These findings are in line with studies that demonstrated that HDAC 3-deficient macrophages were unable to express almost 50% of inflammatory genes studied when stimulated with LPS [27]. Importantly, a study by Ziesché et al. provided similar evidence and indicated that HDAC 3 is involved in deacetylation of the NF-κB p65 transcription factor and acts as a co-activator of this transcription factor in IL-1-signaling [14]. In support of these findings, studies in mice identified HDAC 3 as a mediator of allergic skin inflammation [44]. Taken together, our data on the role of HDAC 3 in macrophages are consistent with literature thus strengthening the evidence that HDAC 3 inhibition has potential to shift inflammatory gene expression toward an anti-inflammatory profile.
In order to get insights into the effects of pharmacological HDAC 3 inhibition mediated by RGFP966 on cell-based model systems that are relevant to inflammatory lung diseases we compared the expression of selected pro- and anti-inflammatory genes in response to LPS/IFNγ stimulation in RAW 264.7 macrophages, HBE cells and hASM cells. When studied in RAW 264.7 macrophages, expression of the IL-1β, IL-6 and IL-12b genes was significantly downregulated in response to RGFP966 treatment, which is in line with the observed downregulation of HDAC 3 expression by siRNA. In contrast, IL-6 was upregulated in HBE cells and not affected in hASM cells (Fig. 2C). This clearly demonstrates that the anti-inflammatory effect of HDAC 3-selective inhibition is cell type dependent and targets macrophages primarily.
In order to determine the overall effects of pharmacological HDAC 3 inhibition on pro- and anti-inflammatory gene expression in lung tissue, mouse PCLS were employed. In PCLS the IL-12b expression was inhibited but in contrast to RAW 264.7 macrophages the IL-1β and IL-6 expression were either unaffected or slightly upregulated. Moreover, the anti-inflammatory gene IL-10 was upregulated in PCLS (Fig. 3B), which was accompanied by upregulation of the M2 maker gene Fizz1 (Fig. 3C) and downregulation of the pro-inflammatory genes iNOS (Fig. 3B). Why effects of RGFP966 on the expression of pro-inflammatory genes iNOS, IL-1β and FIZZ1 are different at 1 μM versus 10 μM is not understood. Overall, analysis of the expression of the investigated genes in LPS/IFNγ-stimulated PCLS indicates a shift from pro- to anti-inflammatory gene expression.
Discrepancies between the cell-based model systems and PCLS can be attributed to differences in cellular composition, cellular communication and cellular microenvironment. Therefore, the studies in PCLS represent an important step toward application of the HDAC 3-selective inhibitor RGFP966 in animal models for airway inflammation. Our data demonstrate the relevance of testing potential novel therapeutics in multiple cell types, and/or tissue of a relevant disease model, to better predict and understand the outcome of drug interventions in an animal model.
The effect of RGFP966 on IL-10 expression levels is remarkable. The important role of IL-10 in inflammatory lung diseases has been previously highlighted in a study that showed diminished IL-10 production in lung tissue of COPD patients after LPS stimulation as compared to lung tissue of patients with normal lung function [45]. In addition, it has been demonstrated that the level of IL-10 and the number of IL-10-positive macrophages in sputum of COPD patients and healthy smokers was decreased as compared to healthy non-smokers [46]. This suggests that macrophages in COPD have lost their ability to produce anti-inflammatory cytokines such as IL-10 and are therefore unable to effectively dampen inflammation, which could be relieved by pharmacological HDAC 3 inhibition. In line with the increased expression of the anti-inflammatory gene IL-10 also the M2 marker gene Fizz1 increases upon RGFP966 treatment. Taken together, both the increased expression of IL-10 and Fizz1 indicate an anti-inflammatory effect for RGFP966 treatment on macrophages in ex vivo studies on PCLS.
A study on the molecular mechanism of action of HDAC 3 inhibitor RGFP966 was performed in LPS/IFNγ-stimulated RAW 264.7 macrophages in order to compare pharmacological inhibition to the current literature on biochemical downregulation of HDAC 3 expression. In our studies we observed that RGFP966 treatment inhibited NF-κB transcriptional activity, which is in line with the literature describing HDAC 3 as an NF-κB p65 co-activator [14], [27], [44]. The co-activator role of HDAC 3 has been reported to involve deacetylation of NF-κB p65 in experiments with overexpression of HDAC 3, which has not been observed in our studies using RGFP966.
It has been described that the duration of the nuclear action of NF-κB p65 is regulated by reversible acetylation and HDAC 3 has been shown to play a role in this process [10], [14]. HDAC 3-mediated deacetylation of NF-κB p65 promotes IκBα binding and nuclear export. Therefore, we investigated the cellular localization of NF-κB p65 upon RGFP966 treatment in RAW 264.7 macrophages. In our experiments we did not observe changes in nuclear localization, which is in line with our observation that RGFP966 did not affect NF-κB p65 acetylation. The observations in our model that pharmacological HDAC 3 inhibition does not lead to NF-κB p65 deacetylation and does not lead to altered NF-κB p65 localization despite the change in NF-κB p65 transcriptional activity point to a mechanism in which HDAC 3 activity plays a co-activator role in NF-κB p65-mediated signaling that is independent from NF-κB p65 deacetylation and localization.
It has been described that HDAC inhibitors also affect the acetylation levels of histone H3 and H4. Analysis of the global acetylation levels of histone H3 and histone H4 using mass spectrometry revealed no changes upon RGFP966 treatment. This indicates that pharmacological HDAC 3 inhibition has no wide-spread effects on histone acetylation. At the mRNA level, RGFP966 did not affect the expression of HDACs 1, 2 and 3 in RAW 264.7 macrophages (Fig. 6A). At the same time, however, the protein levels of HDACs 1 and 2 were slightly but significantly reduced (Fig. 6C). In addition, the localization of HDACs 1, 2 and 3 was not affected in RGFP966-treated RAW 264.7 macrophages compared to vehicle-treated cells. This argues for a model in which pharmacological HDAC 3 inhibition has a pronounced influence on the NF-κB p65 transcriptional activity but does not influence NF-κB p65 acetylation and localization, which was the expected mechanism of action of HDAC 3 inhibition. In addition, no changes in global histone H3 and H4 acetylation levels were detected. Taken together these data indicate that pharmacological HDAC 3 inhibition in RAW 264.7 macrophages inhibits NF-κB p65 transcriptional activity via a mechanism in which the HDAC 3 enzymatic activity plays a key role.
In summary, the results of our study demonstrate that selective pharmacological inhibition of HDAC 3 using RGFP966 has anti-inflammatory effects in response to LPS/IFNγ in RAW 264.7 macrophages and ex vivo in mouse PCLS. Moreover, RGFP966 significantly reduced the transcriptional activity of NF-κB p65, whereas NF-κB p65 acetylation and localization remain unaltered. Our observations confirm the pro-inflammatory role of HDAC 3 in macrophages, and demonstrate for the first time that HDAC 3-selective inhibition in mouse PCLS (a model for inflammatory lung disease) attenuates inflammatory responses. The findings presented in this study justify further optimization and investigation of pharmacological HDAC 3-selective inhibitors such as RGFP966 with the perspective toward development of anti-inflammatory drugs for application in inflammatory lung diseases.
Author contribution
NGJL: Study conception and design; Acquisition, analysis and interpretation of the data; Drafting and revision of the manuscript; Final approval and overall responsibility for the published work. PEvdW: Acquisition, analysis and interpretation of the data. TvdB: Acquisition, analysis and interpretation of the data; Revision of the manuscript. WTRH: Acquisition, analysis and interpretation of the data. MEO: Acquisition, analysis and interpretation of the data; Revision of the manuscript. LEMK: Interpretation of the data; Revision of the manuscript. RB: Interpretation of the data; Revision of the manuscript. RG: Interpretation of the data; Revision of the manuscript. HJH: Interpretation of the data; Revision of the manuscript. FJD: Study conception and design; Interpretation of the data; Revision of the manuscript; Final approval and overall responsibility for the published work.
Acknowledgments
We thank Sophie Bos (Department of Molecular Pharmacology, University of Groningen) for her assistance with preparing mouse precision-cut lung slices. This research was financially supported by the European Research Council with an ERC starting grant (309782) to FJD. Further support was obtained from the Netherlands Organization of Scientific Research (NWO) by a VIDI grant to FJD (723.012.005). We acknowledge COST action ‘epigenetics from bench to bedside (TD0905)’ and ‘epigenetic chemical biology (CM1406)’ for building a European network on epigenetics. Microscopic fluorescence imaging was performed at the UMCG Imaging and Microscopy Center (UMIC), which is supported by NWO-grants 40-00506-98-9021 (TissueFaxs) and 175-010-2009-023 (Zeiss 2p).
References
- 1.Barnes P.J. Immunology of asthma and chronic obstructive pulmonary disease. Nat. Rev. Immunol. 2008;8:183–192. doi: 10.1038/nri2254. [DOI] [PubMed] [Google Scholar]
- 2.Barnes P.J. Corticosteroid resistance in patients with asthma and chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2013;131:636–645. doi: 10.1016/j.jaci.2012.12.1564. [DOI] [PubMed] [Google Scholar]
- 3.Choudhary C., Weinert B.T., Nishida Y., Verdin E., Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 4.Dekker F.J., van den Bosch T., Martin N.I. Small molecule inhibitors of histone acetyltransferases and deacetylases are potential drugs for inflammatory diseases. Drug Discov. Today. 2014;19:654–660. doi: 10.1016/j.drudis.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Yang X.J., Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dekker F.J., Haisma H.J. Histone acetyl transferases as emerging drug targets. Drug Discov. Today. 2009;14:942–948. doi: 10.1016/j.drudis.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Chun P. Histone deacetylase inhibitors in hematological malignancies and solid tumors. Arch. Pharm. Res. 2015;38:933–949. doi: 10.1007/s12272-015-0571-1. [DOI] [PubMed] [Google Scholar]
- 8.Spange S., Wagner T., Heinzel T., Kramer O.H. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 9.Ito K. Impact of post-translational modifications of proteins on the inflammatory process. Biochem. Soc. Trans. 2007;35:281–283. doi: 10.1042/BST0350281. [DOI] [PubMed] [Google Scholar]
- 10.Chen L., Fischle W., Verdin E., Greene W.C. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 11.Chen L.F., Mu Y., Greene W.C. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang B., Yang X.D., Lamb A., Chen L.F. Posttranslational modifications of NF-kappaB: another layer of regulation for NF-kappaB signaling pathway. Cell. Signal. 2010;22:1282–1290. doi: 10.1016/j.cellsig.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buerki C., Rothgiesser K.M., Valovka T., Owen H.R., Rehrauer H., Fey M. Functional relevance of novel p300-mediated lysine 314 and 315 acetylation of RelA/p65. Nucleic Acids Res. 2008;36:1665–1680. doi: 10.1093/nar/gkn003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziesche E., Kettner-Buhrow D., Weber A., Wittwer T., Jurida L., Soelch J. The coactivator role of histone deacetylase 3 in IL-1-signaling involves deacetylation of p65 NF-kappaB. Nucleic Acids Res. 2013;41:90–109. doi: 10.1093/nar/gks916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joshi A.D., Barabutis N., Birmpas C., Dimitropoulou C., Thangjam G., Cherian-Shaw M. Histone deacetylase inhibitors prevent pulmonary endothelial hyper-permeability and acute lung injury, by regulating heat shock protein 90 function. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015 doi: 10.1152/ajplung.00180.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.M.A. Wood, Method and therapeutic for the treatment and regulation of memory formation, 2012, Patent No: WO 2012016081 A2.
- 17.Malvaez M., McQuown S.C., Rogge G.A., Astarabadi M., Jacques V., Carreiro S. HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proc. Natl. Acad. Sci. U.S.A. 2013;110:2647–2652. doi: 10.1073/pnas.1213364110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou C.J., Herman D., Gottesfeld J.M. Pimelic diphenylamide 106 is a slow, tight-binding inhibitor of class I histone deacetylases. J. Biol. Chem. 2008;283:35402–35409. doi: 10.1074/jbc.M807045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rai M., Soragni E., Chou C.J., Barnes G., Jones S., Rusche J.R. Two new pimelic diphenylamide HDAC inhibitors induce sustained frataxin upregulation in cells from Friedreich’s ataxia patients and in a mouse model. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0008825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cozens A.L., Yezzi M.J., Kunzelmann K., Ohrui T., Chin L., Eng K. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 1994;10:38–47. doi: 10.1165/ajrcmb.10.1.7507342. [DOI] [PubMed] [Google Scholar]
- 21.Gkoumassi E., Dekkers B.G., Droge M.J., Elzinga C.R., Schmidt M., Meurs H. Virodhamine and CP55,940 modulate cAMP production and IL-8 release in human bronchial epithelial cells. Br. J. Pharmacol. 2007;151:1041–1048. doi: 10.1038/sj.bjp.0707320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gosens R., Stelmack G.L., Dueck G., McNeill K.D., Yamasaki A., Gerthoffer W.T. Role of caveolin-1 in p42/p44 MAP kinase activation and proliferation of human airway smooth muscle. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006;291:L523–L534. doi: 10.1152/ajplung.00013.2006. [DOI] [PubMed] [Google Scholar]
- 23.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Szymanski W., Ourailidou M.E., Velema W.A., Dekker F.J., Feringa B.L. Light-controlled histone deacetylase (HDAC) inhibitors: towards photopharmacological chemotherapy. Chemistry. 2015;21:16517–16524. doi: 10.1002/chem.201502809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eleftheriadis N., Neochoritis C.G., Leus N.G., van der Wouden P.E., Domling A., Dekker F.J. Rational development of a potent 15-lipoxygenase-1 inhibitor with in vitro and ex vivo anti-inflammatory properties. J. Med. Chem. 2015;58:7850–7862. doi: 10.1021/acs.jmedchem.5b01121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duan Q., Chen H., Costa M., Dai W. Phosphorylation of H3S10 blocks the access of H3K9 by specific antibodies and histone methyltransferase. Implication in regulating chromatin dynamics and epigenetic inheritance during mitosis. J. Biol. Chem. 2008;283:33585–33590. doi: 10.1074/jbc.M803312200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X., Barozzi I., Termanini A., Prosperini E., Recchiuti A., Dalli J. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E2865–E2874. doi: 10.1073/pnas.1121131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boorsma C.E., Draijer C., Melgert B.N. Macrophage heterogeneity in respiratory diseases. Mediators Inflamm. 2013;2013:769214. doi: 10.1155/2013/769214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medzhitov R., Horng T. Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 30.Zhu H., Shan L., Schiller P.W., Mai A., Peng T. Histone deacetylase-3 activation promotes tumor necrosis factor-alpha (TNF-alpha) expression in cardiomyocytes during lipopolysaccharide stimulation. J. Biol. Chem. 2010;285:9429–9436. doi: 10.1074/jbc.M109.071274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin S., Garcia B.A. Examining histone posttranslational modification patterns by high-resolution mass spectrometry. Methods Enzymol. 2012;512:3–28. doi: 10.1016/B978-0-12-391940-3.00001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hersman E., Nelson D.M., Griffith W.P., Jelinek C., Cotter R.J. Analysis of histone modifications from tryptic peptides of deuteroacetylated isoforms. Int. J. Mass Spectrom. 2012;312:5–16. doi: 10.1016/j.ijms.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villar-Garea A., Israel L., Imhof A. Analysis of histone modifications by mass spectrometry. Curr. Protoc. Protein Sci. 2008 doi: 10.1002/0471140864.ps1410s51. (Chapter 14: Unit 14.10) [DOI] [PubMed] [Google Scholar]
- 34.Drogaris P., Villeneuve V., Pomies C., Lee E.H., Bourdeau V., Bonneil E. Histone deacetylase inhibitors globally enhance h3/h4 tail acetylation without affecting h3 lysine 56 acetylation. Sci. Rep. 2012;2:220. doi: 10.1038/srep00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji M.H., Li G.M., Jia M., Zhu S.H., Gao D.P., Fan Y.X. Valproic acid attenuates lipopolysaccharide-induced acute lung injury in mice. Inflammation. 2013;36:1453–1459. doi: 10.1007/s10753-013-9686-z. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Q., Yang F., Li X., Wang L.W., Chu X.G., Zhang H. Trichostatin A protects against experimental acute-on-chronic liver failure in rats through regulating the acetylation of nuclear factor-kappaB. Inflammation. 2015;38:1364–1373. doi: 10.1007/s10753-014-0108-7. [DOI] [PubMed] [Google Scholar]
- 37.Serrat N., Sebastian C., Pereira-Lopes S., Valverde-Estrella L., Lloberas J., Celada A. The response of secondary genes to lipopolysaccharides in macrophages depends on histone deacetylase and phosphorylation of C/EBPbeta. J. Immunol. 2014;192:418–426. doi: 10.4049/jimmunol.1203500. [DOI] [PubMed] [Google Scholar]
- 38.Ashburner B.P., Westerheide S.D., Baldwin A.S., Jr. The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol. 2001;21:7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonneaud A., Gagne J.M., Turgeon N., Asselin C. The histone deacetylase Hdac1 regulates inflammatory signalling in intestinal epithelial cells. J. Inflamm. (Lond.) 2014;11:43. doi: 10.1186/s12950-014-0043-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Q., Zhao K., Shen Q., Han Y., Gu Y., Li X. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 2015;525:389–393. doi: 10.1038/nature15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnes P.J. Role of HDAC2 in the pathophysiology of COPD. Annu. Rev. Physiol. 2009;71:451–464. doi: 10.1146/annurev.physiol.010908.163257. [DOI] [PubMed] [Google Scholar]
- 42.Ahmad T., Barnes P.J., Adcock I.M. Overcoming steroid insensitivity in smoking asthmatics. Curr. Opin. Investig. Drugs. 2008;9:470–477. [PubMed] [Google Scholar]
- 43.Barnes P.J. Reduced histone deacetylase in COPD: clinical implications. Chest. 2006;129:151–155. doi: 10.1378/chest.129.1.151. [DOI] [PubMed] [Google Scholar]
- 44.Kim Y., Kim K., Park D., Lee E., Lee H., Lee Y.S. Histone deacetylase 3 mediates allergic skin inflammation by regulating expression of MCP1 protein. J. Biol. Chem. 2012;287:25844–25859. doi: 10.1074/jbc.M112.348284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hackett T.L., Holloway R., Holgate S.T., Warner J.A. Dynamics of pro-inflammatory and anti-inflammatory cytokine release during acute inflammation in chronic obstructive pulmonary disease: an ex vivo study. Respir. Res. 2008;9:47. doi: 10.1186/1465-9921-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takanashi S., Hasegawa Y., Kanehira Y., Yamamoto K., Fujimoto K., Satoh K. Interleukin-10 level in sputum is reduced in bronchial asthma, COPD and in smokers. Eur. Respir. J. 1999;14:309–314. doi: 10.1034/j.1399-3003.1999.14b12.x. [DOI] [PubMed] [Google Scholar]